Abstract
Doxorubicin (Dox) is a widely used antineoplastic agent that can cause heart failure. Dox cardiotoxicity is closely associated with mitochondrial damage. Mitochondrial fission and mitophagy are quality control mechanisms that normally help maintain a pool of healthy mitochondria. However, unchecked mitochondrial fission and mitophagy may compromise the viability of cardiomyocytes, predisposing them to cell death. Here, we tested this possibility by using Dox-treated H9c2 cardiac myoblast cells expressing either the mitochondria-targeted fluorescent protein MitoDsRed or the novel dual-fluorescent mitophagy reporter mt-Rosella. Dox induced mitochondrial fragmentation as shown by reduced form factor, aspect ratio, and mean mitochondrial size. This effect was abolished by short interference RNA–mediated knockdown of dynamin-related protein 1 (DRP1), a major regulator of fission. Importantly, DRP1 knockdown decreased cell death as indicated by the reduced number of propidium iodide–positive cells and the cleavage of caspase-3 and poly (ADP-ribose) polymerase. Moreover, DRP1-deficient mice were protected from Dox-induced cardiac damage, strongly supporting a role for DRP1-dependent mitochondrial fragmentation in Dox cardiotoxicity. In addition, Dox accelerated mitophagy flux, which was attenuated by DRP1 knockdown, as assessed by the mitophagy reporter mt-Rosella, suggesting the necessity of mitochondrial fragmentation in Dox-induced mitophagy. Knockdown of parkin, a positive regulator of mitophagy, dramatically diminished Dox-induced cell death, whereas overexpression of parkin had the opposite effect. Together, these results suggested that Dox cardiotoxicity was mediated, at least in part, by the increased mitochondrial fragmentation and accelerated mitochondrial degradation by the lysosome. Strategies that limit mitochondrial fission and mitophagy in the physiologic range may help reduce Dox cardiotoxicity.—Catanzaro, M. P., Weiner, A., Kaminaris, A., Li, C., Cai, F., Zhao, F., Kobayashi, S., Kobayashi, T., Huang, Y., Sesaki, H., Liang, Q. Doxorubicin-induced cardiomyocyte death is mediated by unchecked mitochondrial fission and mitophagy.
Keywords: cardiotoxicity, anti-tumor agents, mitochondrial quality control, heart failure
Doxorubicin (Dox) is a potent chemotherapeutic agent widely used to treat a variety of cancers, including leukemias and solid tumors. Unfortunately, Dox treatment can also result in irreversible congestive heart failure (1, 2). This problem is more pronounced when used in childhood cancers, creating a lifelong health burden for cancer survivors (3, 4). Dox is concentrated in the mitochondria and forms complexes with iron that is increased in mitochondria, leading to the production of reactive oxygen species (ROS) (5). Both dexrazoxane, an iron chelator, and ATP binding cassette subfamily B member 8 (ABCB8), a mitochondrial protein that facilitates iron export, are able to decrease mitochondrial iron levels and protect against Dox-induced cardiomyopathy (6), suggesting iron mitochondrial accumulation as a major mechanism of Dox cardiotoxicity. However, although dexrazoxane is the only drug currently approved for reducing Dox-dependent cardiac damage (7, 8), its use has been limited to patients with breast cancer on high doses of Dox because of possible myelosuppression and secondary malignancies (9, 10). Likewise, although ABCB8 can reduce Dox cardiotoxicity, it also confers resistance of tumor cells to Dox chemotherapy (11, 12), suggesting that increasing ABCB8 may not be a viable approach to reducing Dox cardiotoxicity in patients with cancer. Therefore, it is crucial to identify new strategies to protect against Dox-induced heart damage without compromising the antitumor activity of Dox.
Dox cardiotoxicity is closely associated with mitochondrial injury, which is characterized by an early loss of mitochondrial membrane potential followed by dysregulation of mitochondrial quality control mechanisms (13–20). Mitochondria are constantly undergoing fission and fusion, contributing to mitochondrial quality control, but an imbalance between the 2 can lead to cardiac injury. Fusion is controlled by mitofusin (Mfn)1, Mfn2, and optic atrophy 1 (Opa1), whereas fission is controlled by dynamin-related protein 1 (DRP1), mitochondrial fission protein 1 (Fis1), mitochondrial fission factor, and mitochondrial dynamics proteins 49 and 51 (21). Mitochondrial fusion events allow for the mixing of the contents of different mitochondria, thereby diluting injured mitochondrial proteins and DNA. Fission events produce smaller fragmented mitochondrial subunits, which permit even distribution of mitochondria in a cell. Nonetheless, excessive fission has been shown to contribute to cardiac injury under certain conditions, such as pressure-overload, ischemia, and Dox treatment (22–26). Indeed, mitochondrial division inhibitor 1 (Mdivi-1), a chemical inhibitor of mitochondrial fission, and short interference RNA (siRNA)–mediated knockdown of Drp1, a major promoter of mitochondrial fission, were able to attenuate Dox cardiotoxicity in Langendorff-perfused rat hearts (25) and in cultured cardiomyocytes (26), respectively. However, Mdivi-1 activated the prosurvival protein kinase B (Akt) pathway in the heart, suggesting that the cardioprotective effects of Mdivi-1 may not necessarily be mediated solely by the inhibition of mitochondrial fragmentation. Although DRP1 knockdown can inhibit Dox-induced cell injury in neonatal rat cardiomyocytes, it remains to be determined if inactivation of DRP1 can reduce Dox cardiotoxicity in vivo.
Mitophagy is another important mitochondrial quality control mechanism that maintains a healthy mitochondrial network by degrading unwanted or otherwise dysfunctional mitochondria through the autophagy-lysosome pathway. Mitophagy is regulated by a well-established pathway composed of phosphatase and tensin homolog–induced kinase 1 and parkin, an E3 ubiquitin ligase that decorates mitochondria for degradation in the lysosome. Mitophagy is also controlled by a number of adaptors or receptors that are found in cytosol or on mitochondrial membranes (27). Mitophagy is generally believed to play protective roles under various normal and disease conditions (28). However, emerging evidence also suggests that mitophagy can become detrimental, leading to cell death under certain conditions. For example, mitophagy was required for ceramide-induced cancer cell death, which involved microtubule-associated protein light chain 3 (LC3)–ceramide interactions at the mitochondria (29). Cigarette smoke induced mitochondrial dysfunction and triggered mitophagy, causing lung epithelial cell death (30). Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells (31). In the heart, ablation of parkin rescued the cardiac phenotype of DRP1-homozygous knockout mice, suggesting that parkin-mediated mitophagy contributed to cardiac damage in DRP1−/− mice (32). However, it remains controversial whether mitophagy contributes to or protects against Dox cardiotoxicity (33–35). In addition, fission can segregate damaged segments of mitochondria and facilitate their removal by mitophagy (36). Inhibition of fission by inactivating DRP1 often prevents mitophagy in various cell types including cardiomyocytes (37), suggesting a necessary role for fission in mitophagy. However, it is unclear if and how Dox-induced mitochondrial fragmentation would affect mitophagy in cardiomyocytes.
In the present study, we demonstrated that reducing DRP1 expression markedly attenuated Dox-induced cardiotoxic effects both in vitro and in vivo, confirming the role of mitochondrial fragmentation in cardiotoxicity. We also observed that reducing mitochondrial fragmentation effectively decreased mitophagy flux in Dox-treated cardiac myoblast cells, as shown by a novel dual fluorescence mitophagy reporter, suggesting the necessity of mitochondrial fragmentation in Dox-induced mitophagy. Knockdown of parkin, a positive regulator of mitophagy, dramatically diminished Dox-induced cell death, whereas overexpression of parkin had the opposite effect. These results collectively suggested that Dox cardiotoxicity was mediated by the increased mitochondrial fragmentation and accelerated mitochondrial degradation by the lysosome.
MATERIALS AND METHODS
Cell cultures
The H9c2 cardiac myoblast cells were obtained from the American Type Culture Collection (CRL-1446; Manassas, VA, USA) and cultured in DMEM (10-017CV; Corning, Corning, NY, VA, USA) containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin as previously described in Branco et al. (38). Cells were fed every 2–3 d and used for experiment at 80–90% confluence.
Adult mouse cardiomyocyte culture
Ventricular cardiomyocytes from adult mice were isolated as previously described with some adaptations (39). The isolated cardiomyocytes were plated at a density of 50 rod-shaped myocytes/mm2 on laminin-coated coverslips in 35-mm culture dishes and cultured for indicated time periods in a 2% CO2 incubator at 37°C.
Drugs
Dox was purchased from MilliporeSigma (D1515; Burlington, MA, USA). Dox was dissolved in saline to make 1 mM stock solution and then diluted to make a final concentration of 750 nM for H9c2 cells and 3 µM for adult mouse cardiomyocytes upon use. For the whole animal study, mice received a single dose of Dox (15 mg/kg) via intraperitoneal injection. Pepstatin A (PepA) and E64d were purchased from Research Products International (P30100, E57050; Mount Prospect, IL, USA) and dissolved in DMSO (472301; MilliporeSigma).
Western blot analysis
Cardiac tissue and cultured cells were processed for Western blot analysis as previously described (40, 41). H9c2 cells were washed once in PBS and collected in 1× SDS. Samples were boiled for 10 min, loaded onto polyacrylamide gel for electrophoresis, and then transferred to PVDF membranes. After being blocked with 5% milk dissolved in Tris-buffered saline containing 1% Tween 20 for 30 min, the blots were incubated with primary and secondary antibodies in 2.5% milk overnight at 4°C. The blots were then washed in Tris-buffered saline for 45 min and processed for chemiluminescent detection using Lumigen ECL Ultra (TMA-6; Lumigen, Southfield, MI, USA) and the images were acquired using an Amersham Imager 600 (GE Healthcare, Waukesha, WI, USA). Protein abundance on Western blots was quantified with ImageJ [National Institutes of Health (NIH), Bethesda, MD, USA]. The antibodies against DRP-1 (sc-101270), Fis1 (sc-980900), Mfn1 (sc-166644), Mfn2 (sc-100560), and the horseradish peroxidase–conjugated secondary antibodies (sc-2004, sc-2005, sc-2020, and sc-2438) were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). The antibodies against Opa1 (ab42364) and the subunit IV of cytochrome c oxidase (COX; ab14744) were purchased from Abcam (Cambridge, MA, USA). The antibodies against poly (ADP-ribose) polymerase (PARP; 9542), cleaved caspase-3 (cCasp3; 9664), β-Actin (4967), LC3B (3868), pyruvate dehydrogenase (PDH; 2784), phosphorylated (phospho)-DRP1 (Ser616; 4494), and glyceraldehyde 3-phosphate dehydrogenase (5147) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti–phospho-PDHE1-A type I (Ser293) antibody was purchased from MilliporeSigma (ABS204).
Replication-deficient adenoviruses
The human DRP1 cDNA clone was obtained from OriGene Technologies (Rockville, MD, USA). The pLV-mitoDsRed was a gift from Dr. Pantelis Tsoulfas (University of Miami School of Medicine, Miami, FL) (42) (44386; Addgene, Watertown, MA, USA). The plasmid containing the mitophagy reporter mt-Rosella was kindly provided by Dr. Devenish (School of Biomedical Sciences, Monash University, Clayton, VIC, Australia) (43). Rosella is a dual-emission biosensor composed of a pH-stable red fluorescent protein linked to a pH-sensitive green fluorescent protein (GFP). We tagged Rosella with a mitochondrial targeting sequence from the gene that encodes the human COX subunit VIII. To generate the adenoviral vector expressing DRP1, MitoDsRed, or mt-Rosella, we amplified each insert by PCR and subcloned it into the pShuttle-CMV vector via the NotI-XhoI restriction sites. Recombinant adenoviruses expressing DRP1 (AdDRP1s), MitoDsRed (AdDsReds), or mt-Rosella (Admt-Rosellas) were then generated and amplified using the AdEasy Adenoviral Vector System (240009; Stratagene, San Diego, CA, USA). Unless otherwise indicated, H9c2 cardiac blast cells were infected with adenovirus at a multiplicity of infection of 200 for 24 h before drug treatment. The adult mouse cardiomyocytes were infected with AdDsRed at multiplicity of infection of 2000 and cultured for 36 h before Dox treatment.
siRNA gene silencing
siRNAs targeting DRP1 mRNA and a silencer negative control siRNA were obtained from Thermo Fisher Scientific (Waltham, MA, USA). The transfection was performed following the protocol provided by Thermo Fisher Scientific with minor modifications. The culture media were replaced 24 h before transfection with 10% fetal bovine serum in DMEM. Transfections were performed with a mixture of 3 different siRNA oligos targeting DRP1, each with a concentration of 16.67 nM. siRNA oligos were mixed with Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific) and incubated for 15 min. The mixture was then added to the culture dish with media. After 24–36 h, the transfected cells were treated with Dox. Gene silencing efficiency was evaluated with Western blot analysis.
Cell death assay
Propidium iodide (PI), which enters dead cells through disrupted membranes, was used to measure cell death. PI was added directly to the culture medium at a 2 μg/ml working concentration. After 10 min incubation, the cells were examined with a fluorescent microscope and photographed under phase contrast and fluorescent conditions. Five images were obtained per treatment group. The number of PI-positive cells (stained red) were counted and expressed as a percentage against the total number of cells examined per treatment. Apoptotic cell death was determined by Western blot analysis of the apoptotic proteins, cCasp3, and cleaved PARP as described above.
Mitochondrial morphology analysis
Raw images of AdDsRed-infected H9c2 cells or adult mouse cardiomyocytes were obtained with confocal microscopy. Images were optimized for morphometric analysis using methods as previously described (34, 35). Using ImageJ, red-green-blue images were converted to 8 bit and were contrast enhanced. A convolve filter was applied to highlight mitochondrial structures. Small background particles introduced using this method were removed with the “remove outliers” function of ImageJ and by applying a threshold to the image using Otsu’s method.
After optimization, individual cells were outlined, and a particle analysis was performed. The following data was collected with a threshold area of 0.4 μm2 for each cell: number of particles (each representing individual mitochondria), average particle area, perimeter, major axis, and minor axis. A mean form factor (FF) value, which represents both the length and degree of branching of the cell’s mitochondria, was calculated using the formula (Perimeter2)/(4PiArea). A mean aspect ratio (AR) value, a measure of mitochondrial length, was calculated for each cell using the formula (Major Axis)/(Minor Axis). Between 5 and 8 images (totaling between 5 and 15 cells) were analyzed per treatment.
Mitophagy analysis
Dual-fluorescent images of H9c2 cells infected with Admt-Rosella were split into red and green channels, and contrast was optimized. The green channel was subtracted from the red channel using ImageJ’s Image Calculator feature, isolating the “red only” portions of the original image, which represent mitophagy foci. A threshold was applied using the Huang method to exclude small background particles introduced by the channel subtraction.
After optimization, each cell was individually outlined and analyzed. The analysis was performed with a size threshold of 1 μm2 in order to exclude small red noise particles and large red particles such as nuclear Dox aggregates, which were unlikely to represent mitophagy foci. Between 3 and 8 images (totaling between 5 and 15 cells) were analyzed per treatment. The mean numbers of mitophagy foci per cell were compared using 1-way ANOVA. For mitophagy flux analysis, experiments were duplicated with the addition of lysosomal inhibitors PepA (12.5 ng/ml) and E64D (5 ng/ml).
Animals
The DRP1 heterozygous knockout (DRP1+/−) mice were previously described in Kageyama et al. (44). The DRP1 homozygous knockout mice are embryonically lethal, and the heterozygous mice can survive into adulthood without apparent cardiac pathology at baseline as examined by echocardiography and hemodynamic measurements. Both male and female mice of 3–4 mo of age were used in the study. DRP1+/− and their wild-type littermate mice were treated with saline or a single dose of Dox (15 mg/kg body weight) and euthanized 3 d later for all assays. All animal protocols conformed to the Public Health Service Guide for Care and Use of Laboratory Animals and were approved by the New York Institute of Technology College of Osteopathic Medicine Institutional Animal Care and Use Committee.
Measurement of serum lactate dehydrogenase activity and cardiac troponin I in serum
Both lactate dehydrogenase (LDH) and cardiac troponin I (cTnI) were used to evaluate tissue damage caused by various disease conditions; cTnI is more cardiac specific. The serum LDH activity was measured using a kit from MilliporeSigma (MAK066), and the amount of cTnI that was released into the blood after cardiac injury was determined with a Mouse Cardiac Troponin I ELISA Kit (CTNI-1-US; Life Diagnostics, West Chester, PA, USA).
Oxidative injury
Carbonyl groups in the oxidized proteins and lipid peroxidation by-product 4-hydroxynonenal (4-HNE) were measured by Western blot analysis using the OxyBlot Kit from MilliporeSigma and an antibody against 4-HNE (AB5605; MilliporeSigma).
Statistical analysis
Data were presented as the means ± se. One-way ANOVA was used to analyze the differences between experimental groups, followed by Tukey’s Multiple Comparison Test using SPSS software (IBM SPSS, Chicago, IL, USA) or Prism software (GraphPad, La Jolla, CA, USA). A value of P < 0.05 was considered statistically significant.
RESULTS
Dox induced mitochondrial fragmentation in H9c2 cardiac myoblast cells
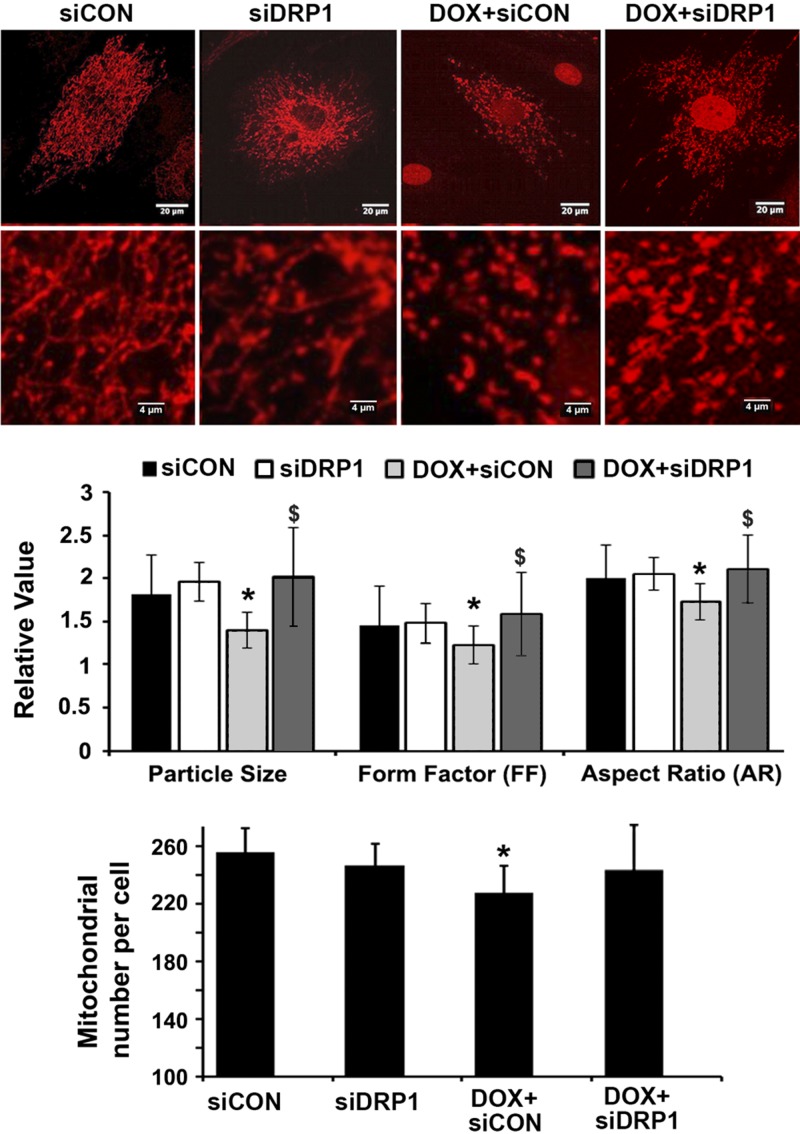
Previous studies reported various pathologic changes in Dox-treated cardiac myocytes including mitochondrial damage (25, 26). To characterize the effects of Dox on mitochondrial morphology in H9c2 cardiac myoblasts, we infected cells with AdDsRed and treated them with 750 nM Dox or saline for 24 h. Mitochondria in saline-treated cells showed elongated rod-like structures, connecting each other to form visible networks. In contrast, mitochondria in Dox-treated cells appeared smaller, shorter, and less connected, demonstrating the ability of Dox to induce mitochondrial fragmentation (Fig. 1) as characterized by the reduced mean mitochondrial particle size (P < 0.01; n = 3), FF (P < 0.01; n = 3), and AR (*P < 0.01; n = 4). Despite the increased mitochondrial fragmentation and reduced size, the total number of mitochondria was not increased by Dox treatment (lower panel of Fig. 1). Instead, Dox led to a small (11%) but significant reduction in the number of mitochondria, probably because of the increased mitochondrial degradation by mitophagy as described in Drp1 knockdown attenuated Dox-induced mitophagic flux. However, this effect was abolished by DRP1 knockdown.
Figure 1.
Drp1 siRNA attenuated Dox-induced mitochondrial fragmentation. H9c2 cells were cultured, infected with AdDsRed, transfected with siDRP1 or control siRNA (siCon), and then exposed to either Dox (750 nM) or saline for 24 h. Mitochondrial morphology was analyzed using ImageJ and the following parameters were calculated: mitochondrial particle number and size, mean FF, and AR. At least 5 images (each containing 5–15 cells) were captured per treatment over 3 separate experiments. Data are expressed as means ± se and were analyzed by ANOVA. *P < 0.01 vs. control (n = 3–4).
Dox altered the protein expressions of genes that control mitochondrial fission and fusion
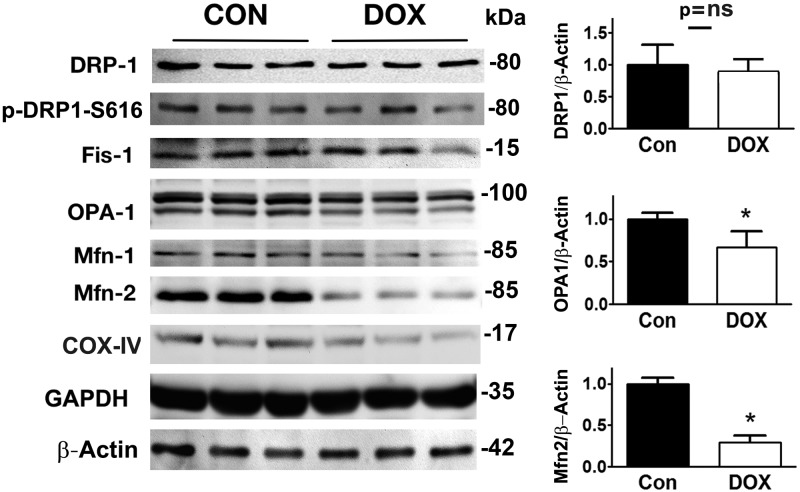
Mitochondrial morphology was determined by the balance between mitochondrial fusion and fission. We screened the factors that control mitochondrial dynamics. Surprisingly, DRP1 and Fis1, 2 major fission factors, were not increased by Dox, suggesting that DRP1 and Fis1 alone were not responsible for Dox-induced mitochondrial fragmentation. However, Dox markedly reduced the protein levels of Opa1 and MFN2, 2 factors that control the fusion process and tilt the balance in the favor of fission, leading to an increased mitochondrial fragmentation (Fig. 2).
Figure 2.
Dox altered the protein expressions of mitochondrial fission and fusion factors. H9c2 cells were treated with either Dox (750 nM) or saline for 24 h. Mitochondrial fission and fusion factors were determined by Western blot analysis. GAPDH, glyceraldegyde 3-phosphate dehydrogenase. Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.05 (n = 6 for Drp1 and n = 3 for Opa1 and Mfn2).
DRP1 knockdown attenuated mitochondrial fragmentation and cell death
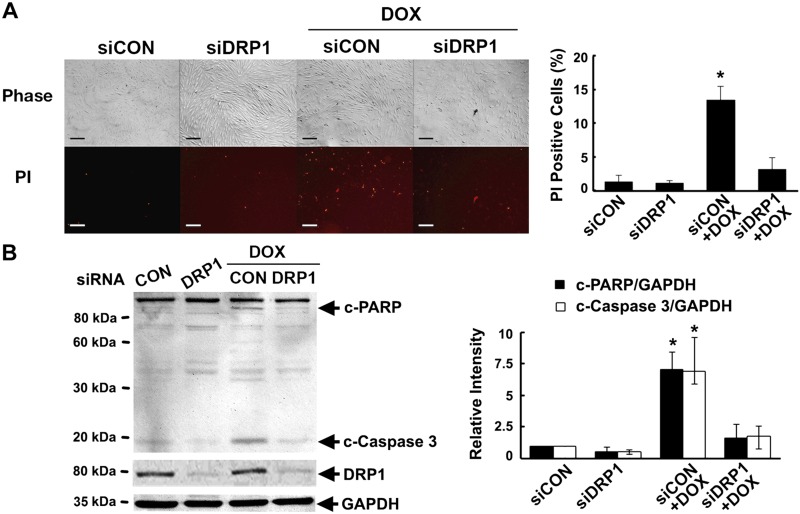
To determine the functional role of fission in Dox-induced cell death, we knocked down DRP1 expression in H9c2 cells with synthetic siRNAs. DRP1 knockdown did not affect mitochondrial morphology by itself but blocked Dox-induced mitochondrial fragmentation (Fig. 1) as demonstrated by the restoration of the elongated rod-shaped structure of the mitochondria. More importantly, DRP1 knockdown dramatically decreased Dox-induced cell death as indicated in Fig. 3 by the number of PI-positive cells (P < 0.05; n = 3) and the levels of cCasp3 (P < 0.01; n = 4) and PARP (P < 0.05; n = 4), suggesting that mitochondrial fragmentation is an essential mechanism that mediates Dox cardiotoxicity.
Figure 3.
DRP1 knockdown protected against Dox-induced cardiomyocyte death. H9c2 cells were pretreated with siDRP1 or control siRNA (siCon) for 24 h and then exposed to either Dox (750 nM) or saline for an additional 24 h. Cell death was determined by PI staining (A) and cleavage of caspase-3 and PARP (B). Scale bars, 200 μm. Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.05 vs. control (n = 3–4).
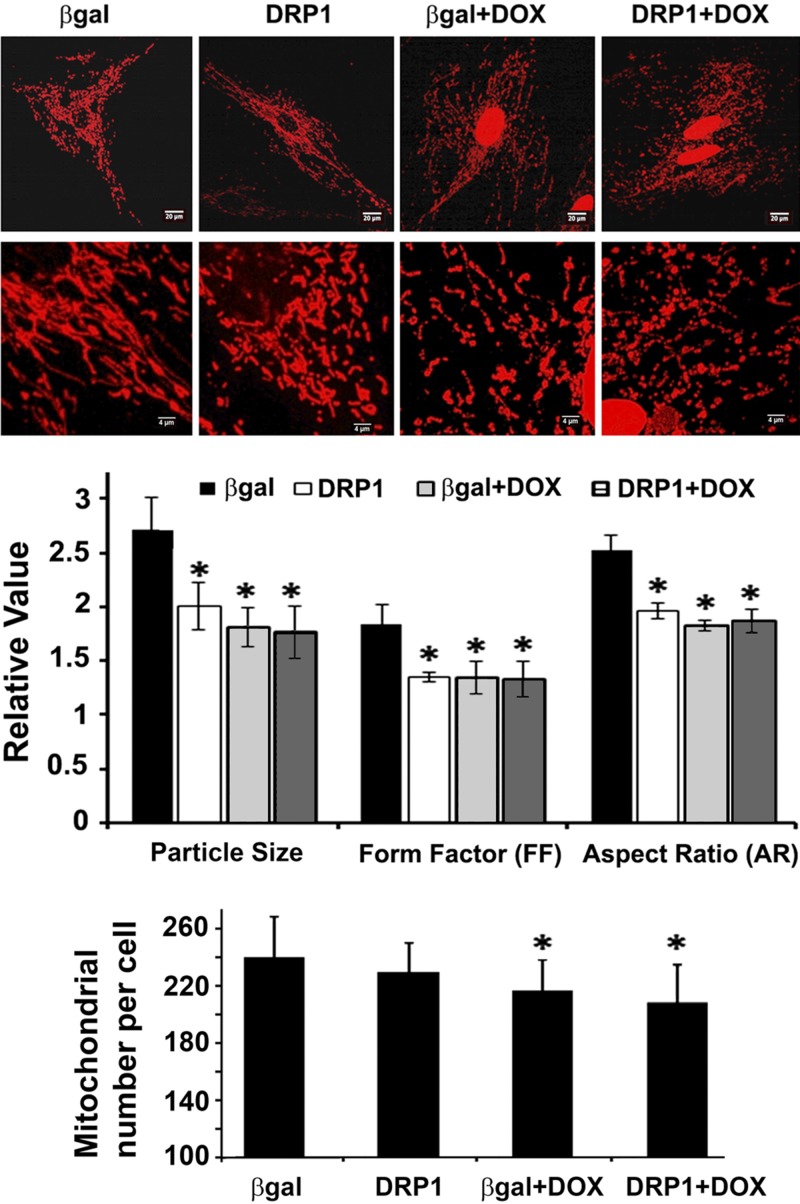
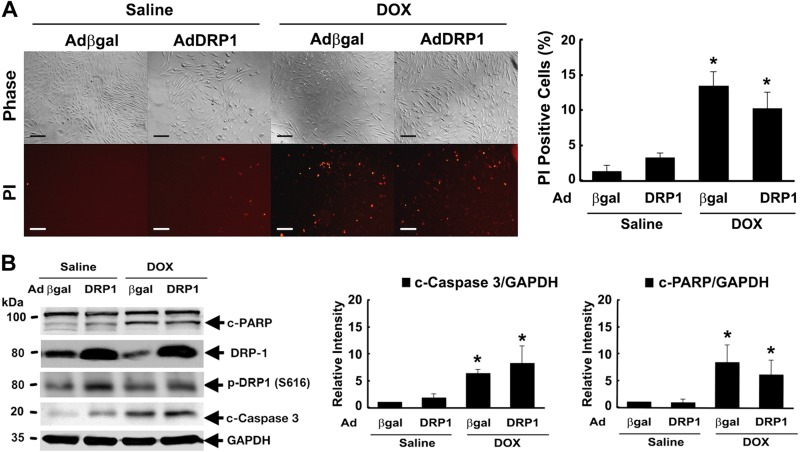
DRP1 overexpression induced mitochondrial fission but did not further increase Dox-induced mitochondrial fragmentation and cell death
We tested the ability of DRP1 to induce mitochondrial fission by using an adenovirus-mediated gene transfer. Expectedly, DRP1 overexpression per se was sufficient to trigger mitochondrial fragmentation as shown by the smaller, shorter, and less interconnected mitochondria (Fig. 4). However, DRP1 did not further increase Dox-induced mitochondrial fragmentation (Fig. 4) nor did it induce cell death by itself or exacerbate Dox-induced cell death (Fig. 5), suggesting a necessity but not sufficiency for DRP1 in Dox cardiotoxicity. Dox induced a small but significant reduction in mitochondrial number (lower panel of Fig. 4), probably because of enhanced mitophagy. However, DRP1 did not exaggerate this effect.
Figure 4.
DRP1 overexpression did not aggravate Dox-induced mitochondrial fragmentation. H9c2 cells were infected with AdDsRed for 48 h, followed by the infection of AdDRP1 or adenovirus expressing β-galactosidase (Adβ-gal) for 48 h. The cells were then exposed to either Dox (750 nM) or saline for 24 h. Mitochondrial morphology was observed with confocal microscopy and analyzed using ImageJ. The following parameters were calculated: mitochondrial particle number and size, mean FF, and AR. At least 5 images (each containing 5–15 cells) were captured per treatment over 3 separate experiments. Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.05 vs. β-gal (n = 3).
Figure 5.
DRP1 overexpression had no further effect on Dox-induced cell death. H9c2 cells were infected with AdDRP1or adenovirus expressing β-galactosidase (Adβ-gal) for 24 h and then exposed to either Dox (750 nM) or saline for 24 h. Cell death was determined by PI staining (A) and cleavage of caspase-3 and PARP (B). Scale bars, 200 μm. Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.05 vs. β-gal (n = 3).
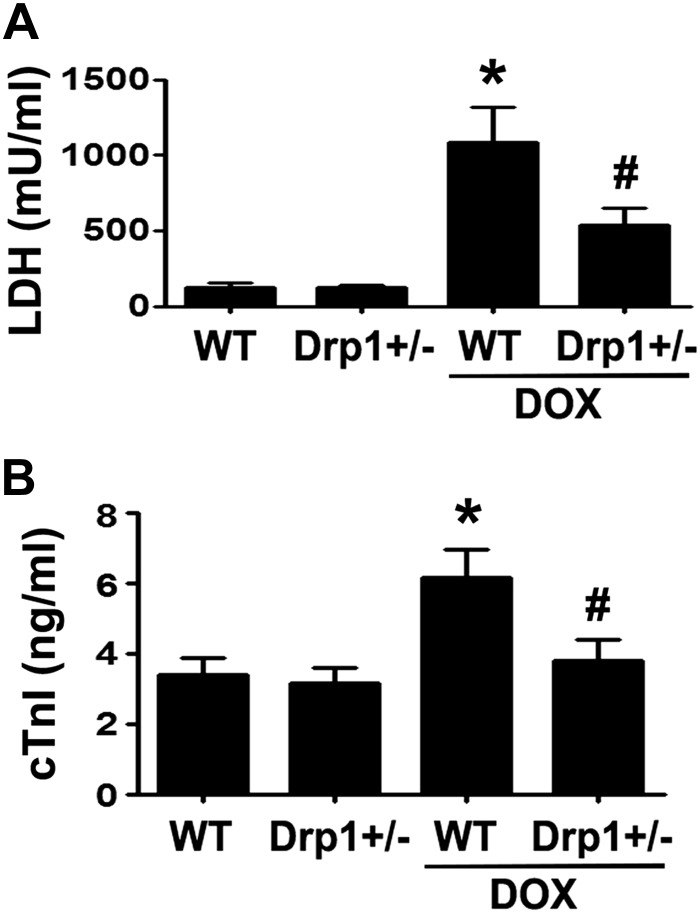
DRP1 haploinsufficiency reduced Dox cardiotoxicity in mice
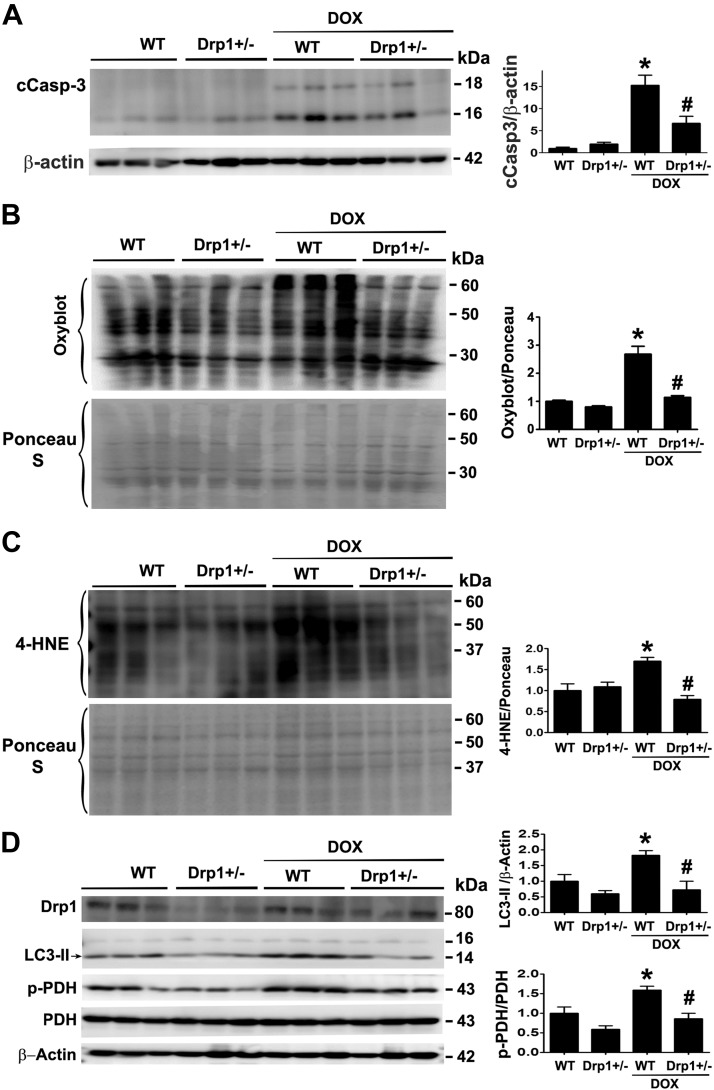
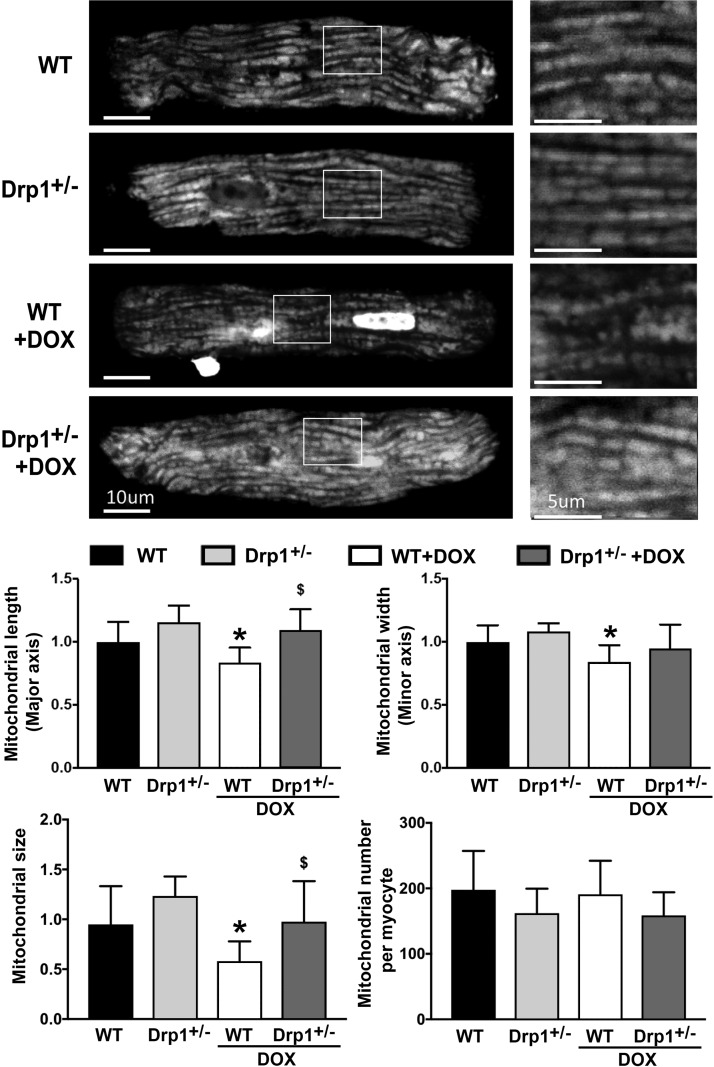
The DRP1 inhibitor Mdivi-1 has been shown to attenuate Dox cardiotoxicity in isolated rat heart (25). However, the role of DRP1 in Dox cardiotoxicity in whole animals remains unclear. Also, Mdivi1 may have nonspecific effects because it can activate Akt (25). Accordingly, we used DRP1-deficient mice to test the necessity of DRP1-dependent mitochondrial fission in Dox cardiotoxicity in vivo. The DRP1 heterozygous knockout mice (DRP1+/−) and their wild-type littermates were treated with saline or a single dose of Dox (15 mg/kg body weight) for 3 d. We measured the serum LDH activity, an indicator of tissue damage. Dox treatment increased serum LDH activity by 8.9-fold in wild-type mice, but only 4.3-fold in Drp1+/− mice (Fig. 6A), demonstrating the ability of DRP1 deficiency to mitigate Dox-induced tissue injury. Because LDH may be released from organs other than the heart, we also measured serum levels of cTnI. Dox increased serum cTnI by 1.84-fold in wild-type mice but this effect was completely abolished in DRP1+/− mice (Fig. 6B). In addition, Dox increased the levels of cCasp3, the protein oxidation products, and the lipid peroxidation by-product 4-HNE in wild-type hearts, but this effect was markedly reduced in DRP1+/− mice (Fig. 7A–C). Changes in enzymes involved in glucose metabolism have been suggested to play an important role in the pathogenesis of Dox-induced cardiotoxicity (45). The PDH complex catalyzes the conversion of pyruvate to acetyl coenzyme A and is composed of 3 enzymatic components including PDHE1 or PDHE1-A type I. The PDHE1-A is inhibited by the phosphorylation of Ser293 by PDH kinase. We found that the PDHE1-A phosphorylation in wild-type mouse heart was increased by Dox treatment, but this increase was greatly attenuated in DRP1+/− mouse heart (Fig. 7D). To determine whether the protective effect of DRP1 deficiency was related to a reduction in mitochondrial fragmentation, we isolated ventricular myocytes from 4-mo-old wild-type and DRP1+/− mice, infected the adult cardiomyocytes with AdDsRed, and treated them with 3 µM Dox for 24 h. Mitochondrial morphology did not significantly differ between wild-type and DRP1+/− cardiomyocytes at the baseline. Dox treatment reduced the major and minor axes as well as the size of mitochondria in wild-type myocytes (Fig. 8), suggesting increased mitochondrial fragmentation. However, this Dox effect was blocked in DRP1+/− cardiomyocytes, indicating an essential role for DRP1 in Dox-induced mitochondrial fragmentation in adult cardiomyocytes. Taken together, these results showed that DRP1 deficiency was sufficient to protect against Dox-induced cardiac damage, strongly supporting the conclusion that DRP1-dependent mitochondrial fragmentation mediates Dox cardiotoxicity.
Figure 6.
DRP1 haploinsufficiency blocked Dox-induced release of LDH and cTnI into the bloodstream. The DRP1 heterozygous knockout mice (DRP1+/−) were treated for 3 d with saline or Dox (15 mg/kg body weight). The serum LDH activity (A) and the serum levels of cTnI (B) were determined by using commercial kits. Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.01 vs. wild-type control, #P < 0.01 vs. Dox-treated wild type (n = 8–12).
Figure 7.
DRP1 haploinsufficiency attenuated Dox-induced oxidative injuries in the mouse heart. The DRP1+/− mice were treated with saline or Dox (15 mg/kg body weight). Mice were euthanized 3 d later, and cardiac tissues were processed for Western blot analysis of cCasp3 (A), oxidized protein (B; Oxyblot), lipid peroxidation by-product 4-HNE (C), and phospho-PDH (D). Data are expressed as means ± se and were analyzed by 1-way ANOVA. *P < 0.01 vs. wild-type control, #P < 0.01 vs. Dox-treated wild-type (n = 6).
Figure 8.
DRP1 haploinsufficiency attenuated Dox-induced mitochondrial fragmentation. Adult ventricular cardiomyocytes were isolated from wild-type and DRP1+/− mice, infected with AdDsRed, and treated with 3 µM Dox for 24 h. Mitochondrial morphology was analyzed using ImageJ. The major and minor axes as well as the size and number of mitochondria were calculated. At least 5 images (each containing 5–15 cells) were captured per treatment. WT, wild type. Data are expressed as means ± se and were analyzed by ANOVA. *P < 0.01 vs. control (n = 3).
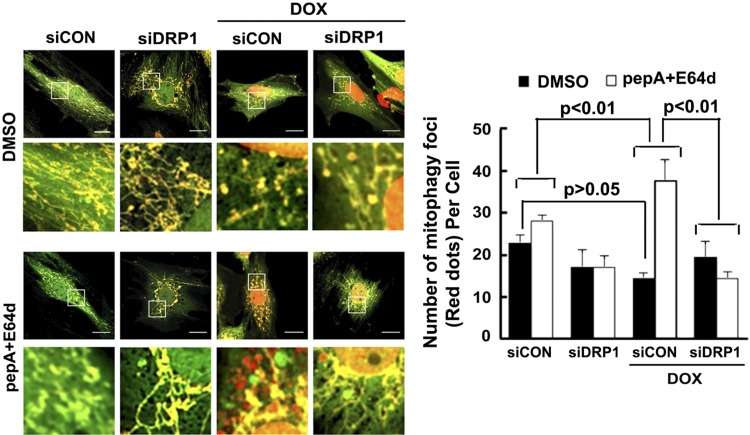
Drp1 knockdown attenuated Dox-induced mitophagic flux
Mitochondrial fission can segregate damaged segments of mitochondria and promote their removal by the autophagy-lysosome pathway, a process termed mitophagy (36). To determine if Dox not only induces mitochondrial fission but also stimulates mitophagy, we constructed an adenovirus encoding the novel mitophagy reporter mt-Rosella. Rosella is a dual-emission biosensor comprising a pH-stable red fluorescent protein linked to a pH-sensitive GFP (43). We tagged Rosella with a mitochondrial targeting sequence from the gene that encodes the human COX subunit VIII. We infected H9c2 cardiac myoblasts with Admt-Rosella. As shown in the merged confocal images (Supplemental Fig. S1A), the mitochondria glowing green or yellow are located in the cytosolic compartments, whereas the fragmented mitochondria fluorescing only red are being degraded within the lysosomes where the pH is low and the GFP is quenched. The red signal or dots colocalized with mitochondrial protein TOM20 (Supplemental Fig. S1A), autophagosomal marker LC3, Supplemental Fig. S1B), and lysosomal protein lysosomal-associated membrane protein 1 (Supplemental Fig. S1C), confirming the specificity of the reporter.
Using the mt-Rosella mitophagy reporter, we assessed the ability of Dox to affect mitophagy in H9c2 cells. A few red dots were found in both saline- and Dox-treated cells (Fig. 9). Compared with saline, Dox caused a small reduction in the number of red dots per cell but it was not significant (saline, 21.4 ± 3.9; Dox, 15.9 ± 2.7; n = 3; P > 0.05), suggesting that Dox might not have any effects on mitophagy. However, mitophagy is a dynamic process and its functional status cannot be determined simply by a snapshot of the number of mitochondrial fragments (red dots) present in the lysosome. Instead, mitophagy flux must be determined in the absence and presence of autophagic degradation inhibitors to reveal the true dynamic changes of the degradation process. Thus, we treated cells with PepA and E64d, 2 lysosomal protease inhibitors. Strikingly, PepA and E64d led to a much larger increase in the number of red dots in the Dox-treated cells than in the saline-treated cells (Fig. 9; saline, 5.3 ± 3.0; Dox, 23.1 ± 6.7; n = 3; P < 0.01), indicating that Dox actually accelerated mitophagy flux. In other words, the trend toward reduced red dots in Dox-treated cells in the absence of a lysosomal inhibitor was caused by a rapid degradation of the mitochondrial fragments in the lysosome. Interestingly, Doxinduced mitophagic flux was prevented by siRNA-mediated DRP1 knockdown (Dox, 23.1 ± 6.7; Dox and DRP1 siRNA (siDRP1; −5.1 ± 3.8; n = 3; P < 0.01), suggesting that DRP1-dependent mitochondrial fragmentation was required for Dox-induced mitophagy. Of note, it was readily observable that DPR1 knockdown blocked Dox-induced mitochondrial fragmentation similar to what was seen in AdDsRed-infected cells.
Figure 9.
DRP1 knockdown inhibited Dox-induced mitophagy flux. H9c2 cells were infected with the Admt-Rosella for 48 h. The cells were then transfected with siDRP1 or control siRNA (siCon) for 24 h and exposed to either Dox (750 nM) or saline for 24 h. Confocal images were captured, mitophagy was analyzed using ImageJ, and the number of mitophagy foci per cell was calculated. Between 3 and 8 images (totaling between 5 and 15 cells) were captured per treatment over 3 separate experiments. Scale bars, 20 µm. For evaluating mitophagy flux, experiments were repeated with lysosomal inhibitors (PepA and E64D) or DMSO 4 h after Dox treatment. Data are expressed as means ± se and were analyzed by 1-way ANOVA (P < 0.01 vs. siCon or siCon+Dox; n = 3).
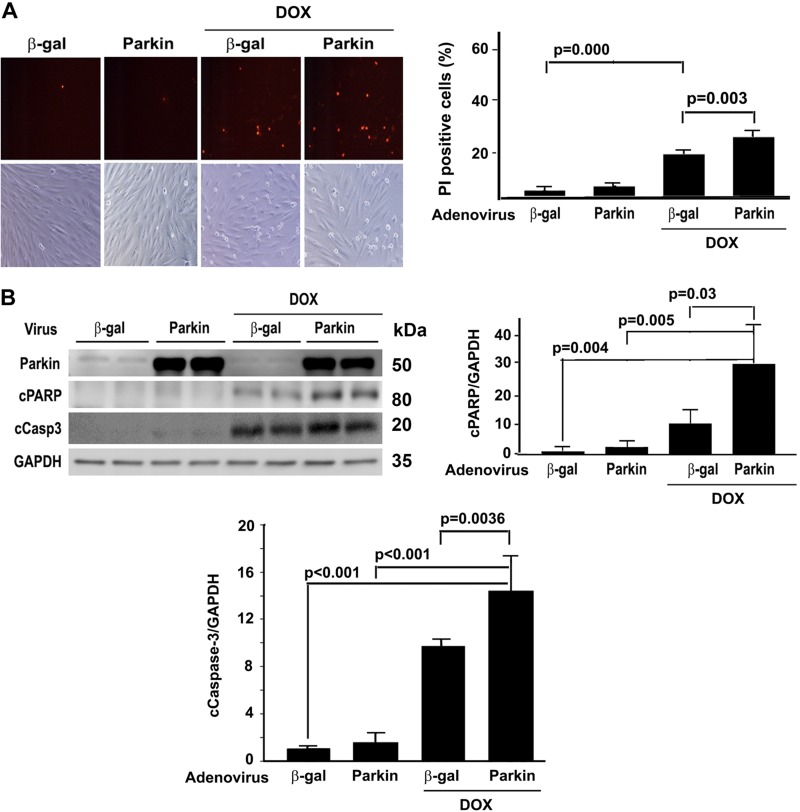
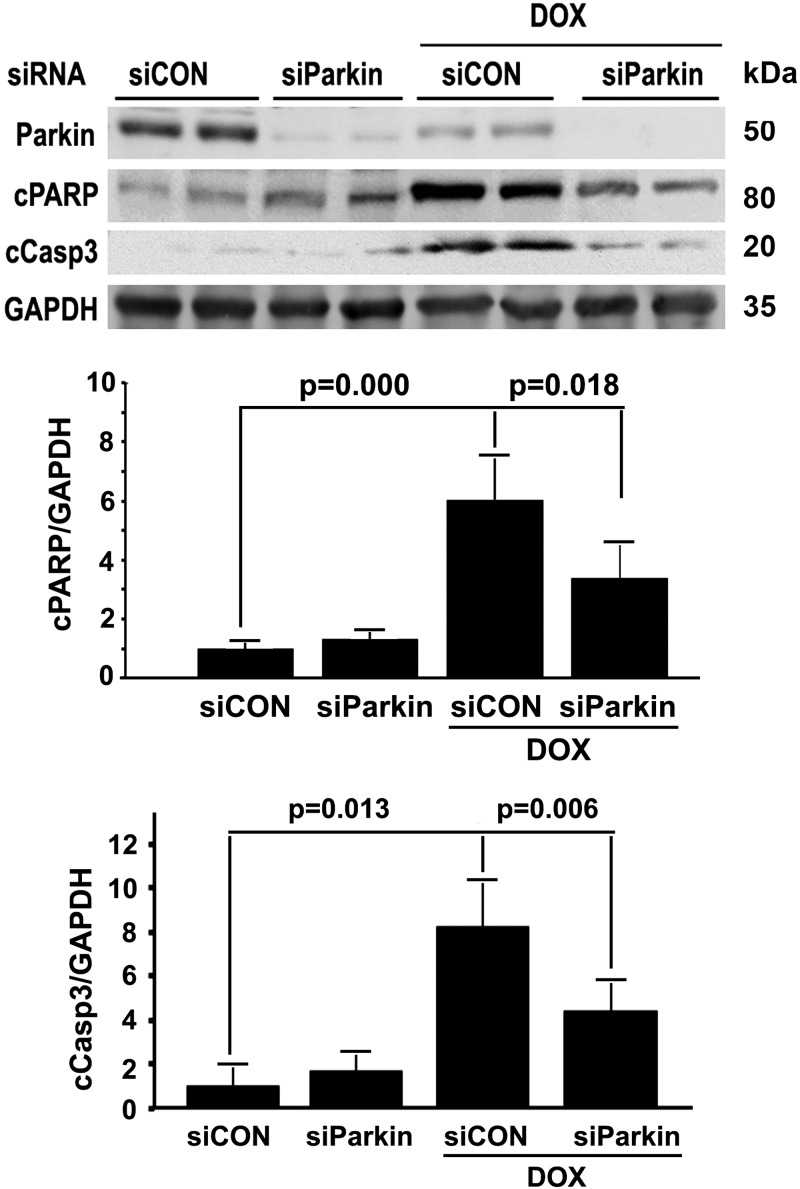
Mitophagy contributed to Dox-induced H9c2 cell death
It is still debatable whether and how mitophagy is involved in Dox cardiotoxicity (33). Parkin is an E3 ubiquitin ligase that ubiquitinates mitochondria to trigger their degradation by mitophagy. We manipulated parkin expression levels in H9c2 cells using adenovirus-mediated gene transfer or siRNA-mediated gene knockdown. Overexpression of parkin was able to increase the number of red dots in the presence of PepA and E64d either with or without Dox treatment. Conversely, knocking down parkin with siRNA attenuated Dox-induced mitophagy flux (Supplemental Fig. S2), confirming the ability of parkin to impact the overall activity of mitophagy. Importantly, increasing mitophagy activity by parkin overexpression exacerbated Dox-induced cell death (Fig. 10), whereas parkin knockdown had the opposite effects (Fig. 11), as measured by the number of PI-positive cells and the levels of cCasp3 and PARP. These results strongly suggested that mitophagy contributed to Dox cardiotoxicity.
Figure 10.
Parkin overexpression exacerbated Dox-induced cell death. H9c2 cells were infected with adenovirus expressing parkin (Adparkin) or adenovirus expressing β-galactosidase (Adβ-gal) for 24 h and then exposed to either Dox (750 nM) or saline for 24 h. Cell death was determined by PI staining (A) and cleavage of caspase-3 and PARP (B). Data were expressed as means ± se and analyzed by 1-way ANOVA (n = 3 for each treatment). GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Figure 11.
Parkin knockdown protected against Dox-induced cardiomyocyte death. H9c2 cells were pretreated with parkin-targeted siRNA (siParkin) or control siRNA (siCon) for 24 h and then exposed to either Dox (750 nM) or saline for an additional 24 h. Cell death was determined by the cleavage of caspase-3 and PARP. Data are expressed as means ± se and were analyzed by 1-way ANOVA (n = 3).
DISCUSSION
Dox cardiotoxicity is closely associated with mitochondrial morphologic changes including mitochondrial fragmentation. The potential contribution of mitochondrial fragmentation to Dox cardiotoxicity has been previously tested in Langendorff-perfused isolated rat heart by using Mdivi-1, a chemical inhibitor of mitochondrial fission. Although Mdivi-1 was able to attenuate Dox cardiotoxicity in isolated rat heart (25), it is not clear if it can achieve the same effect in whole animal studies in vivo. Also, the protective effect may not necessarily be mediated solely by the inhibition of mitochondrial fragmentation because Mdivi-1 activated the prosurvival pathway Akt in the heart, as shown by its phosphorylation levels (25). Although siRNA-mediated knockdown of DRP1 was shown to inhibit Dox-induced cell injury in neonatal rat cardiomyocytes (26), it remains to be determined if inactivation of DRP1 can reduce Dox cardiotoxicity in vivo. In the present study, we showed that DRP1 knockdown not only diminished Dox-induced mitochondrial fragmentation in H9c2 cardiac blast cells (Fig. 1) but also attenuated Dox-induced cell death (Fig. 3), confirming the contribution of DRP1 to Dox-induced cell death. More importantly, we showed that DRP1-deficient mice were protected from Dox-induced cardiac damage (Fig. 7), further supporting a role for DRP1-dependent mitochondrial fragmentation in Dox cardiotoxicity. Interestingly, DRP1 knockdown also attenuated Dox-induced mitophagy flux as assessed with the novel dual-fluorescent mitophagy reporter mt-Rosella, suggesting that mitochondrial fragmentation was required for Dox to induce mitophagy.
Interestingly, we found no difference in the DRP1 levels (Western blot) or activities (phosphorylation at Ser616) between control and Dox groups, suggesting that DRP1 is not the major player in Dox-induced mitochondrial fragmentation and cell death. However, it does not necessarily mean that DRP1 cannot be a drug target from a therapeutic point of view, because mitochondrial morphology is determined by the balance between mitochondrial fusion and fission. Dox markedly reduced the protein levels of Opa1 and Mfn2, 2 factors that control the fusion process and tilt the balance in the favor of fission, leading to an increased mitochondrial fragmentation (Fig. 2). Thus, it is not surprising that reducing Drp1 levels with siRNA reestablished the balance between fission and fusion and blocked Dox-induced mitochondrial fragmentation and cardiotoxicity, which is consistent with a previous study showing the ability of the DRP1 inhibitor Midivi-1 to reduce Dox cardiotoxicity (25). In addition, the abnormal mitochondrial morphology in the hearts of Mfn1 and Mfn2 double-knockout mice was normalized by DRP1 knockout, and Mfn1, Mfn2, and DRP1 cardiac triple-knockout mice survived longer than Mfn1 and Mfn2 double-knockout mice (46), further demonstrating the feasibility of targeting DRP1 to treat cardiac defect caused by reduced fusion proteins as occurred in Dox cardiotoxicity. The fact that DRP1 overexpression did not further increase Dox-triggered mitochondrial fragmentation (Fig. 4) and cell death (Fig. 5) suggests a necessity but not sufficiency for DRP1 in Dox cardiotoxicity.
Mitophagy is an important mitochondrial quality control mechanism that helps maintain a pool of healthy mitochondria by eliminating injured or otherwise dysfunctional mitochondria through the autophagy-lysosome pathway. Although mitophagy is protective in most cases (28), it becomes detrimental under certain conditions (29, 30). The effect of Dox on mitophagy has been previously assessed, but neither mitophagy activity nor its functional role in Dox cardiotoxicity has been unequivocally defined (33). Dox was reported to inhibit cardiac mitophagy because of the sequestration of parkin by p53 that led to decreased mitochondrial translocation of parkin and reduced mitochondria incorporation into autophagic vacuoles (34). In contrast, Dox was also shown to induce mitophagy in cardiomyocytes as evidenced by increased mitochondrial translocation of phosphatase and tensin homolog–induced kinase 1 and parkin and increased colocalization of LC3 with mitochondria (35). These inconsistent results may be in part caused by the methods used to quantify mitophagy. Both studies did not measure mitophagy flux, which reflects the number of mitochondria that are delivered to and degraded in the lysosome. Mitophagy flux can be determined by the difference in the numbers of mitochondria trapped in the lysosome before and after applying mitophagic degradation inhibitors such as PepA and E64d. Using a novel mt-Rosella mitophagy reporter that tracks the fate of mitochondrial fragments, we assessed the ability of Dox to affect mitophagy in H9c2 cells. We found that PepA and E64d led to a much more pronounced lysosomal accumulation of mitochondrial fragments (red dots) in the Dox-treated cells than in the saline-treated cells (Fig. 9), indicating that Dox accelerated mitophagy flux. Interestingly, Dox was previously shown to block autophagic flux in cardiomyocytes (47), suggesting that autophagy and mitophagy are differentially regulated and serve different functions. Further investigation is warranted to elucidate the mechanisms that mediate the differential effects of Dox on autophagy and mitophagy flux. Regardless, increasing mitophagy activity by parkin overexpression exacerbated Dox-induced cell death (Fig. 10), whereas parkin knockdown had the opposite effect (Fig. 11). These results strongly suggested that mitophagy contributed to Dox cardiotoxicity, probably caused by excessive destruction of mitochondria leading to diminished ATP production, which is essential for cardiomyocyte survival.
In summary, we showed that inhibiting mitochondrial fragmentation or mitophagy was able to diminish Dox-induced cardiomyocyte death, suggesting that Dox cardiotoxicity was mediated, at least in part, by the increased mitochondrial fragmentation and accelerated mitochondrial degradation by the lysosome. Accordingly, strategies that limit mitochondrial fission and mitophagy within the physiologic range would be expected to reduce Dox cardiotoxicity. Apparently, further studies are warranted to make sure that these strategies will not compromise the antitumor efficacy of Dox.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
Q.L. was supported by U.S. National Institutes of Health, National Heart, Lung, and Blood Institute Grant 1R15HL137130-01A1, and S.K. was supported by American Heart Association Grant 15SDG25080077. The authors declare no conflicts of interest.
Glossary
- 4-HNE
4-hydroxynonenal
- ABCB8
ATP binding cassette subfamily B member 8
- AdDRP1
adenovirus expressing DRP1
- AdDsRed
adenovirus expressing MitoDsRed
- Admt-Rosella
adenovirus expressing mt-Rosella
- Akt
protein kinase B
- AR
aspect ratio
- cCasp3
cleaved caspase-3
- COX
cytochrome c oxidase
- CtnI
cardiac troponin I
- Dox
doxorubicin
- DRP1
dynamin-related protein 1
- FF
form factor
- Fis1
mitochondrial fission protein 1
- GFP
green fluorescent protein
- LC3
microtubule-associated protein light chain 3
- LDH
lactate dehydrogenase
- Mdivi-1
mitochondrial division inhibitor 1
- Mfn
mitofusin
- Opa1
optic atrophy 1
- PARP
poly (ADP-ribose) polymerase
- PDH
pyruvate dehydrogenase
- PepA
pepstatin A
- phospho
phosphorylated
- PI
propidium iodide
- ROS
reactive oxygen species
- siDRP1
DRP1 siRNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
M. P. Catanzaro, A. Weiner, S. Kobayashi, and Q. Liang designed research and analyzed data; M. P. Catanzaro, A. Weiner, A. Kaminaris, C. Li, F. Cai, F. Zhao, S. Kobayashi, T. Kobayashi, and Y. Huang performed research; M. P. Catanzaro, A. Weiner, and Q. Liang wrote the paper; and H. Sesaki contributed DRP1 gene-targeted mice.
REFERENCES
- 1.Swain S. M., Whaley F. S., Ewer M. S. (2003) Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer 97, 2869–2879 [DOI] [PubMed] [Google Scholar]
- 2.Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. (2004) Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 56, 185–229 [DOI] [PubMed] [Google Scholar]
- 3.Van der Pal H. J., van Dalen E. C., Hauptmann M., Kok W. E., Caron H. N., van den Bos C., Oldenburger F., Koning C. C., van Leeuwen F. E., Kremer L. C. (2010) Cardiac function in 5-year survivors of childhood cancer: a long-term follow-up study. Arch. Intern. Med. 170, 1247–1255 [DOI] [PubMed] [Google Scholar]
- 4.Maeda M. (2008) Late effects of childhood cancer: life-threatening issues. J. Nippon Med. Sch. 75, 320–324 [DOI] [PubMed] [Google Scholar]
- 5.Link G., Tirosh R., Pinson A., Hershko C. (1996) Role of iron in the potentiation of anthracycline cardiotoxicity: identification of heart cell mitochondria as a major site of iron-anthracycline interaction. J. Lab. Clin. Med. 127, 272–278 [DOI] [PubMed] [Google Scholar]
- 6.Ichikawa Y., Ghanefar M., Bayeva M., Wu R., Khechaduri A., Naga Prasad S. V., Mutharasan R. K., Naik T. J., Ardehali H. (2014) Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Invest. 124, 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Injac R., Strukelj B. (2008) Recent advances in protection against doxorubicin-induced toxicity. Technol. Cancer Res. Treat. 7, 497–516 [DOI] [PubMed] [Google Scholar]
- 8.Hasinoff B. B., Herman E. H. (2007) Dexrazoxane: how it works in cardiac and tumor cells. Is it a prodrug or is it a drug? Cardiovasc. Toxicol. 7, 140–144 [DOI] [PubMed] [Google Scholar]
- 9.Swain S. M., Whaley F. S., Gerber M. C., Weisberg S., York M., Spicer D., Jones S. E., Wadler S., Desai A., Vogel C., Speyer J., Mittelman A., Reddy S., Pendergrass K., Velez-Garcia E., Ewer M. S., Bianchine J. R., Gams R. A. (1997) Cardioprotection with dexrazoxane for doxorubicin-containing therapy in advanced breast cancer. J. Clin. Oncol. 15, 1318–1332 [DOI] [PubMed] [Google Scholar]
- 10.Tebbi C. K., London W. B., Friedman D., Villaluna D., De Alarcon P. A., Constine L. S., Mendenhall N. P., Sposto R., Chauvenet A., Schwartz C. L. (2007) Dexrazoxane-associated risk for acute myeloid leukemia/myelodysplastic syndrome and other secondary malignancies in pediatric Hodgkin’s disease. J. Clin. Oncol. 25, 493–500 [DOI] [PubMed] [Google Scholar]
- 11.Elliott A. M., Al-Hajj M. A. (2009) ABCB8 mediates doxorubicin resistance in melanoma cells by protecting the mitochondrial genome. Mol. Cancer Res. 7, 79–87 [DOI] [PubMed] [Google Scholar]
- 12.Mohammad I. S., He W., Yin L. (2018) Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed. Pharmacother. 100, 335–348 [DOI] [PubMed] [Google Scholar]
- 13.Berthiaume J. M., Wallace K. B. (2007) Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol. 23, 15–25 [DOI] [PubMed] [Google Scholar]
- 14.Bianchi C., Bagnato A., Paggi M. G., Floridi A. (1987) Effect of adriamycin on electron transport in rat heart, liver, and tumor mitochondria. Exp. Mol. Pathol. 46, 123–135 [DOI] [PubMed] [Google Scholar]
- 15.Davies K. J., Doroshow J. H. (1986) Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem. 261, 3060–3067 [PubMed] [Google Scholar]
- 16.Doroshow J. H., Davies K. J. (1986) Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J. Biol. Chem. 261, 3068–3074 [PubMed] [Google Scholar]
- 17.Pereira G. C., Silva A. M., Diogo C. V., Carvalho F. S., Monteiro P., Oliveira P. J. (2011) Drug-induced cardiac mitochondrial toxicity and protection: from doxorubicin to carvedilol. Curr. Pharm. Des. 17, 2113–2129 [DOI] [PubMed] [Google Scholar]
- 18.Wallace K. B. (2003) Doxorubicin-induced cardiac mitochondrionopathy. Pharmacol. Toxicol. 93, 105–115 [DOI] [PubMed] [Google Scholar]
- 19.Wallace K. B. (2007) Adriamycin-induced interference with cardiac mitochondrial calcium homeostasis. Cardiovasc. Toxicol. 7, 101–107 [DOI] [PubMed] [Google Scholar]
- 20.Zhou S., Starkov A., Froberg M. K., Leino R. L., Wallace K. B. (2001) Cumulative and irreversible cardiac mitochondrial dysfunction induced by doxorubicin. Cancer Res. 61, 771–777 [PubMed] [Google Scholar]
- 21.Losón O. C., Song Z., Chen H., Chan D. C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Y. W., Chang Y. T., Wang Q., Lin J. J., Chen Y. J., Chen C. C. (2013) Quantitative phosphoproteomic study of pressure-overloaded mouse heart reveals dynamin-related protein 1 as a modulator of cardiac hypertrophy. Mol. Cell. Proteomics 12, 3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong S. B., Subrayan S., Lim S. Y., Yellon D. M., Davidson S. M., Hausenloy D. J. (2010) Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 121, 2012–2022 [DOI] [PubMed] [Google Scholar]
- 24.Sardão V. A., Oliveira P. J., Holy J., Oliveira C. R., Wallace K. B. (2009) Morphological alterations induced by doxorubicin on H9c2 myoblasts: nuclear, mitochondrial, and cytoskeletal targets. Cell Biol. Toxicol. 25, 227–243 [DOI] [PubMed] [Google Scholar]
- 25.Gharanei M., Hussain A., Janneh O., Maddock H. (2013) Attenuation of doxorubicin-induced cardiotoxicity by mdivi-1: a mitochondrial division/mitophagy inhibitor. PLoS One 8, e77713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J. X., Zhang X. J., Feng C., Sun T., Wang K., Wang Y., Zhou L. Y., Li P. F. (2015) MicroRNA-532-3p regulates mitochondrial fission through targeting apoptosis repressor with caspase recruitment domain in doxorubicin cardiotoxicity. Cell Death Dis. 6, e1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Q., Kobayashi S. (2016) Mitochondrial quality control in the diabetic heart. J. Mol. Cell. Cardiol. 95, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nah J., Miyamoto S., Sadoshima J. (2017) Mitophagy as a protective mechanism against myocardial stress. Compr. Physiol. 7, 1407–1424 [DOI] [PubMed] [Google Scholar]
- 29.Sentelle R. D., Senkal C. E., Jiang W., Ponnusamy S., Gencer S., Selvam S. P., Ramshesh V. K., Peterson Y. K., Lemasters J. J., Szulc Z. M., Bielawski J., Ogretmen B. (2012) Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat. Chem. Biol. 8, 831–838; erratum: 1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizumura K., Cloonan S. M., Nakahira K., Bhashyam A. R., Cervo M., Kitada T., Glass K., Owen C. A., Mahmood A., Washko G. R., Hashimoto S., Ryter S. W., Choi A. M. (2014) Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 124, 3987–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basit F., van Oppen L. M., Schöckel L., Bossenbroek H. M., van Emst-de Vries S. E., Hermeling J. C., Grefte S., Kopitz C., Heroult M., Hgm Willems P., Koopman W. J. (2017) Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 8, e2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song M., Gong G., Burelle Y., Gustafsson A. B., Kitsis R. N., Matkovich S. J., Dorn G. W., II (2015) Interdependence of parkin-mediated mitophagy and mitochondrial fission in adult mouse hearts. Circ. Res. 117, 346–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koleini N., Kardami E. (2017) Autophagy and mitophagy in the context of doxorubicin-induced cardiotoxicity. Oncotarget 8, 46663–46680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshino A., Mita Y., Okawa Y., Ariyoshi M., Iwai-Kanai E., Ueyama T., Ikeda K., Ogata T., Matoba S. (2013) Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat. Commun. 4, 2308 [DOI] [PubMed] [Google Scholar]
- 35.Yin J., Guo J., Zhang Q., Cui L., Zhang L., Zhang T., Zhao J., Li J., Middleton A., Carmichael P. L., Peng S. (2018) Doxorubicin-induced mitophagy and mitochondrial damage is associated with dysregulation of the PINK1/parkin pathway. Toxicol. In Vitro 51, 1–10 [DOI] [PubMed] [Google Scholar]
- 36.Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y., Lee H. Y., Hanna R. A., Gustafsson A. B. (2011) Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, H1924–H1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branco A. F., Pereira S. P., Gonzalez S., Gusev O., Rizvanov A. A., Oliveira P. J. (2015) Gene expression profiling of H9c2 myoblast differentiation towards a cardiac-like phenotype. PLoS One 10, e0129303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackers-Johnson M., Li P. Y., Holmes A. P., O’Brien S. M., Pavlovic D., Foo R. S. (2016) A simplified, langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ. Res. 119, 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X., Kobayashi S., Chen K., Timm D., Volden P., Huang Y., Gulick J., Yue Z., Robbins J., Epstein P. N., Liang Q. (2013) Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J. Biol. Chem. 288, 18077–18092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X., Chen K., Kobayashi S., Timm D., Liang Q. (2012) Resveratrol attenuates doxorubicin-induced cardiomyocyte death via inhibition of p70 S6 kinase 1-mediated autophagy. J. Pharmacol. Exp. Ther. 341, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitay B. M., McCormack R., Wang Y., Tsoulfas P., Zhai R. G. (2013) Mislocalization of neuronal mitochondria reveals regulation of Wallerian degeneration and NMNAT/WLD(S)-mediated axon protection independent of axonal mitochondria. Hum. Mol. Genet. 22, 1601–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mijaljica D., Prescott M., Devenish R. J. (2011) A fluorescence microscopy assay for monitoring mitophagy in the yeast Saccharomyces cerevisiae. J. Vis. Exp. 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kageyama Y., Zhang Z., Roda R., Fukaya M., Wakabayashi J., Wakabayashi N., Kensler T. W., Reddy P. H., Iijima M., Sesaki H. (2012) Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J. Cell Biol. 197, 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei S. N., Zhao W. J., Zeng X. J., Kang Y. M., Du J., Li H. H. (2015) Microarray and Co-expression network analysis of genes associated with acute doxorubicin cardiomyopathy in mice. Cardiovasc. Toxicol. 15, 377–393 [DOI] [PubMed] [Google Scholar]
- 46.Song M., Franco A., Fleischer J. A., Zhang L., Dorn G. W., II (2017) Abrogating mitochondrial dynamics in mouse hearts accelerates mitochondrial senescence. Cell Metab. 26, 872–883.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D. L., Wang Z. V., Ding G., Tan W., Luo X., Criollo A., Xie M., Jiang N., May H., Kyrychenko V., Schneider J. W., Gillette T. G., Hill J. A. (2016) Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification. Circulation 133, 1668–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.