Abstract
Background
Sodium MRI (23Na‐MRI)‐derived biomarkers such as total sodium concentration (TSC) have the potential to provide information on tumor cellularity and the changes in tumor microstructure that occur following therapy.
Purpose
To evaluate the repeatability and reproducibility of TSC measurements in the brains of healthy volunteers, providing evidence for the technical validation of 23Na‐MRI‐derived biomarkers.
Study Type
Prospective multicenter study.
Subjects
Eleven volunteers (32 ± 6 years; eight males, three females) were scanned twice at each of two sites.
Field Strength/Sequence
Comparable 3D‐cones 23Na‐MRI ultrashort echo time acquisitions at 3T.
Assessment
TSC values, quantified from calibration phantoms placed in the field of view, were obtained from white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF), based on automated segmentation of coregistered 1H T1‐weighted images and hand‐drawn regions of interest by two readers.
Statistical Tests
Coefficients of variation (CoVs) from mean TSC values were used to assess intrasite repeatability and intersite reproducibility.
Results
Mean GM TSC concentrations (52.1 ± 7.1 mM) were ∼20% higher than for WM (41.8 ± 6.7 mM). Measurements were highly repeatable at both sites with mean scan–rescan CoVs between volunteers and regions of 2% and 4%, respectively. Mean intersite reproducibility CoVs were 3%, 3%, and 6% for WM, GM, and CSF, respectively.
Data Conclusion
These results demonstrate technical validation of sodium MRI‐derived biomarkers in healthy volunteers. We also show that comparable 23Na imaging of the brain can be implemented across different sites and scanners with excellent repeatability and reproducibility.
Level of Evidence: 1
Technical Efficacy: Stage 2
J. Magn. Reson. Imaging 2019;50:1278–1284.
Keywords: sodium MRI, brain, repeatability, reproducibility, biomarker
QUANTIFYING AND IMAGING sodium distribution provides an important biomarker of normal tissue function and of the changes that can occur during different disease processes due to the importance of sodium ions in many aspects of cell physiology. Viable cells maintain a sodium gradient across the cell membrane with a high extracellular sodium concentration (140–150 mM) and a low intracellular sodium concentration (10–15 mM).1 This transmembrane gradient is generated and maintained through the action of the Na+/K+ ATPase (sodium‐potassium pump) that exports three Na+ ions out of the cell in exchange for two K+ ions, and this ionic gradient is then used to drive several other membrane‐bound exchangers and cotransporters.2 The sodium‐23 (23Na) nucleus gives the second strongest nuclear magnetic resonance (NMR) signal from biological tissues after the water proton (1H).3 However, despite the overall sensitivity of 23Na being 10 times lower than that of 1H, sodium distribution has been successfully imaged in vivo using magnetic resonance imaging (MRI).
The lower concentration of 23Na relative to water protons and its short biexponential T2 relaxation time are significant challenges for the implementation of sodium MRI.4 However, advances in MRI hardware and tailored 23Na MRI sequences5 have facilitated higher resolution clinical 23Na MRI as a method to probe tissue biology in health and disease. There is ongoing research into the use of 23Na MRI in many conditions ranging from musculoskeletal and neurodegenerative diseases to stroke.3, 6, 7
Several studies have shown intracellular sodium volume to be significantly higher in malignant tumors compared with benign lesions due to increased cell density, altered metabolism, and changes in cell membrane ion transporters.8, 9 In addition to this increase in intracellular sodium, many malignant tumors also show an increase in the sodium‐rich extravascular–extracellular space, which in some cases may be secondary to an increase in necrosis.10, 11, 12, 13, 14 Combining information on sodium concentration from both the intracellular and the extracellular compartments provides complementary information that can be used to probe tissue structure, cell density, and membrane integrity in more detail, which could be used in the assessment of tumor progression and treatment response.15 23Na MRI has been shown to be sensitive to therapy‐induced changes in both breast and central nervous system tumors.16 In a recent study, 23Na MRI outperformed an invasive histological correlate for brain tumor prognostication.17 23Na MRI may therefore allow better assessment of tissue cellularity and may allow treatment to be more personalized as a result of documenting the variety of responses seen within and between tumors.
The most widely reported metric in 23Na MRI studies is the total sodium concentration (TSC), which is the combination of both the intracellular and extracellular contributions. Before such biomarkers can be clinically utilized, they require both technical and biological validation to establish their accuracy and precision, as well as their relationship to cancer biology. Repeatability and reproducibility need to be demonstrated at an early stage of imaging biomarker development. While a number4, 18, 19, 20, 21 of studies report low standard deviations in normal tissue sodium concentrations, absolute TSC results and acquisition methods vary substantially between different studies and different centers, making comparisons difficult. As such, the purpose of this work was to implement comparable 23Na MRI acquisition protocols at different sites, and evaluate the repeatability (intrasite variation) and reproducibility (intersite variation) of TSC measurements, providing data contributing to the technical validation of 23Na MRI‐derived biomarkers.
Materials and Methods
Participants
Local research Ethics Committee approval was granted, and volunteers provided informed consent. The brains of 11 healthy volunteers (32 ± 6 years; eight males, three females), with no history of neurological disease, were scanned at two sites (A and B) with different MRI scanners. At site A, all the volunteers were scanned twice; at site B, seven were scanned twice and four once. The two scans at a given site were <1 hour apart and were used to assess repeatability (intrasite variation); scans for a given volunteer at both sites ranged from 1–41 days apart and were used to assess reproducibility (intersite variation).
Calibrants
Two batches of sodium calibration phantoms were made from a stock of 20 mM and 80 mM sodium chloride (NaCl) in water with 4% agar and 1% nickel sulfate (NiSO4) and distributed to the two sites. Each of the phantoms were cylindrical tubes of 30 ml capacity (physical dimensions 2.45 × 8.9 cm; Dutscher, France), filled with the stock solution. These were placed beside the ear defenders/headphones worn by the subjects to include them in the imaging field of view (FOV). An in‐house script written in MatLab v. 9.1 (MathWorks, Natick, MA) was used to automatically outline the phantoms and erode the voxels at the periphery, which are most likely to suffer from partial volume based on intensity thresholding.
MRI Acquisition
The sodium acquisition was performed using a matched protocol 3D‐Cones UTE sequence. The 3D‐Cones trajectory is attractive for this type of study: the code for waveform generation is freely available; the trajectory is utilizable with both 3D and UTE imaging, therefore increasing signal‐to‐noise ratio (SNR) over standard 2D gradient echo sequences; and it has a high sampling efficiency, allowing for shorter scans compared with 3D radial UTE sequences.22 The two 3 T systems were from different MR manufacturers (GE MR750, GE Healthcare, Waukesha, WI; and Achieva, Philips Medical Systems, Best, The Netherlands). Both systems used the same model of 1H/23Na dual‐tuned head coil (RAPID Biomedical, Rimpar, Germany). The 3D‐Cones trajectory was computed offline and imported into the respective scanner environments, making the gradient waveforms and the main sequence parameters identical on each system: 4 mm isotropic nominal resolution, 24 × 24 cm FOV, 10 msec readout, 2184 readouts, 3 averages, 166 kHz full readout bandwidth, 30 mT/m maximum gradient amplitude, 120 mT/s maximum slew rate. The echo time (TE) and repetition time (TR) were 0.5 msec and 100 msec for both systems.
To account for gradient timing errors, phantom imaging at both sites was undertaken to optimize the line profile through a homogenous phantom. The difference between actual and theoretical waveform was corrected by convolving the reconstruction trajectory with an exponential hysteresis function that shifts the k‐space samples in time. An experimentally determined delay of 5 μs provided the best line profile (straight through the phantom, abrupt at phantom edges) for both sites.
A volumetric T1‐weighted (T1W) fast spoiled gradient echo (FSPGR) sequence was acquired at both sites with 1 mm isotropic resolution, 24 × 24 cm FOV, TE/TR = 2.6/7 msec, 62.5 kHz full readout bandwidth for segmentation and region of interest (ROI) analysis.
Data Analysis
Images were reconstructed in MatLab v. 9.1.23 A sample density weighted apodization24 was used to match the SNR between the sites. TSC maps were generated based on the calibration phantoms placed in the FOV.25 TSC values were obtained from white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF), based on automated segmentation of coregistered 1H T1W images (SPM12, UCL, London, UK). To reduce the effect of partial volume, only voxels with a tissue probability larger than 95% were included in the masks. In addition, a hand‐drawn ROI analysis was performed using Osirix 8.5 (Pixmeo SARL, Switzerland) by two observers (6 and 12 years of experience with radiological imaging) to assess interobserver agreement.
The selection of the anatomical structures included in the analysis were performed by two observers in consensus to reduce bias. The caudate and the putamen were selected as representative regions of deep GM; the centrum semiovale was chosen to represent WM. Care was taken to diminish the potential partial volume effect of the ventricles on the ROIs and all the ROIs were drawn bilaterally. Coefficients of variation (CoVs) of mean values were used to assess repeatability (between scans 1 and 2 at a given site) and reproducibility (between the first scan at each site).
Results
The phantoms appeared on imaging with a diameter of at least 6 voxels over 20 slices and each individual 2D phantom ROI that was obtained contained 10–30 voxels.
Example TSC maps from one subject scanned twice at both sites are shown in Fig. 1a, with WM, GM, and CSF values averaged over the first scan from all subjects using the segmentation masks is shown in Fig. 1b. Qualitatively, TSC values in segmented regions were consistent across sites, with mean GM concentrations slightly higher than WM, and CSF values generally higher than WM and GM. Minor qualitative differences between sites A and B are shown in Fig. 1a.
Figure 1.
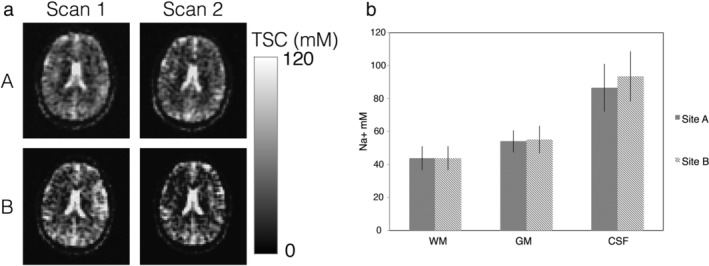
Mean total sodium concentrations across brain regions from the two sites: (a) Example TSC maps from one subject scanned twice at sites A and B. (b) Mean TSC ± SD for the first scan and all volunteers in segmented WM, GM, and CSF for sites A and B.
The repeatability and reproducibility of segmented TSC values are presented with Bland–Altman plots in Fig. 2a,b, respectively. Measurements were more repeatable at site A than B, with mean repeatability coefficients of 4.7 mM and 8.5 mM, respectively. The limits of agreement at the 95% confidence interval (CI) were –3.3 to +5.3 mM and –6.1 to +9.7 mM at sites A and B, respectively. The repeatability CoVs for WM, GM, and CSF were 1.8, 2.0, and 1.9% for site A and 4.2, 4.7, and 4.1%, respectively, for site B. The reproducibility CoVs between site A and B were 2.7, 3.2, and 6.3% for WM, GM, and CSF. At the 95% CI, the limits of agreement of reproducibility between site A and B were –9.3 to +12.7 mM with greater variation in CSF (Fig. 2b). Mean sodium concentration estimates for the segmentation analysis for each scan at the two sites are given in Table 1.
Figure 2.
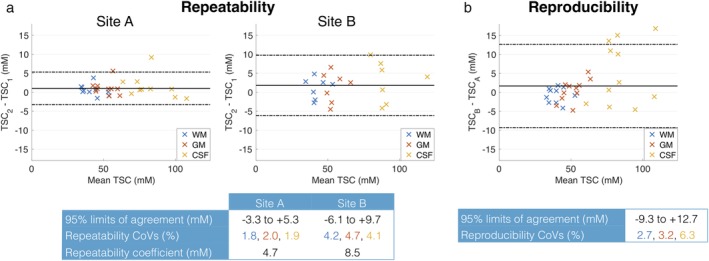
Bland–Altman plots of repeatability and reproducibility for automated segmentation. (a) Repeatability of TSC in segmented brain regions, comparing scans 1 and 2 at each site. Individual data points represent the difference in mean TSC between scans for a given subject, plotted against the mean of the two TSC measurements. (b) Reproducibility of TSC in segmented brain regions, comparing the first scan at each site.
Table 1.
Mean Total Sodium Concentrations (mM) From the Segmentation Analysis for All Subjects Given for Each Site and Each Scan
| Site A | Site B | |||||||
|---|---|---|---|---|---|---|---|---|
| Scan 1 | Scan 2 | Mean | Scan 1 | Scan 2 | Mean | Site A & B | Mean | |
| WM | 41.6 ± 7.3 | 42.4 ± 7.1 | 42.0 ± 6.9 | 41.1 ± 7.3 | 43.0 ± 6.3 | 41.8 ± 6.7 | WM | 41.8 ± 6.7 |
| GM | 51.0 ± 6.5 | 52.3 ± 6.7 | 51.8 ± 6.5 | 51.6 ± 8.6 | 54.6 ± 6.9 | 52.7 ± 7.8 | GM | 52.1 ± 7.1 |
| CSF | 81.8 ± 14.3 | 83.7 ± 13.9 | 82.7 ± 13.8 | 86.9 ± 15.7 | 93.0 ± 12.8 | 89.2 ± 14.5 | CSF | 85.8 ± 14.3 |
The repeatability and reproducibility of TSC in hand‐drawn ROIs are shown in Fig. 3a,b, respectively, for one observer. As with the segmentation analysis, repeatability was better at site A than B, with repeatability coefficients of 5.4 mM and 9.1 mM, respectively (Fig. 3a). The limits of agreement at the 95% CI were –4.0 to +6.1 mM and –8.0 to +10.0 mM at sites A and B, respectively. The repeatability CoVs for centrum semiovale, putamen, and caudate were 2.9, 3.2, and 4.6% for site A and 4.6, 6.3, and 7.8%, respectively, for site B. The reproducibility CoVs between site A and B were 5.2, 5.2, and 7.6% for centrum semiovale, putamen, and caudate. At the 95% CI, interval, the limits of agreement of reproducibility between site A and B were –8.1 to +9.3 mM (Fig. 3b).
Figure 3.
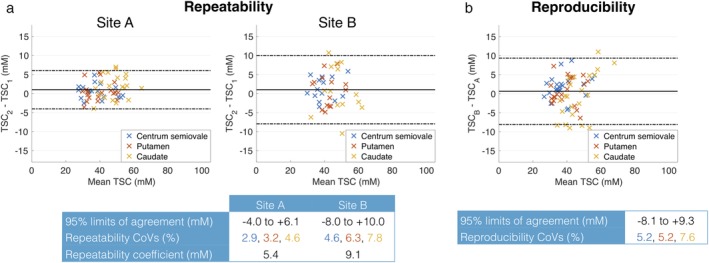
Bland–Altman plots of repeatability and reproducibility for manual segmentation. (a) Repeatability of TSC in three manually defined ROIs, comparing scans 1 and 2 at each site. Individual data points represent the difference in mean TSC between scans for a given subject, plotted against the mean of the mean TSCs. (b) Reproducibility of TSC in manually defined ROIs, comparing subjects' first scan at each site.
The results of the hand‐drawn ROI analysis described above come from one observer. Similar results were obtained from the second observer, with intraclass correlation coefficients (ICCs) used to formally assess interobserver agreement. ICCs calculated separately for each ROI in the first scan at each site generally indicated good interobserver agreement (Fig. 4), with ICC point estimates >0.85 in all cases. Mean sodium concentration estimates for the ROI analysis for each observer are given in Table 2.
Figure 4.
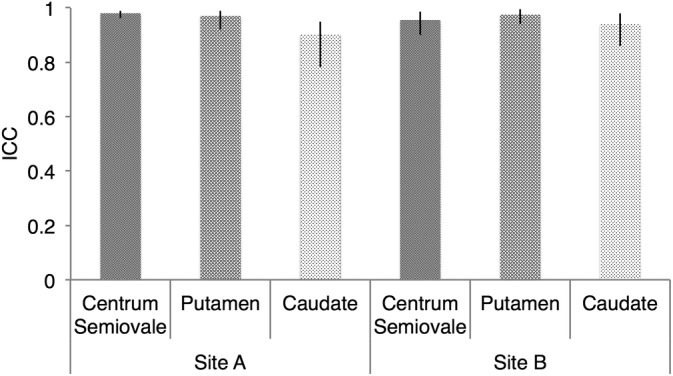
Assessment of interobserver agreement using ICCS. Data points and error bars represent the ICC and 95% CI for TSC values in ROIs drawn by the two observers. ICCs were calculated separately for each ROI for the first scan at sites A and B.
Table 2.
Mean Sodium Concentrations (mM) From the ROI Analysis for All Subjects at Each Site and for Each Observer
| Site A | Site B | ||||||
|---|---|---|---|---|---|---|---|
| Observer 1 | Observer 2 | Mean | Observer 1 | Observer 2 | Mean | Site A & B Mean |
|
| Caudate | 48.1 ± 7.1 | 47.8 ± 6.3 | 47.9 ± 6.7 | 47.2 ± 8.0 | 46.6 ± 7.5 | 47.0 ± 7.6 | 47.6 ± 7.1 |
| Centrum Semiovale | 37.6 ± 8.0 | 37.2 ± 8.0 | 37.4 ± 8.0 | 39.5 ± 8.2 | 39.9 ± 8.4 | 39.6 ± 11.3 | 38.6 ± 8.0 |
| Putamen | 39.3 ± 6.3 | 39.9 ± 7.1 | 39.5 ± 7.1 | 41.1 ± 6.9 | 42.4 ± 7.0 | 41.6 ± 6.9 | 40.5 ± 7.1 |
Discussion
This study assessed the repeatability and reproducibility of total brain tissue sodium as well as the first to utilize scanners from different vendors. Furthermore, it is the first to compare the use of a 3D Cones acquisition that has been harmonized across platforms.
The total sodium concentration measurements acquired in this study from automated segmentation of white and gray matter analysis are consistent with previous studies,19, 21, 26 eg, 41.8 ± 6.9 vs. 45.9 ± 3.1 mM, respectively, for WM and 52.1 ± 7.1 vs. 52.7 ± 3.6 mM for GM.26 WM measurements from the hand‐drawn ROI analysis here are also in agreement with previous studies: for example, Qian et al27 showed 37.4 ± 2.9 vs. 38.6 ± 8.0 mM here. GM values also showed good agreement with the literature: 47.6 ± 7.1 for the caudate here vs. 52.0 ± 6.0 mM28; and 40.9 ± 7.1 for the putamen here vs. 43.0 ± 3.0 mM.28 In general, differences between overall GM and the putamen are unsurprising due to cellular differences between deep GM and cortical GM: for example, the basal ganglia is permeated by a small amount of WM fiber bundles which would give a TSC that is a composite between WM and GM. Cortical GM, however, is more likely to be affected by partial volume effects due to the proximity of the CSF. The caudate GM values also demonstrate lower reproducibility, possibly due to partial volume effects, as it is bordering on WM and CSF.
Within our study cohort, measured CSF concentration values demonstrated poorer reproducibility compared with WM and GM measurements, which may be due to partial volume effects from surrounding tissue, real differences in sodium CSF concentration, or potential signal loss due to flow and pulsation.29 The CSF spaces are typically small in this young healthy cohort (average age 32 ± 6 years), leading to a higher risk of partial volume that may impact the absolute concentration measurement. In healthy volunteers, CSF flows at a rate of 11.1 ± 4.9 ml/min and changes in Na+ ion concentration have been reported in relation to migraine onset,30 suggesting that the CSF may be more subject to short‐term changes in sodium concentration.
Absolute sodium ion concentration within the CSF from studies that use nonimaging methods has been reported in the range of 135–155 mM,30 but exact values are rarely reported in 23Na MRI studies due to the partial T1‐recovery saturation issues at short repetition time (the T1 of CSF is ∼47 msec, compared with the T1 of brain tissue, which is ∼22 msec).31 Nevertheless, in a study undertaken by Ouwerkerk et al,32 the mean sodium concentration of CSF, derived using 23Na MRI at 1 .5T in a cohort of healthy volunteers (age range 22–63 years), was 135 ± 15 mM. While this value is higher than that reported in our study, the standard deviation in CSF values was large and is not inconsistent with the range of CSF measurement values (69.2–121.3) seen across our patient cohort. Furthermore, while Ouwerkerk et al's32 result was calculated from a hand‐drawn ROI placed within the lateral ventricles, a segmentation mask that included contributions from all CSF containing regions was used in our study. Due to the lack of appreciable brain atrophy within the young, healthy cohort studied, the cortical CSF spaces and lateral ventricles are often small, increasing the risk for partial volume effects, and potentially thereby lowering measured CSF concentration values within the subarachnoid and ventricular space respectively.
Qualitative differences in the appearance of the images between sites A and B such as blurring and contrast between brain parenchyma and CSF could be due to variations in eddy current behavior. Blurring of images related to eddy currents in spiral‐like imaging has previously been shown.33 Blurring may increase the risk of partial volume due to a poorer point spread function; however, in this study site A did not perform less well than site B in reproducibility and repeatability.
The small CoVs of our results confirm that TSC is highly reproducible between sites and observers. Madelin et al35 assessed repeatability of fluid‐suppressed sodium concentration in the brain by scanning 11 subjects twice on the same system using the FLORET sequence34 and found CoVs for estimated parameters between 10–20%.35 The higher CoVs in that study may be due to the lower SNR nature of fluid suppressed imaging and the advent of 23Na imaging as compared with the total sodium imaging utilized in our study. In a different study, Madelin et al36 looked at reproducibility and repeatability of total sodium imaging and fluid suppressed sodium concentration in the articular cartilage of six volunteers utilizing two different systems (3 T and 7 T, same vendor) and found CoVs over both systems to be between 7.5–13.6% using a radial sequence.36 Newbould et al and Jordan et al37, 38 both looked at repeatability of total sodium MRI measures in articular cartilage by scanning the same subjects using a 3D Cones sequence and found CoVs of 3.2% and 11.3%, respectively. Given the small size of articular cartilage and the involved potential partial volume issues compared with the comparatively larger brain structures investigated in this study, the smaller CoVs observed in our study are to be expected.
Previous authors have demonstrated increased TSC within CNS neoplasms such as high‐grade glioma,17, 32, 39 and that 23Na MRI can be used as a predictive marker of both tumor grade32 and prognosis.17 Within glioma, cellular proliferation and changes in cell membrane ion transporters17 contribute to elevated intracellular sodium concentrations, but these tumors also display increases in the sodium‐rich extravascular–extracellular space secondary to necrosis and loss of normal cellular packing.10, 11, 12, 13, 14 Evidence from in vivo animal studies of temporal 23Na MRI changes in glioma following chemotherapy treatment suggest that TSC may also serve as an early marker of progression and treatment response in this tumor group.40 While nonresponsive tumors in this study displayed a progressive increase in measured TSC, within responsive glioma regions there was an early sharp rise in TSC, an effect they hypothesized was due to therapy‐induced loss of membrane integrity and later cellular necrosis.40 Further studies should be undertaken to assess treatment and tumor growth‐related TSC changes in human glioma, but our study findings that these measurements demonstrate good repeatability and reproducibility in normal brain represent an early important step in the development of TSC as an imaging biomarker in CNS tumors.
The present study has a number of limitations, including technical aspects and study design. The true resolution of non‐Cartesian imaging studies is also difficult to define due to the nonuniform sampling and blurring introduced by dephasing of short T2 species during readout. The real resolution obtained is likely to be larger than the nominal resolution. However, by adapting the readout length to be shorter than the average expected sodium T2s, the effect of blurring was minimized in our study. Modeling of the image point‐spread‐function and correction in postprocessing was not used in our study and may improve estimates in future studies. B0 ± B1 correction are not a major concern at our field strength due to the low gyromagnetic ratio of sodium and the use of a volume resonator. Studies investigating 23Na at higher field strengths and/or using phased arrays will benefit from such corrections. In terms of technical limitations, an important consideration is the potential influence of relaxation on TSC measurements. Although a short TE was used to reduce T2* weighting, changes in transverse relaxation and/or tissue compartments may confound TSC measurements. Similarly, while the chosen TR matches that used in previous studies and provides a reasonable compromise between SNR, scan time, and T1‐weighting, TSC comparisons between tissues with different and long (>20 msec) T1 may be confounded. These limitations may be addressed in future studies by incorporating 23Na relaxation time measurements. In terms of study design, limitations include the small number of subjects, and their reasonably narrow age range. While including more subjects over a broader age range would be beneficial, we do not think the conclusions would be substantially altered, especially given the evidence that TSC shows little variation in cognitively normal aging.43 The restriction of recruiting only healthy volunteers is another limitation, with the present study providing results against which pathology‐specific repeatability measurements can be compared. Finally, comparisons among more sites and scanners is crucial if 23Na MRI is to yield robust biomarkers.
In conclusion, this study demonstrated good intersite and intrasite reproducibility and repeatability for 23Na MRI of the normal human brain. This work can be used to inform biologically meaningful differences in sodium concentration that could be used in the future as a biomarker in multisite studies. This is a key step in establishing sodium MRI‐derived metrics of tissue microstructure in cancer and other diseases.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by a research agreement between Philips Healthcare and the University of Manchester. The authors acknowledge the contributions of Christian Stehning from Philips Research, Hamburg, Germany, Matthew Clemence and Dave Higgins from Philips Healthcare, UK, and Rolf Schulte from GE Global Research, Munich, Germany.
The first two and last two authors contributed equally to this work.
Contract grant sponsor: Cancer Research UK; Contract grant numbers: C8742/A18097 and C19212/A16628. This is a contribution from the Cancer Imaging Centre in Cambridge & Manchester, which is funded by the Engineering and Physical Sciences Research Council and Cancer Research UK. The project was supported by the Addenbrooke's Charitable Trust, the National Institute for Health Research (NIHR) [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] in partnership with the University of Cambridge, Evelyn Trust, the MS Society, Cambridge Experimental Cancer Medicine Centre, and a research agreement between Philips Healthcare and the University of Manchester. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Data Accessibility
All data are available on reasonable request from the authors.
References
- 1. Madelin G, Kline R, Walvick R, Regatte RR. A method for estimating intracellular sodium concentration and extracellular volume fraction in brain in vivo using sodium magnetic resonance imaging. Sci Rep 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaplan JH. Biochemistry of Na,K‐ATPase. Annu Rev Biochem 2002;71:511–535. [DOI] [PubMed] [Google Scholar]
- 3. Madelin G, Regatte RR. Biomedical applications of sodium MRI in vivo. J Magn Reson Imaging 2013;38:511–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ouwerkerk R, Bleich KB, Gillen JS, Pomper MG, Bottomley PA. Tissue sodium concentration in human brain tumors as measured with 23Na MR imaging. Radiology 2003;227–2:529–537. [DOI] [PubMed] [Google Scholar]
- 5. Konstandin S, Schad L. 30 years of sodium/X‐nuclei magnetic resonance imaging. Magn Reson Mater Phy 2014;27:1–4. [DOI] [PubMed] [Google Scholar]
- 6. Hussain MS, Stobbe RW, Bhagat YA, et al. Sodium imaging intensity increases with time after human ischemic stroke. Ann Neurol 2009;66:55–62. [DOI] [PubMed] [Google Scholar]
- 7. Jones SC, Kharlamov A, Yanovski B, et al. Stroke onset time using sodium {MRI} in rat focal cerebral ischemia. Stroke 2006;37:883–888. [DOI] [PubMed] [Google Scholar]
- 8. Ng KH, Bradley DA, Looi LM. Elevated trace element concentrations in malignant breast tissues. Br J Radiol 1997;70:375–382. [DOI] [PubMed] [Google Scholar]
- 9. Zs.‐Nagy I, Lustyik G, Zs.‐Nagy V, Lukacs G, Balazs G. Correlation of malignancy with the intracellular Na+:K+ ratio in human thyroid tumors. Cancer Res 1983;43:5395–5402. [PubMed] [Google Scholar]
- 10. Jakubovic R, Zhou S, Heyn C, et al. The predictive capacity of apparent diffusion coefficient (ADC) in response assessment of brain metastases following radiation. Clin Exp Metastasis 2016;33:277–284. [DOI] [PubMed] [Google Scholar]
- 11. Price SJ, Jena R, Burnet NG, et al. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: An image‐guided biopsy study. AJNR Am J Neuroradiol 2006;27:1969–1974. [PMC free article] [PubMed] [Google Scholar]
- 12. Zámecník J, Vargová L, Homola A, Kodet R, Syková E. Extracellular matrix glycoproteins and diffusion barriers in human astrocytic tumors. Neuropathol Appl Neurobiol 2004;30:338–350. [DOI] [PubMed] [Google Scholar]
- 13. Mills S, Soh C, Rose C, et al. A domparison of DCE‐MRI derived measure of extracellular volume and ADC in glioblastoma multiforme. In: Proc 16th Sci Meet Int Soc Magn Reson Med 2008; Toronto; p 3484.
- 14. Thulborn KR. Quantitative sodium MR imaging: A review of its evolving role in medicine. Neuroimage 2018. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lüdemann L, Grieger W, Wurm R, Budzisch M, Hamm B, Zimmer C. Comparison of dynamic contrast‐enhanced MRI with WHO tumor grading for gliomas. Eur Radiol 2001;11:1231–1241. [DOI] [PubMed] [Google Scholar]
- 16. Schepkin V, Ross B, Chenevert T, et al. Sodium magnetic resonance imaging of chemotherapeutic response in a rat glioma. Magn Reson Med 2005;53:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biller A, Badde S, Nagel A, et al. Improved brain tumor classification by sodium MR imaging: Prediction of IDH mutation status and tumor progression. Am J Neuroradiol 2016;37:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inglese M, Madelin G, Oesingmann N, et al. Brain tissue sodium concentration in multiple sclerosis: A sodium imaging study at 3 Tesla. Brain 2010;133(Pt 3):847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paling D, Solanky BS, Riemer F, et al. Sodium accumulation is associated with disability and a progressive course in multiple sclerosis. Brain 2013;136. [DOI] [PubMed] [Google Scholar]
- 20. Thulborn K, Lui E, Guntin J, et al. Quantitative sodium MRI of the human brain at 9.4T provides assessment of tissue sodium concentration and cell volume fraction during normal aging. NMR Biomed 2016;29:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zaaraoui W, Konstandin S, Audoin B, et al. Distribution of brain sodium accumulation correlates with disability in multiple sclerosis: A cross‐sectional 23Na MR imaging study. Radiology 2012;264:859–867. [DOI] [PubMed] [Google Scholar]
- 22. Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med 2006;55:575–582. [DOI] [PubMed] [Google Scholar]
- 23. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min‐max interpolation. IEEE Trans Signal Process 2003;51:560–574. [Google Scholar]
- 24. Zwart NR, Johnson KO, Pipe JG. Efficient sample density estimation by combining gridding and an optimized kernel. Magn Reson Med 2012;67:701–710. [DOI] [PubMed] [Google Scholar]
- 25. Christensen JD, Barrere BJ, Boada FE, Vevea JM, Thulborn KR. Quantitative tissue sodium concentration mapping of normal rat brain. Magn Reson Med 1996;36:83–89. [DOI] [PubMed] [Google Scholar]
- 26. Maarouf A, Audoin B, Pariollaud F, et al. Increased total sodium concentration in gray matter better explains cognition than atrophy in MS. Neurology 2017;88:289–295. [DOI] [PubMed] [Google Scholar]
- 27. Qian Y, Zhao T, Zheng H, Weimer J, Boada FE. High‐resolution sodium imaging of human brain at 7T. Magn Reson Med 2012;68:227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reetz K, Romanzetti S, Dogan I, et al. Increased brain tissue sodium concentration in Huntington's disease — A sodium imaging study at 4 T. Neuroimage 2012;63:517–524. [DOI] [PubMed] [Google Scholar]
- 29. Alperin N, Vikingstad EM, Gomez‐Anson B, Levin DN. Hemodynamically independent analysis of cerebrospinal fluid and brain motion observed with dynamic phase contrast MRI. Magn Reson Med 1996;35:741–754. [DOI] [PubMed] [Google Scholar]
- 30. Stoquart‐ElSankari S, Balédent O, Gondry‐Jouet C, Makki M, Godefroy O, Meyer M‐E. Aging effects on cerebral blood and cerebrospinal fluid flows. J Cereb Blood Flow Metab 2007;27:1563–1572. [DOI] [PubMed] [Google Scholar]
- 31. Solanky BS, Riemer F, Wheeler‐Kingshott CAM, Golay X. Evaluation of cerebrospinal fluid suppression techniques in sodium MRI at 3T. In: Proc 20th Sci Meet Int Soc Magn Reson Med, Melbourne; 2012:1702.
- 32. Ouwerkerk R, Bleich KB, Gillen JS, Pomper MG, Bottomley PA. Tissue sodium concentration in human brain tumors as measured with 23Na MR imaging. Radiology 2003;227:529–537. [DOI] [PubMed] [Google Scholar]
- 33. Duyn JH, Yang Y, Frank JA, Van Der Veen JW. Simple correction method for k‐space trajectory deviations in MRI. J Magn Reson 1998;132:150–153. [DOI] [PubMed] [Google Scholar]
- 34. Pipe JG, Zwart NR, Aboussouan EA, Robison RK, Devaraj A, Johnson KO. A new design and rationale for 3D orthogonally oversampled k‐space trajectories. Magn Reson Med 2011;66:1303–1311. [DOI] [PubMed] [Google Scholar]
- 35. Madelin G, Babb J, Xia D, Regatte RR. Repeatability of quantitative sodium magnetic resonance imaging for estimating pseudo‐intracellular sodium concentration and pseudo‐extracellular volume fraction in brain at 3 T. PLoS One 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Madelin G, Babb JS, Xia D, Chang G, Jerschow A, Regatte RR. Reproducibility and repeatability of quantitative sodium magnetic resonance imaging in vivo in articular cartilage at 3 T and 7 T. Magn Reson Med 2012;68:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newbould RD, Miller SR, Tielbeek JAW, et al. Reproducibility of sodium MRI measures of articular cartilage of the knee in osteoarthritis. Osteoarthr Cartil 2012;20:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordan CD, McWalter EJ, Monu UD, et al. Variability of CubeQuant T1rho, quantitative DESS T2, and cones sodium MRI in knee cartilage. Osteoarthr Cartil 2014;22:1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagel AM, Bock M, Hartmann C, et al. The potential of relaxation‐weighted sodium magnetic resonance imaging as demonstrated on brain tumors. Invest Radiol 2011;46:539–547. [DOI] [PubMed] [Google Scholar]
- 40. Schepkin VD, Ross BD, Chenevert TL, et al. Sodium magnetic resonance imaging of chemotherapeutic response in a rat glioma. Magn Reson Med 2005;53:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available on reasonable request from the authors.


