A group of cytoprotective genes is regulated by heterodimers composed of the cap’n’collar (CNC) family member Nrf2 and one of the small Maf (sMaf) proteins (MafF, MafG, or MafK) through the antioxidant response element (ARE, also referred to as the CNC-sMaf binding element [CsMBE]). Many lines of evidence support this model; however, a direct and specific evaluation of the Nrf2-sMaf heterodimer remains to be executed.
KEYWORDS: CsMBE, Nrf2, transcription factor, small Maf, tethered molecule
ABSTRACT
A group of cytoprotective genes is regulated by heterodimers composed of the cap’n’collar (CNC) family member Nrf2 and one of the small Maf (sMaf) proteins (MafF, MafG, or MafK) through the antioxidant response element (ARE, also referred to as the CNC-sMaf binding element [CsMBE]). Many lines of evidence support this model; however, a direct and specific evaluation of the Nrf2-sMaf heterodimer remains to be executed. To address this issue, we constructed a tethered Nrf2-MafG (T-N2G) heterodimer using a flexible linker peptide. We then introduced the T-N2G construct into cells lacking all three sMaf proteins to specifically evaluate the function of the tethered heterodimer without interference from other endogenous CNC-sMaf heterodimers or sMaf homodimers. In response to an Nrf2 activator, diethyl maleate, the T-N2G protein can widely activate the target genes of Nrf2 but not those of Nrf1, such as proteasome subunit genes. Genome-wide binding analysis showed that the T-N2G protein preferentially bound to the CsMBE motifs in the regulatory regions of the Nrf2 target genes. These results provide direct evidence that the Nrf2-MafG heterodimer acts as a transcriptional activator of Nrf2-dependent genes and show that this assay system will be a powerful tool to specifically examine the function of other CNC-sMaf heterodimers.
INTRODUCTION
Basic region-leucine zipper (bZIP)-type transcription factors form various combinations of homodimers or heterodimers that differentially regulate the expression of their target genes (1). Functional evaluation of individual dimer complexes is an essential step in understanding regulatory axes between transcription factors (trans factors) and cis-acting elements (cis elements). However, due to the conserved structure of the leucine zipper domain, each bZIP factor shows a different affinity to different bZIP factors (2). Thus, multiple combinations of heterodimer models are often proposed along with individual homodimers, and the combinations and functional contributions of various dimers remain to be evaluated.
Well-studied examples of bZIP transcription factors are the cap’n’collar (CNC) proteins. The CNC family consists of four members, p45 nuclear factor-erythroid 2 (NF-E2) and NF-E2-related factors (Nrf1, Nrf2, and Nrf3), as well as two distantly related members, Bach1 and Bach2, and plays differential roles in gene regulation (3–6). Among them, Nrf2 is the first CNC protein that was found to be stress inducible (7). Nrf2 is constantly subjected to ubiquitination by the Keap1-Cul3 E3 ligase complex and is degraded through the proteasome system (8–10). Oxidative and electrophilic stresses inactivate Keap1, allowing Nrf2 to accumulate in nuclei and to activate the expression of their target genes. On the other hand, a sibling transcription factor in the CNC family, Nrf1, regulates proteasome subunit genes and some related genes in response to proteolytic stress (11–13). While it is possible that these CNC family factors regulate their target genes in a partially overlapping manner, available lines of evidence support the notion that each CNC factor regulates a distinct and specific set of target genes (14–16). More importantly, the CNC factors cannot bind to DNA as a monomer but need to form heterodimers with other bZIP transcription factors to exert their functions.
The cellular counterpart of the v-Maf oncogene consists of a small transcription factor family with a bZIP structure (17). Of the members of the Maf family transcription factors, small Maf (sMaf) proteins have been shown to serve as bZIP heterodimeric partners of the CNC family factors (18, 19). sMafs consist of three functionally redundant members, i.e., MafF, MafG, and MafK, and these members lack any canonical transcriptional activation domain (20). Accordingly, the functions of CNC-sMaf heterodimers rely on those of the CNC proteins. However, an intriguing observation is that each sMaf can also form a homodimer by itself and can act as a negative regulator (21).
Other bZIP factors also heterodimerize with CNC factors. For instance, Nrf1 and Nrf2 are reported to form heterodimers with the Jun family members c-Jun, JunB, and JunD (22). In addition, Nrf2 is reported to form a heterodimer with ATF4 (23). Comprehensive interaction data based on a protein array with 49 bZIP factors revealed that the CNC factors show wide-ranging affinities for multiple bZIP factors, including sMaf, c-Jun, and ATF4 (2), suggesting the possible existence of heterodimers composed of CNC and bZIP factors other than sMaf proteins.
To prove the biological significance of CNC-sMaf heterodimers, we and other groups have been trying to obtain solid lines of experimental evidence. For instance, in vitro DNA-protein binding analyses, such as electrophoretic gel mobility shift assays, showed that the CNC-sMaf heterodimers can bind to certain types of cis-acting consensus binding sequences, as discussed below. The binding sequences of CNC-sMaf heterodimers have been called an NF-E2 binding element (24), an antioxidant response element (ARE) (25), or an electrophile response element (EpRE) (26), depending on their locations and functional roles. These NF-E2-related sequences harbor a common consensus sequence, RTGASNNNGC (IUPAC code: R is A or G and S is G or C), that partially overlaps the Maf recognition element (MARE; TGCTGACTCAGCA) to which sMaf homodimers preferentially bind (27). Structural analysis revealed that the underlined GC in the NF-E2-related sequences is recognized by the sMaf protein (28).
R (underlined) in the NF-E2-related sequences (RTGASNNNGC) is also important to preferentially recruit CNC proteins rather than sMaf. A tyrosine (Y) residue in the sMaf basic regions, which corresponds to alanine (A) in the sequences of the CNC and c-Jun/c-Fos family members, is responsible for the GC recognition (29). We reported that mutant Nrf2, whose A was replaced with Y, such as sMaf, preferentially recognized MARE, showing that the CNC-sMaf heterodimers structurally distinguish their binding sites from those of MARE (27, 29). Taking all these observations into consideration, we have proposed the CNC-sMaf binding element (CsMBE) as a common designation for these NF-E2-related sequences (27). Next-generation sequencing technologies have also provided us supportive lines of evidence, such that chromatin immunoprecipitation sequencing (ChIP-Seq) analysis revealed genome-wide Nrf2 and MafG cooccupying sites that harbor CsMBEs and are located in the regulatory regions of the Nrf2 target genes (30).
We have been working extensively on mouse genetics to prove the cooperativity of the CNC and sMaf proteins. Our rationale of the analyses is that if CNC proteins and sMaf act by forming heterodimers, similar phenotypes should be observed upon introducing mutations into mouse genes in vivo. In fact, we generated loss-of-function mutant mouse lines of each sMaf as well as double or triple mutant mouse lines and compared their phenotypes with those of the CNC mutant mice (31–33). Both p45 NF-E2 and sMaf mutant mice show thrombocytopenia, supporting the notion that p45 NF-E2 and sMaf cooperatively regulate genes responsible for platelet production (31, 34). Both Nrf1 and sMaf liver-specific conditional knockout mice display a fatty liver (14, 16, 35). sMaf triple knockout mice die in utero, showing a very good coincidence with Nrf1 and Nrf2 double mutant mice (36, 37), and primary cultured mouse embryonic fibroblasts (MEFs) established from sMaf triple mutants show the impaired induction of Nrf2 target genes (38).
Whereas these studies support the importance of the CNC-sMaf heterodimer model, we wish to obtain much more direct and conclusive verifications of this model. To comprehensively clarify the molecular basis of the CNC-sMaf factor activity, in this study, we challenged two elaborate means of evaluation and incorporated them into our analyses of the CNC-sMaf heterodimer model. We constructed a tethered Nrf2-MafG heterodimer (T-N2G) and all three sMaf-deficient cells. These two means enabled us to perform a functional evaluation of the T-N2G protein in vivo, completely avoiding interferences from the other CNC-sMaf heterodimers. Our data revealed that T-N2G widely and specifically activated the Nrf2 target genes through CsMBEs, whereas T-N2G did not activate the proteasome subunit genes, known as the target genes of Nrf1.
RESULTS
Construction of the tethered Nrf2-MafG heterodimer.
Because the direct evaluation of the Nrf2-sMaf heterodimer has been critically important, we decided to adopt the protein tethering system, which has been used for heterodimeric transcription factors (39). For instance, a flexible polypeptide linker consisting of 22 amino acids [RS(GGGS)4GGRS] was used to connect p45 NF-E2 and MafK (40). In this study, we used the same-length polypeptide linker and generated a tethered Nrf2-MafG (T-N2G) molecule. Since the amino acid sequence after the leucine zipper of Nrf2 is only one amino acid longer than that of p45 NF-E2, we hypothesized that the linker size of p45-MafK might also work well for the Nrf2-MafG-tethered molecule to preserve the binding specificity and each molecule function.
As shown in Fig. 1A and B, we fused mouse FLAG-tagged Nrf2 cDNA without a stop codon with MafG cDNA by the linker and inserted the tethered dimer cDNA into an expression vector. An important point here is how strictly we can evaluate the binding and function of the T-N2G molecule, avoiding the influences of the endogenous sMaf proteins. If the tethered protein is introduced into wild-type cells, this protein competes with various endogenous CNC-sMaf heterodimers and sMaf homodimers, depending on their expression levels (Fig. 1C, upper panel). We believe that it is ideal to evaluate the T-N2G-tethered molecule in the absence of sMaf proteins (Fig. 1C, lower panel).
FIG 1.
Construction of an assay system for a functional evaluation of the T-N2G protein in the absence of sMaf proteins. (A) Structure of the T-N2G expression vector. A cDNA encoding FLAG-Nrf2 fused to MafG with a flexible polypeptide linker was inserted into the PiggyBac dual promoter vector. (B) Amino acid sequences of the T-N2G protein from the bZIP region of Nrf2 to that of MafG are shown. bZIP regions are underlined. Leucine residues of the Nrf2 leucine zipper motif are shown with a black background. The linker is boxed. (C) A schematic diagram showing that in the absence of sMaf proteins, the T-N2G protein can exert its function while the other CNC factors cannot.
Establishment of immortalized sMaf-deficient MEFs.
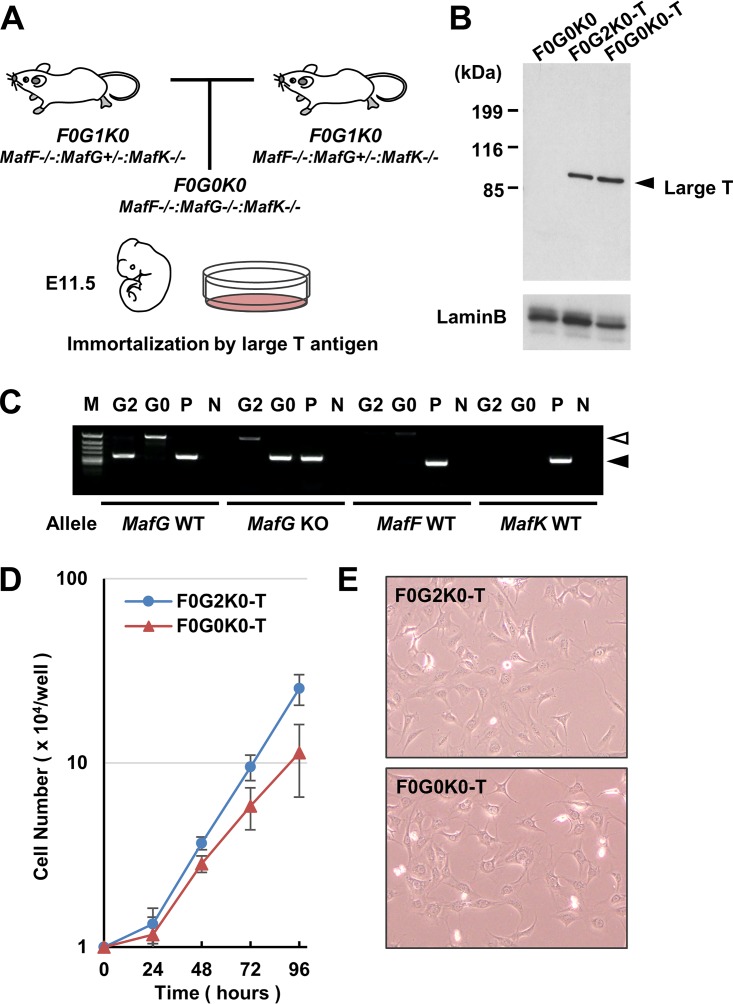
To rigorously verify the binding and function of the T-N2G molecule while avoiding the influences of the endogenous sMaf proteins, we were then challenged to establish immortalized triple sMaf knockout MEFs. Since the MafF−/− MafG−/− MafK−/− (F0G0K0) mice lacking all three sMaf proteins die in utero around embryonic day 11.5 (37), we established MEFs from F0G0K0 embryos as well as from MafF−/− MafG+/+ MafK−/− (F0G2K0) embryos by intercrossing the MafF−/− MafG+/– MafK−/− (F0G1K0) mice (Fig. 2A).
FIG 2.
Establishment of immortalized sMaf-deficient MEFs. (A) Mating strategy for the production of sMaf triple knockout (F0G0K0) embryos. (B) Immunoblot analysis showing that large T antigen is expressed in both F0G2K0-T and F0G0K0-T cells. Lamin B1 was detected as a loading control. (C) Genotyping PCR with genomic DNA from F0G2K0-T (G2) and F0G0K0-T (G0) cells. P and N, positive and negative controls, respectively. Note that MafF and MafK wild-type (WT) alleles are not detected in either cell. The MafG WT allele was detected only in F0G2K0-T cells, while the MafG KO allele was detected only in F0G0K0-T cells. A closed arrowhead indicates specific PCR products. An open arrowhead indicates nonspecific products. (D) Growth curves of F0G2K0-T and F0G0K0-T cells. (E) Phase-contrast images of F0G2K0-T and F0G0K0-T cells.
To obtain MEFs that can be stably expanded, we introduced large T antigens into the MEFs from F0G0K0 and F0G2K0 embryos and induced the immortalization of these MEFs (Fig. 2B, F0G2K0-T and F0G0K0-T). After culturing over 30 passages, we reconfirmed by PCR genotyping that the F0G0K0-T cells lacked the wild-type alleles of the MafG, MafK, and MafF genes (Fig. 2C).
We found that the F0G0K0-T cells grew slightly slower than the F0G2K0-T cells did (Fig. 2D). The doubling times of the F0G0K0-T cells and F0G2K0-T cells were approximately 24 and 17 h, respectively. Inspection of the cell morphology revealed that these two types of cells showed fibroblast morphology and appeared to be like each other (Fig. 2E). These results thus demonstrate that we have successfully prepared immortalized sMaf-null cells and support our contention that triple sMaf knockout MEFs can be used for further characterizations of the T-N2G molecule.
T-N2G activates the CsMBE-dependent reporter gene.
We next asked whether the T-N2G molecule retained binding specificity and transactivation function. To address this question, we first performed a reporter cotransfection-transactivation assay. When we transfected the expression vectors of either Nrf2, MafG, or both with a CsMBE-dependent luciferase (Luc) reporter into the established F0G0K0-T cells, single Nrf2 or MafG transfection failed to activate the CsMBE-dependent reporter gene expression (Fig. 3A). Notably, however, Nrf2 and MafG cotransfection markedly activated the Luc reporter, depending on the amount of MafG plasmid.
FIG 3.

T-N2G protein activates CsMBE-dependent reporter genes in the absence of sMaf proteins. (A) The pNQO1-ARE-luc reporter construct was transfected with an Nrf2 expression vector with or without various amounts (micrograms) of the MafG expression vector as shown. (B) The pNQO1-ARE-luc reporter was transfected with the T-N2G expression vector as well as the Nrf2 and MafG expression vectors. The total amounts (micrograms) of Nrf2, MafG, and T-N2G plasmids transfected into three wells are shown. Firefly luciferase activity in the absence of any effector plasmids was set to 1 as the relative luciferase unit (RLU). The error bars represent the standard deviations (SDs) (n = 3). Representative data from two independent experiments are shown.
While it has been shown that Nrf2 alone activates the CsMBE-dependent reporter in many cultured cells (e.g., see reference 41), we interpreted that the activation utilized heterodimerization with endogenous sMaf proteins. However, since there were no experimental systems that directly verify this interpretation, this notion remained to be rigorously tested. In this study, we showed for the first time that Nrf2 transfection alone did not give rise to such transactivation of the reporter (Fig. 3). As the present experiments were conducted in the absence of any endogenous sMaf molecules, the results unequivocally demonstrate that Nrf2 transactivates reporter gene transcription by forming heterodimers with sMaf, excluding the hypothesis that some of the endogenous molecules other than sMaf may be able to heterodimerize and activate transcription from the CsMBE sites.
Another important observation here was that T-N2G greatly activated the transcription of the CsMBE reporter by itself in the sMaf-deficient MEFs (Fig. 3B). The transfection vectors for Nrf2, MafG, and T-N2G utilized different promoter systems, so we could not directly compare the transactivation efficiencies of Nrf2 plus MafG and T-N2G in the experiment shown in Fig. 3B. Nonetheless, this result unequivocally demonstrates that the T-N2G molecule is a functional protein that is capable of activating transcription through CsMBEs.
Nrf2 target genes are activated in stable T-N2G-expressing cells.
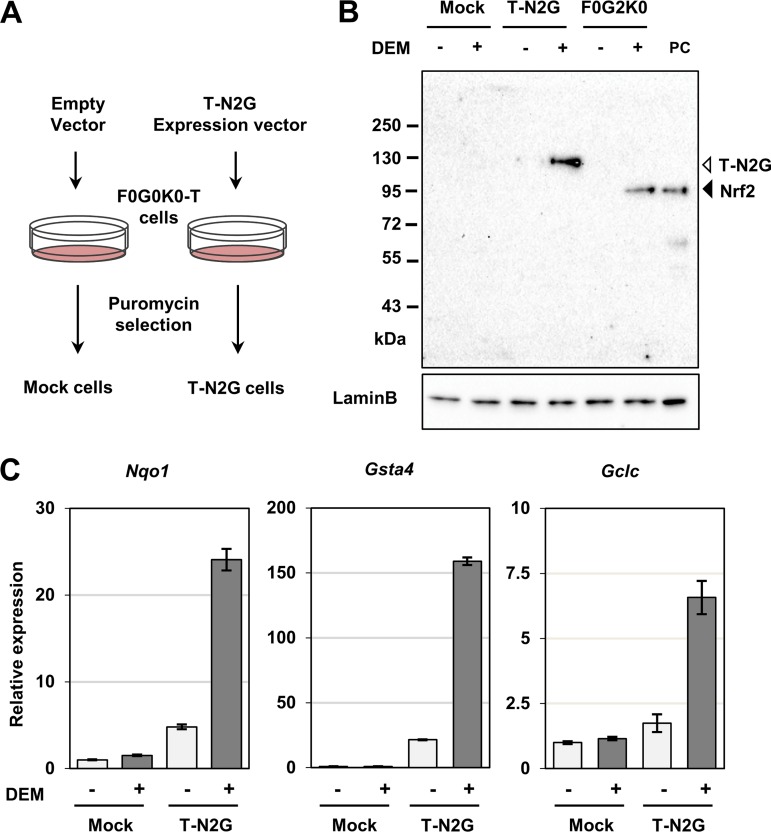
To address the functions of the T-N2G protein, we introduced the T-N2G expression vector into sMaf-deficient MEFs and established T-N2G-expressing MEFs (Fig. 4A, T-N2G cells). We also established mock cells that were sMaf-deficient MEF cells with an empty vector. To ascertain whether the T-N2G protein was also degraded by the proteasome system through the Keap1-mediated ubiquitination of Nrf2, we examined the expression of T-N2G proteins in diethyl maleate (DEM)-treated cells as well as in nontreated cells (Fig. 4B). The T-N2G protein was detected at the expected molecular size, which was larger than the size of endogenous Nrf2 observed in F0G2K0-T cells as well as exogenous Nrf2 loaded as a positive control.
FIG 4.
Establishment of sMaf-deficient MEFs expressing the T-N2G protein. (A) Schematic diagram of the protocol for obtaining stable cell lines. After transfection followed by puromycin selection, mock and T-N2G MEFs were established. (B) Mock, T-N2G, and F0G2K0-T MEFs were treated with 100 μM DEM for 5 h, and their nuclear lysates were analyzed by immunoblot analysis using an anti-Nrf2 antibody. Lamin B1 was detected as a loading control. (C) T-N2G-mediated induction of typical Nrf2 target genes, Nqo1, Gsta4, and Gclc, was examined by qPCR. The expression level of each mRNA was normalized to that of Hprt mRNA. The data represent the means ± SDs (n = 3). The gene expression levels in vehicle-treated mock cells were set to 1.
Notably, the T-N2G protein was expressed at a very low level in T-N2G cells, whereas expression was significantly increased by DEM treatment (Fig. 4B), indicating that the Keap1-mediated degradation machinery retains the ability to degrade the T-N2G protein. This observation is in very good agreement with our previous observation that Nrf2 protein fused with beta-galactosidase in the N-terminal domain is degraded by the Keap1 pathway (42). Indeed, the latter fusion protein has been utilized as an excellent reporter to monitor Nrf2 activity (43).
We next examined whether the T-N2G protein could activate Nrf2 target genes or exhibit different capabilities against the wild-type Nrf2 protein. To this end, we treated the mock and T-N2G cells with DEM and assessed the induction of representative Nrf2 target genes, those coding for NAD(P)H quinone dehydrogenase 1 (Nqo1), glutathione S-transferase A4 (Gsta4), and glutamate-cysteine ligase catalytic subunit (Gclc), by means of quantitative PCR (qPCR). As a result, DEM significantly induced these three genes in T-N2G cells (Fig. 4C), indicating that the T-N2G protein retains transactivating activity similar to that of the Nrf2-sMaf heterodimer. Notably, all these genes were slightly activated in the T-N2G cells even without the DEM treatment (Fig. 4C), indicating that these Nrf2 target genes have already been activated by the basal expression level of the T-N2G protein. On the other hand, DEM treatment failed to induce the expression of these genes in the mock cells, supporting our hypothesis that the DEM-mediated induction of the Nrf2 target genes observed in the T-N2G cells is indeed T-N2G dependent. These wide-ranging results thus demonstrate that the T-N2G protein has the capability to activate Nrf2 target genes within cells.
T-N2G activates a wide range of Nrf2 target genes.
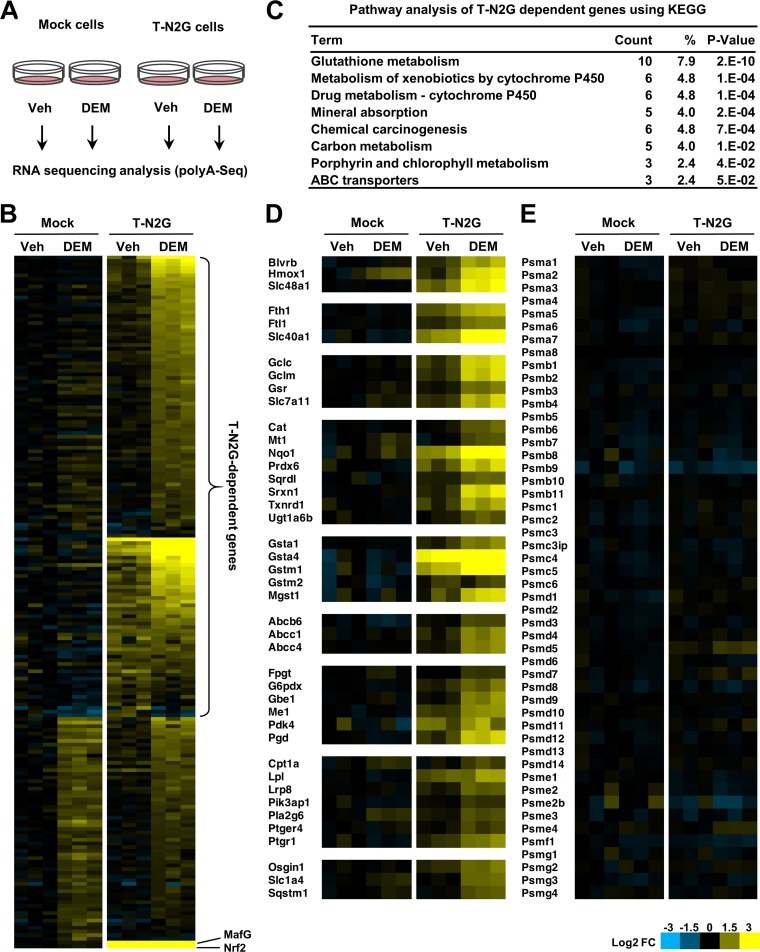
To assess how widely T-N2G activates Nrf2 target genes, we performed transcriptome sequencing (RNA sequencing [RNA-Seq]) analysis. To this end, we prepared RNA samples from the mock and T-N2G cells with or without DEM treatment (Fig. 5A). Based on differential gene expression analysis, we first identified genes whose expression was significantly induced by DEM in the T-N2G cells and genes whose basal expression in the T-N2G cells was already significantly higher than that in mock cells (Fig. 5B). These included Nrf2 and Mafg, which were derived from exogenous T-N2G cDNA.
FIG 5.
T-N2G activates a wide range of Nrf2 target genes. (A) Schematic diagram of the protocol for RNA-Seq. RNA was extracted from the mock and T-N2G cells treated with 100 μM DEM or the vehicle (Veh) for 12 h and subjected to RNA-Seq. (B) Heatmap of relative expression levels of genes whose expression was significantly induced by DEM in T-N2G cells and genes whose basal expression in T-N2G cells was significantly higher than that in mock cells (false discovery rate [FDR] < 0.05, log2 fold change [FC] > 0.7). Genes whose expression was induced by DEM, even in mock cells, are regarded as T-N2G-independent genes. The expression levels of MafG and Nrf2, shown at the bottom, are derived from the T-N2G vector. (C) Pathway analysis of T-N2G-dependent genes using KEGG data sets. Count represents the number of genes involved in each pathway, and the percent represents the percentage of genes involved in the total genes in each pathway. P values represent modified Fisher’s exact P values. (D) Heatmap of relative expression levels of genes induced in a T-N2G-dependent manner. Genes are categorized into groups based on their functions. (E) Heatmap of relative expression levels of proteasome subunit genes. Heatmap colors indicate the log2 FC values relative to the median expression level of each gene in mock cells without DEM treatment (B, D, and E).
Notably, there was a group of genes that were induced, albeit mildly, by the DEM treatment in mock cells (Fig. 5B, lower area of the left heatmaps). This finding indicates that the inductions of these genes are T-N2G independent or independent from the Nrf2-sMaf system. This group of genes includes, for instance, heat shock protein genes, implying that DEM activates signaling pathways other than the Nrf2-sMaf system. We therefore excluded these T-N2G-independent genes, plus Nrf2 and Mafg, from our current analyses and worked on the remaining 126 genes as T-N2G-dependent genes (Fig. 5B).
To obtain an overview of T-N2G-dependent genes, we then performed a pathway analysis using the KEGG database (44). Pathways involved in glutathione metabolism, xenobiotic metabolism, chemical carcinogenesis, carbon metabolism, and ABC transporters were significantly enriched in this analysis (Fig. 5C), which have been shown by the Nrf2-regulated pathways (45).
We then manually categorized the T-N2G-dependent genes according to their function. A portion of these categorizations is shown in Fig. 5D. Confirming our expectation, these T-N2G-dependent genes heavily overlap various types of Nrf2 target genes, such as genes involved in heme and iron metabolism, glutathione synthesis, and oxidative and xenobiotic-stress responses. In contrast, T-N2G did not significantly activate the proteasome subunit genes (Fig. 5E), which are known to be regulated mainly by Nrf1, demonstrating that the T-N2G protein possesses strict specificity toward the Nrf2 target genes.
Genome-wide binding of T-N2G proteins.
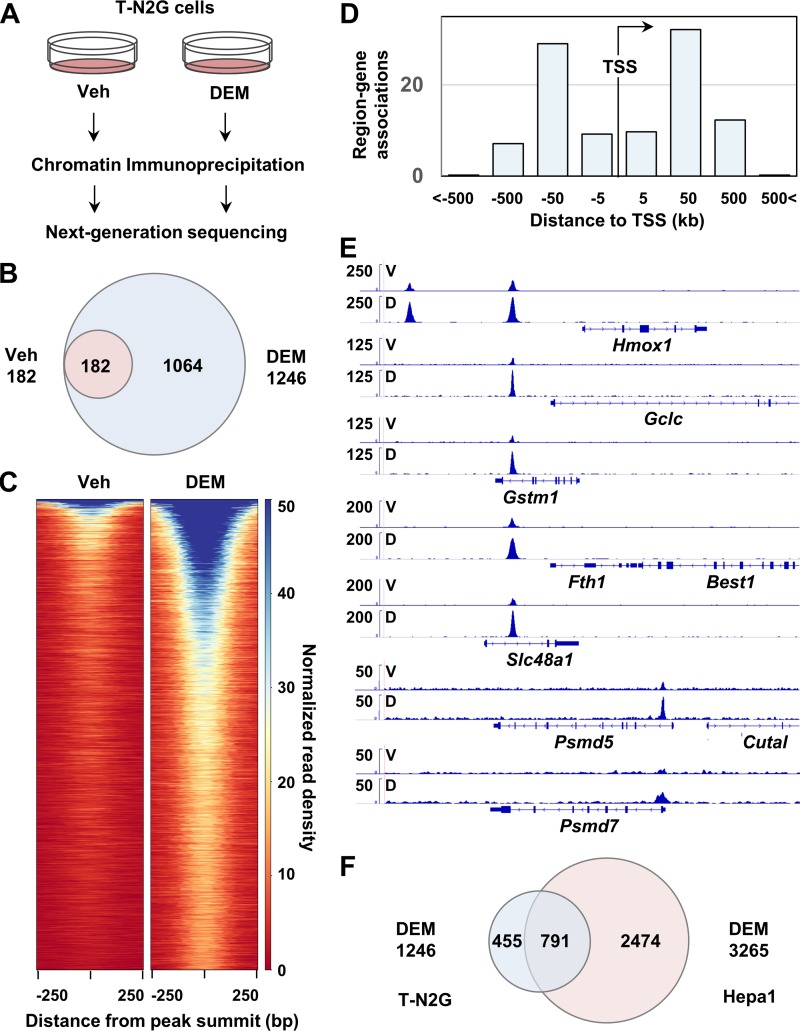
To characterize the transcriptional properties of the T-N2G protein, we determined the genome-wide binding sites of the T-N2G protein. Chromatin extracts from the T-N2G cells treated with or without DEM were subjected to immunoprecipitation, followed by next-generation deep sequencing analysis (Fig. 6A). In vehicle-treated control T-N2G cells, we identified 182 genomic regions as T-N2G binding sites by a peak calling algorithm (Fig. 6B). In contrast, as many as 1,246 regions were identified as the T-N2G binding sites in the DEM-treated T-N2G cells. The latter sites included all 182 regions identified in the vehicle-treated T-N2G cells (Fig. 6B). The fold enrichment values in the overlapped regions increased in the range from 1.3- to 6.7-fold after DEM treatment, except for one region whose value decreased by 0.6-fold (data not shown). These data indicate that virtually most of the T-N2G binding sites are inducible, while inducibility varies depending on the site. Showing very good agreement, the heatmaps of normalized ChIP-Seq signals clearly demonstrated that the majority of the T-N2G binding sites were DEM inducible (Fig. 6C). The increase in the number of positive sites reflects the increase in cellular T-N2G protein binding.
FIG 6.
Genome-wide analysis of T-N2G binding sites. (A) Schematic diagram of the protocol for ChIP-Seq. Chromatin extracts from the T-N2G cells treated with 100 μM DEM or the vehicle (Veh) for 4 h were subjected to immunoprecipitation, followed by next-generation deep sequencing analysis. (B) Venn diagram showing the overlap of the T-N2G binding sites between vehicle (Veh)-treated cells and DEM-treated cells. (C) Heatmap of normalized read density of ChIP-Seq data from Veh-treated cells and DEM-treated cells. (D) Relative positions of the T-N2G binding sites relative to the TSS. (E) Representative T-N2G binding sites in Hmox1, Gclc, Gstm1, Fth1, Slc48a1, Psmd5, and Psmd7 gene loci in the T-N2G cells treated with the vehicle (V) and DEM (D). (F) Overlap of the T-N2G binding sites between DEM-treated T-N2G cells and DEM-treated Hepa1 cells (30).
An intriguing observation was that more than 80% of the T-N2G binding sites in DEM-treated T-N2G cells were located within 50 kbp, both upstream and downstream, of transcription start sites (TSS); approximately 19% of them were in the promoter-proximal regions, i.e., within 5 kbp of TSS (Fig. 6D). We next tried to identify the nearest gene to each T-N2G binding site by exploiting the Genomic Regions Enrichment of Annotations Tool (GREAT) (46). We identified 916 genes to which the T-N2G binding sites were located within 100 kbp. These genes included typical Nrf2 target genes. Actual peaks for several of them, including those for heme oxygenase 1 (Hmox1), Gclc, glutathione S-transferase M1 (Gstm1), ferritin heavy chain 1 (Fth1), and solute carrier family 48 (heme transporter), member 1 (Slc48a1) genes, are shown in Fig. 6E.
Notably, the T-N2G binding sites were found in the possible regulatory regions of some of the proteasome subunit genes, such as Psmd5 and Psmd7 (Fig. 6E), which are known to be Nrf1 target genes. However, the bindings were relatively weak compared with those in the regulatory regions of canonical Nrf2 target genes. These genes were not induced strongly by DEM in the T-N2G cells (Fig. 5E). Based on these findings, we surmise that either the contribution of T-N2G recruited to the proteasome subunit gene regulatory regions is marginal or that the T-N2G protein has not been recruited efficiently to the regulatory regions of the genes.
We also asked whether the binding sites of T-N2G overlapped those previously identified as Nrf2-sMaf binding sites. Therefore, we compared the T-N2G binding sites identified in this study with the sites previously identified in DEM-treated Hepa1 cells as the Nrf2-MafG binding sites (30). As shown in Fig. 6F, from this comparison, we found that more than 60% of the T-N2G binding sites in MEFs (this study) overlapped the Nrf2-MafG binding sites in Hepa1 cells (30). These results strongly argue that the T-N2G protein substantially recapitulates the binding profile of the Nrf2-MafG heterodimer identified in other cell types.
CsMBEs are enriched in the T-N2G binding regions.
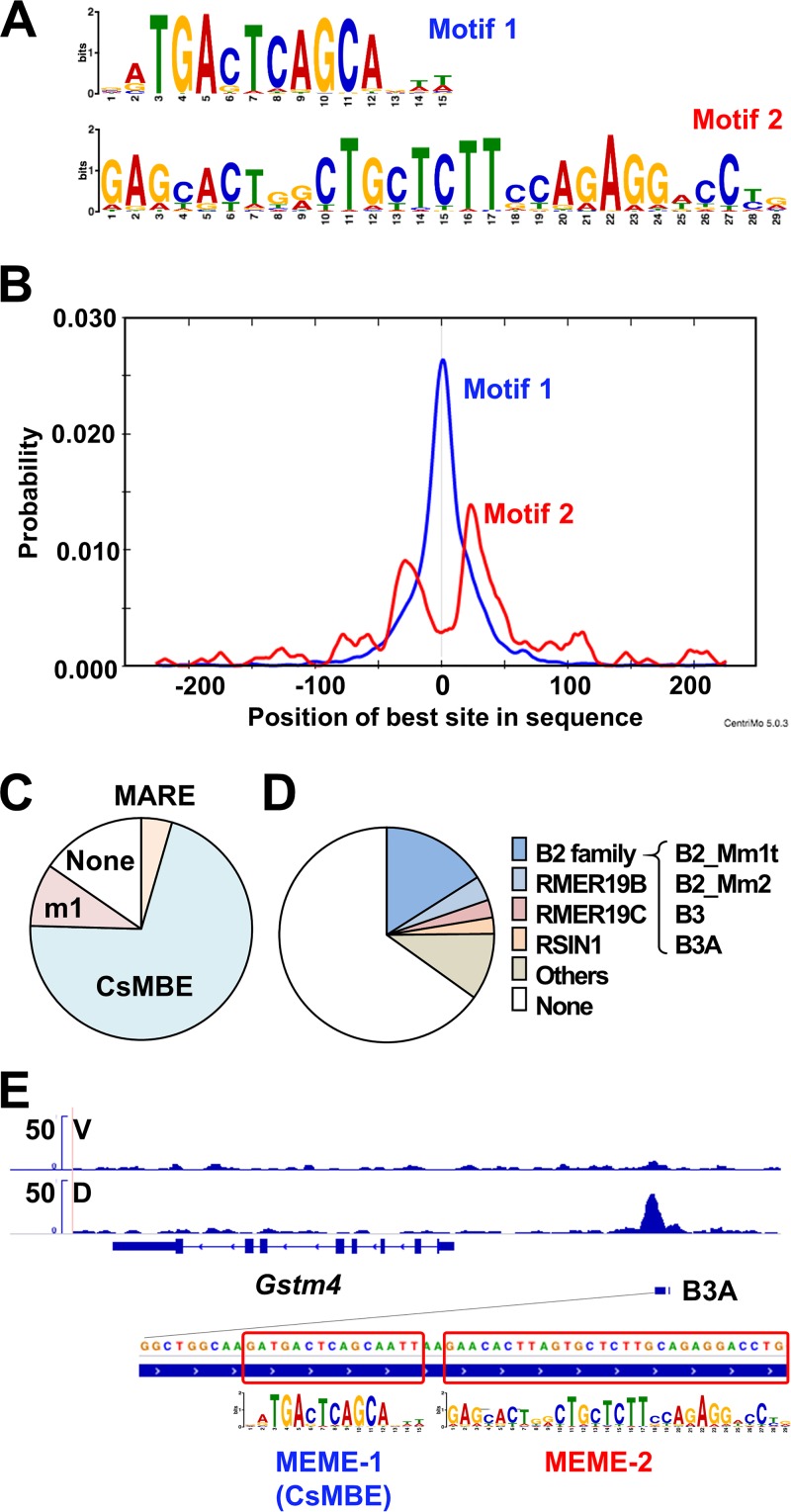
Finally, we asked whether CsMBEs are enriched in the T-N2G binding regions. To answer this question, we first performed de novo motif analysis using a web-based motif analysis tool, MEME-ChIP (47). The 500-bp regions centered on the peak summits of the T-N2G binding sites were subjected to the analysis. As a result, typical consensus CsMBE sequences were significantly enriched at the center of the tested regions (Fig. 7A and B, motif 1) with a highly significant E value, 2.4e–2935 (for definition of E value, see reference 47). We then searched for the presence of CsMBEs (RTGASNNNGC) throughout the T-N2G binding regions and found that more than 70% of the regions include perfect CsMBEs, in which underlined sequences are ideal sequences for sMaf binding (RTGASTCAGC) (Fig. 7C). We allowed one mismatch in the underlined regions of perfect CsMBEs, and we found that approximately 80% of the T-N2G binding regions include CsMBEs (Fig. 7C, CsMBE plus m1). We also searched for MAREs based on the sequence GCTGACTCAGC and found that only 4% of the regions contain MAREs, showing that CsMBE is the dominant element identified in the T-N2G binding regions. Taken together, these results support the notion that T-N2G preferentially recognizes CsMBEs in native in vivo chromatin.
FIG 7.
De novo motif analysis of T-N2G binding sites. (A) Two motifs identified by the de novo motif discovery algorithm MEME-ChIP in the T-N2G binding sites are shown. (B) Average profiles of the enriched motifs in the T-N2G binding sites determined by MEME-ChIP. (C) Pie charts showing the proportions of MARE (GCTGASTCAGC), CsMBE (RTGASTCAGC), and m1 (one mismatch allowed in CsMBEs) in the T-N2G binding sites. (D) Pie charts showing the proportions of repeats that overlapped the T-N2G binding regions. Repeats deposited in the RepeatMasker data were used. (E) A B3A element, which belongs to the B2 SINE family and harbors a CsMBE, in the regulatory region of the Gstm4 gene. The locations of two enriched motifs are boxed.
Interestingly, de novo motif analysis also showed another motif spanning relatively long regions with an E value of 3.0e–346 (Fig. 7A, motif 2). This second motif is obviously enriched at regions approximately 25 bp away from the CsMBE, suggesting this motif’s relevance to CsMBEs (Fig. 7B). We suspected that the second motif may be a part of repeat elements, such as retrotransposable elements. To test this hypothesis, we searched for repeat sequences overlapping CsMBEs in T-N2G binding regions by referring to the RepeatMasker data set (48). We found that approximately 35% of the CsMBEs in the T-N2G binding regions overlapped repeats in the data set and that 16% of the CsMBEs were embedded with the SINE B2 family (Fig. 7D). Figure 7E shows an example of a CsMBE embedded in a SINE B3A element, which belongs to the SINE B2 family, located upstream of the Gstm4 gene. In this case, a CsMBE is located at the edge of the B3A element, followed by motif 2. These results suggest that the SINE B2 elements contribute, albeit partially, to the expansion of CsMBEs over the course of molecular evolution.
DISCUSSION
Our best working hypothesis is that the CNC factors utilize sMaf factors as an obligatory heterodimeric partner referred to here as the CNC-sMaf hypothesis (20). Whereas we have been providing multiple lines of evidence supporting this hypothesis through biochemical and molecular biological analyses and mouse in vivo genetics, there still remains uncertainty in the hypothesis; therefore, a direct elucidation of this hypothesis has been necessary for a long time. In this study, we challenged the direct demonstration of the CNC-sMaf hypothesis by constructing an Nrf2-sMaf-tethered molecule, T-N2G, and a triple sMaf knockout MEF cell line, F0G0K0-T. These two means enabled us to evaluate the specific heterodimer functions of Nrf2-MafG among various heterodimer combinations without interfering with the contributions of the other CNC-sMaf dimers. The results unequivocally demonstrate that the T-N2G protein widely transactivates Nrf2 target gene expression through binding to CsMBEs localized in their regulatory regions. Thus, the Nrf2-sMaf heterodimer is the authentic dimer acting as a specific transcriptional activator for a battery of Nrf2-dependent cytoprotective genes, and our data strongly support the CNC-sMaf hypothesis.
In this study, we tested MafG as a representative partner molecule of Nrf2 because MafG is the most widely and abundantly expressed sMaf in various tissues (20, 32, 33). Since no major functional difference has been recognized in terms of their bZIP structures, we think that the current conclusion can be generalized to other Nrf2-sMaf heterodimers, such as Nrf2-MafF and Nrf2-MafK. However, it is still possible that each sMaf protein differentially contributes to Nrf2-mediated regulation. For instance, it is known that the expression of each sMaf gene is under differential regulation in various tissues (20). This observation suggests that the type and amount of active Nrf2-sMaf heterodimer are finely and tissue specifically tuned.
An important outcome of this study is the identification of the strict target gene specificity of the Nrf2-sMaf heterodimers. Indeed, T-N2G did not significantly activate the proteasome subunit genes, which are representative Nrf1 target genes. Our present ChIP-Seq data strongly agree with this conclusion and show that T-N2G is not efficiently recruited to most of the regulatory regions of proteasome subunit genes. These observations suggest that the stable binding of the T-N2G protein is allowed only in certain regulatory regions of the genes. It seems possible that the Nrf2-sMaf heterodimer forms stable complexes only when it interacts with particular neighboring transcription factors or when it interacts with the promoter complexes through appropriate transcriptional cofactors. Since it has been reported that Nrf1 and Nrf2 recruit different transcriptional cofactors to exert their transactivation activities (49–51), these cofactors may also confer target gene specificity to these CNC factors.
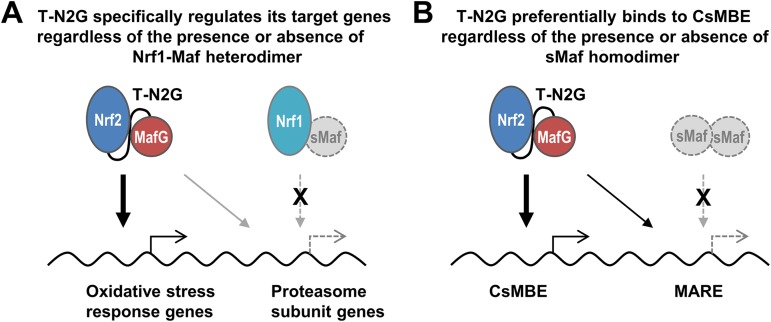
Upon evaluation of the target gene specificity of the CNC-sMaf heterodimers, it is critical to examine whether there is interference from the other endogenous CNC-sMaf heterodimers. Our results clearly exclude the possibility of interference from the other factors, since the T-N2G protein has been detected in the absence of sMaf proteins in this study. As shown in Fig. 8A, this study has excluded the possibility that endogenous Nrf1-sMaf heterodimers prevent the T-N2G protein from binding to the regulatory regions of proteasome subunit genes. We believe that this is one of the most important features of this assay, as this assay allows us to examine the function of each CNC-sMaf heterodimer in an environment that is free from interference from the other sMaf-containing dimers.
FIG 8.
Schematic models of two advantages of the evaluation system of tethered heterodimers in sMaf-deficient cells. (A) The possible interference of endogenous CNC-sMaf heterodimers can be excluded. In this case, it is suggested that the T-N2G protein preferentially regulates oxidative stress response genes even in the circumstance where endogenous Nrf1 cannot work. (B) The possible interference of endogenous sMaf homodimers can be excluded. In this case, it is suggested that the T-N2G protein preferentially binds to a CsMBE rather than to a MARE, even in circumstances where sMaf homodimers do not exist.
The current ChIP-Seq data have nicely demonstrated that asymmetric cis-acting binding sequences for the CNS-sMaf heterodimers (CsMBEs or AREs/EpREs) are clearly enriched in the T-N2G binding regions. While MARE, a palindromic Maf homodimer binding sequence, has also been found in the T-N2G binding sites, it is a minor fraction, indicating that the T-N2G protein preferentially recognizes a CsMBE. This observation strongly agrees with the results of Nrf2-MafG heterodimer analysis, in which Nrf2 and MafG are individually expressed (30). When interpreting these results, we reaffirm the importance of evaluating tethered molecules in the absence of sMafs. As shown in Fig. 8B, if endogenous sMaf proteins are present, then sMaf homodimers may prevent the T-N2G protein from binding to MARE, affecting the apparent T-N2G binding specificity. This study is the first to evaluate the binding properties of the Nrf2-sMaf heterodimer in the in vivo chromatin configuration context and to conclusively demonstrate that the Nrf2-sMaf heterodimer preferentially recognizes CsMBEs.
We identified a part of a CsMBE in the repeats of the SINE B2 family, which is a transposable element family that has expanded in rodents. Transposable elements have been reported to contribute to the evolution of transcriptional regulation by expanding the cis-acting elements (52). Our data also suggest that a portion of CsMBEs has expanded with the help of transposable elements. Since the SINE B2 family is rodent specific, additional studies are needed to clarify whether transposable elements have also contributed to the expansion of CsMBE-mediated regulation in humans.
In summary, we conducted a direct evaluation of Nrf2 and MafG heterodimer function utilizing tethered heterodimer T-N2G and sMaf-deficient test cells. These two means allowed us to evaluate the functions of the tethered molecule without any interference from the other sMaf-containing dimers. This study opens a new pathway to examine the function of each CNC-sMaf heterodimer specifically, which is indispensable to comprehending the CNC-sMaf heterodimer networks.
MATERIALS AND METHODS
Plasmid constructs.
A T-N2G expression vector was generated by two successive PCRs. In the first PCR, Flag-Nrf2 and MafG cDNAs were fused with a portion of linker peptide sequences. Using these PCR products as the templates, T-N2G cDNA was amplified by the second PCR with primers containing sequences homologous to the following PiggyBac vector (see below). By using the In-Fusion system (TaKaRa Bio), the resulting PCR product was inserted into the PiggyBac dual promoter vector containing red fluorescent protein (RFP) and a puromycin selection marker (PB514B-2; System Biosciences). Primer sequences are available upon request.
Establishment of immortalized MEFs and MEFs stably expressing T-N2G.
MEFs were prepared from MafF−/− MafG−/− MafK−/− and MafF−/− MafG+/+ MafK−/− embryos at embryonic day 11.5. After removal of the head and the internal organs, the torso was minced and dispersed into 0.25% trypsin–EDTA and maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and antibiotics. To immortalize MEFs, the simian virus 40 (SV40) large T antigen expression vector was introduced into the cells by electroporation (42). To obtain MEFs stably expressing the T-N2G protein and/or the RFP marker, the T-N2G expression vector or its empty vector was cotransfected with a transposase expression vector into sMaf-deficient MEFs. Then, at 48 h posttransfection, the cells were treated with puromycin (5 μg/ml) for the selection of stably transfected cells. The expression of RFP was confirmed in both cell lines.
Luciferase assay.
sMaf-deficient MEFs were seeded at a density of 1 × 105 cells per well into a 12-well plate. The pNQO1-ARE-Luc luciferase vector (53) was cotransfected with the empty vector or the Nrf2, MafG, or T-N2G expression vector using ViaFect transfection reagent (Promega). At 24 h posttransfection, the luciferase activities were measured by the Dual-Luciferase reporter assay system (Promega) using a luminometer (Lumat LB 9507; Berthold). The firefly luciferase activity was normalized to Renilla luciferase activity (pRL-TK; Promega). All samples were prepared in triplicate.
Immunoblot analysis.
Nuclear lysates were prepared by NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific) solubilized in Laemmli SDS sample buffer and subjected to SDS-PAGE, followed by semidry electroblotting onto an Immobilon-P membrane (Millipore, Bedford, MA). The expression of both T-N2G and endogenous Nrf2 proteins was detected using an anti-Nrf2 antibody (clone 103) (54). An anti-Lamin B1 antibody (Zymed; 33-2000) was used as a loading control for the nuclear protein.
RNA purification and qPCR analyses.
Total RNA was extracted using a Qiagen RNeasy mini kit (Qiagen) and was reverse transcribed into cDNA using a SuperScript VILO cDNA synthesis kit (Life Technologies, Carlsbad, CA). qPCR was performed with a PCR master mix using a TaqMan probe or SYBR green (Life Technologies, Carlsbad, CA). The primers and probes for Nqo1, Gsta4, and Gclc and for hypoxanthine-guanine phosphoribosyl transferase (Hprt) were described previously (38).
RNA-Seq.
Sequencing libraries were prepared from 2 μg of total RNA by a SureSelect strand-specific RNA sample prep kit (Agilent). The resulting libraries were quantified by the quantitative MiSeq (qMiSeq) method (55) and sequenced on a HiSeq 2500 sequencing system (Illumina) generating 76-bp single-end reads. Mapping and expression analyses were performed with TopHat (version 2.0.8b) and Cufflinks (version 2.1.1). Pathway analysis was conducted using the KEGG databases (44) on the DAVID platform (56).
ChIP-qPCR and ChIP-Seq analysis.
Cross-linking was performed with 1% formaldehyde for 10 min at room temperature and was subsequently quenched with 0.125 M glycine for 5 min. The samples were then lysed, and the nuclear fractions were extracted and sonicated with a focused ultrasonicator (S220; Covaris) to generate 100- to 300-bp DNA fragments. The ChIP reaction was performed with an anti-MafG antibody (30) by following the ENCODE Snyder lab’s protocol (available at http://www.encodeproject.org). Purified DNA subjected to ChIP and input DNA were used for preparing the sequencing libraries by a NEBNext Ultra II DNA library prep kit (NEB). The resulting libraries were quantified by qMiSeq and sequenced on a HiSeq 2500 sequencing system (Illumina) generating 101-bp single-end reads. Mapping and peak calling analyses were performed with Bowtie2 (version 2.2.9) and MACS2 (version 2.1.1.20160309). Heatmaps of normalized read density were generated by deepTools (version 3.1.2) (57). De novo motif analysis was performed using MEME-ChIP (47). A motif search was performed using FIMO (58).
Data availability.
All sequencing data are present in the NCBI GEO Superseries under accession no. GSE134353.
ACKNOWLEDGMENTS
We thank Nozomi Hatanaka for her excellent technical support.
This study was supported in part by the MEXT/JSPS KAKENHI, Grant-in-Aid for Scientific Research (C) 17K08617 (to F.K.), and Grant-in-Aid for Scientific Research (S) 19H05649 (to M.Y.).
We declare no conflicts of interest.
REFERENCES
- 1.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22:6321–6335. doi: 10.1128/mcb.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman JR, Keating AE. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 3.Sykiotis GP, Bohmann D. 2010. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal 3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T, Yamamoto M. 2015. Molecular basis of the Keap1-Nrf2 system. Free Radic Biol Med 88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M, Kensler TW, Motohashi H. 2018. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol Rev 98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem 280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. 2010. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HM, Han JW, Chan JY. 2016. Nuclear factor erythroid-2 like 1 (NFE2L1): structure, function and regulation. Gene 584:17–25. doi: 10.1016/j.gene.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird L, Tsujita T, Kobayashi EH, Funayama R, Nagashima T, Nakayama K, Yamamoto M. 2017. A homeostatic shift facilitates endoplasmic reticulum proteostasis through transcriptional integration of proteostatic stress response pathways. Mol Cell Biol 37:e00439-16. doi: 10.1128/MCB.00439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. 2008. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem 283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotsu Y, Hataya N, Katsuoka F, Yamamoto M. 2012. NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1beta genes. Mol Cell Biol 32:2760–2770. doi: 10.1128/MCB.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsuoka F, Yamazaki H, Yamamoto M. 2016. Small Maf deficiency recapitulates the liver phenotypes of Nrf1- and Nrf2-deficient mice. Genes Cells 21:1309–1319. doi: 10.1111/gtc.12445. [DOI] [PubMed] [Google Scholar]
- 17.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. 1989. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci U S A 86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara KT, Kataoka K, Nishizawa M. 1993. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene 8:2371–2380. [PubMed] [Google Scholar]
- 19.Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH. 1993. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc Natl Acad Sci U S A 90:11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsuoka F, Yamamoto M. 2016. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene 586:197–205. doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. 2000. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 22.Venugopal R, Jaiswal AK. 1998. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 23.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, Alam J. 2001. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 24.Mignotte V, Eleouet JF, Raich N, Romeo PH. 1989. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci U S A 86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rushmore TH, Morton MR, Pickett CB. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266:11632–11639. [PubMed] [Google Scholar]
- 26.Friling RS, Bensimon A, Tichauer Y, Daniel V. 1990. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc Natl Acad Sci U S A 87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M. 2016. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med 91:45–57. doi: 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa H, Motohashi H, Sueno S, Kimura M, Takagawa H, Kanno Y, Yamamoto M, Tanaka T. 2009. Structural basis of alternative DNA recognition by Maf transcription factors. Mol Cell Biol 29:6232–6244. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M, Yamamoto T, Zhang J, Itoh K, Kyo M, Kamiya T, Aburatani H, Katsuoka F, Kurokawa H, Tanaka T, Motohashi H, Yamamoto M. 2007. Molecular basis distinguishing the DNA binding profile of Nrf2-Maf heterodimer from that of Maf homodimer. J Biol Chem 282:33681–33690. doi: 10.1074/jbc.M706863200. [DOI] [PubMed] [Google Scholar]
- 30.Hirotsu Y, Katsuoka F, Funayama R, Nagashima T, Nishida Y, Nakayama K, Engel JD, Yamamoto M. 2012. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shavit JA, Motohashi H, Onodera K, Akasaka J, Yamamoto M, Engel JD. 1998. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev 12:2164–2174. doi: 10.1101/gad.12.14.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onodera K, Shavit JA, Motohashi H, Katsuoka F, Akasaka JE, Engel JD, Yamamoto M. 1999. Characterization of the murine mafF gene. J Biol Chem 274:21162–21169. doi: 10.1074/jbc.274.30.21162. [DOI] [PubMed] [Google Scholar]
- 33.Onodera K, Shavit JA, Motohashi H, Yamamoto M, Engel JD. 2000. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J 19:1335–1345. doi: 10.1093/emboj/19.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. 2005. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci U S A 102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung L, Kwong M, Hou S, Lee C, Chan JY. 2003. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem 278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki H, Katsuoka F, Motohashi H, Engel JD, Yamamoto M. 2012. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol Cell Biol 32:808–816. doi: 10.1128/MCB.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. 2005. Genetic evidence that small Maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol 25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhold LA, Wold B. 1993. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell 74:1033–1042. doi: 10.1016/0092-8674(93)90725-6. [DOI] [PubMed] [Google Scholar]
- 40.Kotkow KJ, Orkin SH. 1995. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol 15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirotsu Y, Katsuoka F, Itoh K, Yamamoto M. 2011. Nrf2 degron-fused reporter system: a new tool for specific evaluation of Nrf2 inducers. Genes Cells 16:406–415. doi: 10.1111/j.1365-2443.2011.01496.x. [DOI] [PubMed] [Google Scholar]
- 43.Higashi C, Kawaji A, Tsuda N, Hayashi M, Saito R, Yagishita Y, Suzuki T, Uruno A, Nakamura M, Nakao K, Furusako S, Yamamoto M. 2017. The novel Nrf2 inducer TFM-735 ameliorates experimental autoimmune encephalomyelitis in mice. Eur J Pharmacol 802:76–84. doi: 10.1016/j.ejphar.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 44.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 46.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma W, Noble WS, Bailey TL. 2014. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc 9:1428–1450. doi: 10.1038/nprot.2014.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter 4:Unit 4.10. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Hosoya T, Maruyama A, Nishikawa K, Maher JM, Ohta T, Motohashi H, Fukamizu A, Shibahara S, Itoh K, Yamamoto M. 2007. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem J 404:459–466. doi: 10.1042/BJ20061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekine H, Okazaki K, Kato K, Alam MM, Shima H, Katsuoka F, Tsujita T, Suzuki N, Kobayashi A, Igarashi K, Yamamoto M, Motohashi H. 2018. O-GlcNAcylation signal mediates proteasome inhibitor resistance in cancer cells by stabilizing NRF1. Mol Cell Biol 38:e00252-18. doi: 10.1128/MCB.00252-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han JW, Valdez JL, Ho DV, Lee CS, Kim HM, Wang X, Huang L, Chan JY. 2017. Nuclear factor-erythroid-2 related transcription factor-1 (Nrf1) is regulated by O-GlcNAc transferase. Free Radic Biol Med 110:196–205. doi: 10.1016/j.freeradbiomed.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katsuoka F, Motohashi H, Engel JD, Yamamoto M. 2005. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J Biol Chem 280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- 54.Maruyama A, Tsukamoto S, Nishikawa K, Yoshida A, Harada N, Motojima K, Ishii T, Nakane A, Yamamoto M, Itoh K. 2008. Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch Biochem Biophys 477:139–145. doi: 10.1016/j.abb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Katsuoka F, Yokozawa J, Tsuda K, Ito S, Pan X, Nagasaki M, Yasuda J, Yamamoto M. 2014. An efficient quantitation method of next-generation sequencing libraries by using MiSeq sequencer. Anal Biochem 466:27–29. doi: 10.1016/j.ab.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 56.Huang dW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing data are present in the NCBI GEO Superseries under accession no. GSE134353.