Abstract
Systems biology uses computational approaches to integrate diverse data types to understand cell and organ behavior. Data derived from complementary technologies, for example transcriptomic and proteomic analyses, are providing new insights into development and disease. We compared mRNA and protein profiles from purified endothelial, epithelial, immune, and mesenchymal cells from normal human infant lung tissue. Signatures for each cell type were identified and compared at both mRNA and protein levels. Cell-specific biological processes and pathways were predicted by analysis of concordant and discordant RNA-protein pairs. Cell clustering and gene set enrichment comparisons identified shared versus unique processes associated with transcriptomic and/or proteomic data. Clear cell-cell correlations between mRNA and protein data were obtained from each cell type. Approximately 40% of RNA-protein pairs were coherently expressed. While the correlation between RNA and their protein products was relatively low (Spearman rank coefficient rs ~0.4), cell-specific signature genes involved in functional processes characteristic of each cell type were more highly correlated with their protein products. Consistency of cell-specific RNA-protein signatures indicated an essential framework for the function of each cell type. Visualization and reutilization of the protein and RNA profiles are supported by a new web application, “LungProteomics,” which is freely accessible to the public.
Keywords: human lung, integrated omics, proteome, sorted cells, transcriptome
INTRODUCTION
The lung is a complex organ consisting of a diversity of distinct cell types derived from ectodermal, mesodermal, and endodermal compartments. A diversity of specialized cells mediates gas exchange, host defense, and ion transport, all required for lung structure and function (35). The gas exchange functions of the lung are mediated by the alveoli, wherein extensive epithelial surfaces are in close apposition to the capillary network enabling efficient transfer of oxygen and carbon dioxide (5, 24, 27). Alveolarization of the human lung begins before the time of birth and continues through adolescence. Most of the alveoli are formed in the first and second year of life (5, 24). Alveologenesis is a highly regulated process tat requires cooperative interactions among interstitial, epithelial, mesenchymal, and vascular compartments of the lung (4). The lung is continuously exposed to microorganisms, allergens, particulates, and toxicants. Thus a multilayered innate and acquired host defense system has evolved to protect the lung and to maintain homeostasis (28, 34). Understanding the molecular and cellular processes forming and maintaining the pulmonary structures has important implications for the treatment of lung diseases. Elucidation of mechanisms controlling normal lung formation will provide insight into the pathogenesis of common, chronic pulmonary disorders and the mechanisms underlying normal repair (4, 24).
High-throughput technologies now enable precise measurement of thousands of mRNA and proteins simultaneously. In general, each data type, whether lipid, protein, DNA, or RNA have utilized a single “omic” data type for analysis (4, 5, 24, 28, 34). Relationships among cell-selective gene expression and protein concentration have not yet been correlated in the human lung. Recent studies demonstrate a general lack of correlation between mRNA and protein (14, 15, 17, 19, 31). In the present study, we directly compared mRNA and protein using sorted cells from same human lungs to minimize technical variability and to ensure that differences observed in matching RNA and protein data represent mostly the biological effects. Identification of quantified cell selective RNAs and proteins will be useful in identifying the diverse cell types within complex organs like the lung. We established an analytic pipeline to integrate transcriptomic and proteomic data to provide insights that are not accessible using mRNA or protein expression data alone. Signature genes/proteins from four sorted lung cell types were identified using the same analytic pipeline. mRNA and protein correlations were assessed at both the cell level (global scale) and gene level (individual gene-protein pair). Both coherent and noncoherent signatures of mRNA and protein were identified, and their potential impact on functional processes was addressed. To facilitate access to this human protein/mRNA data resource, we developed a new web application, LungProteomics, as a functional component of the Lung Gene Expression Analysis (LGEA) web portal (12, 13) supported by the LungMAP consortium (1). LungProteomics provides a user-friendly web interface for users to query proteins of interest or protein signatures for each cell type. The web tool provides side-by-side comparisons of the relative expression of protein and mRNA pairs and their profile correlation. The website will be naturally extended to ongoing data generated from normal lung tissue and cells from both human and mouse at additional developmental time points.
MATERIALS AND METHODS
Fluorescence Activated Cell Sorting, mRNA, and Protein Profiling
Human lung tissue was obtained from the International Institute for the Advancement of Medicine and the National Disease Research Interchange. Consent has been given for the use of tissue in research and is considered to be exempt from human subject regulation (since the tissue was recovered from the deceased and provided as deidentified samples to investigators), as outlined by the University of Rochester Research Subjects Review Board protocol (RSRB00056775).
Human lung endothelial cells (CD45−/CD326−/CD31+/144+), epithelial cells (CD45−/CD326+/CD31−/CD144−), mixed immune cells (CD45+/CD326−/CD31−/CD144−) and mesenchymal cells (CD45−/CD326−/CD31−/CD144−) were isolated from three female donors (D001, D008, and D011) who died at 20 mo of age from nonrespiratory causes. The organs were recovered by standard organ procurement protocols following determination of brain death. D001 identified as black, while D008 and D011 were white. Past medical history, last blood gases, histologic assessment of lung growth and structure, expected-to-observed lung weight ratios, and radial alveolar counts all supported normal lung development and lung function. However, histopathology performed by pediatric pathologists after case selection demonstrated “acute bronchopneumonia, with moderate, focal bacteria” in D001, while D008 and D011 were described as “normal structure and development with patchy mild atelectasis and mild macrophage accumulation.” Cells were isolated using the FACSAria II (Becton Dickinson, San Jose, CA) instrument. The workflow for isolation of each cell type by FACS is depicted in Supplemental Fig. S1 (all Supplemental Material is available at https://doi.org/10.6084/m9.figshare.8066276). High throughput RNA sequencing of sorted cell populations was performed essentially as we previously described (2). GENCODE database (https://www.gencodegenes.org/) was used for mRNA annotation. Counts per million were calculated for mRNA abundances analysis.
Proteomics Sample Processing
Proteins were extracted using a modified Folch extraction as previously described (19). After extraction, the protein pellet was dried in a speedvac and then reconstituted in 30 µl of 8 M urea containing 50 mM ammonium bicarbonate. Then, the proteins were reduced with DTT (5 mM for 30 min at 60°C) and alkylated with iodoacetamide (400 mM for 1 h at 37°C in the dark), diluted 10 times in 50 mM ammonium bicarbonate containing 1 mM CaCl2 before digestion. Resulting peptides were desalted using C18 SPE cartridges (Discovery C18, 1 mL, 50 mg; Sulpelco, Belfonte, PA). The peptide concentrations were measured by BCA assay (Thermo Scientific). Five microliters of 0.1 μg/μl of peptides were analyzed via tandem mass spectrometry (LC-MS/MS) using a label-free relative quantification approach (8, 21, 23, 29). Identification and quantification of the proteins were performed using MaxQuant software (8) as previously described (7, 23, 29). The expression values of protein were log2 transformed and median normalized.
Immunofluorescence and Confocal Microscopy
Human lungs were inflation fixed with diethyl pyrocarbonate (DEPC)-treated 4% paraformaldehyde (PFA), followed by washing in DEPC-treated 1× PBS and cryoprotection in 30% DEPC-treated sucrose. Tissue was then frozen in OCT at −80°C. Immunofluorescence staining was performed on 7-µm-thick tissue sections rehydrated in 1× PBS fixed in 4% PFA for 5 min and washed in 1× PBS. Antigen retrieval was performed in 0.1 M citrate buffer (pH 6.0) by microwaving. Slides were blocked for 2 h at room temperature using 4% normal goat or donkey serum in 1× PBS containing 0.2% Triton X-100 and then incubated with primary antibodies diluted in blocking buffer for ~16 h at 4°C. Primary antibodies included ACTA2 (1:2,000; A5228; Sigma-Aldrich, St. Louis, MO), AGER (1:100; R&D Systems, Minneapolis, MN), ABCA3 (1:100; WMAM-17G524; Seven Hill Bioreagents, Cincinnati, OH), CD68 (1:100; Abcam, Cambridge, MA), FN1 (1:100; Abcam), FOXF1 (1:100; R&D Systems), HOPX (1:100, Santa Cruz Biotechnology, Santa Cruz, CA), and NKX2.1 (1:1,000; RB TTF-1 1231; Seven Hills Bioreagents). Appropriate secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 633 were used at a dilution of 1:200 in blocking buffer for 1 h at room temperature. Nuclei were counterstained with DAPI (1 μg/mL; Thermo Fisher Scientific, Waltham, MA). Sections were mounted using ProLong Gold (Thermo Fisher Scientific) mounting medium and coverslipped. Tissue sections stained by immunofluorescence were imaged on an inverted Nikon A1R confocal microscope (×60 magnification) 1.27-NA objective using a 1.2-AU pinhole. Maximum intensity projections of multilabeled Z stack images obtained sequentially using channel series across the 7-μm-thick sections were generated using Nikon NIS-Elements software.
Human lung tissue sections (6 µm) from paraffin blocks were deparaffinized and rehydrated. In situ hybridization was performed using the RNAscope multiplex fluorescent v2 assay (cat. no. 323110; ACD Bio) following the manufacturer’s instruction. Tissues were slightly digested using Protease plus provided in the RNAscope assay then hybridized with RNA probes for COL6A1 (cat. no. 482461; ACD Bio), EMCN (cat. no. 549721; ACD Bio), FOXF1 (cat. no. 505741-C3; ACD Bio), NKX2.1 (ACD Bio, cat. no.468991), and SFTPC (cat. no. 452561; ACD Bio). In situ hybridization was followed by immunofluorescent staining using anti-ACTA2 (C6198; Sigma-Aldrich) (9), anti-CD31 antibody (RB-10333-P1, NeoMarkers; Thermo Scientific) (10), anti-CDH1 antibody (610181; BD Biosciences) anti-COL6A1 antibody (Abcam; ab151422), anti-EMCN antibody (eBioV.7C7; Invitrogen), anti-FOXF1 antibody (R&D Systems; AF4798), anti-NKX2.1 (WRAB-1231; Seven Hills), and anti-SFTPC antibody (LS-B10952; Lifespan Biosciences). Supplemental Table S1 summarizes the antibodies used in the present study and their specificity.
Proteomic and Transcriptomic Data Analyses
In this study, 3,320 proteins were detected through mass spectrometry (MS) and 58,723 mRNA entries were generated through RNA-seq sequencing. The Uniprot Retrieve/ID mapping tool (https://www.uniprot.org/uploadlists/) was used to join two data sets, and the combined data set contains 3,320 mRNA-protein pair expression records. One hundred percent of proteins have matched mRNAs found in corresponding mRNA data set. Data were further standardized (z-scored) with mean as zero and standard deviation as one in all genes for mRNA and protein separately before hierarchical clustering and principal component analysis (PCA). Hierarchical clustering analysis and PCA were performed using Partek Genomic Suite 6.6 (http://www.partek.com/). Donor D001 was identified as an outlier in PCA analysis. Data from this tissue were removed from the correlation analyses but included in the signature gene identification since the outlier largely influences the sample correlation but not the signature genes identification. The genome-wide correlation between mRNA and protein expression was measured by Spearman correlation coefficient for all conditions. Differentially expressed genes and proteins between one cell type and the other three cell types were identified by modified one way ANOVA analysis using REML (restricted maximum likelihood) model (16) to accommodate the low sample numbers (n = 3 per condition), with the cutoff as: P < 0.05; fold change > 2 between the average expression of a gene in a given cell and the average expression of all other cells; and the average expression of a gene in a given cell type >1.2 of the maximal expression of this gene in any other cell types. Gene set enrichment analysis was performed using ToppGene Suite (6).
To better understand potential factors influencing mRNA and protein coherent and noncoherent expression, chi-square test and logistic regression analysis were conducted using packages of car, gmodels, and ggplot2 in R (https://www.r-project.org/). mRNA and protein signatures identified in the same cell type were considered as coherently expressed (n = 765). mRNA and protein signatures were considered as noncoherently expressed when the signature represents a different cell type or is not detected in proteomics profiling (n = 6276). Considering the remarkable group size difference, we compared each group to the whole human genome and estimate relative enrichment of individual factors between the two groups. The potential factors of interest influencing protein-mRNA expression difference include cellular component [plasma membrane GO:0005886, cytoplasm GO:0005737, nucleus GO:0005634, cell surface GO:0009986, extracellular matrix (ECM) GO:0031012, and cell junction GO:0030054], and protein type/function [transcription factor (Ingenuity Pathway Analysis, Genomatix, and CIS-BP database), cell surface receptor (Ingenuity Pathway Analysis), and secreted protein (Human Protein Atlas)]. Other properties including mRNA/protein abundance, mRNA/protein half-life, translation rate, and transcription rate were collected from previous publications (3, 25) and tested using Wilcoxon/Kruskal-Wallis tests (rank sums). Bivariate associations were assessed using cross tabulation and chi-square test (discrete) and loess fits on untransformed and log scales (continuous). The type I error probability applied for statistical significance tests was α = 0.05, and all tests were two sided. A logistic regression model was initially fitted with coordination (1 = coherent, 0 = noncoherent) as the dependent variable and the six protein subcellular location terms (1–0) as the predictor variables (n = 7,041 UniProt entry names). Next, we removed the nonsignificant predictors determined by the initial model assessment and added other categorical variables (secreted proteins, cell surface receptors, and transcription factors) back to the model one-at-a-time; none reached the level of statistical significance. Since protein properties (half-life, turnover rate, copy number, translation rate, transcription rate, etc.) information was only available for ~25% of the data, association of these (continuous) variables with coordination was assessed separately. A data set composed of the subset of records with complete information for all the continuous variables was created (n = 903). A logistic regression model was fit with coordination (1 = coherent, 0 = noncoherent) as the dependent variable and all continuous variables as the predictor variables. Factor was considered statistically significant if P < 0.05 was obtained from analysis.
LungProteomics in LGEA Web Portal
The web query functions were developed in Eclipse (https://www.eclipse.org), an integrated development environment. Oracle Database was used for data storage and data retrieval. The web interface was written using HTML, CSS, and JavaScript. Java servlets were created on server side to respond to request from the web page, to extract and process data retrieved from database, and to return formatted data to client browser for display. The Javascript-based charting library “Highcharts” (https://www.highcharts.com) provides dynamic web page construction, data visualization and interaction effects. The LGEA web portal (13) hosts access links through newly developed component LungProteomics on its home page (https://research.cchmc.org/pbge/lunggens/mainportal.html).
Data Sharing
The RNA-seq data are available in the LungMAP web resource (https://lungmap.net/breath-omics-experiment-page/?experimentTypeId=LMXT0000000018&experimentId=LMEX0000000295&analysisId=LMAN0000000059&view=allEntities). Proteomics data are available in the MassIVE data repository (ID: MSV000081973; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) as well as in the LungMAP web resource (https://lungmap.net/breath-omics-experiment-page/?experimentTypeId=LMXT0000000015&experimentId=LMEX0000000661&analysisId=LMAN0000000096&view=allEntities). The analytic and interpreted results are available in the “LungProteomics” web portal of the LGEA Web Portal database (https://research.cchmc.org/pbge/lunggens/lungProteomics/human_sorted_profiles.html).
RESULTS
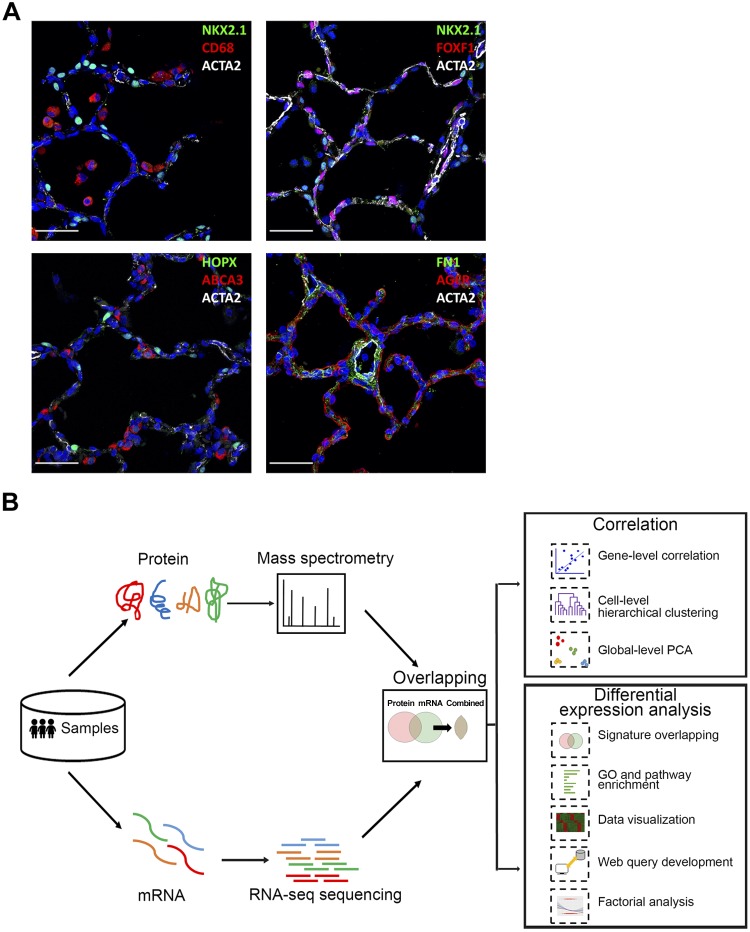
Multiple factors, including differences in sample preparation, protein/mRNA stability, and posttranscriptional regulation may contribute to the lack of correlation between transcriptomic and proteomic data seen experimentally (11, 30). In the present study, we generated mRNA and protein data from four abundant cell types (epithelial, endothelial, mesenchymal, and immune cells) from infant lungs at 20 mo of age (Supplemental Table S2). At this age, lung morphology is characteristic of the alveolar stage during which alveolar epithelial growth occurs in parallel with microvascular maturation. Immunofluorescence confocal microscopy was utilized to define alveolar structures at 20 mo of age (Fig. 1A). mRNA profiling and protein profiling were compared in each sample to identify relationships between protein and mRNA as illustrated in Fig. 1B. Genes and proteins coherently or noncoherently expressed in each cell type were identified at this stage of alveolarization.
Fig. 1.
Anatomic location of human cell types and schematic of analytic workflow. A: confocal microscopy was utilized to define major lung cell types by immunofluorescence staining for NKX2-1, ABCA3 (AT2 cells), HOPX, AGER (AT1 cells), CD68 (immune cells), ACTA2 (smooth muscle/myofibroblast), FOXF1 (endothelial-mesenchymal cells), and FN1 (fibroblast) in donor lungs (D011 and D08) from infants of 20 mo of age. Scale bars, 100 μm. B: workflow of integrative transcriptomic and proteomic profiling analysis. Human lung samples underwent same sorting procedures to isolate endothelial, epithelial, immune, and mesenchymal cells for RNA-seq and proteomic profiling. 3,320 mRNA-protein pairs were obtained for downstream analyses (n = 3/cell type). Sample-level and gene-level correlations were calculated to assess the overall correlation of transcriptomic and proteomic data; differential RNA/protein expression analyses were performed using one-way ANOVA with REML (restricted maximum likelihood) model and compared to identify coherent vs. noncoherent signatures for major cell types; functional enrichment analyses were performed to identify unique and shared biological processes and pathways; and chi-square test and logistic regression analysis were conducted to determine potential factors influencing mRNA-protein expression coherency. Data visualization and web query tools were developed to facilitate data sharing and reutilization. PCA, principle component analysis; GP, gene ontology.
Genome-Wide mRNA-Protein Correlation at Both Sample and Gene Level
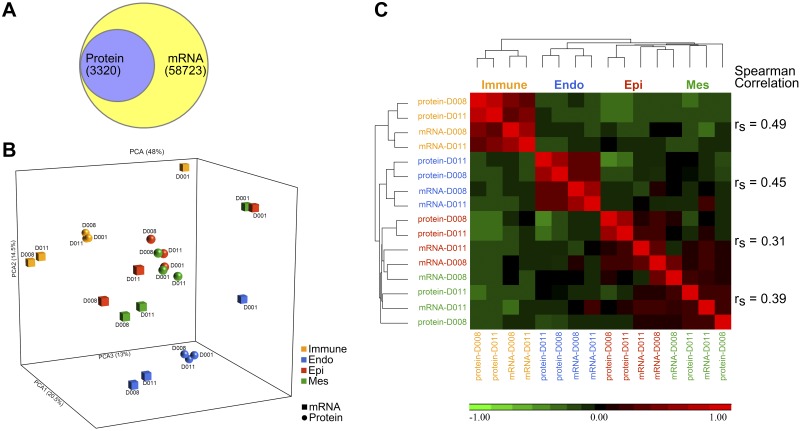
Transcriptomic and proteomic data from all samples were combined into a single data set, shown in a Venn diagram in Fig. 2A, indicating the overlap of 3,320 mRNA-protein pairs (Supplemental Table S3). A principal component analysis (PCA) was performed to visualize the relatedness of mRNA and protein data from each of the cell types (Fig. 2B). Cell type relatedness represents the largest variance in the data (~21% of variance). Purified mRNA and protein data from each cell type clustered together. Immune and endothelial cells clustered into cell-specific modules. Mesenchymal and epithelial cells were more closely related with some degree of overlap in gene expression and protein profiles (Fig. 2B). Pair-wised mRNA/protein cell-cell correlations were calculated using the Pearson correlation on each of the four cell types. Cell-cell correlation heatmap based on hierarchical clustering of all four cell types was generated using mRNA and protein data (Fig. 2C). In general, mRNA/protein from the same cell types were positively correlated and clustered together. Consistent with PCA, cell level correlations were stronger in endothelial and immune cells and weaker in epithelial and mesenchymal cells. We calculated correlations based on individual mRNA-protein pairs in each of the four cell types. In general, protein and mRNA were moderately correlated with Spearman correlation (rs) ranging from 0.31 to 0.49 (Fig. 2C).
Fig. 2.
Overview and correlation of transcriptomic and proteomic data sets. A: overlap of 3,320 pairs of mRNA/proteins is shown from combined human lung transcriptomic and proteomic data [n = 3, human lung at 20 mo, sorted endothelial (Endo), epithelial (Epi), immune, and mesenchymal (Mes) cells] using a Venn diagram. B: principle component analysis (PCA) of human lung transcriptomic and proteomic data (color by cell types and shape by omics types). C: hierarchical clustering of transcriptomic and proteomic data and cell-cell correlation heatmap. rs, Spearman's rank correlation coefficient.
Cell Type-Specific mRNA-Protein Signatures
Epithelial cells.
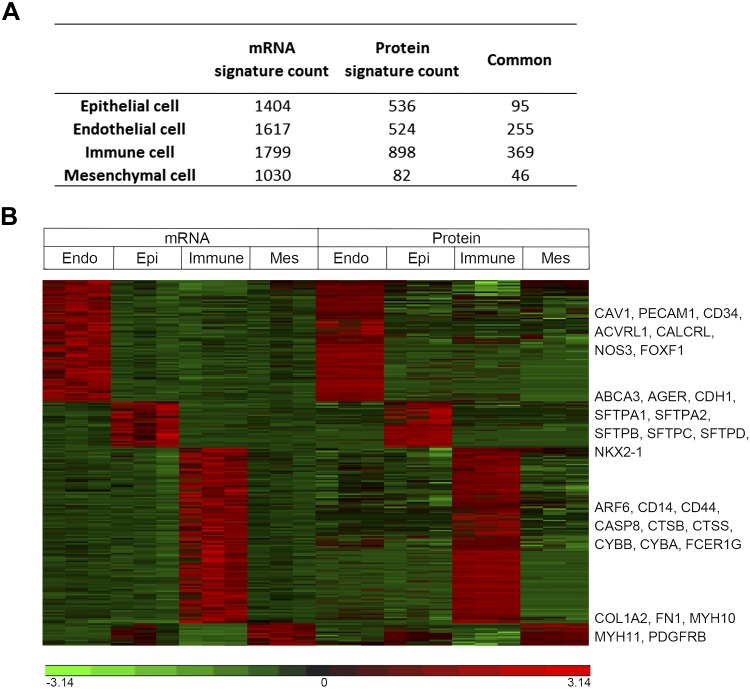
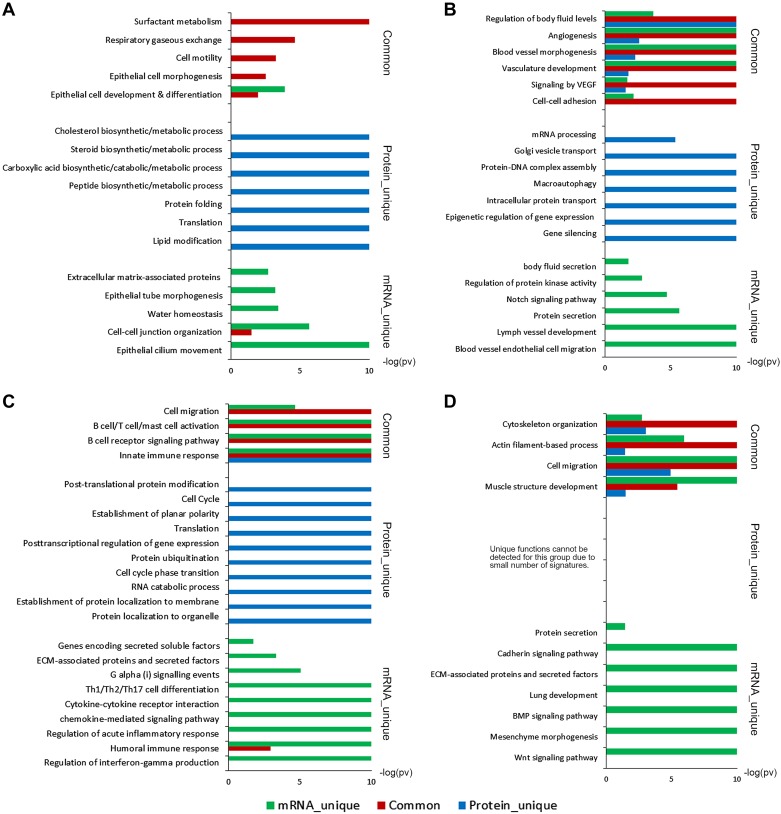
mRNA and protein signatures for lung epithelial cells are shown in Fig. 3; 1,404 mRNAs and 536 proteins provided the epithelial signature (Supplemental Table S3). Of these, 95 included protein and mRNA pairs. Functional enrichment analysis revealed that “surfactant metabolism” and “respiratory gaseous exchange” (including SFTPB, SFTPC, SFTPD, SLC34A2, and ABCA3) and “epithelial cell development/morphogenesis/differentiation” (including AGR2, CDH1, DSP, NKX2–1, and SCEL) were highly conserved in both mRNA and protein analyses (Fig. 4A and Supplemental Table S4). Proteins involved in metabolism and biosynthetic processes, including steroid, cholesterol, carboxylic acid and peptide “biosynthesis/metabolism,” “lipid modification,” and “translation” were enriched in epithelial protein signatures but were not well correlated with the mRNA (Fig. 4A, Supplemental Table S4). Among 1,404 epithelial signature genes, 1,291 (92%) of the encoded protein products were not detectable (Fig. 3A). mRNAs involved in “epithelial tube morphogenesis” (including ETV5, FGFR2, FOXA1, FOXA2, and SOX2), “water homeostasis” (including AQP3, AQP5, SCNN1A, and SCNN1G), and genes encoding of “extracellular matrix-associated proteins” (including LAMA3, LAMC2, WNT3, WNT4, and WNT7A) were highly enriched but were not detected by proteomics profiling (Fig. 4A and Supplemental Table S4), perhaps related to differences in the sensitivity of the two analytical platforms or due to the secretory nature of the EMC gene products.
Fig. 3.
Summary of mRNAs and proteins and coherently and noncoherently expressed mRNA-protein pairs of each cell type. A: the numbers of mRNA signatures, protein signatures and conserved signatures identified for epithelial, endothelial, immune, and mesenchymal cells. B: heatmap depicts z-score-normalized transcriptomic and proteomic profiling of epithelial (Epi), endothelial (Endo), immune and mesenchymal (Mes) cells using common signatures.
Fig. 4.
Gene set enrichment analyses predicted enriched functions for coherently and noncoherently expressed mRNA-protein pairs of each cell type. Functional enrichment analysis predicted enriched bio-processes and pathways of epithelial cells (A), endothelial cells (B), immune cells (C), and mesenchymal cells (D), which are commonly or independently regulated at mRNA and protein levels. The x-axis represents the –log10 transformed enrichment P value.
Endothelial cells.
Endothelial cells play key roles in lung morphogenesis and function, secreting morphogens, providing nutrients, and mediating gas exchange (32). Signature mRNAs and proteins expressed by endothelial cells are shown in Fig. 3; 1,617 mRNA and 524 proteins defined the endothelial cell signature. Among these, 255 mRNA/protein pairs (including PECAM1, FOXF1, CDH5 and NOS3) were coherently expressed (Supplemental Table S3). Genes involved in “regulation of body fluid levels” (including AQP1, CAV1, CD34, FLI1, and NOS3), “signaling by VEGF” (including CAV1, CDH5, JAK1, NOS3, and WASF2), and “blood vessel morphogenesis” and “angiogenesis” (including ACVRL1, CALCRL, FOXF1, NOS3, and PECAM1) were conserved at mRNA and protein levels (Fig. 4B and Supplemental Table S4). Endothelial mRNA signature genes were enriched in “blood vessel endothelial cell migration” (including DLL4, KDR, NOTCH1, SOX18, and VEGFC), “lymph vessel development” (including PROX1, EPHA2, FLT4, FOXC1, and PTPN14), and signaling molecules important for vascular branching and differentiation (including BMP6, NOS1, NOS2, NOTCH1, and VEGFC) (Fig. 4B, Supplemental Table S4). While a large group genes regulating or directly involved in Notch signaling (including DLL4, HES1, JAG2, NOTCH1, and NOTCH4) were enriched in endothelial cells, their corresponding protein products were not detectable. Endothelial cell protein signatures that were not correlated with mRNAs were enriched in “intracellular protein transport,” histone family members involved in “gene silencing,” “epigenetic regulation of gene expression,” and “protein-DNA complex assembly” (Fig. 4B and Supplemental Table S4).
Immune cells.
The extensive surface of the lung is constantly exposed to environment pathogens and toxicants. An intricate innate and acquired host defense system has evolved to protect lung from infection and injury (28, 33, 37). Immune responses of the lung are highly active after birth and during the first years of life to make distinct immune responses appropriate for early life (26, 34). 1,799 mRNA and 898 proteins identified as CD45+ lung immune cell signatures. Among these, 369 correlated mRNA-protein pairs were identified (Fig. 3 and Supplemental Table S3). Biological functions of the conserved immune signatures were enriched in “innate immune response” (including ARF6, CASP8, CTSB, FCER1G, and MAPKAPK3) and B cell/T-cell/mast cell activation (including BTK, FCER1G, LYN, PTPRC, and SYK; Fig. 4C and Supplemental Table S4). While genes encoding “ECM-associated proteins and secreted factors” involved in “cytokine-cytokine receptor interaction,” “Th1/Th2/Th17 cell differentiation,” regulation of “interferon-gamma production,” and “acute inflammatory response” were enriched in the mRNA immune cell signature, their protein products were generally not detectable (Fig. 4C, Supplemental Table S4). Protein signatures that were not correlated with mRNAs were primarily involved in “protein localization to organelle,” “protein ubiquitination,” and “posttranslational protein modification” (including USP14, ADRM1, PSME1/2/3, and PSMD14), “posttranscriptional regulation of gene expression” (including YBX1, SERBP1, YWHAB, proteasomal subunits and eukaryotic translation initiation factor subunits), and “establishment of planar polarity” (including CDC42, PFN1, RHOA, and multiple proteasome subunits; Fig. 4C and Supplemental Table S4).
Mesenchymal cells.
mRNA and protein signatures were identified in CD45−/CD326−/CD31−/CD144− cells, which could not be assigned to any of the other (epithelial, endothelial, or immune) cell types. mRNA and protein signature genes in these mixed mesenchymal cells are shown in Fig. 3A. We identified 1,030 mRNA and 82 protein signatures, among the 82 protein signatures, 46 were correlated mRNA expression (Fig. 3 and Supplemental Table S3). Coherent mRNA-protein signatures, including known mesenchymal selective markers FN1, MYH10, MYH11, and PDGFRB, were enriched in genes mediating “cytoskeleton organization,” “muscle structure development,” and “actin filament-based process” (Fig. 4D, Supplemental Table S4). Remarkably, protein products for 89% of mesenchymal mRNA signature genes were not detected in the corresponding protein data set, as shown in Fig. 3A. Genes encoding “ECM-associated proteins and secreted factors” involved in “protein secretion” and “BMP and WNT signaling pathways” were unique mRNA signatures (Fig. 4D and Supplemental Table S4). Since mesenchymal cells produce much of the matrix supporting the alveoli, it is likely that secreted proteins encoded by the signature mRNAs are lost during proteolytic separation of cells from the lung stroma.
Experimental Validations of Selective Candidates of Coherent and Noncoherent Expressed mRNA-Protein Pairs
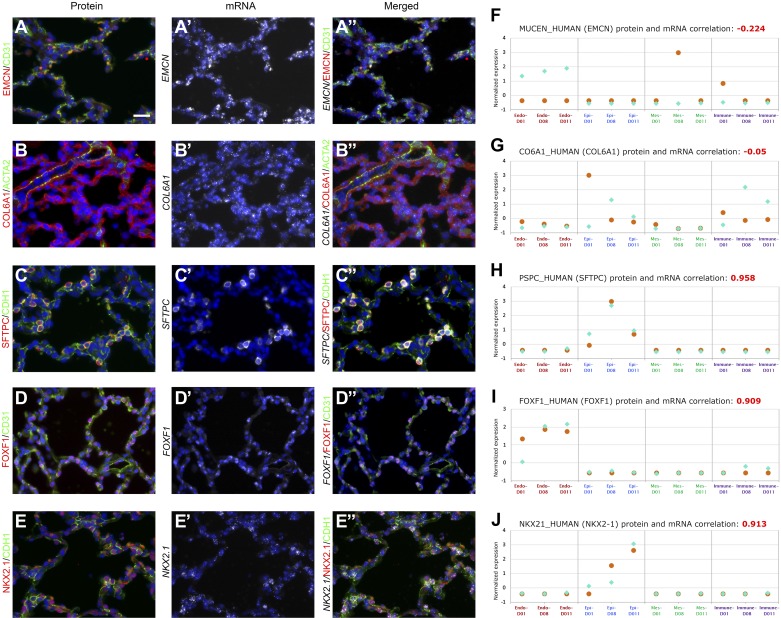
We applied the combination of in situ hybridization (RNAscope multiplex fluorescent) and immunofluorescence staining to the same donor samples to validate coherent and noncoherent expressed mRNA-protein pairs in different cell types. The coimmunofluorescence/in situ hybridization of a given protein/mRNA was performed in conjunction with an additional marker stain to demonstrate markers of corresponding cell types. As shown in Fig. 5, noncoherent expression of RNA and protein expression of EMCN and COL6A1were observed (Fig. 5, A, A’, A”, B, B’, and B”). EMCN RNA (Fig. 5A’) was highly expressed throughout the lung tissue, whereas EMCN protein (Fig. 5A) was only detected in a select number of cells. Costaining with CD31 demonstrated that very few endothelial cells express the EMCN protein while many endothelial cells express the RNA for EMCN. This finding was consistent with the transcriptomics and proteomics data in this cell type (Fig. 5, A” and F). Alternatively, COL6A1 protein were highly expressed in the lung tissue, with evident expression in the vascular smooth muscle cells, as determined by ACTA2 staining (Fig. 5B), while COL6A1 RNA were detected in a few mesenchymal cells only (Fig. 5, B’ and B”). In contrast, in situ and immunofluorescence stain for SFTPC showed strong coexpression of RNA and protein in CDH1-positive epithelial cells (Fig. 5, C, C’, and C”). Comparable protein and RNA coexpression patterns were also observed for transcription factors FOXF1 (Fig. 5, D, D’, and D”) and NKX2.1 (Fig. 5, E, E’, and E”), critical for lung epithelial and endothelial development. FOXF1 colocalization with CD31 suggests its presence in the endothelial cells, whereas the coexpression of NKX2.1 with CDH1 indicates its strong presence in epithelial cells. Comparable results of mRNA/protein pair expression pattern were observed in two additional 20-mo-old lungs (Supplemental Fig. S2). Figure 5, F–J, shows the corresponding mRNA/protein expression profiles of EMCN, COL6A1, SFTPC, FOXF1, and NKX2.1 and their correlations in sorted cells. mRNA and protein expressions of SFTPC, FOXF1, and NKX2.1 were correlated (r = 0.958, 0.909, and 0.913) while EMCN and COL6A1 expressions were not correlated (r = −0.05 and −0.224, respectively).
Fig. 5.
Correlation between immunofluorescence staining and RNA expression patterns. Protein and RNA localization was assessed in lung sections from infants of 20 mo of age by coimmunofluorescence antibody and in situ hybridization staining with RNAscope. A–E: indicated proteins are shown in red and costaining cell-specific markers CD31 (endothelial), CDH1 (epithelial), or ACTA2 (smooth muscle myofibroblasts) are shown in green. A’–E’: indicated RNA is shown in white, performed simultaneously on the same sections. A”–E”: combination of coimmunofluorescent staining of proteins (red/green) and RNA expression (white) are shown. Scale bars, 25 μm. F–J: abundance of each RNA and protein in each cell type and their correlation (red). Correlation was calculated using the Pearson correlation coefficient.
Factors Influencing mRNA-Protein Expression Coherency
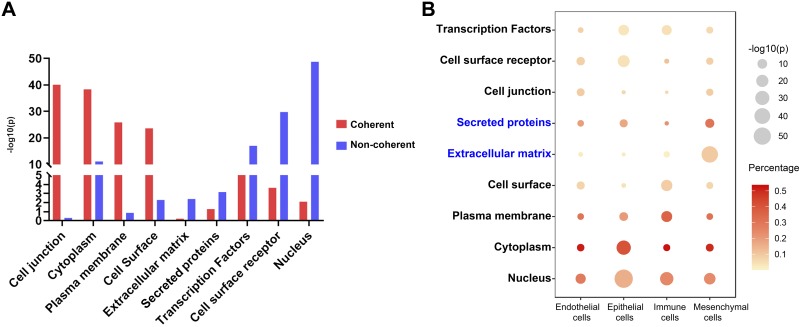
The present study suggested the overall mRNA-protein correlation is ~0.4 with ~50% signatures noncoherently expressed between mRNAs and their protein products. This correlation is comparable to previous studies (3, 11, 17, 18, 25, 30). Little is known about the mechanism behind the poor correlation of protein-mRNA expression. To address this issue, we employed chi-square test and logistic regression to a collection of potential factors influencing protein-mRNA expression differences including protein subcellular localizations, protein types/functions, and protein properties (Supplemental Table S5). Both chi-square tests and logistic regression model predicted protein subcellular localization is the most important variable influencing protein-mRNA expression difference (chi-square test P = 5.05E-36), among these, proteins located in “plasma membrane,” “cytoplasm,” “cell surface,” and “cell junction” were more coherently expressed while proteins located in “extracellular matrix” and “nucleus” were more noncoherently expressed (Fig. 6A). Proteins of “transcription factor,” “secreted protein,” and “cell surface receptor” were positively associated with noncoherent expression (Fig. 6A). Logistic regression model produced consistent results that protein subcellular localization has the greatest impact on protein-mRNA expression differences as shown in Table 1. Protein subcellular location variables of “cytoplasm,” “cell junction,” “plasma membrane,” and “cell surface” are statistically significant to predict protein and mRNA expression coherence. Protein properties data (half-life, turnover rate, copy number, translation rate, and transcription rate) was collected from previous proteomics publications (3, 25). Since only 25% of the current data mapped to these collected properties, we assessed these factors (continuous variables) separately from the protein subcellular localization data (discrete variables). Among these continuous protein properties, Wilcoxon/Kruskal-Wallis test (rank sums) predicted that protein/mRNA half-life and protein turnover had moderate but a statistically significant influence on the group mean difference of coherently and noncoherently expressed protein-mRNA pairs (Table 2). Protein turnover rate and half-life (measured in HeLa cell) were positively associated with noncoherent expression, while mRNA half-life (measured in NIH3T3 mouse fibroblasts) was positively associated coherent expression (Table 2). Logistic regression was used to fit these protein properties, demonstrating that only protein turnover rate (measured in HeLa cell) was positively associated with noncoherent expression (Table 3). Both methods predicted that noncoherently expressed protein-RNA pairs are likely to have higher protein turnover rate. We noticed a difference of protein properties between HeLa and NIH3T3 cell lines; there are two possible reasons: 1) the protein properties collected from HeLa cell (3) and NIH3T3 cell (25) were in a range of 4,000–5,000 of proteins, which only cover small and different portions of the genome after overlapping with data in present study; or 2) HeLa and NIH3T3 cell lines grew under different culture conditions and were measured using different method, which could influence protein stability and cause difference.
Fig. 6.
Analyses of potential factors influencing mRNA and protein expression coherency. A: bar graph showing the significance of potential factors influencing coherently (n = 765) and noncoherently expressed mRNA-protein pairs (n = 6276) using chi-square test. P values are in –log10 transformed. B: enrichment of potential factors in endothelial, epithelial, immune, and mesenchymal cells. Node size is proportional to the significance (–log10 transformed) of factor in each cell type. Node color is proportional to percentage of genes associated with the factor in a given cell type.
Table 1.
Logistic regression analysis on protein subcellular localizations
| 95% CI of OR |
||||||
|---|---|---|---|---|---|---|
| Variable | Coefficient | Z Value | P Value | OR | Lower | Upper |
| Intercept | −3.222 | −35.668 | 1.25E-278 | 0.04 | 0.033 | 0.047 |
| Cell junction | 0.65 | 5.61 | 2.03E-08 | 1.916 | 1.523 | 2.401 |
| Cell surface | 0.486 | 3.593 | 3.26E-04 | 1.626 | 1.244 | 2.113 |
| Plasma membrane | 0.475 | 5.225 | 1.74E-07 | 1.608 | 1.344 | 1.919 |
| Cytoplasm | 1.248 | 12.636 | 1.34E-36 | 3.483 | 2.878 | 4.242 |
| Nucleus | −0.006 | −0.071 | 0.943 | 0.994 | 0.837 | 1.177 |
| Extracellular matrix | −0.261 | −1.062 | 0.288 | 0.77 | 0.462 | 1.218 |
OR, odds ratio; CI, confidence interval.
Table 2.
Wilcoxon test (rank sums) on protein properties
| P Value | S | Z | Rank Sum (Coherent) | Rank Sum (Noncoherent) | Mean Rank (Coherent) | Mean Rank (Noncoherent) | Median (Coherent) | Median (Noncoherent) | |
|---|---|---|---|---|---|---|---|---|---|
| Protein half-life (HeLa cell) | 0.030 | 159,328 | 2.170 | 159,328 | 762,075 | 634.773 | 689.037 | 999 | 999 |
| Turnover (HeLa cell) | 0.008 | 277,693 | 2.674 | 277,693 | 1,310,960 | 824.015 | 907.239 | 19.747 | 20.527 |
| Protein half-life (NIH3T3 mouse fibroblasts) | 0.965 | 272,986 | 0.044 | 272,986 | 1,177,970 | 853.08 | 851.75 | 57.385 | 56.190 |
| mRNA half-life (NIH3T3 mouse fibroblasts) | 0.002 | 271,450 | 3.072 | 271,450 | 1,017,370 | 875.645 | 785.61 | 11.780 | 11.100 |
| mRNA transcription rate (NIH3T3 mouse fibroblasts) | 0.306 | 212,488 | 1.023 | 212,488 | 914,763 | 727.699 | 756.628 | 1.605 | 1.870 |
| Protein translation rate (NIH3T3 mouse fibroblasts) | 0.200 | 196,151 | 1.280 | 196,151 | 867,460 | 700.539 | 736.384 | 173.765 | 181.280 |
| Protein copy number (NIH3T3 mouse fibroblasts) | 0.167 | 246,271 | 1.381 | 246,271 | 1,134,020 | 796.994 | 838.772 | 75167.4 | 89686.8 |
| mRNA copy number (NIH3T3 mouse fibroblasts) | 0.810 | 199,870 | 0.240 | 199,870 | 856,462 | 721.551 | 728.284 | 18.590 | 20.375 |
Table 3.
Logistic regression analysis on protein turnover
| 95% CI of OR |
||||||
|---|---|---|---|---|---|---|
| Variable | Coefficient | Z Value | P Value | OR | Lower | Upper |
| Intercept | −0.78 | −2.616 | 0.009 | 0.458 | 0.253 | 0.816 |
| Turnover | −0.035 | −2.395 | 0.017 | 0.966 | 0.939 | 0.994 |
Protein properties including mRNA/protein abundance, mRNA/protein half-life, translation rate, and transcription rate were collected from previous publications (25, 26). A logistic regression model was fit with coordination (1 = coherent, 0 = noncoherent) as the dependent variable and all continuous protein property variables as the predictor variables. After sequentially removing nonsignificant predictors, only turnover remained statistically significant. OD, odds ratio; CI, confidence interval.
From the proteomic expression profiling analysis, we detected fewer protein signatures and the most unmatched mRNA signatures were in mesenchymal cells compared with epithelial, endothelial and mixed immune cell types. We assessed the potential factors influencing the differences among the four cell types. Compared with other cell types, mesenchymal mRNA signatures were significantly enriched in “secreted proteins” (P = 8.49E-09) and proteins located in “extracellular matrix” (P = 9.11E-45) (Fig. 6B). Thus these “ECM-associated proteins and secreted proteins” may not be stored intracellularly and may be lost from stromal-matrixes during proteolytic procedures used to isolate the cells.
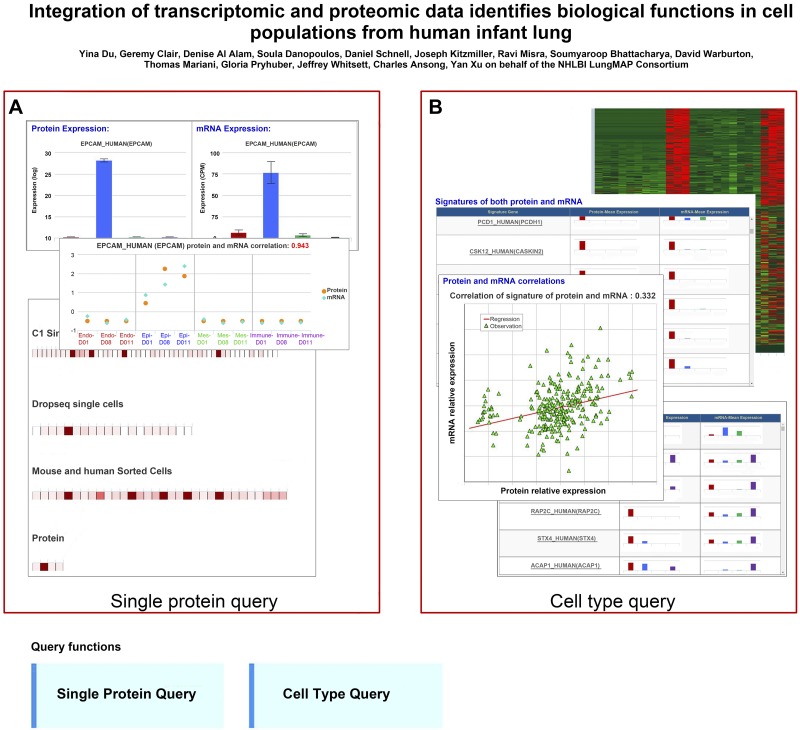
Web-Based Query Functions in LGEA Web Portal
To facilitate queries of proteomic and transcriptomic data, we developed LungProteomics as a functional component of the LGEA web portal (12, 13). LungProteomics (https://research.cchmc.org/pbge/lunggens/lungProteomics/human_sorted_profiles.html) provides two protein-centered query functions: “protein query” and “cell type query.” “Protein query” retrieves expression information and provides comparisons of protein and mRNA pairs in conjunction with associated statistical summaries (Fig. 7A), enabling users to quantitatively examine the consistency or discordancy of protein-mRNA pairs and the selectivity of queried proteins for each cell type. The “protein query” provides an expression overview of proteins/genes of interest across all data sets within the LGEA database using a monocolor heatmap (Fig. 7A). The “cell type query” retrieves signature genes from each cell type and displays signature proteins and mRNAs using an interactive heatmap (Fig. 7B). Protein signatures are paired into coherent and noncoherent protein signatures in downloadable data tables. The “cell type query” provides cell-level correlations based on expression of coherent mRNA/protein signatures and displays correlations with a scatter plot and regression line (Fig. 7B). Both the database and web portal are readily expandable to accommodate new proteomic data.
Fig. 7.
Screenshots of LungProteomics web application. “LungProteomics” provides two protein-centered query functions, “protein query” and “cell type query,” for users to query a protein of interest or protein signatures of a given cell type. LungProteomics is a functional component of the Lung Gene Expression Analysis (LGEA) web portal freely available at https://research.cchmc.org/pbge/lunggens/lungProteomics/human_sorted_profiles.html and is readily expendable for new proteomic data from other lung developmental times in human and mouse.
DISCUSSION
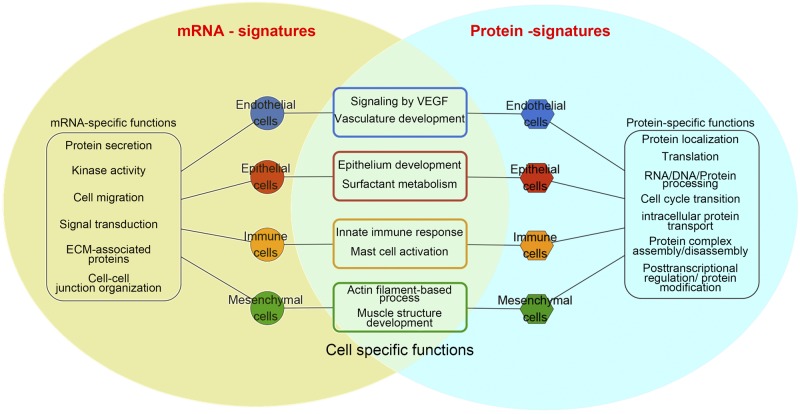
We performed an integrative analysis of transcriptomic and proteomic expression profiling of major cell types in the human lung. Transcriptomic and proteomic data from all four cell types are shown in Fig. 8. Cell type-specific biological functions of mRNA/protein signatures were generated for each cell type. Fundamental cell selective functions of each cell type were represented by coherently expressed genes and their protein products with an average of correlation between RNA and their protein products up to 0.8. For example, epithelial mRNA-protein signatures that mediate “epithelial cell morphogenesis/differentiation,” “surfactant metabolism,” and “respiratory gaseous exchange” were predicted; mRNA-protein signatures involved in “immune cell activation” and “innate immune response” were coherently expressed in mixed immune cells; and mRNA-protein signatures involved in “VEGF signaling,” “regulation of body fluid levels,” and “vasculature development” were selectively expressed in endothelial cells.
Fig. 8.
Summary of transcriptomic and proteomic profiling of human major cell types. The Venn diagram compares signatures identified from transcriptomic and proteomics analyses. The common bioprocesses and pathways represented by coherently expressed signatures are shown in the overlap area. Functional annotations related to noncoherent mRNA signatures are represented in the yellow portion of the Venn diagram and predicted functions derived from noncoherent protein signatures are depicted in the blue portion of the Venn diagram. Overall correlations between mRNA and protein levels were relatively low (~0.4), but cell-specific functions and processes serving as characteristics of each cell type are most represented by coherently expressed mRNA/protein signatures. ECM, extracellular matrix.
Cell-cell correlation and PCA analyses demonstrated that protein-mRNA correlations were better in endothelial and immune cells and weaker in epithelial and mesenchymal cells. From PCA and correlation analyses, endothelial and immune cells were clearly separated from other cell types, while the separation of epithelial and mesenchymal cells was less clear (Fig. 2, B and C). At gene level, a number of mesenchymal markers including ACTA2, COL1A1, FN1, and MYL9 were detected in epithelial cells at relatively high levels. Confocal microscopy of lung tissue from these samples did not show staining for ACTA2, FN1, or COL1A1 in epithelial cells. Whether this observation has biological meaning or is caused by cross contamination of cells or RNAs during sample preparation is unclear at present.
While RNA analyses identified most of the coding genes in the human genome, current proteomic data sets are relatively incomplete due to the imperfect identification of coding sequences and relatively limited sensitivity of current peptide detection technologies (20, 38). Low abundance of the proteins, high turnover rate, or technical limitations may contribute to this observation (20). We and others have shown that it is possible to detect >8,000 proteins from whole lung tissue homogenates utilizing a two-dimensional LC-MS analysis strategy. In the present study, we utilized a one-dimensional LC-MS analysis strategy due to resource restrictions, which may account for lower detection levels of proteins. Many of the unique mRNA signatures identified were important for functions typical of each cell type. In contrast, many protein products were not detected by proteomic profiling, likely due to platform sensitivity, proteolytic loss, and other technical issues. There is a small portion of proteins (n = 117) that are detected by mass spectrometry without detectable mRNA expressions, 32 out of 117 of these belong to histone families. This group of genes is mostly involved in “gene silencing,” “chromatin silencing,” and “epigenetic regulation of gene expression” (Supplemental Table S6). Approximately 45~60% of protein signatures were not coherently expressed with their corresponding mRNAs. These protein signatures were highly enriched in the fundamental housekeeping bioprocesses, including protein localization and transport, translation, RNA/DNA/protein processing, cell cycle transition, and protein complex assembly/disassembly/degradation. Lack of mRNA-protein expression correlation may be influenced at multiple posttranslational levels, including stability of RNAs and proteins, posttranscriptional, translational, and protein degradation processes controlling steady-state protein abundances (11, 22, 30). The analyses of potential factors influencing protein-mRNA expression difference suggested that protein subcellular localization was an important variable influencing protein-mRNA expression difference (chi-square test P = 5.05E-36). Among these, proteins located in “plasma membrane,” “cell surface,” “cell junction,” and “cytoplasm” were more coherently expressed while proteins located in “extracellular matrix” and “nucleus” were more noncoherently expressed. Transcription factors, cell surface receptors, and secreted proteins were positively associated with noncoherent expression. Other protein properties play less significant roles. Based on our analyses, noncoherently expressed protein-RNA pairs tend to have longer half-life, higher turnover rate and translation rate, and more abundant protein expression. Considering that the collected protein properties (i.e., protein half-life, turnover rate, etc.) were in range of 4,000–5,000 proteins, which only cover a small portion of the entire genome after overlapping with data in present study, and each property was measured in different cell lines and by different methods, caution is needed in interpreting these findings.
The present study is limited by low sample size (n = 3). With the increasing awareness of the pediatric origins of adult disease, it is especially important to demonstrate the feasibility of multi-omics studies in pediatrics, in comparison with the mature lung. Variability in present data was demonstrated by PCA among the three donors, highlighting that variability may be related to areas of inflammation, patient heterogeneity, or technical issues related to sample handling. Nevertheless, the present study comprised samples from infants of the same age, representing the first integrative analysis of multiomics data from major pulmonary cell types, identified cell type-specific gene/protein signatures, bioprocesses and signaling pathways that are coherently or noncoherently regulated in this pediatric population.
In summary, we correlated mRNA and protein profiles in cells isolated from normal human infant lung tissue. Clearly, each methodological approach (transcriptomics, proteomics) has strengths and weaknesses in identifying key cell features. Consideration of both transcriptomic and proteomic data, independently and together, provides a better representation of the molecular status of each of the major pulmonary cell types. mRNA abundance explained less than 40% of the variation in protein abundance, suggesting that the overall correlations between mRNA and protein levels were largely dependent on translation and posttranslational activities and not determined by mRNA abundance alone (31, 36). Protein levels provide insights useful in identifying cells by immunochemical and chemical approaches. In spite of the relatively low global correlation (~0.4) between proteomic and mRNA profiles, sorted cells from the same types clustered together, demonstrating that cell type identification is supported by either analysis. Cell-specific signature genes and genes involved in the essential functions of each major human lung cell type were highly correlated with their protein products (>0.8). Conserved mRNA/protein signatures for each cell type may serve as solid biomarkers for cell identification and sorting for study of human lung. To facilitate access and utilization of these data, a web portal consisting of tools for protein query, cell type query, and protein-mRNA comparisons was developed. These web tools are freely available (https://research.cchmc.org/pbge/lunggens/lungProteomics/human_sorted_profiles.html).
GRANTS
This research was supported in part by National Heart, Lung, and Blood Institute LungMAP Grants U01 HL122642 (to J. A. Whitsett and Y. Xu), U01 HL122700 (to T. J. Mariani and G. Pryhuber), U01 HL122681 (to D. Warbuton and D. Al Alam), and U01 HL122703 (to C. Ansong). Proteomics analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy and located at Pacific Northwest National Laboratory (PNNL) in Richland, WA. PNNL is a multiprogram national laboratory operated by Battelle for the Department of Energy under Contract DE-AC05-76RLO 1830.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
G.C.C., D.A.A., D.W., T.J.M, G.S.P., J.A.W., and Y.X. conceived and designed research; G.C.C., D.A.A., S.D., J.A.K., R.S.M., S.B., D.W., T.J.M., G.S.P., J.A.W., and C.A. performed experiments; Y.D. and D.S. analyzed data; Y.D. and Y.X. interpreted results of experiments; Y.D., D.A.A., S.D., J.A.K., and D.W. prepared figures; Y.D., J.A.W., and Y.X. drafted manuscript; Y.D., G.C.C., D.A.A., S.D., D.S., J.A.K., R.S.M., S.B., D.W., T.J.M., G.S.P., J.A.W., C.A., and Y.X. edited and revised manuscript; Y.D., G.C.C., D.A.A., S.D., D.S., J.A.K., R.S.M., S.B., D.W., T.J.M., G.S.P., J.A.W., C.A., and Y.X. approved final version of manuscript.
REFERENCES
- 1.Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, Hagood JS, Kaminski N, Mariani TJ, Potter SS, Pryhuber GS, Warburton D, Whitsett JA, Palmer SM, Ambalavanan N; LungMAP Consortium . LungMAP: The Molecular Atlas of Lung Development Program. Am J Physiol Lung Cell Mol Physiol 313: L733–L740, 2017. doi: 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay G, Huyck HL, Misra RS, Bhattacharya S, Wang Q, Mereness J, Lillis J, Myers JR, Ashton J, Bushnell T, Cochran M, Holden-Wiltse J, Katzman P, Deutsch G, Whitsett JA, Xu Y, Mariani TJ, Pryhuber GS. Dissociation, cellular isolation, and initial molecular characterization of neonatal and pediatric human lung tissues. Am J Physiol Lung Cell Mol Physiol 315: L576–L583, 2018. doi: 10.1152/ajplung.00041.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert FM, Ahmad Y, Gierliński M, Charrière F, Lamont D, Scott M, Barton G, Lamond AI. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics 11: M111.011429, 2012. doi: 10.1074/mcp.M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbon J, Boucherat O, Chailley-Heu B, Delacourt C. Control mechanisms of lung alveolar development and their disorders in bronchopulmonary dysplasia. Pediatr Res 57: 38R–46R, 2005. doi: 10.1203/01.PDR.0000159630.35883.BE. [DOI] [PubMed] [Google Scholar]
- 5.Burri PH. Structural aspects of postnatal lung development - alveolar formation and growth. Biol Neonate 89: 313–322, 2006. doi: 10.1159/000092868. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, Web Server: W305-11, 2009. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clair G, Piehowski PD, Nicola T, Kitzmiller JA, Huang EL, Zink EM, Sontag RL, Orton DJ, Moore RJ, Carson JP, Smith RD, Whitsett JA, Corley RA, Ambalavanan N, Ansong C. Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci Rep 6: 39223, 2016. doi: 10.1038/srep39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372, 2008. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 9.Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, Al Alam D. Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am J Physiol Lung Cell Mol Physiol 314: L144–L149, 2018. doi: 10.1152/ajplung.00379.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danopoulos S, Krainock M, Toubat O, Thornton M, Grubbs B, Al Alam D. Rac1 modulates mammalian lung branching morphogenesis in part through canonical Wnt signaling. Am J Physiol Lung Cell Mol Physiol 311: L1036–L1049, 2016. doi: 10.1152/ajplung.00274.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512–1526, 2009. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Y, Guo M, Whitsett JA, Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70: 1092–1094, 2015. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, Pryhuber GS, Mariani TJ, Bhattacharya S, Guo M, Potter SS, Dexheimer P, Aronow B, Jobe AH, Whitsett JA, Xu Y. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72: 481–484, 2017. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4: 117, 2003. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haider S, Pal R. Integrated analysis of transcriptomic and proteomic data. Curr Genomics 14: 91–110, 2013. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyle RH, Gottfredson NC. Sample size considerations in prevention research applications of multilevel modeling and structural equation modeling. Prev Sci 16: 987–996, 2015. doi: 10.1007/s11121-014-0489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep 5: 10775, 2015. doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Wei Y, To C, Zhu CQ, Tong J, Pham NA, Taylor P, Ignatchenko V, Ignatchenko A, Zhang W, Wang D, Yanagawa N, Li M, Pintilie M, Liu G, Muthuswamy L, Shepherd FA, Tsao MS, Kislinger T, Moran MF. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat Commun 5: 5469, 2014. doi: 10.1038/ncomms6469. [DOI] [PubMed] [Google Scholar]
- 19.Moghieb A, Clair G, Mitchell HD, Kitzmiller J, Zink EM, Kim YM, Petyuk V, Shukla A, Moore RJ, Metz TO, Carson J, McDermott JE, Corley RA, Whitsett JA, Ansong C. Time-resolved proteome profiling of normal lung development. Am J Physiol Lung Cell Mol Physiol 315: L11–L24, 2018. doi: 10.1152/ajplung.00316.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie L, Wu G, Brockman FJ, Zhang W. Integrated analysis of transcriptomic and proteomic data of Desulfovibrio vulgaris: zero-inflated Poisson regression models to predict abundance of undetected proteins. Bioinformatics 22: 1641–1647, 2006. doi: 10.1093/bioinformatics/btl134. [DOI] [PubMed] [Google Scholar]
- 21.Piehowski PD, Zhao R, Moore RJ, Clair G, Ansong C. Quantitative proteomic analysis of mass limited tissue samples for spatially resolved tissue profiling. Methods Mol Biol 1788: 269–277, 2018. doi: 10.1007/7651_2017_78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP, Wang R. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics 25: 1397–1403, 2009. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rindler TN, Stockman CA, Filuta AL, Brown KM, Snowball JM, Zhou W, Veldhuizen R, Zink EM, Dautel SE, Clair G, Ansong C, Xu Y, Bridges JP, Whitsett JA. Alveolar injury and regeneration following deletion of ABCA3. JCI Insight 2: 97381, 2017. doi: 10.1172/jci.insight.97381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schittny JC. Development of the lung. Cell Tissue Res 367: 427–444, 2017. doi: 10.1007/s00441-016-2545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 473: 337–342, 2011. [Erratum in Nature 495: 126–127, 2013.] doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 26.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci 282: 20143085, 2015. doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surate Solaligue DE, Rodríguez-Castillo JA, Ahlbrecht K, Morty RE. Recent advances in our understanding of the mechanisms of late lung development and bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 313: L1101–L1153, 2017. doi: 10.1152/ajplung.00343.2017. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Chow CW, Downey GP. Role of innate immune cells and their products in lung immunopathology. Int J Biochem Cell Biol 40: 1348–1361, 2008. doi: 10.1016/j.biocel.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Snowball JM, Xu Y, Na CL, Weaver TE, Clair G, Kyle JE, Zink EM, Ansong C, Wei W, Huang M, Lin X, Whitsett JA. EMC3 coordinates surfactant protein and lipid homeostasis required for respiration. J Clin Invest 127: 4314–4325, 2017. doi: 10.1172/JCI94152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13: 227–232, 2012. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Wu G, Chen L, Zhang W. Integrated analysis of transcriptomic and proteomic datasets reveals information on protein expressivity and factors affecting translational efficiency. Methods Mol Biol 1375: 123–136, 2016. doi: 10.1007/7651_2015_242. [DOI] [PubMed] [Google Scholar]
- 32.Wang T, Gross C, Desai AA, Zemskov E, Wu X, Garcia AN, Jacobson JR, Yuan JX, Garcia JG, Black SM. Endothelial cell signaling and ventilator-induced lung injury: molecular mechanisms, genomic analyses, and therapeutic targets. Am J Physiol Lung Cell Mol Physiol 312: L452–L476, 2017. doi: 10.1152/ajplung.00231.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitsett JA. Intrinsic and innate defenses in the lung: intersection of pathways regulating lung morphogenesis, host defense, and repair. J Clin Invest 109: 565–569, 2002. doi: 10.1172/JCI0215209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 16: 27–35, 2015. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev 99: 513–554, 2019. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu G, Nie L, Zhang W. Integrative analyses of posttranscriptional regulation in the yeast Saccharomyces cerevisiae using transcriptomic and proteomic data. Curr Microbiol 57: 18–22, 2008. doi: 10.1007/s00284-008-9145-5. [DOI] [PubMed] [Google Scholar]
- 37.Zaas AK, Schwartz DA. Innate immunity and the lung: defense at the interface between host and environment. Trends Cardiovasc Med 15: 195–202, 2005. doi: 10.1016/j.tcm.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Culley DE, Gritsenko MA, Moore RJ, Nie L, Scholten JC, Petritis K, Strittmatter EF, Camp DG 2nd, Smith RD, Brockman FJ. LC-MS/MS based proteomic analysis and functional inference of hypothetical proteins in Desulfovibrio vulgaris. Biochem Biophys Res Commun 349: 1412–1419, 2006. doi: 10.1016/j.bbrc.2006.09.019. [DOI] [PubMed] [Google Scholar]