Abstract
Background
Severe asthma in horses, known as severe equine asthma (SEA), is a prevalent, performance‐limiting disease associated with increased allergen‐specific immunoglobulin E (IgE) against a range of environmental aeroallergens.
Objective
To develop a protein microarray platform to profile IgE against a range of proven and novel environmental proteins in SEA‐affected horses.
Animals
Six SEA‐affected and 6 clinically healthy Warmblood performance horses.
Methods
Developed a protein microarray (n = 384) using protein extracts and purified proteins from a large number of families including pollen, bacteria, fungi, and arthropods associated with the horses, environment. Conditions were optimized and assessed for printing, incubation, immunolabeling, biological fluid source, concentration techniques, reproducibility, and specificity.
Results
This method identified a number of novel allergens, while also identifying an association between SEA and pollen sensitization. Immunolabeling methods confirmed the accuracy of a commercially available mouse anti‐horse IgE 3H10 source (R 2 = 0.91). Biological fluid source evaluation indicated that sera and bronchoalveolar lavage fluid (BALF) yielded the same specific IgE profile (average R 2 = 0.75). Amicon centrifugal filters were found to be the most efficient technique for concentrating BALF for IgE analysis at 40‐fold. Overnight incubation maintained the same sensitization profile while increasing sensitivity. Reproducibility was demonstrated (R 2 = 0.97), as was specificity using protein inhibition assays. Arthropods, fungi, and pollens showed the greatest discrimination for SEA.
Conclusions and Clinical Importance
We have established that protein microarrays can be used for large‐scale IgE mapping of allergens associated with the environment of horses. This technology provides a sound platform for specific diagnosis, management, and treatment of SEA.
Keywords: allergen, horse, IgE, protein microarray, severe equine asthma
Abbreviations
- BALF
bronchoalveolar lavage
- BSA
bovine serum albumin
- IgE
immunoglobulin E
- PBS
phosphate buffered saline
- PBST
phosphate buffered saline with Tween 20
- RAST
radioallergosorbent test
- SEA
severe equine asthma
- sIgE
specific immunoglobulin E
1. INTRODUCTION
Severe asthma in horses, known as severe equine asthma (SEA), is a performance‐limiting, allergic response to inhaled allergens in genetically predisposed horses that affects approximately 14% of the equine population in the United Kingdom.1, 2, 3 Allergen exposure in affected horses results in small airway inflammation, mucus hypersecretion, and bronchoconstriction, altering pulmonary resistance, dynamic compliance, and pleural pressure.4, 5, 6 The predominant source of these aeroallergens is the organic dust portion of forage and bedding, which contains fungi, bacteria, pollen, and arthropods.7, 8, 9, 10, 11 Removal from the aeroallergen‐rich stabling environment results in some degree of remission,12 but owner compliance is limited because of seasonality, competition schedule, health issues, and nutritional demands. Treatments with corticosteroids and bronchodilators provide short‐term relief, but such therapeutic approaches have been associated with undesirable adverse effects, and their use is prohibited under Fédération Équestre Internationale and Jockey Club rules.13, 14 Allergen avoidance is the cornerstone of prevention and effective treatment, but the efficacy of the latter approach relies on identification of causal allergens.15 Currently, the main obstacles to diagnostic and therapeutic developments include major limitations in the number of allergens screened to establish causal agents and lack of a clinically applicable in vitro test.
The pathogenesis of this condition remains unclear, but several studies have implicated immunoglobulin E (IgE) by in vitro histamine release assays,16, 17, 18 natural hay and straw challenges,19 intradermal testing,20 and specific IgE (sIgE) analysis of bronchoalveolar lavage fluid (BALF) and sera.21, 22 The sIgE assays suggest that Aspergillus fumigatus, Alternaria alternate, rAsp f 8, Tyrophagus putrescentiae, Saccharopolyspora rectivirgula, Asp f 1/a, Aspergillus terreus, Eurotium amstelodami, Geotrichum candidum, and Wallemia sebi are implicated in the etiology of SEA.8, 21, 23, 24, 25 Although many recombinant proteins are available,26 advances in causal allergen identification have been limited because of the practicality of testing with classical methods, such as ELISA, which are time‐consuming, expensive, and require large quantities of samples and reagents.27
In recent years, protein microarrays have been gaining popularity in allergy diagnostic testing because of their ability to assess the interaction of thousands of proteins with specific immunoglobulin isotypes using techniques such as fluorescence on a miniaturized scale, a technique known as microarray profiling.28 This technique circumvents the aforementioned limitations associated with techniques such as ELISA, enabling multiallergen testing to assess complex sensitization profiles. Furthermore, with specific allergens, these tests show similar sensitivity to standard laboratory methods, including ELISA, UniCAP, radioallergosorbent test (RAST), ImmunoCAP, and immunoblot testing.29, 30, 31 Previously published sensitivity and specificity values using protein microarrays have indicated the high discriminatory power of the protein extracts and pure recombinant Culicoides proteins associated with insect bite hypersensitivity in the horse.22
Our aim was to develop a widespread allergen profiling technique using microarray methods that would enable rapid and accurate IgE profiling of SEA‐affected horses. Furthermore, we wanted to analyze the correlation between BALF and sera‐specific IgE profiles, a crucial consideration with respect to diagnostic sample requirements. Profiling data allows for advances in diagnostic testing and treatment.
2. MATERIALS AND METHODS
2.1. BALF and sera samples
Clinical assessment including physical examination, pulmonary function tests, and BALF cytology was used to define the inclusion and exclusion criteria for the selection of 6 horses with SEA and 6 control horses.5 Bronchoalveolar lavage fluid was collected as previously described,32 filtered through a 100‐mL syringe filter (Biocomma, Shenzhen, China), and decanted into 10‐mL aliquots in 15‐mL centrifuge tubes with the addition of Thermo Scientific Pierce Mini‐Protease Inhibitor Tablets—EDTA free (product # 13437766). The mixture was gently agitated and incubated at 4°C for 10 minutes before addition of 2.5 mL of glycerol (Fisher Scientific, Leicestershire, United Kingdom) and stored at −80°C until analysis. To concentrate, BALF was thawed, maintained at 4°C, and filtered using a Sartorius Stedim 0.45‐μm filter syringe (product # 17598). The BALF samples then were concentrated in an Amicon Ultra‐15 centrifugal filter (product # UFC905024) and used immediately. Blood was collected, and sera were prepared as previously described,33 before storage at −80°C until analysis, at which time samples were thawed at room temperature and placed on ice.
2.2. Proteins, printing and hybridization
To maximize utility, the microarray was designed to be as comprehensive as possible by including extracts and pure proteins from a wide range of protein families derived predominantly from fungi, bacteria, pollen, and arthropods. The extracts and pure proteins were obtained from commercial suppliers, produced in house, and from donations. Because of the limited commercial availability of some bacterial and fungal protein extracts, it was necessary to produce them in house. Lyophilized purified samples of the desired strain were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (https://www.dsmz.de) and grown in 150 mL of liquid media according to the supplier's recommendation (250‐mL Erlenmeyer flask). Once grown, the media were centrifuged in 50‐mL tubes at 4000g for 10 minutes, and the supernatant was carefully removed before washing the individual pellets in 5 mL of phosphate buffered saline (PBS). The tubes were centrifuged at 4000g for 10 minutes, the supernatant was removed, and 1 mL of lysis buffer solution was added to each tube (PBS, 0.5% TritonX‐100 wt/vol and Thermo Scientific EDTA free protease inhibitor mini‐tablet). The resuspended pellets were pooled into a single 50‐mL centrifuge tube, placed on ice, and sonicated using an MSE Soniprep 150 (15 seconds sonication with 30‐second cooling periods in between for 10 cycles). Subsequently, the solution was filtered through a Nalgene 0.45‐μm syringe filter (product # 190‐25‐45), and the protein content quantified using a Pierce Bicinchoninic Acid Protein Assay Kit (product # 23225). The remaining solution underwent lyophilization, was resuspended in MiliQ water with 10% glycerol (filtered through a 0.02 μm syringe filter), and was normalized to 1 mg/mL protein and stored at −80°C.
Bronchoalveolar lavage concentration work initially was conducted using slides described previously and consisting of extracts (n = 240) and pure proteins (n = 120) from a range of protein families, including fungi, pollens, and arthropods, to establish the optimal BALF concentration to be utilized in subsequent development work.22 In house extracts not present in the initial array setup were tested by printing normalized samples (1 mg/mL protein) onto 16‐pad FAST slides (Whatman Schleicher & Schuell, Dassel, Germany) using a QArraylite arrayer (Genetix, United Kingdom). After sample selection, a new set of 384 proteins (see Supporting Information Appendix) was printed in a professional setting using a Marathon microarrayer (ArrayJet, Roslin, Scotland) printer as previously described22 with an approximate spot size of 200 μm diameter and replicated with even spacing 2 times across each of the individual 16 pads into 2 identical blocks to final spot density of 12 288 spots/slide. For alignment and quality control, spots of Cy3, Cy5, and PBS were printed onto each slide. Once printed, slides were blocked for 3 hours at 37°C in 3% bovine serum albumin (BSA; wt/vol) in PBS inside a Corning 5 slide holder (product # 40082) using a mini hybridization oven (Appligene, USA), washed 3 times for 2 minutes in PBS containing 0.05% (wt/vol) Tween‐20, followed by five 1‐minute washes with MiliQ water, and dried by centrifugation (MSE Mistral 3000i, Sanyo, United Kingdom) at 1000g for 10 minutes at room temperature.
Slides were fitted with Proplate slide modules (Grace Bio‐Labs, product # 204862) and washed 3 times (60‐second dwell time) with PBS with Tween 20 (PBST; Tween at 0.2%). Samples (BALF/sera) were diluted in a ratio of 1:2 with 4% BSA in PBST (Tween at 0.4% wt/vol) containing Thermo Scientific Pierce Mini‐Protease Inhibitor Tablets—EDTA free (product # 13437766; 1 tablet in 5 mL), which previously had been passed through a Whatman 13‐mm, 0.45‐μm filter syringe (product # 6784‐1304). One hundred microliters of the prepared sample was added to each well, excluding well 4, which was used as control and filled with 100 μL of the dilution solution (1:2) in PBS. The Proplate was fitted with an adhesive seal strip and incubated for 16 hours at 4°C on the Stuart mini see‐saw rocker (SSM4) at 13 oscillations/minute. Slides were washed 3 times with PBST (Tween at 0.05%) using the BioTek plate washer and incubated for 2 hours at 37°C in a ThermoHybaid (HyPro 20) at vibration setting 3 with 100 μL per well of mouse anti‐horse IgE (BioRad, product # MCA5982GA) in a ratio of 1:400 in 1% BSA in PBST (Tween at 0.2% w/v), washed 3 more times with PBST (Tween at 0.05%) and incubated for 1 hour at 37°C in the ThermoHybaid with 100 μL per well of DyLight 649 conjugated anti‐mouse IgG1 (Rockland, product # 610‐443‐040) in a ratio of 1:400 in 1% BSA in PBST (Tween at 0.2% wt/vol). The slide then was washed 3 times in PBST (Tween at 0.05%) followed by 3 washes with Milli‐Q water, and dried by centrifugation at 300×g for 10 minutes (Mistral 3000i, rotor 43124‐708).
2.3. Data analysis
Processed slides were scanned in a GenePix 4000B (Molecular devices, USA) with the photomultiplier tube settings at 440 and 310 at 635 and 532 nm, respectively, and saved as TIF files. Images were processed in GenePix Pro software v6.0.1.27 (Axon Instruments) and saved as comma‐delimited text files. Fluorescence values were calculated for each spot by subtracting local background from the median fluorescence value of the spot. One pad per slide contained all reagents with addition of PBS instead of serum for control purposes; these results were deducted from samples on the same slide to account for any protein autofluorescence and nonspecific binding. Further analysis and data presentation were carried out using Microsoft Excel. Average fluorescence values for each protein were compared between SEA and control groups using a conventional Z‐test in Excel (Microsoft, USA). The Benjamini‐Hochberg method was used to account for false discovery at a rate of 0.05. Benjamini‐Hochberg corrected values were considered significant at P < .05. Linear regression (coefficient of determination) of IgE fluorescence values for all proteins (n = 384) was used to establish the relationship between BALF and sera, reproducibility of results, and various mouse anti‐horse IgE sources. Bronchoalveolar lavage fluid concentration techniques and concentrations were tested by 1‐way analysis of variance. Tukey's honestly significant difference test for multiple comparisons was performed if significant differences were found (P < .05).
3. RESULTS
3.1. Optimizing sera incubation conditions
3.1.1. BALF concentration techniques
Bronchoalveolar lavage fluid concentrations employing Amicon and PD10/lyophilizing methods were compared using a BALF pool from 6 horses (n = 3 SEA and 3 control). Total IgE for each protein group was used to compare concentration methods (Figure 1A) and indicated no significant difference (P > .05) between concentration techniques. The Amicon concentration method was used to evaluate optimal BALF concentration by total IgE fluorescence for each protein group, indicating that a plateau was reached at 40‐fold concentration (Figure 1B). Therefore, all subsequent BALF concentrations were carried out by Amicon filtration to a 40‐fold final concentration.
Figure 1.
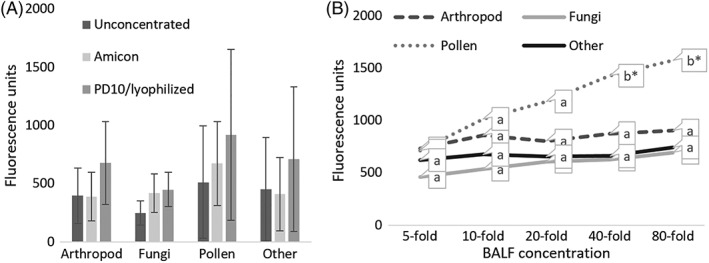
A, Bronchoalveolar lavage concentration optimization. Average BALF IgE fluorescence results for each protein group demonstrating concentration efficacy for unconcentrated, and concentrated (10‐fold) using either Amicon filter or PD10 columns/lyophilizing, A 1‐way ANOVA for each protein group using a BALF pool from 6 horses (n = 3 SEA; n = 3 control) demonstrated there was no significant difference (p > .05). Means that have no superscript in common are significantly different from each other. B, Average BALF pool horses (n = 3 SEA; n = 3 control) IgE fluorescence results for the main protein groups at various concentrations using Amicon filtration. Groupings included arthropod, fungi, pollen and other; which largely consisted of food and environmental proteins. Significant differences of each protein group were calculated individually by 1‐way ANOVA with Tukey's HSD (* = p < .05). Means that have no superscript in common are significantly different from each other
3.1.2. Incubation time
Two conditions were tested for optimal sera incubation times using a sera pool from 6 horses (n = 3 SEA and 3 control): 3 hours at 37°C as previously used for equine sera22 and overnight (16 hours) at 4°C, which previously has been shown to be more sensitive in studies performed in humans.34 As shown in Figure 2, the IgE profile between the 2 incubation times was significantly correlated (R 2 = 0.76). However, when the serum was incubated for 16 hours at 4°C, it was more sensitive with 28.1% of proteins showing positive reactions, compared to the 4 hours incubation, which showed 16.4% of proteins with positive reactions (data not shown). Therefore, subsequent serological incubations were conducted overnight at 4°C to increase sensitivity.
Figure 2.
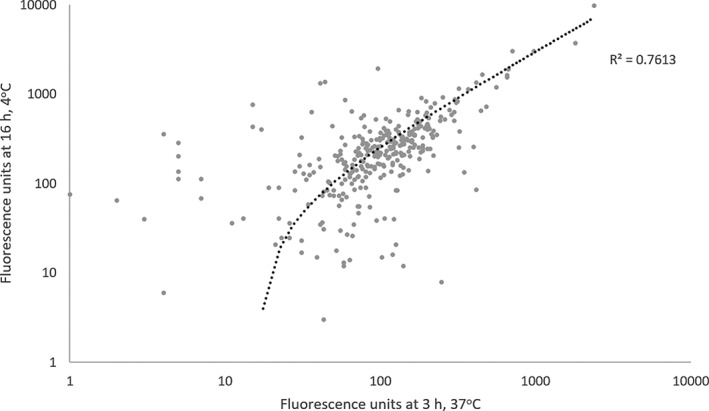
Comparison of incubation methods. Linear regression of microarray immunoglobulin E fluorescence results (n = 384 proteins) of pooled sera (n = 3 severe equine asthma; n = 3 control) incubated for 3 hours, 37°C, and 16 hours, 4°C, on a log scale
3.1.3. Comparison of specific IgE in BALF and sera
Bronchoalveolar lavage samples concentrated by Amicon (40‐fold) were compared with sera from 6 horses (n = 3 SEA; n = 3 control), and correlation coefficients were calculated for each separate protein group. Strong correlations were found between BALF and sera (Table 1). Thus, all subsequent incubations were conducted with serum because it is far easier to obtain, less invasive, more economical, and stable to transport. Horse 5, a clinically healthy horse, showed poor BALF/sera correlations across all protein groups, which was thought to be a result of the horse recently changing to a different barn on the same yard and, therefore, localized IgE production in the lung because of environmental allergen correlated poorly with serological IgE.23, 35
Table 1.
Regression coefficients between total specific immunoglobulin E fluorescence results between protein groups in serum and bronchoalveolar lavage fluid (n = 6)
| Pollen | Arthropod | Fungus | Bacteria | |
|---|---|---|---|---|
| Horse 1 | 0.63 | 0.82 | 0.71 | 0.74 |
| Horse 2 | 0.79 | 0.96 | 0.73 | 0.91 |
| Horse 3 | 0.65 | 0.92 | 0.54 | 0.88 |
| Horse 4 | 0.91 | 0.99 | 0.56 | 0.72 |
| Horse 5 | 0.44 | 0.58 | 0.33 | 0.89 |
| Horse 6 | 0.87 | 0.85 | 0.59 | 0.98 |
3.2. Reproducibility
3.2.1. Printing lot variation
The effect of printing lot on reproducibility of the protein microarray was assessed using 2 microarray slides printed on the same day. Sera from 3 SEA and 3 control horses was hybridized on the 2 slides simultaneously. Fluorescence results from replicate arrays were evaluated using linear regression and indicated that fluorescence results from the array were repeatable between printing lots (R 2 = 0.97).
3.2.2. Comparison between monoclonal mouse anti‐horse 3H10 sources
Two mouse anti‐horse IgE monoclonal antibodies (derived from the 3H10 clone) were compared using linear regression of the fluorescence results using a sera pool from 6 horses (n = 3 SEA; n = 3 control), which included the original 3H10 from a previous study36 and the commercially available BioRad 3H10 (product # MCA5982GA).
As shown in Figure 3, the fluorescence results from the array had a correlation coefficient of R 2 = 0.91, indicating a significantly similar IgE profiles.
Figure 3.
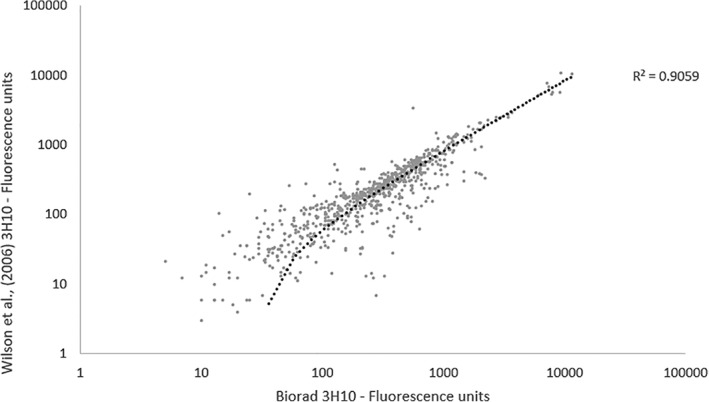
Comparison of secondary antibodies. Linear regression comparing protein microarray (n = 384) immunoglobulin E (IgE) fluorescence results on log scale from pooled sera (n = 3 severe equine asthma; n = 3 control) using two mouse anti‐horse IgE 3H10 clones; Wilson et al,36 and BioRad (product # MCA5982GA)
3.3. Specificity—protein inhibition assay
To test the IgE specificity of IgE‐protein binding, a protein inhibition assay was performed to assess cross‐reactivity, in which pooled sera were spiked with several proteins in serial dilution and the effect on related and neighboring proteins evaluated. A protein inhibition assay enables the confirmation of specificity of an antibody against the target protein and usually is conducted using several proteins to confirm both the antibody's specificity to the target protein and to assess potential cross‐reactivity.34 Two different protein inhibition groups were used, each containing 2 different proteins. Group 1 consisted of Blattella germanica (Bla g 1) and Rumex crispus (Rum cr), and group 2 consisted of Penicillium notatum (Pen ch) and Acinetobacter gerneri (Aci g). Interestingly, decreased fluorescence from proteins other than those targeted also was observed, indicating either nonspecific inhibition or some similarity among the allergenic components of the proteins. Group 1 showed no nonspecific binding in the bacteria, arthropod, or fungi groups, but cross‐reactivity was seen among grass pollens, most notably Cynodon dactylon, R crispus, Zea mays, and Anthoxanthum odoratum.
Group 2 demonstrated no nonspecific binding in the bacteria, arthropod, and pollen groups, but cross‐reactivity was seen among Aspergillus niger, Aspergillus versicolor, Penicillium expansum, P notatum, Aspergillus nidulans and A fumigatus (Figure 4).
Figure 4.
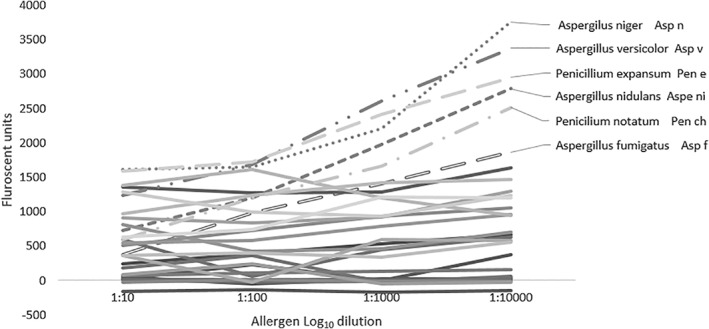
Group 2 inhibition group using pooled sera (n = 3 severe equine asthma; n = 3 control). Group 2 proteins (target protein: Penicilium notatum Pen ch) deominstrated altered immunoglobulin E fluorescence results between serial dilutions for Aspergillus niger, Aspergillus versicolor, Penicillium expansum, P notatum, Aspergillus nidulans and Aspergillus fumigatus, indicating cross‐reactivity/nonspecific binding
3.4. Allergen comparison
Whether or not the prototype array was able to identify novel allergens associated with SEA was assessed using Z‐tests with Benjamini‐Hochberg adjusted P‐values among 6 SEA and 6 control sera samples. The results shown in Table 2 confirmed the ability to conduct IgE profiling and identify potential SEA allergens using microarray methods. As expected, it confirmed the presence of fungi and mite as main reactants in the SEA population, while also identifying an association with pollen which has not been implicated previously.
Table 2.
Z‐test results with Benjamini‐Hochberg corrected P‐values showing all statistically significant allergen between the severe equine asthma (n = 6) and control group (n = 6; P = .05)
| Name | Benjamini‐Hochberg P‐value | |
|---|---|---|
| Dermatophagoides farinae | Der f 2 | .0004 |
| Blattella germanica | Bla g 5 | .0004 |
| Aspergillus restrictus | Asp r 1 | .002 |
| Linum usitatissimum | Lin us [pollen] | .009 |
| Dermatophagoides pteronyssinus | Der p 7 | .01 |
| B germanica | Bla g 5 | .01 |
| Hevea brasiliensis | Hev b 11 | .01 |
| Triticum polonicum | Tri tp | .02 |
| H brasiliensis | Hev b 5.0101 | .02 |
| Penicillium notatum | Pen ch | .02 |
| Actinidia chinensis | Act c 5 | .02 |
| Malassezia pachydermatis | Mala p | .03 |
| Actinidia deliciosa | Act d 11 | .04 |
| Olea europaea | Ole e 2 | .04 |
| Anthoxanthum odoratum | Ant o [pollen] | .04 |
| H brasiliensis | Hev b 6.02 | .04 |
| Parietaria judaica | Par j 1 | .04 |
| Alternaria alternata | Alt a 1 | .04 |
| Triticum turgidum ssp. durum | Tri td | .05 |
| Aspergillus fumigatus | Asp f | .05 |
4. DISCUSSION
We previously have determined the sensitivity and specificity of microarrays in the diagnosis of insect bite hypersensitivity,22 confirming the high discriminatory power of complex extracts and pure recombinant Culicoides proteins associated with the allergy. Based on these principles, an array was constructed to enable multiallergen testing and assess the complex sensitization of profiles associated with SEA in a single assay, based on proteins in the environment of the horse. The use of protein extracts was essential because very few proteins have been assessed in relation to SEA to date. Therefore, in the early stages of development, we used a range of extracts to maximize coverage in combination with pure proteins, where available or where the allergen has been previously associated, thus simultaneously maintaining specificity. Furthermore, we observed comparable accuracy between natural extracts and recombinant allergens, whereas some studies have indicated that the use of the recombinant component alone may be insufficient for some allergens.37 Although the eventual goal will be to move toward component‐resolved diagnostics utilizing individual allergen molecules for increased sensitivity and minimizing cross‐reactivity, the genus and species must be identified to enable the production of pure proteins. Component‐resolved diagnostics offers markedly increased accuracy over routine diagnostic tests (eg, skin prick and specific IgE determination),38 and enables the accurate selection of allergens to be used for allergen immunotherapy.39 Moreover, the identification of sensitization to pure proteins will assist in development of diagnostic tests and treatments. This microarray approach has several advantages. It allows substantial allergen profiling with minimal sample, collection of less invasive readily obtained in vivo samples, permits automation, and enables the generation of mathematical predictive models to assist in clinical allergy diagnosis.
Protein microarray methods primarily consist of 2 steps: first, the printing of proteins onto the nitrocellulose slides, and second, the profiling of equine IgE. Printing methods are well established; therefore, the latter was optimized to enable analysis of SEA BALF and sera samples. Although it has been suggested that developing technology means sensitivity is such that many immunoglobulin isotypes in BALF can be assessed unconcentrated to the nanogram or microgram, it is often not possible to detect allergen‐specific IgE in BALF because of the low concentration of the isotype present.23, 40 Therefore, concentration techniques often must be employed to assess BALF IgE. Lyophilization41 and centrifugal filtration methods42, 43 have been successfully utilized to concentrate BALF for immunoglobulin analysis, but certain techniques, such as ammonium sulfate precipitation, can result in denaturation of the liable epitopes.44 Similarly, lyophilizing BALF samples without desalting results in high concentrations of sodium chloride, which can denature proteins. The most commonly utilized BALF concentration technique is the centrifugal filter (Amicon Ultra‐15 or Centricon‐10) to a 10‐fold concentration of immunoglobulin,42, 45, 46, 47, 48, 49 but, even this technique results in a 20% loss of specific IgE and IgG.46
On collection of BALF for immunoglobulin analysis, protease inhibitors must be added to prevent proteolytic cleavage of proteins, which would otherwise leave the immunoglobulin unviable. Similarly, immunoglobulin in BALF is markedly affected by freeze–thaw cycles, and the inclusion of a cryopreservative in the form of glycerol has been shown to be effective.50 Therefore, glycerol was added as a cryoprotectant to a final concentration of 20% to help stabilize the proteins and prevent formation of ice crystals during freezing that destroy protein structure. Unfortunately, samples containing cryopreservative tend to thaw during lyophilization, meaning it was not possible to achieve a 40‐fold concentration using this technique. To avoid this problem and remove sodium chloride, samples underwent a buffer exchange using a PD‐10 column before lyophilization. Bronchoalveolar lavage concentrations and techniques were trialed, varying hybridization temperatures and times were assessed, and BALF/sera analyzed. The highest binding capacity was observed with overnight incubation at 4°C. When using BALF, the highest binding was seen using the Amicon filter and the PD‐10 elution column/lyophilizing at a 40‐fold final concentration. The Amicon filter technique was quicker, easier, decreased the risk of contamination, and had previously been utilized, and, therefore, this technique was selected. Interestingly, during optimization, a biased response toward pollens was observed. Plants are polyploids and show a large number of gene duplications; hence, high cross‐reactivity among species generally is observed. It is our experience that pollen response in humans and other animals is commonly amplified.51
Because of the significant correlation of BALF and sera (average R 2 = 0.75), sera were used in subsequent analysis because of the ease of use. Previous studies comparing the specific IgE profiles of BALF and sera using ELISA techniques have been limited and contradictory to date. Previous authors concluded that although BALF may be valuable for analysis, sera was of little clinical relevance.21, 23 Here, we demonstrated the ability to profile unconcentrated sera instead of BALF to assess potential allergens. This approach has several advantages because serum is far easier to collect, store, and prepare for analysis in comparison to BALF. Collection also is less stressful for the horse. Moreover, it holds further potential in the use of diagnostic microarrays because serum is far more stable to ship for analysis.52
Repeatability is an essential factor in the development of new diagnostic tests. Therefore, the effects of printing lot and mouse anti‐horse IgE 3H10 sources were assessed. These results confirmed reproducibility among printing lots (average R 2 = 0.97). The original mouse anti‐horse IgE 3H10 used was that from a previous study.40 The commercial availability of antibodies is essential in diagnostic tests; therefore, the 3H10 clone40 was compared with the commercially available BioRad 3H10 (product # MCA5982GA), confirming reproducibility with the commercially available clone (R 2 = 0.90).
Specificity is an important aspect of protein microarrays, which was confirmed by a protein inhibition assay. In this assay, some cross‐reactivity was seen, predominantly with grass pollens, as well as with Aspergillus and Penicillium. A previous study reported that pollens from grasses (Poaceae) often had high immunological cross‐reactivity, potentially indicating common antigenic/allergenic component(s).53 Furthermore, cross‐reactivity was identified between the genus Penicillium and Aspergillus, which is expected because taxonomically, the genera Penicillium and Aspergillus have many similarities, because both produce and contain galactomannans with similar galactofuranosyl and immunogenic side chains. Cross‐reactivity in the fungi group was seen only with whole protein extracts, emphasizing the importance of including pure proteins. Analysis of human sera in a variety of assays has indicated that A fumigatus contains determinants in common with Cladosporium, Candida, Alternaria, Trichophyton, and Epidermophyton,54 but cross‐reactivity was not identified here.
Several allergens of interest identified in our study were consistent with those previously identified as SEA‐associated by ELISA, Western blot, and RAST methods (A fumigatus, A alternate, E amstelodami, and G candidum). Ours was the largest panel of proteins tested with a controlled SEA group to date and thus identified new and relevant allergens. Several SEA‐associated allergens identified in our study previously have been associated with allergic asthma in humans (eg, Dermatophagoides farina, B germanica, Aspergillus restrictus, Dermatophagoides pteronyssinus). The novel SEA‐associated allergens identified in our study strongly implicate fungi and mite as the main reactants, as well as identifying previously unrecognized reaction with pollens. This observation confirms the future potential of specific IgE as a biomarker for the serological diagnosis of SEA.
Our results have established a reliable protein microarray for large‐scale IgE profiling of environmental proteins of horses, confirming identified SEA‐associated allergens and elucidating a range of previously unidentified allergens. The technique is sufficiently sensitive and specific to differentiate between sensitized allergens in SEA and in control horses. Furthermore, the developed serological assay enables accurate identification of an individual horse's sensitization profile. This information provides a reliable, fast, and repeatable method for screening a wide variety of potential allergens found in the stable environment in a miniaturized and affordable format, while offering a platform to support management and treatment of this debilitating respiratory disorder in horses.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Approval by the Royal Agricultural University's Animal Welfare Committee.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Appendix S1: Supplementary Information
White S, Moore‐Colyer M, Marti E, et al. Development of a comprehensive protein microarray for immunoglobulin E profiling in horses with severe asthma. J Vet Intern Med. 2019;33:2327–2335. 10.1111/jvim.15564
Funding information Fred And Marjorie Sainsbury Charitable Trust; HAYGAIN; The Royal Agricultural University
REFERENCES
- 1. Hotchkiss JW, Reid SWJ, Christley RM. A survey of horse owners in Great Britain regarding horses in their care. Part 1: horse demographic characteristics and management. Equine Vet J. 2007;39(4):294‐300. [DOI] [PubMed] [Google Scholar]
- 2. Moran G, Folch H. Recurrent airway obstruction in horses—an allergic inflammation: a review. Vet Med (Praha). 2011;56(1):1‐13. [Google Scholar]
- 3. Gerber V, Swinburne JE, Blott SC, et al. Genetics of recurrent airway obstruction (RAO). Dtsch Tierarztl Wochenschr. 2008;115(7):271‐275. [PubMed] [Google Scholar]
- 4. Miskovic M, Couëtil LL, Thompson CA. Lung function and airway cytologic profiles in horses with recurrent airway obstruction maintained in low‐dust environments. J Vet Intern Med. 2007;21(5):1060‐1066. [DOI] [PubMed] [Google Scholar]
- 5. Pirie RS. Recurrent airway obstruction: a review. Equine Vet J. 2014;46(3):276‐288. [DOI] [PubMed] [Google Scholar]
- 6. Leclere M, Lavoie‐Lamoureux A, Joubert P, et al. Corticosteroids and antigen avoidance decrease airway smooth muscle mass in an equine asthma model. Am J Respir Cell Mol Biol. 2012;47(5):589‐596. [DOI] [PubMed] [Google Scholar]
- 7. Bogacka E, Jahnz‐Rózyk K. Allergy to fungal antigens. Pol Merkur Lekarski. 2003;14(83):381‐384. [PubMed] [Google Scholar]
- 8. Künzle F, Gerber V, Van Der Haegen A, Wampfler B, Straub R, Marti E. IgE‐bearing cells in Bronchoalveolar lavage fluid and allergen‐specific IgE levels in sera from RAO‐affected horses. J Vet Med A. 2007;54(1):40‐47. [DOI] [PubMed] [Google Scholar]
- 9. Morán G, Ojeda G, Diedrichs K, Ortloff A, Barria M, Folch H. Inhalation of Aspergillus fumigatus spores induces airway inflammation in mice in a similar manner as observed in recurrent airway obstruction in horses. Arch Med Vet. 2011;43(2):163‐171. [Google Scholar]
- 10. Pirie RS, Dixon PM, McGorum BC. Endotoxin contamination contributes to the pulmonary inflammatory and functional response to Aspergillus fumigatus extract inhalation in heaves horses. Clin Exp Allergy. 2003;33(9):1289‐1296. [DOI] [PubMed] [Google Scholar]
- 11. Tahon L, Baselgia S, Gerber V, et al. In vitro allergy tests compared to intradermal testing in horses with recurrent airway obstruction. Vet Immunol Immunopathol. 2009;127(1–2):85‐93. [DOI] [PubMed] [Google Scholar]
- 12. Jackson CA, Berney C, Jefcoat AM, Robinson NE. Environment and prednisone interactions in the treatment of recurrent airway obstruction (heaves). Equine Vet J. 2000;32(5):432‐438. [DOI] [PubMed] [Google Scholar]
- 13. Cornelisse CJ, Robinson NE. Glucocorticoid therapy and the risk of equine laminitis. Equine Vet Educ. 2013;25(1):39‐46. [Google Scholar]
- 14. Thomson JR, McPherson EA. Chronic obstructive pulmonary disease in the horse. 2: therapy. Equine Vet J. 1983;15(3):207‐210. [DOI] [PubMed] [Google Scholar]
- 15. Couëtil LL, Chilcoat CD, DeNicola DB, Clark SP, Glickman NW, Glickman LT. Randomized, controlled study of inhaled fluticasone propionate, oral administration of prednisone, and environmental management of horses with recurrent airway obstruction. Am J Vet Res. 2005;66(10):1665‐1674. [DOI] [PubMed] [Google Scholar]
- 16. Gerber H, Hockenjos P, Lazary S, Kings M, de Weck A. Histamine release from equine leucocytes provoked by fungal allergens. Dtsch Tierarztl Wochenschr. 1982;89(7):267‐270. [PubMed] [Google Scholar]
- 17. Dirscherl P, Grabner A, Buschmann H. Responsiveness of basophil granulocytes of horses suffering from chronic obstructive pulmonary disease to various allergens. Vet Immunol Immunopathol. 1993;38(3–4):217‐227. [DOI] [PubMed] [Google Scholar]
- 18. Hare JE, Viel L, Conlon PD, Marshall JS. In vitro allergen‐induced degranulation of pulmonary mast cells from horses with recurrent airway obstruction (heaves). Am J Vet Res. 1999;60(7):841‐847. [PubMed] [Google Scholar]
- 19. McGorum BC, Dixon PM, Halliwell RE. Quantification of histamine in plasma and pulmonary fluids from horses with chronic obstructive pulmonary disease, before and after “natural (hay and straw) challenges”. Vet Immunol Immunopathol. 1993;36(3):223‐237. [DOI] [PubMed] [Google Scholar]
- 20. McPherson EA, Lawson GHK, Murphy JR, Nicholson JM, Breeze RG, Pirie HM. Chronic obstructive pulmonary disease (COPD) in horses: aetiological studies: responses to intradermal and inhalation antigenic challenge. Equine Vet J. 1979;11(3):159‐166. [DOI] [PubMed] [Google Scholar]
- 21. Halliwell REW, McGorum BC, Irving P, Dixon PM. Local and systemic antibody production in horses affected with chronic obstructive pulmonary disease. Vet Immunol Immunopathol. 1993;38(3–4):201‐215. [DOI] [PubMed] [Google Scholar]
- 22. Marti E, Wang X, Jambari NN, et al. Novel in vitro diagnosis of equine allergies using a protein array and mathematical modelling approach: a proof of concept using insect bite hypersensitivity. Vet Immunol Immunopathol. 2015;167(3–4):171‐177. [DOI] [PubMed] [Google Scholar]
- 23. Eder C, Crameri R, Mayer C, et al. Allergen‐specific IgE levels against crude mould and storage mite extracts and recombinant mould allergens in sera from horses affected with chronic bronchitis. Vet Immunol Immunopathol. 2000;73(3–4):241‐253. [DOI] [PubMed] [Google Scholar]
- 24. Niedzwiedz A, Jaworski Z, Kubiak K. Serum concentrations of allergen‐specific IgE in horses with equine recurrent airway obstruction and healthy controls assessed by ELISA. Vet Clin Pathol [Internet]. 2015;44(3):391‐396. [DOI] [PubMed] [Google Scholar]
- 25. Schmallenbach K, Rahman I, Sasse HH, et al. Studies on pulmonary and systemic Aspergillus fumigatus‐specific IgE and IgG antibodies in horses affected with chronic obstructive pulmonary disease (COPD). Vet Immunol Immunopathol. 1998;66(3–4):245‐256. [DOI] [PubMed] [Google Scholar]
- 26. Crameri R, Garbani M, Rhyner C, Huitema C. Fungi: the neglected allergenic sources. Allergy. 2014;69(2):176‐185. [DOI] [PubMed] [Google Scholar]
- 27. Fall BI, Nießner R. Detection of known allergen‐specific IgE antibodies by immunological methods Methods in Molecular Biology. Clifton, NJ: Springer. 2009;509:107‐122. 10.1007/978-1-59745-372-1_7. [DOI] [PubMed] [Google Scholar]
- 28. Jambari NN, Wang X, Alcocer M. Protein Microarray‐Based IgE Immunoassay for Allergy Diagnosis. New York, NY: Humana Press; 2017:129‐137. [DOI] [PubMed] [Google Scholar]
- 29. Jahn‐Schmid B, Harwanegg C, Hiller R, et al. Allergen microarray: comparison of microarray using recombinant allergens with conventional diagnostic methods to detect allergen‐specific serum immunoglobulin E. Clin Exp Allergy. 2003;33(10):1443‐1449. [DOI] [PubMed] [Google Scholar]
- 30. Renault NK, Gaddipati SR, Wulfert F, et al. Multiple protein extract microarray for profiling human food‐specific immunoglobulins a, M, G and E. J Immunol Methods. 2011;364(1–2):21‐32. [DOI] [PubMed] [Google Scholar]
- 31. Jeon H, Jung JH, Kim Y, Kwon Y, Kim ST. Allergen microarrays for in vitro diagnostics of allergies: comparison with ImmunoCAP and AdvanSure. Ann Lab Med. 2018;38(4):338‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ivester KM, Couëtil LL, Moore GE, Zimmerman NJ, Raskin RE. Environmental exposures and airway inflammation in young thoroughbred horses. J Vet Intern Med. 2014;28(3):918‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eder C, Curik I, Brem G, et al. Influence of environmental and genetic factors on allergen‐specific immunoglobulin‐E levels in sera from Lipizzan horses. Equine Vet J. 2001;33(7):714‐720. [DOI] [PubMed] [Google Scholar]
- 34. Lin J, Bardina L, Shreffler WG, et al. Development of a novel peptide microarray for large‐scale epitope mapping of food allergens. J Allergy Clin Immunol. 2009;124(2):315‐322, 322.e1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Froidure A, Mouthuy J, Durham SR, Chanez P, Sibille Y, Pilette C. Asthma phenotypes and IgE responses. Eur Respir J. 2016;47(1):304‐319. [DOI] [PubMed] [Google Scholar]
- 36. Wilson AD, Harwood L, Torsteinsdottir S, Marti E. Production of monoclonal antibodies specific for native equine IgE and their application to monitor total serum IgE responses in Icelandic and non‐Icelandic horses with insect bite dermal hypersensitivity. Vet Immunol Immunopathol. 2006;112(3–4):156‐170. [DOI] [PubMed] [Google Scholar]
- 37. Smoldovskaya O, Feyzkhanova G, Arefieva A, et al. Allergen extracts and recombinant proteins: comparison of efficiency of in vitro allergy diagnostics using multiplex assay on a biological microchip. Allergy Asthma Clin Immunol. 2016;12(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dodig S, Čepelak I. The potential of component‐resolved diagnosis in laboratory diagnostics of allergy. Biochem Med. 2018;28(2):020501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matricardi P, Stringari G, Caffarelli C, Asero R, Dondi A, Tripodi S. The impact of component resolved diagnosis on allergen‐specific immunotherapy prescription in children with pollen‐related allergic rhinitis. J Allergy Clin Immunol. 2014;133(2):AB45. [DOI] [PubMed] [Google Scholar]
- 40. Wilson DR, Merrett TG, Varga EM, et al. Increases in allergen‐specific IgE in BAL after segmental allergen challenge in atopic asthmatics. Am J Respir Crit Care Med. 2002;165(1):22‐26. [DOI] [PubMed] [Google Scholar]
- 41. Wilkie BN, Markham RJ. Sequential titration of bovine lung and serum antibodies after parenteral or pulmonary inoculation with Pasteurella haemolytica. Am J Vet Res. 1979;40(12):1690‐1693. [PubMed] [Google Scholar]
- 42. Peebles RS, Hamilton RG, Lichtenstein LM, et al. Antigen‐specific IgE and IgA antibodies in bronchoalveolar lavage fluid are associated with stronger antigen‐induced late phase reactions. Clin Exp Allergy. 2001;31(2):239‐248. [DOI] [PubMed] [Google Scholar]
- 43. Koh YY, Park Y, Lee HJ, Kim CK. Levels of interleukin‐2, interferon‐gamma, and interleukin‐4 in bronchoalveolar lavage fluid from patients with mycoplasma pneumonia: implication of tendency toward increased immunoglobulin E production. Pediatrics. 2001;107(3):E39. [DOI] [PubMed] [Google Scholar]
- 44. Marti E, Peveri P, Griot‐Wenk M, et al. Chicken antibodies to a recombinant fragment of the equine immunoglobulin epsilon heavy‐chain recognising native horse IgE. Vet Immunol Immunopathol. 1997;59(3–4):253‐270. [DOI] [PubMed] [Google Scholar]
- 45. Bartlett JA, Albertolle ME, Wohlford‐Lenane C, et al. Protein composition of bronchoalveolar lavage fluid and airway surface liquid from newborn pigs. Am J Physiol Lung Cell Mol Physiol. 2013;305(3):L256‐L266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peebles RS, Liu MC, Adkinson NF, Lichtenstein LM, Hamilton RG. Ragweed‐specific antibodies in bronchoalveolar lavage fluids and serum before and after segmental lung challenge: IgE and IgA associated with eosinophil degranulation. J Allergy Clin Immunol. 1998;101(2 Pt 1):265‐273. [DOI] [PubMed] [Google Scholar]
- 47. Lamkhioued B, Renzi PM, Abi‐Younes S, et al. Increased expression of eotaxin in bronchoalveolar lavage and airways of asthmatics contributes to the chemotaxis of eosinophils to the site of inflammation. J Immunol. 1997;159(9):4593‐4601. [PubMed] [Google Scholar]
- 48. Richard EA, Depecker M, Defontis M, et al. Cytokine concentrations in Bronchoalveolar lavage fluid from horses with neutrophilic inflammatory airway disease. J Vet Intern Med. 2014;28(6):1838‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Foster MW, Thompson JW, Que LG, et al. Proteomic analysis of human bronchoalveolar lavage fluid after subsgemental exposure. J Proteome Res. 2013;12(5):2194‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kneidinger N, Warszawska J, Schenk P, et al. Storage of bronchoalveolar lavage fluid and accuracy of microbiologic diagnostics in the ICU: a prospective observational study. Crit Care. 2013;17(4):R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang X, Tsilochristou O, Perna S, et al. Evolution of the IgE and IgG repertoire to a comprehensive array of allergen molecules in the first decade of life. Allergy. 2018;73(2):421‐430. [DOI] [PubMed] [Google Scholar]
- 52. Rodríguez‐Capote K, Schnabl KL, Maries OR, Janzen P, Higgins TN. Stability of specific IgE antibodies to common food and inhalant allergens. Clin Biochem. 2016;49(18):1387‐1389. [DOI] [PubMed] [Google Scholar]
- 53. Sridhara S, Singh BP, Kumar L, Verma J, Gaur SN, Gangal SV. Antigenic and allergenic relationships among airborne grass pollens in India. Ann Allergy Asthma Immunol. 1995;75(1):73‐79. [PubMed] [Google Scholar]
- 54. Vojdani A. Cross‐reactivity of aspergillus, Penicillium, and Stachybotrys antigens using affinity‐purified antibodies and immunoassay. Arch Environ Health. 2004;59(5):256‐265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Information


