Abstract
Loss of functional metabolic muscle mass remains a strong and consistent predictor of mortality among people living with human immunodeficiency virus (PLWH). PLWH have a higher incidence of alcohol use disorder (AUD), and myopathy is a significant clinical comorbidity due to AUD. One mechanism of skeletal muscle (SKM) mass maintenance and repair is by differentiation and fusion of satellite cells (SCs) to existing myofibers. Previous studies demonstrated that chronic binge alcohol (CBA) administration decreases SC differentiation potential, myogenic gene expression, and miR-206 expression in simian immunodeficiency virus (SIV)-infected male rhesus macaques and that miR-206 targets the Class IIA histone deacetylase, HDAC4. The aim of this study was to determine whether alcohol-induced increases in Class IIA HDACs mediate the observed decrease in differentiation potential of SCs. Data show that CBA dysregulated HDAC gene expression in SKM and myoblasts of SIV-infected macaques. CBA and antiretroviral therapy increased HDAC activity in SKM and this was positively correlated with HDAC4 gene expression. In vitro ethanol (ETOH) treatment increased HDAC expression during differentiation and decreased differentiation potential of myoblasts. HDAC expression was negatively correlated with fusion index and myotube formation, indicators of differentiation potential. Treatment with a Class II HDAC inhibitor, TMP195, restored differentiation in ETOH-treated myoblasts. MEF2C expression at day 3 of differentiation was positively correlated with fusion index and myotube formation. These findings suggest that an alcohol-mediated increase in Class IIA HDAC expression contributes to decreased myoblast differentiation by downregulating MEF2C, a transcription factor critical for myogenesis.
Keywords: chronic alcohol, histone deacetylases, stem cell differentiation
INTRODUCTION
Loss of lean body mass has been associated with poor health outcomes, opportunistic infections, and decreased survival in persons living with human immunodeficiency virus (PLWH) (11, 12, 16) and remains a consistent predictor of mortality (48). Antiretroviral therapy (ART) has decreased patient mortality and increased life expectancy among PLWH, leading to an increased need to study nonacquired immunodeficiency syndrome (AIDS) comorbidities in this population (4, 7). The prevalence of alcohol use disorder is significantly higher in PLWH than in the general population, and at-risk alcohol consumption increases the risk for comorbid clinical complications including alcoholic myopathy (14, 15). Myopathy is a significant clinical concern in PLWH, as it is a predictor of mortality even in the absence of overt muscle wasting (37, 40). Alcoholic myopathy occurs in up to 60% of chronic alcoholics as a result of an imbalance between anabolic and catabolic mechanisms that maintain skeletal muscle (SKM) mass (38, 44, 50), and at-risk alcohol use accentuates SKM atrophy in human immunodeficiency virus (HIV)/AIDS (5, 51). Our studies have examined the underlying mechanisms of accentuated SKM atrophy in a nonhuman primate model of HIV infection. Our data show that chronic binge alcohol (CBA) exacerbates loss of SKM mass and accelerates time to end-stage disease in non-ART-treated, simian immunodeficiency virus (SIV)-infected male rhesus macaques through mechanisms that include increased proteasomal activity, a profibrotic milieu, decreased antioxidant capacity, mitochondrial dyshomeostasis, and dysregulation of gene networks that regulate muscle homeostasis and satellite cell (SC) activation (8, 10, 19, 43). The latter are crucial initial steps in myogenesis.
Myogenesis occurs in response to muscle injury or atrophy and is orchestrated by muscle regulatory factors (MRFs) and transcription factors that coordinate the proliferation and fusion of muscle SCs to restore normal muscle fiber size (1, 3, 56). Our studies have shown that CBA-mediated impaired differentiation potential was associated with decreased myogenic gene expression (45). The studies also showed that cultured myoblasts retained gene expression changes resulting from in vivo CBA administration, suggesting an epigenetic memory that could mediate impaired myogenesis. The three major epigenetic modifications are DNA methylation, noncoding RNA interference of transcription and translation, and posttranslational modifications of histones (24). Histone modification alters chromatin structure by either making DNA more accessible and thus increasing gene transcription and expression, or condensation of chromatin decreasing expression (24). Our data demonstrated alterations in microRNA (miR) and HDAC associated with alcohol-mediated impaired myogenesis (42, 43). Myoblasts isolated from CBA/SIV/ART+ macaques showed decreased myogenic capability after up to four in vitro passages in the absence of ethanol (ETOH) or SIV viral protein exposure. Moreover, CBA significantly decreased miR-206 expression in SKM and myoblasts of SIV-infected macaques, and this was correlated with decreased myogenic gene expression (42). Skeletal muscle-specific miR-206 targets HDAC4 (42), a class IIA histone deacetylase that represses muscle differentiation (46, 53, 55).
HDACs regulate gene expression by two mechanisms. Classically, HDACs remove acetyl groups from lysine residues on histone tails, leading to tightly packed chromatin and a decrease in gene transcription due to a lack of accessibility by transcription factors. HDACs can also modulate transcription factor by activating or deactivating transcription factors via deacetylation or by binding to transcription factors and preventing them from transcribing target genes (20). Class IIA HDACs are highly expressed in SKM, heart, thymus, and brain (53). They have repression and deacetylase domains and mainly function as transcriptional repressors by binding directly to transcription factors or acting through other corepressors (55). HDACs 4, 5, and 7 contain a nuclear export sequence and a nuclear localization sequence and exhibit a dynamic trafficking equilibrium between the nucleus and cytoplasm (25, 26, 54). Perturbation of this equilibrium has been implicated in impaired myogenesis (9, 28), and upregulation of these HDACs can alter equilibrium. Class IIA HDACs also contain an MEF2 binding site allowing for repression of Mef2-mediated transcription. Mef2 is required for MRF-induced myogenic conversion and myotube formation in C2C12 myoblasts (34), and overexpression of Class IIA HDACs inhibits muscle differentiation partially by inhibiting Mef2 activity (23).
Taking together our previous data and published literature, we hypothesized that alcohol decreases myoblast differentiation by upregulating Class IIA histone deacetylases and that the impaired differentiation could be ameliorated using a selective Class IIA HDAC inhibitor. Results indicate that CBA increased Class IIA HDAC expression in SKM and SC of SIV-infected, ART-treated male rhesus macaques and that ETOH treatment of naïve myoblasts increased Class IIA HDAC expression during differentiation. Administration of a selective Class IIA HDAC inhibitor restored differentiation in ETOH-treated myoblasts, with an increase in MEF2C expression. These studies suggest that ETOH impairs myoblast differentiation via a Class IIA HDAC-mediated mechanism that decreases MEF2C expression, thus hindering progression through myogenic programming.
METHODS
Animal Experiments
All experiments described in this study were approved by the Institutional Animal Care and Use Committee at Louisiana State University Health Sciences Center (LSUHSC, New Orleans, LA) and Tulane National Primate Research Center (TNPRC, Covington, LA) and adhered to the National Institutes of Health guidelines for the care and use of experimental animals. Detailed experimental design including virological, immune, and metabolic data collected from the parent longitudinal study have been previously published (2, 13, 29). Briefly, adult (4–6 yr old) male rhesus macaques (Macaca mulatta) were obtained from TNPRC. Animals were surgically implanted with gastric catheters and administered alcohol daily for 30 min at a dose of 13–14g ETOH/kg body weight/week. Macaques achieved a blood alcohol level of 50–60 mM, 2 h after the start of alcohol initiation. Daily CBA or isocaloric sucrose (VEH) administration continued throughout the study. After 3 mo of CBA or VEH administration, animals were inoculated intrarectally with 1,000 times the TCID50 of SIVmac251 and SIV disease progression monitored throughout the study. After 2.5 mo of SIV infection, at viral set-point, animals were randomized to receive daily subcutaneous injections of 20 mg/kg tenofovir {9-[R-2-(phosphonomethoxy) propyl] adenine} and 30 mg/kg emtricitabine, provided by Gilead Sciences (Foster City, CA) for a total of four treatment groups: VEH/SIV/ART− (n = 4), VEH/SIV/ART+ (n = 7), CBA/SIV/ART− (n = 6), and CBA/SIV/ART+ (n = 7). This ART protocol suppresses viral load in VEH and CBA animals and does not have renal or hepatic toxicity (29). Following 11.5 mo of SIV infection and 14.5 mo of CBA or VEH administration, all macaques were euthanized according to the American Veterinary Medical Association’s guidelines. Three non-ART-treated macaques met criteria for euthanasia before study end point.
Myoblast Isolation and Culture
Quadriceps muscle biopsy samples were excised from fasted animals at baseline or at study end point. Primary myoblasts were isolated from quadriceps femoris muscle according to previously published protocols (45). In brief, SKM tissue (~100 mg) was enzymatically dissociated with 0.05% trypsin during two 1 h treatments. Cells were plated for 4–5 h for fibroblast separation. Nonadhered cells were collected and cultured in Ham’s F-12 medium (Thermo Fisher, Waltham, MA) with 10% fetal bovine serum (FBS, Thermo Fisher) and 2.5 ng/ml human fibroblast growth factor (R & D Systems, Minneapolis, MN) in 100 mm dishes and grown to 70% confluence (passage 0). Myoblasts were cultured on collagen type I-coated plates (Corning, Corning, NY) and frozen at each passage in FBS + 10% DMSO. More than 90% of cells from all primary macaque myoblast lines are positive for PAX7 and α7-integrin by flow cytometry (PAX7+/ α7-integrin+) (45). Myoblasts used in experiments in this study were at passage 3 (P3) or passage 4 (P4).
HDAC Activity Assay
SKM tissue collected at baseline and study end point was homogenized in tissue protein extraction (T-PER) buffer (Pierce Thermo Scientific, Rockford, IL) with HALT protease and phosphatase inhibitor cocktail (Thermo Fisher) and centrifuged at 4°C at 12,000 g for 15 min, and supernatants were removed for cytoplasmic protein. The pellet was homogenized in RIPA Lysis Buffer 1X (Millipore Sigma, Burlington, MA) with a syringe and needle followed by vortexing and centrifuged at 4°C at 12,000 g for 15 min, and supernatants containing nuclear protein were used for analyses. Protein concentration was determined with the Pierce bicinchoninic acid protein assay kit (Thermo Fisher). HDAC activity was determined with the HDAC Activity Fluorometric Assay kit (BioVision, Milpitas, CA) using 20 µg nuclear protein according to manufacturer’s instructions. Relative fluorescent units (RFU) were read following 60 min incubation with a lysine developer at excitation/emission wavelength 370 nm/450 nm.
In Vitro Ethanol Treatment
Naïve myoblasts (P4) were plated 80,000 cells/well in six-well collagen-coated plates (Corning) in growth media (Ham’s F-12, 10% FBS, 2% L-glutamine, 1% P/S/F). In vitro ethanol treatment was by supplementing media with 50 mM ETOH during proliferation and differentiation and cultured in a 75 mM ETOH atmosphere at 37°C and 5% CO2 to prevent evaporation of ETOH. The incubator was supplemented with 75 mM ETOH water bath every 48 h. Myoblasts were grown to 80% confluence (~3 days, proliferation phase) then changed to differentiation media (Ham’s F-12, 2% horse serum, 2% L-glutamine, 1% P/S/F). Media were changed every 48 h during proliferation and differentiation. Cells were harvested at the end of proliferation and after 5 days of differentiation by trypsinization (0.25% trypsin-EDTA, GIBCO, Thermo Fisher) for RNA extraction and reverse transcription-quantitative PCR (RT-qPCR). After 5 days of differentiation, one well of each cell line and each treatment group was fixed with cold methanol for 10 min and air-dried for quantification of differentiation.
Treatment with Class IIA HDAC Inhibitor, TMP195
TMP195, a pan-Class IIA HDAC inhibitor, was used to determine whether HDAC inhibition would rescue impaired myoblast differentiation. TMP195 inhibits HDACs both in vivo and in vitro (17) and targets HDACs 4, 5, and 7 at concentrations below 100 nM (22). Its efficacy was confirmed in our studies using the HDAC activity assay in which TMP195 at 100 nM reduced HDAC activity in nuclear protein extracts from SKM tissue at levels comparable to that of the pan-HDAC inhibitor Trichostatin A. Using trichostatin, we found a 96.9% decrease in HDAC activity, and for TMP195, there was a 96.7% decrease in activity as measured by RFU. Standard curves generated using the deacetylated substrate indicate these activity levels correspond to ~1.4 μmol of deacetylated substrate compared with ~17 μmol deacetylated substrate in controls. Naïve P4 macaque myoblasts were plated 80,000 cells/well in six-well collagen-coated plates with growth media. Myoblasts were supplemented with 50 mM ETOH in media (ETOH) or control (CTRL) and grown as stated above. Myoblasts were grown to 80% confluence (~3 days, proliferation phase) then changed to differentiation media. During differentiation, media were supplemented with 50 mM ETOH and 100 nM TMP195 or VEH (0.1% DMSO) yielding four groups, n = 5–7 cell lines for each (CTRL/VEH, CTRL/TMP195, ETOH/VEH, ETOH/TMP195). Media were changed every 24 h. Cells were harvested after 3 days of differentiation and 5 days of differentiation by trypsinization for RNA extraction and RT-qPCR, and wells were fixed with cold methanol for 10 min and air-dried at end of differentiation.
qPCR for Gene Expression
RNA extraction.
Total RNA was extracted from SKM (25–35 mg) and myoblast (500,000–1,000,000 cells) samples using the miRNeasy Mini kit (Qiagen Sciences; Germantown, MD). For the in vitro studies, RNA was extracted from myoblasts (5 × 105–1 × 106 cells) using RNeasy Mini kit (Qiagen Sciences). For SKM, cDNA was synthesized from 1 μg of the resulting total RNA with the QuantiTect Reverse Transcriptase kit (Qiagen Sciences), and for myoblasts 250 or 500 ng total RNA was used.
qPCR.
Primers (Table 1) were designed to span exon-exon junctions (Integrated DNA Technologies, Coralville, IA) and used at a concentration of 500 nmol. HDACs 4, 5, and 7 and myogenic genes PAX7, MEF2C, and MYF5 were assessed. The final reactions were made to a total volume of 20 µl with QuantiTect SYBR Green PCR kit (Qiagen). All reactions were carried out in duplicate on a CFX96 real-time PCR system (Bio-Rad Laboratories, Hercules, CA). qPCR data were analyzed by the comparative CT method (ΔΔCT). Target genes were compared with the endogenous control ribosomal protein S13 (RPS13). We have previously shown RPS13 does not change among different treatments and tissues in SIV-infected macaques and serves as a reliable reference gene for normalization of mRNA (41). Results from in vivo SKM and myoblast experiments were normalized to control values obtained from quadriceps muscle biopsies of naïve male macaques. In vitro ETOH and ETOH/TMP195 myoblast experiments were normalized to CTRL or CTRL/VEH group, respectively.
Table 1.
List of primers used for qPCR analysis
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| HDAC4 | GGA GTG TCA ACC TCC TAT AAC | GTC TTT CGG CCA CTT TCT |
| HDAC5 | GCA GGA GAG CTC AAG AAT G | CAG TGA TGG CTA CAG AGT TG |
| HDAC7 | TGG ACA CTG ACA CCA TCT | CCA CAG CGA AAC CAT TCT |
| PAX7 | TCC TGG AAG AAG GTG GTT GAA TGC | CGT GTG CAG GTC TGG TTC AGT AA |
| MEF2C | GTC AAT TGG GAG CTT GCA CTA GCA | TGG TAC GGT CTC TAG GAG GAG AAA |
| MYF5 | AGC CCT ACC TCC AAC TGT TCT GAT | AGG TTG CTC TGA GGA GGT GAT CC |
| RPS13 | TCT GAC GAC GTG AAG GAG CAG ATT | TCT CTC AGG ATC ACA CCG ATT TGT |
Primers are from IDT Technologies.
Jenner-Giemsa Staining for Assessment of Differentiation
Jenner-Giemsa staining was performed as previously described (42, 45, 52). In brief, methanol-fixed cells were exposed to Jenner staining solution (Fisher Scientific, Hampton, NH) for 10 min, rinsed, and then exposed to Giemsa stain (Fisher Scientific) for 10 min. The stained cells were air-dried, and photomicrographs were taken using an Olympus BX51 microscope in a blinded fashion. Fusion index was determined by counting the total number of nuclei incorporated into myotubes (≥2 nuclei in a single cell) and dividing by the total number of nuclei present. The number of myotubes was measured as the total number of myotubes with ≥3 nuclei per field of measurement.
Statistical Analysis
All data are presented as means ± SE, where n = 4–7 for each animal treatment group at baseline and study end-point (specific n for each group presented in figure legends). For in vitro studies, all data are presented as means ± SE, where n = 5–7 for each treatment group. Two-way analysis of variance (ANOVA) was used to compare all measures containing four treatment groups. For ETOH and TMP195 experiments, gene expression and differentiation at day 3 and day 5 are expressed as % CTRL, to reflect that each treatment group was normalized to CTRL/VEH values. This is due to variability among the primary macaque cell lines. One-way ANOVA was used to analyze gene expression and HDAC activity in 11 mo post-SIV myoblasts, and Tukey’s post hoc analysis was used when one-way ANOVA revealed statistical group differences. Student’s t test was used to compare gene expression and differentiation measures in ETOH in vitro experiments (Prism GraphPad version 5; La Jolla, CA). Statistical significance was established at P < 0.05.
RESULTS
CBA Increases HDAC4 Expression in SKM of SIV-infected Male Macaques
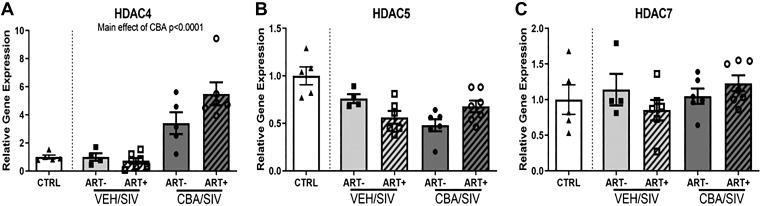
Expression of Class IIA histone deacetylases 4, 5, and 7, enriched in SKM and implicated in myogenesis, were determined in the quadriceps muscle. HDAC4 expression was significantly (P < 0.05) higher in muscle of the CBA/SIV macaques irrespective of ART status (Fig. 1A). There were no statistically significant differences in the mRNA expression of HDAC5 and HDAC7 due to CBA, SIV or ART (Fig. 1, B and C).
Fig. 1.
Relative gene expression of Class IIA histone deacetylases, HDAC4, HDAC5, and HDAC7 in skeletal muscle. A: there was a main effect of chronic binge alcohol (CBA) to significantly increase HDAC4 expression. Control (white bars); sucrose (VEH)/simian immunodeficiency virus (SIV)/antiretroviral therapy (ART)− (light gray bars), VEH/SIV/ART+ (light gray diagonal hatched bars), chronic binge alcohol (CBA)/ SIV/ ART− (dark gray bars), and CBA/SIV/ART+ (dark gray diagonal hatched bars). B: there was no significant effect of CBA, SIV, and ART on HDAC5 expression in skeletal muscle. C: there was no significant effect of CBA, SIV, and ART on HDAC7 expression in skeletal muscle. 2-way ANOVA. Values are means ± SE.
CBA and ART increased nuclear HDAC Activity, which correlates positively with HDAC4 expression
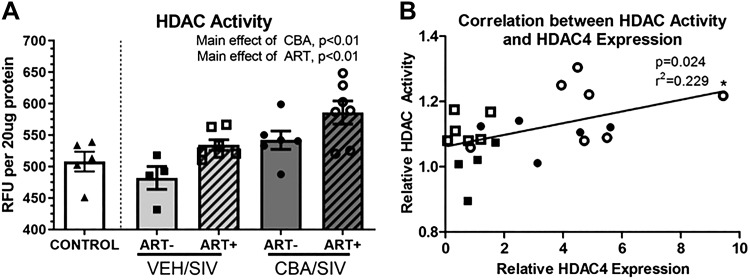
Since HDAC mRNA expression was increased in the quadriceps muscle, total HDAC activity was determined. There was a main effect of CBA and ART to significantly increase HDAC activity in quadriceps muscle (Fig. 2A). HDAC activity was not affected by SIV. HDAC activity was positively correlated HDAC4 expression (Fig. 2B), but not with HDAC5 or 7 expression.
Fig. 2.
Histone deacetylase (HDAC) activity in skeletal muscle of SIV-infected macaques. A: there was a main effect of chronic binge alcohol (CBA) and antiretroviral therapy (ART) to significantly increase total nuclear HDAC activity in skeletal muscle. Control (white bars); sucrose (VEH)/simian immunodeficiency virus (SIV)/ART− (light gray bars), VEH/SIV/ART+ (light gray diagonally hatched bars), CBA/ SIV/ ART− (dark gray bars) and CBA/SIV/ART+ (dark gray diagonally hatched bars). B: there was a significant positive correlation of HDAC4 expression with total nuclear HDAC activity. RFU, relative fluorescence units. 2-way ANOVA and Pearson correlation. Values are means ± SE.
CBA Increases HDAC4 Expression, and SIV Increases HDAC5 Expression in Proliferating Myoblasts
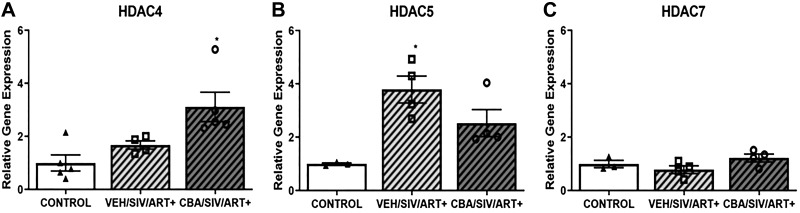
Expression of histone deacetylases 4, 5, and 7 was determined in myoblasts during the proliferation phase. CBA administration significantly increased HDAC4 expression compared with control myoblasts (Fig. 3A). VEH/SIV/ART+ myoblasts had significantly higher HDAC5 expression than control myoblasts (Fig. 3B). Although CBA increased HDAC5 expression, it failed to achieved statistical significance (P = 0.051). CBA or SIV did not alter the expression of HDAC7 expression in myoblasts.
Fig. 3.
Relative gene expression of Class IIA histone deacetylases, HDAC4, HDAC 5, and HDAC7 in myoblasts. A: there was significant increase in HDAC4 expression in myoblasts of chronic binge alcohol (CBA)/simian immunodeficiency virus (SIV)/antiretroviral (ART)+ (dark gray hatched bars) macaques compared with VEH/SIV/ART+ (light gray hatched bars) and CONTROL (white bars) myoblasts during proliferation. B: there was a significant increase in HDAC5 expression in myoblasts of VEH/SIV/ART+ compared with CONTROL macaques. C: there were no significant differences in HDAC7 expression in myoblasts of CONTROL, VEH/SIV/ART+, and CBA/SIV/ART+ macaques. *P < 0.05 (1-way ANOVA). Values are means ± SE.
ETOH Increases Class IIA HDAC Expression in Naïve Myoblasts during Differentiation
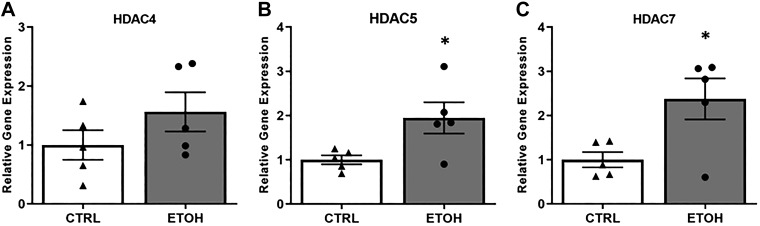
Naïve myoblasts were cultured in growth media supplemented with 50 mM ETOH for 3 days and differentiated in media supplemented with 50 mM ETOH for 5 days. HDAC4 expression measured during differentiation was 1.6 ± 0.3 (P = 0.196, Fig. 4A) in ETOH-treated myoblasts compared with CTRL. HDAC5 (1.8 ± 0.5, P < 0.05, Fig. 4B) and HDAC7 (2.3 ± 0.46, P < 0.05, Fig. 4C) expression was significantly increased in ETOH-treated compared with CTRL myoblasts.
Fig. 4.
Class IIA HDAC expression in ethanol (ETOH)-treated myoblasts. A: there was no significant change in HDAC4 expression in ETOH-treated (dark gray bar) compared with CTRL (white bars) myoblasts at day 5 of differentiation. B: there was a significant increase in HDAC5 expression in ETOH-treated compared with CTRL myoblasts. C: there was a significant increase in HDAC7 expression in ETOH-treated compared with CTRL myoblasts. *P < 0.05 (t test). Values are means ± SE.
ETOH Decreases and TMP195 Restores Differentiation in Naïve Myoblasts
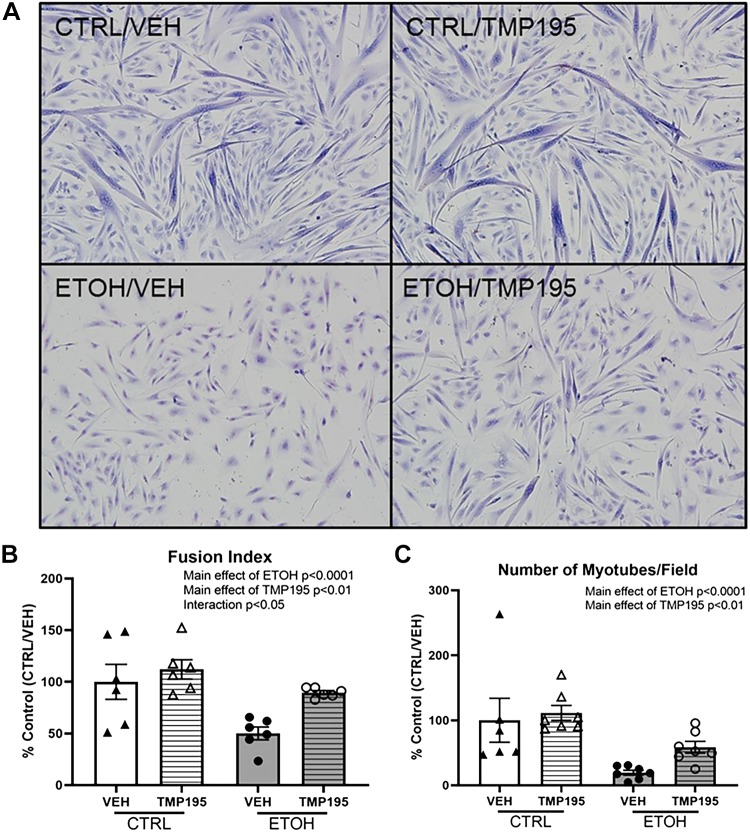
Since a negative relationship was observed between Class IIA HDACs and differentiation, we predicted that targeting these enzymes might restore myogenic potential. Myoblasts were differentiated in the presence of ETOH for 5 days. There was a significant decrease in differentiation in the ETOH-treated myoblasts compared with CTRL myoblasts, as measured by Jenner-Giemsa staining and quantification (Fig. 5A). ETOH significantly decreased the fusion index (Fig. 5B), and number of myotubes formed (Fig. 5C). Selective Class IIA HDAC inhibitor, TMP195, had no significant effect on CTRL myoblasts but significantly restored myotube formation and fusion index (Fig. 5, B and C) in ETOH-treated myoblasts.
Fig. 5.
In vitro differentiation potential of ethanol (ETOH) and Class IIA histone deacetylase inhibitor, TMP195-treated myoblasts. A: representative images of Jenner-Giemsa stained myoblast cultures from CTRL/VEH, CTRL/TMP195, ETOH/VEH, and ETOH/TMP195 at day 5 of differentiation. B: there was a main effect of ETOH to decrease and TMP195 to increase fusion index. CTRL/VEH (white bars), CTRL/TMP195 (white horizontal hatched bars), ETOH/VEH (dark gray bars), and ETOH/TMP195 (dark gray horizontal hatched bars). C: there was a main effect of ETOH to decrease and a main effect of TMP195 to increase number of myotubes per field at day 5 of differentiation. For statistical purposes, treatment group myoblasts are normalized to their respective controls due to variability between primary macaque cell lines. 2-way ANOVA. Values are means ± SE.
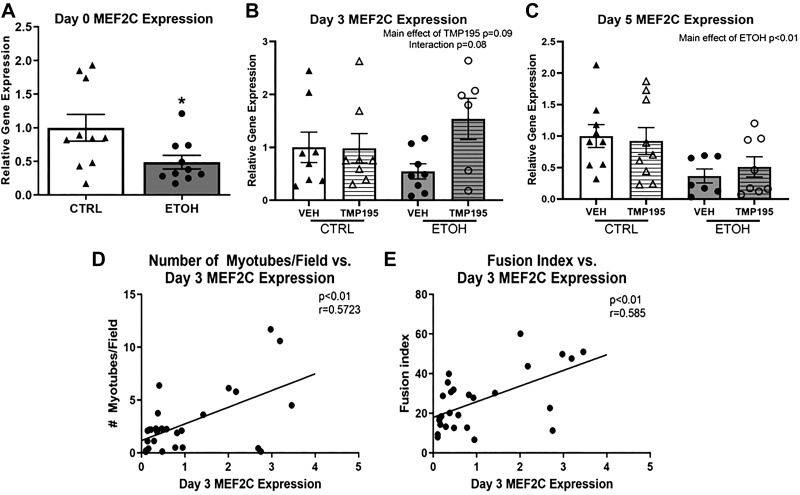
ETOH Decreases and TMP195 Restores MEF2C Expression, which Correlates Positively with Differentiation
Since TMP195 restored myoblast differentiation, myogenic gene expression was determined during differentiation. Ethanol significantly decreased MEF2C expression at day 0 of differentiation (Fig. 6A). At day 3 of differentiation, there was a main effect of ETOH to decrease MEF2C expression (P = 0.06) and a main effect of TMP195 to increase MEF2C expression (P = 0.09) (Fig. 6B). MEF2C expression at this time point (day 3) significantly correlated with fusion index and number of myotubes per field quantified at day 5 of differentiation (Fig. 6, D and E). At day 5 of differentiation, there was a main effect of ETOH to decrease MEF2C expression (Fig. 6C).
Fig. 6.
Relative gene expression of MEF2C in ethanol (ETOH) and Class IIA histone deacetylase inhibitor, TMP195-treated myoblasts. A: there was a significant decrease in MEF2C expression during proliferation in ETOH-treated (dark gray bars) compared with CTRL (white bars) myoblasts. B: MEF2C expression at day 3 of differentiation in CTRL/VEH (white bars), CTRL/TMP195 (white horizontally hatched bars), ETOH/VEH (dark gray bars), ETOH/TMP195 (dark gray horizontally hatched bars). Main effect of TMP195, P = 0.09, Interaction of ETOH and TMP195, P = 0.08. C: there was a significant decrease in MEF2C expression at day 5 of differentiation in ETOH/VEH and ETOH/TMP195 groups compared with CTRL/VEH and CTRL/TMP195 groups. D, E: there was a significant positive correlation of MEF2C expression at day 3 of differentiation with number of myotubes per field and fusion index in all treatment groups. Values are means ± SE 2-way ANOVA and Pearson correlation.
DISCUSSION
This study examined alcohol-mediated mechanisms of epigenomic dysregulation causing impaired muscle stem cell differentiation. Our results indicate in vivo CBA administration increased HDAC4 expression in SKM, which was correlated with an increase in total nuclear HDAC activity. Increased Class IIA HDAC expression in myoblasts isolated from CBA/SIV was consistent with our previous findings of decreased miR-206 expression in myoblasts from CBA-administered animals, as HDAC4 is a target of miR-206 (42). CBA effects were observed in myoblasts after isolation, passaging, and culturing with no further exposure to in vitro alcohol and led us to hypothesize that epigenetic mechanisms contribute to this “memory.” Epigenetic dysregulation was confirmed in both SKM tissue and myoblasts. Using an in vitro approach, we showed ETOH treatment to impair differentiation of naïve macaque myoblasts and to increase Class IIA HDAC expression. HDAC inhibition with TMP195 partially restored impaired differentiation in ETOH-treated myoblasts, and this was positively correlated with MEF2C expression at day 3 of differentiation. These findings suggest that temporal MEF2C expression may predict differentiation potential. The functional relationship between Class IIA HDACs and MEF2C warrants further exploration.
Previous studies demonstrated that CBA, SIV, and ART decrease muscle metabolic function in male rhesus macaques (30, 32). In addition, we have shown that CBA exacerbates muscle wasting at end-stage disease (31). Gene regulatory network studies showed dysregulation of a number of pathways affecting muscle function, including satellite cell function and myogenesis (43). The primary focus of this study was to investigate the role of Class IIA HDACs 4, 5, and 7, which are enriched in SKM and have been studied in myoblast differentiation. HDACs interact with MEF2 family of transcription factors, and MEF2C is a crucial driver of myogenesis (23, 27, 34, 49). HDACs 4, 5, and 7 also participate in a carefully orchestrated shuttling between the nucleus and cytoplasm through the process of differentiation (28, 46, 55). HDACs decrease gene expression in several tissues (6). Alterations in Class IIA HDAC expression can impair myogenesis by several mechanisms including skewing HDAC localization, inhibiting MEF2 activity, and decreasing myogenic gene expression (46, 47). HDAC4 expression was increased in both SKM and myoblasts of CBA-administered macaques, also previously reported by our group (42). HDAC4 expression in SKM was positively correlated with total HDAC activity, which is consistent with previous findings that HDAC4 mRNA expression is directly related to protein expression (21). HDAC5 and HDAC7 were not significantly changed due to CBA, SIV, or ART in SKM, but HDAC5 was increased in VEH/SIV/ART+ myoblasts (and approached a nonsignificant increase in CBA/SIV/ART+, Fig. 3B). The inconsistency in HDAC5 expression due to SIV, ART, and CBA in the muscle and myoblasts is potentially due to specific effects of HDAC5 on myoblast function, the effect being diluted when considering the expression in whole muscle tissue.
Alcohol increases histone deacetylase expression in the brain (35, 36, 39), indicating alcohol is impacting HDAC expression in other tissues, though the exact mechanism has yet to be elucidated. Naïve myoblasts treated with ETOH in vitro show increased Class IIA HDAC expression, and this corresponded to a decrease in differentiation (Supplemental Fig. 1: https://figshare.com/articles/Adler_Supplemental_Figue_1_jpg/8135456).
Alterations in HDAC expression and significant changes in differentiation suggest epigenomic modulation of myogenic gene expression leading to decreased differentiation. The programming of myogenic gene expression is crucial in determining if an SC leaves the quiescent state to proliferate (58) or if proliferation yields nonidentical daughter cells that are primed to differentiate (57). This can cause committed myoblasts to return to a quiescent state rather than fuse into myofibers (33, 57). Increased Class IIA HDAC expression at day 5 of differentiation may correspond to decreased expression of downstream genes and repression of MEF2C. ETOH caused a significant decrease in MEF2C, a positive regulator of further downstream MRFs (1, 3). Class IIA HDAC inhibitor, TMP195, did not significantly alter differentiation in CTRL myoblasts but did restore fusion index and increased the number of myotubes formed in ETOH-treated myoblasts when compared with their respective CTRL.
Taken together, these results suggest that TMP195-mediated increase in MEF2C expression partially restored myogenic capacity of ETOH-treated myoblasts. MEF2C expression is decreased during proliferation in ETOH-treated myoblasts. At day 3 of differentiation, the expression in ETOH/TMP-treated is comparable to CTRL myoblasts. MEF2C expression also correlates with differentiation (Fig. 6, B and C). Class IIA HDACs repress MEF2C during myogenesis, and inhibition of these HDACs increases MEF2C expression and ability to drive myogenesis. We speculate that MEF2C at day 3 of differentiation could be directing cells to terminal differentiation and stimulating the activity of downstream MRFs, accounting for increased fusion in these groups. MEF2C was decreased in the ETOH and ETOH/TMP groups at day 5 of differentiation.This may be due to the significant decrease in cell numbers (Supplemental Fig. S2: https://figshare.com/articles/Supplemental_Figure_2/8135504) decreasing the ability of fusion of myoblasts due to decreased cell-to-cell contact (18). There are two potential mechanisms that lead to decreased cell numbers, ethanol-mediated cell death or decrease in the proliferative ability of myoblasts, and these mechanisms warrant further investigation.
In conclusion, evidence from both the in vivo and in vitro alcohol administration models suggests a Class IIA HDAC-mediated impairment of SC differentiation. Our data provide evidence that a Class IIA HDAC inhibitor improves in vitro differentiation of ethanol-treated myoblasts. The data also support the hypothesis that inhibition of MEF2C and potentially histone hypoacetylation contributes to the decreased differentiation ability of ethanol-treated myoblasts (Fig. 7).
Fig. 7.
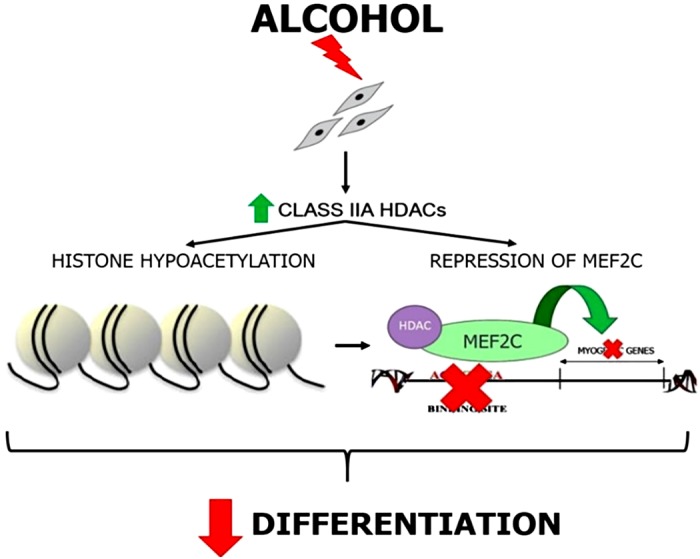
Working model of how alcohol-mediated dysregulation of Class IIA histone deacetylases decreases differentiation potential of satellite cells.
GRANTS
Research reported in this publication was supported by the National Institutes of Health Grants K01 AA-024494, P60 AA-009803, and P51 RR-000164.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.A., P.E.M., and L.S. conceived and designed research; K.A. and L.S. performed experiments; K.A. and L.S. analyzed data; K.A., P.E.M., and L.S. interpreted results of experiments; K.A. and L.S. prepared figures; K.A. and L.S. drafted manuscript; K.A., P.E.M., and L.S. edited and revised manuscript; K.A., P.E.M., and L.S. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge Drs. Gregory Bagby and Steve Nelson, Comprehensive Alcohol-HIV/AIDS Research Center, LSUHSC, for administrative support and design of the original animal study; Dr. Jason Dufour, Tulane National Research Primate Center, for veterinary expertise. The authors are grateful for excellent technical assistance from Larissa Devlin, Wayne A. Cyprian, and Nancy Dillman at TNPRC Pathology Laboratory. From LSUHSC-NO, we are grateful for the technical support of Rhonda R. Martinez, Curtis Vande Stouwe, Paul Berner, Jane Schexnayder, Amy B. Weinberg, and Jean Carnal.
REFERENCES
- 1.Asfour HA, Allouh MZ, Said RS. Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp Biol Med (Maywood) 243: 118–128, 2018. doi: 10.1177/1535370217749494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagby GJ, Zhang P, Purcell JE, Didier PJ, Nelson S. Chronic binge ethanol consumption accelerates progression of simian immunodeficiency virus disease. Alcohol Clin Exp Res 30: 1781–1790, 2006. doi: 10.1111/j.1530-0277.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- 3.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol 4: a008342, 2012. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregenzer A, Conen A, Knuchel J, Friedl A, Eigenmann F, Näf M, Ackle P, Roth M, Fux CA. Management of hepatitis C in decentralised versus centralised drug substitution programmes and minimally invasive point-of-care tests to close gaps in the HCV cascade. Swiss Med Wkly 147: w14544, 2017. [DOI] [PubMed] [Google Scholar]
- 5.Clary CR, Guidot DM, Bratina MA, Otis JS. Chronic alcohol ingestion exacerbates skeletal muscle myopathy in HIV-1 transgenic rats. AIDS Res Ther 8: 30, 2011. doi: 10.1186/1742-6405-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370: 737–749, 2003. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet 382: 1525–1533, 2013. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd T, Simon L, LeCapitaine NJ, Zabaleta J, Mussell J, Berner P, Ford S, Dufour J, Bagby GJ, Nelson S, Molina PE. Chronic binge alcohol administration accentuates expression of pro-fibrotic and inflammatory genes in the skeletal muscle of simian immunodeficiency virus-infected macaques. Alcohol Clin Exp Res 38: 2697–2706, 2014. doi: 10.1111/acer.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J Biol Chem 276: 17007–17013, 2001. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 10.Duplanty AA, Simon L, Molina PE. Chronic Binge Alcohol-Induced Dysregulation of Mitochondrial-Related Genes in Skeletal Muscle of Simian Immunodeficiency Virus-Infected Rhesus Macaques at End-Stage Disease. Alcohol Alcohol 52: 298–304, 2017. doi: 10.1093/alcalc/agw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlandson KM, Li X, Abraham AG, Margolick JB, Lake JE, Palella FJ Jr, Koletar SL, Brown TT. Long-term impact of HIV wasting on physical function. AIDS 30: 445–454, 2016. doi: 10.1097/QAD.0000000000000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlandson KM, Perez J, Abdo M, Robertson K, Ellis RJ, Koletar SL, Kalayjian R, Taiwo B, Palella FJ Jr, Tassiopoulos K. Frailty, Neurocognitive Impairment, or Both in Predicting Poor Health Outcomes Among Adults Living With Human Immunodeficiency Virus. Clin Infect Dis 68: 131–138, 2019. doi: 10.1093/cid/ciy430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford SM Jr, Simon L, Vande Stouwe C, Allerton T, Mercante DE, Byerley LO, Dufour JP, Bagby GJ, Nelson S, Molina PE. Chronic binge alcohol administration impairs glucose-insulin dynamics and decreases adiponectin in asymptomatic simian immunodeficiency virus-infected macaques. Am J Physiol Regul Integr Comp Physiol 311: R888–R897, 2016. doi: 10.1152/ajpregu.00142.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvan FH, Bing EG, Fleishman JA, London AS, Caetano R, Burnam MA, Longshore D, Morton SC, Orlando M, Shapiro M. The prevalence of alcohol consumption and heavy drinking among people with HIV in the United States: results from the HIV Cost and Services Utilization Study. J Stud Alcohol 63: 179–186, 2002. doi: 10.15288/jsa.2002.63.179. [DOI] [PubMed] [Google Scholar]
- 15.Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991-1992 and 2001-2002. Drug Alcohol Depend 74: 223–234, 2004. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Grinspoon S, Mulligan K; Grinspoon S, Mulligan K. Weight loss and wasting in patients infected with human immunodeficiency virus. Clin Infect Dis 36: S69–S78, 2003. doi: 10.1086/367561. [DOI] [PubMed] [Google Scholar]
- 17.Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, Schad S, Johnson SF, Carrasco RD, Lazo S, Bronson RT, Davis SP, Lobera M, Nolan MA, Letai A. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 543: 428–432, 2017. doi: 10.1038/nature21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauss RS, Joseph GA, Goel AJ. Keep Your Friends Close: Cell-Cell Contact and Skeletal Myogenesis. Cold Spring Harb Perspect Biol 9: a029298, 2017. doi: 10.1101/cshperspect.a029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeCapitaine NJ, Wang ZQ, Dufour JP, Potter BJ, Bagby GJ, Nelson S, Cefalu WT, Molina PE. Disrupted anabolic and catabolic processes may contribute to alcohol-accentuated SAIDS-associated wasting. J Infect Dis 204: 1246–1255, 2011. doi: 10.1093/infdis/jir508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Sun Z. Mechanism of Action for HDAC Inhibitors-Insights from Omics Approaches. Int J Mol Sci 20: 1616, 2019. doi: 10.3390/ijms20071616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Pore N, Kim M, Voong KR, Dowling M, Maity A, Kao GD. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol Biol Cell 17: 585–597, 2006. doi: 10.1091/mbc.e05-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, Murray-Thompson M, Schwartz B, Chakravorty S, Wu Z, Mander PK, Kruidenier L, Reid RA, Burkhart W, Turunen BJ, Rong JX, Wagner C, Moyer MB, Wells C, Hong X, Moore JT, Williams JD, Soler D, Ghosh S, Nolan MA. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol 9: 319–325, 2013. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 23.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell 6: 233–244, 2000. doi: 10.1016/S1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 24.Mahnke AH, Miranda RC, Homanics GE. Epigenetic mediators and consequences of excessive alcohol consumption. Alcohol 60: 1–6, 2017. doi: 10.1016/j.alcohol.2017.02.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408: 106–111, 2000. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol 21: 6312–6321, 2001. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J 18: 5099–5107, 1999. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res 29: 3439–3447, 2001. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molina PE, Amedee AM, Veazey R, Dufour J, Volaufova J, Bagby GJ, Nelson S. Chronic binge alcohol consumption does not diminish effectiveness of continuous antiretroviral suppression of viral load in simian immunodeficiency virus-infected macaques. Alcohol Clin Exp Res 38: 2335–2344, 2014. doi: 10.1111/acer.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molina PE, Amedee AM, Winsauer P, Nelson S, Bagby G, Simon L. Behavioral, Metabolic, and Immune Consequences of Chronic Alcohol or Cannabinoids on HIV/AIDs: Studies in the Non-Human Primate SIV Model. J Neuroimmune Pharmacol 10: 217–232, 2015. doi: 10.1007/s11481-015-9599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina PE, Lang CH, McNurlan M, Bagby GJ, Nelson S. Chronic alcohol accentuates simian acquired immunodeficiency syndrome-associated wasting. Alcohol Clin Exp Res 32: 138–147, 2008. doi: 10.1111/j.1530-0277.2007.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina PE, McNurlan M, Rathmacher J, Lang CH, Zambell KL, Purcell J, Bohm RP, Zhang P, Bagby GJ, Nelson S. Chronic alcohol accentuates nutritional, metabolic, and immune alterations during asymptomatic simian immunodeficiency virus infection. Alcohol Clin Exp Res 30: 2065–2078, 2006. doi: 10.1111/j.1530-0277.2006.00252.x. [DOI] [PubMed] [Google Scholar]
- 33.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 275: 375–388, 2004. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ornatsky OI, Andreucci JJ, McDermott JC. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J Biol Chem 272: 33271–33278, 1997. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 35.Palmisano M, Pandey SC. Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol 60: 7–18, 2017. doi: 10.1016/j.alcohol.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey SC, Kyzar EJ, Zhang H. Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology 122: 74–84, 2017. doi: 10.1016/j.neuropharm.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto Neto LF, Sales MC, Scaramussa ES, da Paz CJ, Morelato RL. Human immunodeficiency virus infection and its association with sarcopenia. Braz J Infect Dis 20: 99–102, 2016. doi: 10.1016/j.bjid.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preedy VR, Adachi J, Ueno Y, Ahmed S, Mantle D, Mullatti N, Rajendram R, Peters TJ. Alcoholic skeletal muscle myopathy: definitions, features, contribution of neuropathy, impact and diagnosis. Eur J Neurol 8: 677–687, 2001. doi: 10.1046/j.1468-1331.2001.00303.x. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez-Roman ME, Billini CE, Ghezzi A. Epigenetic Mechanisms of Alcohol Neuroadaptation: Insights from Drosophila. J Exp Neurosci 12: 1179069518779809, 2018. doi: 10.1177/1179069518779809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rees HC, Meister E, Mohler MJ, Klotz SA. HIV-Related Frailty Is Not Characterized by Sarcopenia. J Int Assoc Provid AIDS Care 15: 131–134, 2016. doi: 10.1177/2325957414553848. [DOI] [PubMed] [Google Scholar]
- 41.Robichaux S, Lacour N, Bagby GJ, Amedee AM. Validation of RPS13 as a reference gene for absolute quantification of SIV RNA in tissue of rhesus macaques. J Virol Methods 236: 245–251, 2016. doi: 10.1016/j.jviromet.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon L, Ford SM Jr, Song K, Berner P, Vande Stouwe C, Nelson S, Bagby GJ, Molina PE. Decreased myoblast differentiation in chronic binge alcohol-administered simian immunodeficiency virus-infected male macaques: role of decreased miR-206. Am J Physiol Regul Integr Comp Physiol 313: R240–R250, 2017. doi: 10.1152/ajpregu.00146.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon L, Hollenbach AD, Zabaleta J, Molina PE. Chronic binge alcohol administration dysregulates global regulatory gene networks associated with skeletal muscle wasting in simian immunodeficiency virus-infected macaques. BMC Genomics 16: 1097, 2015. doi: 10.1186/s12864-015-2329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon L, Jolley SE, Molina PE. Alcoholic Myopathy: Pathophysiologic Mechanisms and Clinical Implications. Alcohol Res 38: 207–217, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon L, LeCapitaine N, Berner P, Vande Stouwe C, Mussell JC, Allerton T, Primeaux SD, Dufour J, Nelson S, Bagby GJ, Cefalu W, Molina PE. Chronic binge alcohol consumption alters myogenic gene expression and reduces in vitro myogenic differentiation potential of myoblasts from rhesus macaques. Am J Physiol Regul Integr Comp Physiol 306: R837–R844, 2014. doi: 10.1152/ajpregu.00502.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sincennes MC, Brun CE, Rudnicki MA. Concise Review: Epigenetic Regulation of Myogenesis in Health and Disease. Stem Cells Transl Med 5: 282–290, 2016. doi: 10.5966/sctm.2015-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sousa-Victor P, Muñoz-Cánoves P, Perdiguero E. Regulation of skeletal muscle stem cells through epigenetic mechanisms. Toxicol Mech Methods 21: 334–342, 2011. doi: 10.3109/15376516.2011.557873. [DOI] [PubMed] [Google Scholar]
- 48.Tang AM, Jacobson DL, Spiegelman D, Knox TA, Wanke C. Increasing risk of 5% or greater unintentional weight loss in a cohort of HIV-infected patients, 1995 to 2003. J Acquir Immune Defic Syndr 40: 70–76, 2005. [DOI] [PubMed] [Google Scholar]
- 49.Taylor MV, Hughes SM. Mef2 and the skeletal muscle differentiation program. Semin Cell Dev Biol 72: 33–44, 2017. doi: 10.1016/j.semcdb.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 50.Urbano-Márquez A, Fernández-Solà J. Effects of alcohol on skeletal and cardiac muscle. Muscle Nerve 30: 689–707, 2004. doi: 10.1002/mus.20168. [DOI] [PubMed] [Google Scholar]
- 51.Vagenas P, Azar MM, Copenhaver MM, Springer SA, Molina PE, Altice FL. The Impact of Alcohol Use and Related Disorders on the HIV Continuum of Care: a Systematic Review : Alcohol and the HIV Continuum of Care. Curr HIV/AIDS Rep 12: 421–436, 2015. doi: 10.1007/s11904-015-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veliça P, Bunce CM. A quick, simple and unbiased method to quantify C2C12 myogenic differentiation. Muscle Nerve 44: 366–370, 2011. doi: 10.1002/mus.22056. [DOI] [PubMed] [Google Scholar]
- 53.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet 19: 286–293, 2003. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 54.Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol 21: 5992–6005, 2001. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang XJ, Grégoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol 25: 2873–2884, 2005. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 72: 19–32, 2017. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 57.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol 166: 347–357, 2004. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119: 1824–1832, 2006. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]