Abstract
Background
Grading of meningiomas is important in the choice of the most effective treatment for each patient.
Purpose
To determine the diagnostic accuracy of a deep convolutional neural network (DCNN) in the differentiation of the histopathological grading of meningiomas from MR images.
Study Type
Retrospective.
Population
In all, 117 meningioma‐affected patients, 79 World Health Organization [WHO] Grade I, 32 WHO Grade II, and 6 WHO Grade III.
Field Strength/Sequence
1.5 T, 3.0 T postcontrast enhanced T1 W (PCT1W), apparent diffusion coefficient (ADC) maps (b values of 0, 500, and 1000 s/mm2).
Assessment
WHO Grade II and WHO Grade III meningiomas were considered a single category. The diagnostic accuracy of the pretrained Inception‐V3 and AlexNet DCNNs was tested on ADC maps and PCT1W images separately. Receiver operating characteristic curves (ROC) and area under the curve (AUC) were used to asses DCNN performance.
Statistical Test
Leave‐one‐out cross‐validation.
Results
The application of the Inception‐V3 DCNN on ADC maps provided the best diagnostic accuracy results, with an AUC of 0.94 (95% confidence interval [CI], 0.88–0.98). Remarkably, only 1/38 WHO Grade II–III and 7/79 WHO Grade I lesions were misclassified by this model. The application of AlexNet on ADC maps had a low discriminating accuracy, with an AUC of 0.68 (95% CI, 0.59–0.76) and a high misclassification rate on both WHO Grade I and WHO Grade II–III cases. The discriminating accuracy of both DCNNs on postcontrast T1W images was low, with Inception‐V3 displaying an AUC of 0.68 (95% CI, 0.59–0.76) and AlexNet displaying an AUC of 0.55 (95% CI, 0.45–0.64).
Data Conclusion
DCNNs can accurately discriminate between benign and atypical/anaplastic meningiomas from ADC maps but not from PCT1W images.
Level of evidence: 2
Technical Efficacy: Stage 2
J. Magn. Reson. Imaging 2019;50:1152–1159.
Keywords: meningioma, deep learning, apparent diffusion coefficient, postcontrast, grading
MENINGIOMAS account for 33.8% of all the primary intracranial neoplasms in the USA.1, 2 Different treatment options, based on the histopathological grading of the lesions, are recommended for meningiomas.1 Nevertheless, no widely accepted methods to predict the histopathological grading of these neoplasms by means of magnetic resonance imaging (MRI), using either standard MRI sequences or advanced MRI sequences such as diffusion‐weighted imaging, have been available to date. The possibility to accurately predict the grading of meningiomas from MR images could enable a more targeted, and therefore more effective, treatment plan for each patient.
Machine learning has grown increasing popular in radiology research in recent years, mostly driven by the perspective of creating greater interconnection between radiologists and machines.3, 4 Currently, the main scope for applying machine learning in medical imaging is the creation of an integrated environment where the machines support, speed up, and supervise the radiologists' work.3 The diffusion of machine‐learning algorithms is currently hindered by the scarcity of large and well‐annotated databases, which are difficult and expensive to produce. Moreover, the introduction of such algorithms in clinical practice requires integration with preexisting workflows, along with an actual demonstration of their value in terms of cost reduction and outcome improvement. The possible applications of machine learning to assist the radiologist during routine clinical activity range from the automatic creation of study protocols5 to the hanging of study protocols, and to the improvement of computed tomography (CT) image quality; among the various advantages is also a reduction in the radiation dose.6, 7 In addition, many recent articles have highlighted the ability to use deep convolutional neural networks (DCNNs) to assist radiologist interpretation of radiographic images.8, 9
Deep learning is a branch of machine learning that uses patterns gained directly from data, through a general‐purpose learning procedure instead of human‐engineered features, to make predictions.10 DCNNs automatically use filters to create feature maps depicting the distribution of such features in the images. These maps are then analyzed to create increasingly more complex and abstract representation of the items represented in the images. A broad number of applications of DCNNs in neuroradiology are being studied, and the most thoroughly investigated are: 1) automatic brain segmentation11, 12; 2) automatic detection of Alzheimer's disease‐associated lesions from functional MRI13; 3) automatic detection of stroke‐related lesions from CT images14; 4) prediction of genetic mutation15; and 5) grading of gliomas.16, 17
Recently, the scope to predict the grading of spontaneously occurring meningiomas from routine MRI sequences by means of DCNNs has been explored in dogs; an 82% accuracy in discrimination between benign, atypical, and malignant lesions was achieved on a small‐sized dataset (56 patients).18
Thus, the aim of our study was to determine whether application of DCNNs on apparent diffusion coefficient (ADC) maps and postcontrast T1‐weighted (PCT1W) images could enable accurate discrimination of the histopathological grading of human meningiomas.
Materials and Methods
Patients
This study was approved by the institutional Ethical Committee. As this is a retrospective study and retrieving the informed consent from all the patients would have been very difficult, in agreement with the Ethical Committee of the University of Padua, all the MRI scans were anonymized prior to the analysis. In all, 117 meningioma cases (79 World Health Organization [WHO] Grade I, 32 WHO Grade II and 6 WHO Grade III; 80 women and 37 men; mean age 58.6 years ±14.9 [standard deviation]), who were admitted to our Neuroradiology and Neurosurgery units between January 2012 and December 2016 and who underwent an MRI scan and received a final histopathological diagnosis of meningioma, were retrospectively selected. Subtypes of benign lesion included: meningothelial (n = 30), transitional (n = 22), fibroblastic (n = 11), psammomatous (n = 5), angiomatous (n = 4), secretory (n = 3), microcystic (1), not specified (n = 2), and cordoid (n = 1). Due to the relatively low number of anaplastic lesions, WHO Grade II and WHO Grade III lesions were combined as a single category.
Imaging
All the MRI scans included were deidentified prior to analysis in compliance with the requirements of the institutional Ethical Committee. Image quality, in particular the presence of the most common MRI artifacts, of all the MRI scans was assessed by three neuroradiologists (F.C., A.D.P., F.C. with 26, 24, and 14 years of experience, respectively) prior to the beginning of the study. Twenty‐four of the 117 MRI scans included in the present study were performed at our institution with a 3.0 T Ingenia system (Philips Medical Systems, Best, the Netherlands) using the following parameters: diffusion‐weighted (DW) single‐shot echo planar imaging sequence (field of view [FOV], 230 × 230 mm; matrix size, 256 × 256; slice thickness, 3 mm; gap thickness 0 mm; repetition time [TR] greater than 3000 msec / echo time [TE] minimum; b values of 0 and 1000 s/mm2; ADC maps were automatically generated by the embedded software) and a 3D sagittal T1‐weighted postcontrast fast field echo sequence (FOV, 240 × 240 mm; matrix size, 448 × 448; reduction factor 1 to 2,5; TR minimum / TE minimum; 5' after i.v. administration of Gadovist 0.1 mmol/kg). Eighty‐five of the 117 MRI scans were performed at our institution with an Avanto 1.5 T (Siemens Medical Solutions, Erlangen, Germany) MRI scanner equipped with 16 multichannel receiver head coils using the following settings: DW single‐shot echo planar imaging sequence (FOV, 270 × 270 mm; slice thickness, 5 mm; gap thickness 0 mm; TR 3000 msec / TE 89; b values of 0, 500, and 1000 s/mm2; ADC maps were automatically generated by the embedded software) and 3D sagittal T1‐weighted postcontrast gradient echo sequence (FOV, 256 × 256 mm; TR 1200 / TE 3.9; 5' after i.v. administration of Gadovist 0.1 mmol/kg) Further images from MRI scans of patients referred from external facilities were included in the study. The other MRI scanners utilized were: Achieva 1.5 T (Philips Medical Systems) (n = 3), Signa 1.5 T (GE Medical Systems, Milwaukee, WI) (n = 2), and Espree 1.5 T (Siemens Medical Solutions) (n = 3).
Image Processing
All the MR images were exported in an 8‐bit Joint Photographic Experts Group (JPEG) format using a freely available image visualization and analysis software (Horos; Nimble, Purview, Annapolis, MD). The lesions were manually selected (carefully maintaining a square format) by one of the authors (T.B.) on the PCT1W images and then the same cropping was applied on the ADC maps. Following recommendations in the available literature,16 an adaptive contrast filter was applied to the images using ImageJ (NIH, Bethesda, MD). Thereafter, the images were resized to a 299 × 299 pixel format. Lastly, the images were saved into different folders according to the histopathological grading assigned by the pathologist.
Deep Learning
The deep‐learning analysis was performed on a workstation with a Linux operating system (Ubuntu 16.04; Canonical, London, UK) equipped with two Titan XP (NVIDIA, Santa Clara, CA) graphic processing units, a Xeon E5‐2640 v4 2.1 GHz (Intel, Santa Clara, CA) processor, and 32 GB of random access memory. The deep‐learning toolbox included in MatLab (v. 2017b, MathWorks, Natick, MA) was used for image analysis. The Inception‐V3 and Alexnet deep neural networks pretrained on a large‐scale database (ImageNet‐www.image-net.org) of everyday color images were used for image classification. ADC maps and PCT1W images were analyzed separately. A custom MatLab function that augmented the image database by randomly rotating, cropping, flipping, and/or creating mirror images was then used on the same models to increase diagnostic accuracy. Network hyperparameters were set as follows: adaptive moment estimation (Adam) loss function, an adaptive learn rate with an initial learn rate of 0.0001, a squared‐gradient decay factor of 0.99. The diagnostic accuracy of the DCNN was tested using a leave‐one‐out cross‐validation procedure performed at a patient level. For the leave‐one‐out cross‐validation procedure the training database, containing n –1 cases (where n = number of cases), was divided into a training and a validation set comprising 90% and 10% of the images, respectively. The predicted class of each case was recorded. Cases were classified as WHO Grade II–III based on the modal‐predicted class of the images. To avoid overfitting, the DCNNs were initialized prior to each iteration of the leave‐one‐out cross‐validation and no layers of the network were frozen because we considered basic features (such as shape or texture, which are typically extracted by the first layers of the network) to be highly relevant for our classification problem.
Statistics and Data Analysis
A contingency table with the results of the leave‐one‐out cross‐validation was created and the area under the curve (AUC), sensitivity, specificity, and accuracy were calculated using commercially available software (MedCalc v. 15.05; Ostend, Belgium; and MatLab and Statistics Toolbox Release 2018a). The 95% confidence intervals were calculated as exact Clopper‐Pearson confidence intervals. The AUC value as a criterion of discrimination accuracy was classified as low (0.5–0.7), moderate (0.7–0.9), or high (>0.9).15 The AUC of the individual models was compared with the DeLong method.19 For a thorough evaluation of the accuracy of our test, and to account for the strong imbalance between benign and atypical/anaplastic meningiomas in our database, the Matthews Correlation Coefficient was calculated using the Multi Class Confusion Matrix function embedded in MatLab. The Matthews Correlation Coefficient takes values in the interval [–1, 1], with 1 showing a complete agreement, –1 a complete disagreement, and 0 showing that the prediction was uncorrelated with the ground truth.20 A schematic representation of the workflow used in this retrospective study is reported in Fig. 1.
Figure 1.
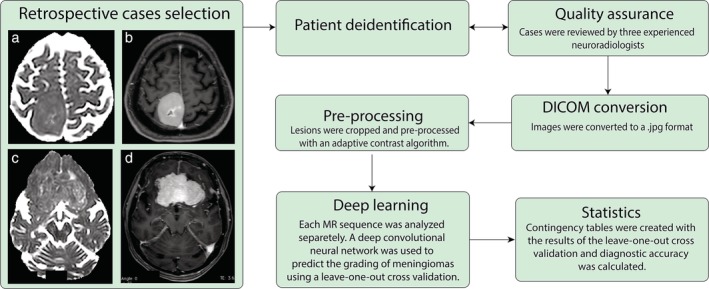
Schematic representation of the workflow used in this retrospective study. Axial (a) ADC Maps, (b) PCT1W images obtained in a 58‐year‐old woman showing a WHO Grade I right fronto‐parietal meningioma. Axial (c) ADC Maps, (d) PCT1W images obtained in a 72‐year‐old woman showing a WHO Grade II bilateral fronto‐basal meningioma.
Results
The contingency tables resulting from the leave‐one‐out cross‐validation of the combinations between Inception‐V3 and ADC maps (IncV3‐ADC), Inception‐V3, and PCT1W images (IncV3‐PCT1W), AlexNet and ADC maps (Alex‐ADC), and AlexNet and PCT1W images (Alex‐PCT1W) are reported in Fig. 2.
Figure 2.
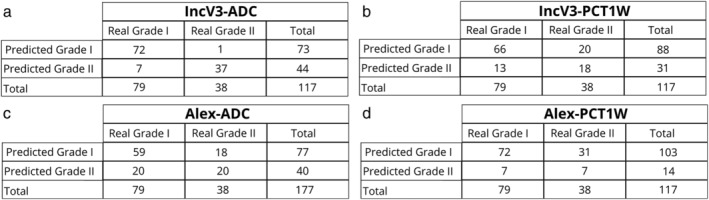
Contingency tables of the results of the leave‐one‐out cross‐validation for the ADC and the PCT1W images.
The IncV3‐ADC model displayed the highest discriminating accuracy, with an AUC of 0.94 (95% confidence interval [CI], 0.88–0.98). Remarkably, only one WHO Grade II was misclassified. The discrimination accuracy of AlexNet on ADC maps was low, with an AUC of 0.63 (95% CI, 0.54–0.72). In particular, 15 WHO grade II and 5 WHO grade III were misclassified.
The application of DCNNs on PCT1W images did not provide satisfactory results and the discriminating accuracy for both DCNN architectures was low. In particular, the IncV3‐PCT1W model had an AUC of 0.68 (95% CI, 0.59–0.76). In particular, 16 WHO grade II and 4 WHO grade III were misclassified. The Alex‐PCT1W model had an even lower performance, with an AUC of 0.55 (95% CI, 0.45–0.64). In particular, 25 WHO grade II and 6 WHO grade III were misclassified.
The complete results of the ROC curve analysis are reported in Table 1. The comparisons between ROC curves are reported in Fig. 3. There was a significant difference between the ROC curve of the IncV3‐ADC model and all the remaining models (P < 0.0001), whereas no statistically significant differences between IncV3‐PCT1W and Alex‐ADC (P = 0.733), between Alex‐ADC and Alex‐PCT1W(P = 0.134), and between Alex‐PCT1W and IncV3‐PCT1W (P = 0.051) were evident. Two examples of misclassified lesions, one WHO Grade I lesion classified as WHO Grade II–III and the WHO Grade II–III misclassified by the IncV3‐ADC model, are reported in Figs. 4, 5, respectively.
Table 1.
Results of the ROC Curve Analysis, Along With Their 95% Confidence Intervals, for the MR Datasets
| Dataset | Sensitivity,% | Specificity,% | AUC | Accuracy,% | PPV,% | NPV,% | MCC |
|---|---|---|---|---|---|---|---|
| IncV3‐ADC | 97.4 (96.2–99.9) | 91.1 (82.6–96.4) | 0.94 (0.88–0.98) | 93.1 (87–97) | 84.1 (69.9–93.4) | 98.6 (92.6–99.9) | 0.86 |
| IncV3‐PCT1W | 52.6 (35.8–69) | 83.5 (73.5–90.9) | 0.68 (0.59–0.76) | 71.8 (62.7–79.7) | 60.6 (42.1–77.1) | 76.7 (66.4–85) | 0.37 |
| Alex‐ADC | 52.6 (35.8–69) | 74.6 (63.6–83.8) | 0.63 (0.54–0.72) | 76.6 (69.6–82.4) | 50 (33.8–66.2) | 76.6 (65.6–85.5) | 0.14 |
| Alex‐PCT1W | 18.4 (7.7–34.3) | 91.1 (82.6–96.4) | 0.55 (0.45–0.64) | 67.5 (58.4–75.9) | 50 (23–77) | 69.9 (60–78.6) | 0.42 |
ROC: receiver operator curve; AUC: area under the curve; MCC: Matthews Correlation Coefficient; PPV: positive predictive value; NPV: negative predictive value.
Figure 3.
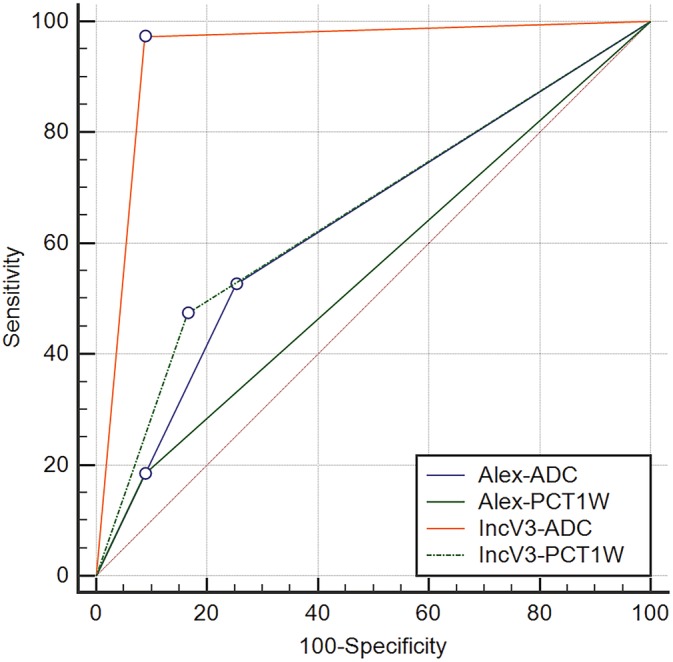
ROC curves of the IncV3‐ADC, the IncV3‐PCT1W, the Alex‐ADC, and the Alex‐PCT1W models, with their corresponding AUCs.
Figure 4.
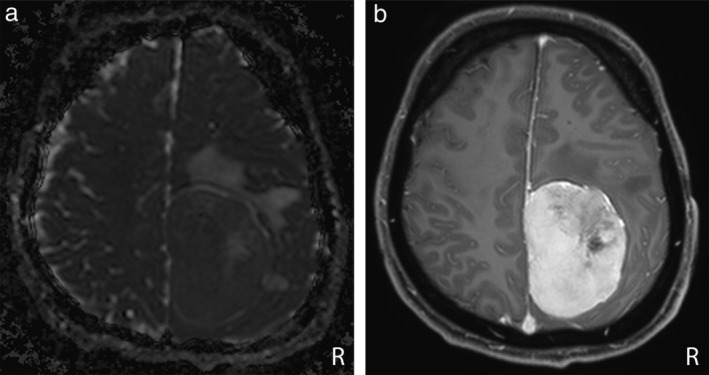
Axial ADC map (a) and PCT1W MR images (b) in a 36‐year‐old man with a WHO Grade II parietal falcine meningioma. The lesion was misclassified by the IncV3‐ADC model, whereas it was correctly classified by the IncV3‐PCT1W model.
Figure 5.
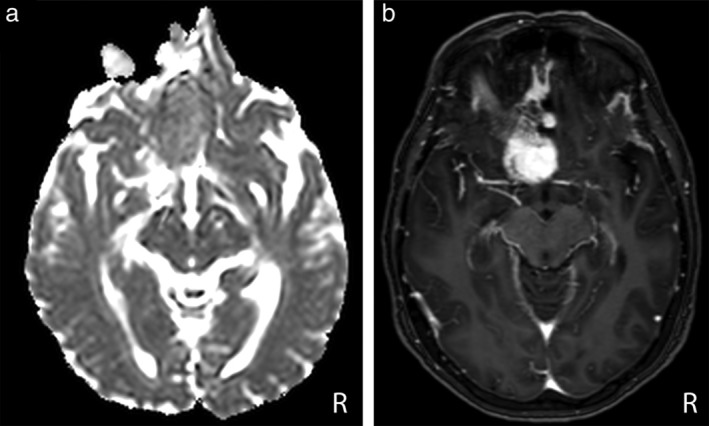
Axial ADC map (a) and PCT1W images (b) in a 59‐year‐old woman with a Grade I anterior clinoid meningioma. The lesion was misclassified as a WHO Grade II–III lesion by the IncV3‐ADC model, whereas it was correctly classified as a WHO Grade I lesion by the IncV3‐PCT1W model.
Discussion
ADC maps and postcontrast T1W images were included in the present study mainly for two reasons: 1) because most of the previous studies trying to determine meningioma grading from MR images were focused on these two imaging sequences; 2) because the combination between semantic and radiomic features on postcontrast T1W images has recently shown promising results with an AUC of 0.86.21 The possibility to determine the grading of human meningiomas using diffusion‐tensor imaging has been explored by several authors,22, 23, 24, 25, 26 with differing results. In particular, a study by Surov et al22 including 389 meningioma‐affected patients reported an ADC value below 0.85 × 10‐3 mm2 s‐1 having a 73% accuracy (72.9% sensitivity and 73.1% specificity) in the distinction between benign and atypical/malignant meningiomas. By contrast, Santelli et al26 and Sanverdi et al24 found no statistically significant differences between the ADC calculated on benign and on atypical/malignant lesions in 177 patients.
The use of histogram analysis of diffusion‐tensor MRI metrics, including tensor shape measurements, showed promising results in the determination of the grading of human meningiomas.25 The study by Wang et al25 reported an 88% accuracy (in a leave‐one‐out cross‐validated procedure) in the distinction between typical and atypical/anaplastic meningiomas. Remarkably, we obtained a higher diagnostic accuracy (94%) on a much larger study population (117 vs. 49 cases). In the previous studies using texture analysis,21, 25 some predetermined texture features, mainly describing the homogeneity of the signal in a determined region of interest, were extracted from the MR images and then combined to develop a predictive model (logistic regression or multivariate analysis). Although this approach might provide accurate classification results, the number of steps required for the analysis (image selection, image preprocessing, feature extraction, feature selection, model development, and model testing) limits their clinical applicability. Moreover, most of the previously mentioned steps (except for the model development and testing) are required also to predict the grading of new cases. One of the main advantages of the approach we are proposing here compared with the texture analysis‐based model put forward by Wang et al25 is that once the DCNN has been retrained on our dataset, it can be directly used to predict the histopathological grading of new cases without any further steps (feature extraction, model calculation). DCNNs are capable of detecting features that are learned directly from data using a general‐purpose learning procedure,10 thus overcoming the need to "manually" extract the features from the images. The other advantage of DCNNs is that the learning process is "embedded" in the algorithm, and hence there is no need to develop and test statistical or machine‐learning models to classify the results. The main disadvantage of DCNNs is that these algorithms act like black‐boxes, and thus the structure of the networks does not provide any useful information from the database being analyzed.27 Moreover, DCNNs are extremely easy to implement, simply by adding new labeled images and retraining the model to include the new cases, thus potentially increasing model accuracy.
The application of DCNNs on PCT1W and ADC maps showed a markedly different classification accuracy. For the above‐described limitations of DCNNs, it is not possible to determine the exact reason for such differences. It is the authors' opinion that the large variability in the PCT1W features of meningiomas, regardless of the histopathological grading, may have limited the classification performance of both the DCNNs tested. In particular, even if a heterogeneous enhancement is more commonly associated with WHO Grade II and III lesions, several WHO Grade I lesions have a cystic, and therefore heterogeneous, appearance.21 Indeed, other studies using texture analysis (DCNNs are capable to analyze thoroughly the texture of the images) to predict the grading of meningiomas from PCT1W images reported a similar diagnostic accuracy.21 On the other hand, the application of DCNNs on ADC maps provided a very high diagnostic accuracy. WHO Grade II meningiomas are characterized by a higher degree of tissue disruption (and therefore a higher heterogeneity of the distribution of cells in the lesion) compared with WHO Grade I lesions. ADC maps depict a representation of the directionality of the cells within a lesion and, therefore, this MRI sequence could inherently reveal more information regarding the histopathological grade of meningiomas. In fact, previous studies on the possible applications of texture analysis to predict the grading of meningiomas described a lower anisotropy (a measure of the signal alignment) in WHO Grade II meningiomas.25
Another limitation of the present study is that the clinical translatability of CNNs in general has yet to be proven.3 The two CNN architectures used in the present study (AlexNet and Inception V3: most previous studies used GoogleNet, an earlier version of Inception V3) have been successfully applied in different research scenarios in the medical literature,8, 30, 31 but, to the best of the authors' knowledge, no commercially available medical products based on these two architectures are currently available.
According to the literature, disease prevalence affects the probability predictions of DCNNs.28 However, the prevalence of benign against atypical/anaplastic lesions in our database closely resembles the prevalence in the general population.1 Therefore, the high sensitivity (97.4%) and positive predictive value (PPV) (84%) of the IncV3‐ADC model suggest that the methodology proposed here is potentially applicable in clinical environments.29 On the other hand, the relatively high false‐positive rate of the IncV3‐ADC model (n = 7) might suggest that increasing the number of atypical/malignant cases in the database could enable the PPV of our model to increase.
A further limitation of this study is that, due to the relatively low number of anaplastic lesions present in the database, atypical and anaplastic meningiomas were considered as a single category. To the best of the authors' knowledge, no clinical trials focused on anaplastic lesions alone have been made available, and such lesions are grouped together with atypical meningiomas in most of the available studies.1 Interestingly, the diagnostic accuracy obtained in the present study for the standard MRI sequences (PCT1W) is comparable to the diagnostic accuracy reported for the same MRI sequences in dogs.18 The inclusion of ADC maps, which were not available in the canine study, enabled the development of a noticeably more accurate test in humans than in dogs.
The generalization ability of our DNN was not completely assessed due to both the relatively low number of MRI devices included in the project (n = 5) and the premise that most of the studies were performed using the same two MRI scanners installed at the Neuroradiology unit of our institution. Interestingly, all the cases misclassified by the IncV3‐ADC model were acquired with either one of the two MRI scanners (3.0 T Ingenia system; Philips Medical Systems; and, Avanto 1.5 T; Siemens Medical Solutions) supplied to our institution, suggesting that factors other than MRI scanner type can influence the analysis results. Given the low influence of the MRI scanner on the analysis, the inclusion of a larger number of facilities appears as a feasible solution for incorporating a greater number of cases within the model.
The histopathological grading score of lesions was considered as the DCNN target even if the choice between different treatment options for each patient depended on several factors, such as: lesion location, the presence or absence of brain edema, the presence or absence of clinical signs, patient age, etc. However, the 5‐year recurrence rate of atypical and anaplastic meningiomas is five to ten times higher than that of benign meningiomas29; therefore, the scope to accurately predict the histopathological grading of meningiomas may prospectively help the clinician in deciding on the best treatment for each patient (observation, surgery, radiotherapy, embolization, etc.) when surgical removal of the lesion is not mandatory.
In this study the lesion‐containing images were manually cropped and then resized to fit the requirements of DCNNs. In such a procedure some of the information regarding the lesion size is likely to be lost. Larger lesions are reported to be associated with a greater likelihood of a meningioma being WHO grade II.25 Further studies, possibly preserving the lesions size on the DCNN, might provide better classification results.
In conclusion, this study shows that the application of DCNNs on ADC maps provides a high diagnostic accuracy in discriminating between benign and atypical/anaplastic meningiomas, whereas the same application of DCNNs on PCT1W gives rise to inaccurate results. Additional studies, ideally including a larger number of cases from different institutions, are needed in order to fully assess the generalization ability of this test and to understand the safety and possible implications of machine learning as an adjunctive tool in clinical practice and decision‐making algorithms for patients, and particularly those with WHO Grade II and III meningiomas.
Acknowledgment
Contract grant sponsor: University of Padua, Italy; Contract grant numbers: Junior Research Grant from the UniPD (2015), Supporting Talents in Research@University of Padua, with a project entitled: “Prediction of the histological grading of human meningiomas using MR image texture and deep learning: a translational application of a model developed on spontaneously occurring meningiomas in dogs.”
The authors thank the NVIDIA Corp. (CA, USA) for donation of the GPU cards used in this study.
References
- 1. Goldbrunner R, Minniti G, Preusser M, et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol 2016;17:383–391. [DOI] [PubMed] [Google Scholar]
- 2. Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol 2010;99:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dreyer KJ, Geis JR. When machines think: Radiology's next frontier. Radiology 2017;285:713–718. [DOI] [PubMed] [Google Scholar]
- 4. Zaharchuk XG, Gong XE, Wintermark XM, Rubin XD, Langlotz XCP. Deep learning in neuroradiology. Am J of Neuroradiol 10.3174/ajnr.A5543. 2018. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown AD, Marotta TR. Using machine learning for sequence‐level automated MRI protocol selection in neuroradiology. J Am Med Inform Assoc 2018;25:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H, Zhang Y, Kalra MK, et al. Low‐dose CT with a residual encoder‐decoder convolutional neural network (RED‐CNN). IEEE Trans Med Imaging Ithaca, NY: Cornell University Library. http://ieeexplore.ieee.org/document/7947200/ 2018 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 7. Lakhani P, Prater AB, Hutson RK, et al. Machine learning in radiology: Applications beyond image interpretation. J Am Coll Radiol 2017;1–10. [DOI] [PubMed] [Google Scholar]
- 8. Cicero M, Bilbily A, Colak E, et al. Training and validating a deep convolutional neural network for computer‐aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol 2017;52:281–287. [DOI] [PubMed] [Google Scholar]
- 9. Lakhani P, Sundaram B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017;284:574‐582. [DOI] [PubMed] [Google Scholar]
- 10. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 11. Havaei M, Davy A, Warde‐Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal 2017;35:18–31. [DOI] [PubMed] [Google Scholar]
- 12. Mehta R, Majumdar A, Sivaswamy J. BrainSegNet: A convolutional neural network architecture for automated segmentation of human brain structures. J Med Imaging 2017;4:024003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage 2017;145:137–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scherer M, Cordes J, Younsi A, et al. Development and validation of an automatic segmentation algorithm for quantification of intracerebral hemorrhage. Stroke 2016;47:2776–2782. [DOI] [PubMed] [Google Scholar]
- 15. Chang P, Grinbad BD, Weinberg M, et al. Deep‐learning convolutional neural networks accurately classify genetic mutations in gliomas. Am J Neruroradiol 2018;39:1201–1207, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khawaldesh S, Perviaiz U, Rafiq A, et al. Noninvasive grading of glioma tumor using magnetic resonance imaging with convolutional neural networks. Apll Sci 2018;8:27. [Google Scholar]
- 17. Chang K, Bai HX, Zhou H, et al. Residual convolutional neural network for the determination of IDH status in low‐ and high‐grade gliomas from MR imaging. Clin Cancer Res 2017;24:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banzato T, Cherubini GB, Atzori M, Zotti A. Development of a deep convolutional neural network to predict grading of canine meningiomas from magnetic resonance images. Vet J 2018;235:90–92. [DOI] [PubMed] [Google Scholar]
- 19. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988;44:837–845. [PubMed] [Google Scholar]
- 20. Boughorbel S, Jarray F, El‐Anbari M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS One 2017;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coroller TP, Bi LW, Huynh E, et al. Radiographic prediction of meningioma WHO Grade by semantic and radiomic features. PLoS One 2017;12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Surov A, Gottschling S, Mawrin C, et al. Diffusion‐weighted imaging in meningioma: Prediction of tumor WHO Grade and association with histopathological parameters. Transl Oncol 2015;8:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Surov A, Ginat DT, Sanverdi E, et al. Use of diffusion weighted imaging in differentiating between malignant and benign meningiomas. A multicenter analysis. World Neurosurg 2016;88:598–602. [DOI] [PubMed] [Google Scholar]
- 24. Sanverdi SE, Ozgen B, Oguz KK, et al. Is diffusion‐weighted imaging useful in grading and differentiating histopathological subtypes of meningiomas? Eur J Radiol 2012;81:2389–2395. [DOI] [PubMed] [Google Scholar]
- 25. Wang S, Kim S, Zhang Y, et al. Determination of WHO grade and subtype of meningiomas by using histogram analysis of diffusion‐tensor imaging metrics. Radiology 2012;262:584–592. [DOI] [PubMed] [Google Scholar]
- 26. Santelli L, Ramondo G, Della Puppa A, et al. Diffusion‐weighted imaging does not predict histological grading in meningiomas. Acta Neurochir 2010;152:1315–1319. [DOI] [PubMed] [Google Scholar]
- 27. Prieto A, Prieto B, Ortigosa EM, et al. Neural networks: An overview of early research, current frameworks and new challenges. Neurocomputing 2016;214:242–268. [Google Scholar]
- 28. Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018;283:800–809. [DOI] [PubMed] [Google Scholar]
- 29. Rogers CL, Perry A, Pugh S, et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro Oncol 2016;18:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noguchi T, Higa D, Asada T, et al. Artificial intelligence using neural network architecture for radiology (AINNAR): Classification of MR imaging sequences. Jns J Radiol 2018:5–11. [DOI] [PubMed] [Google Scholar]
- 31. Chi J, Walia E, Babyn P, Wang J, Groot G, Eramian M. Thyroid nodule classification in ultrasound images by fine‐tuning deep convolutional neural network. J Digit Imaging 2017;30:477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]


