Abstract
Drug-like molecules are known to contain many different building blocks with great potential as pharmacophores for drug discovery. The continued search for unique scaffolds in our laboratory led to the isolation of a novel Ghanaian soil bacterium, Streptomyces sp. MA37. This strain produces many bioactive molecules, most of which belong to carbazoles, pyrrolizidines, and fluorinated metabolites. Further probing of the metabolites of MA37 has led to the discovery of a new naphthacene-type aromatic natural product, which we have named accramycin A 1. This molecule was isolated using an HPLC-photodiode array (PDA) guided isolation process and MS/MS molecular networking. The structure of 1 was characterized by detailed analysis of LC-MS, UV, 1D, and 2D NMR data. Preliminary studies on the antibacterial properties of 1 using Group B Streptococcus (GBS) produced a minimum inhibitory concentration (MIC) of 27 µg/mL. This represents the first report of such bioactivity amongst the naphthacene-type aromatic polyketides, and also suggests the possibility for the further development of potent molecules against GBS based on the accramycin scaffold. A putative acc biosynthetic pathway for accramycin, featuring a tridecaketide-specific type II polyketide synthase, was proposed.
Keywords: accramycin, naphthacene, type II polyketide, Streptomyces sp. MA37, Group B Streptococcus
1. Introduction
Natural products continue to provide highly diverse chemical scaffolds, which have facilitated the research and development of many currently marketed drugs and viable clinical candidates [1]. Excellent examples of these natural scaffolds that have aided in the design of several compound libraries include the β-lactams (penicillin G), macrolactones (erythromycin), glycopeptides (vancomycin), tetracylines, and modified nucleosides [1,2].
Bacterial aromatic polyketides are prolific sources of many diverse scaffolds, some of which constitute a major class of antibiotics known to be generated on enzymatic assembly lines. For example, fasamycins and naphthacemycins contain 1-phenylnaphthacene moiety, which is known to have a rare carbon skeleton, and is a potent scaffold [3]. The gem-dimethyl tetracyclic carbon framework in these compounds has been reported to show potent anti-methicillin-resistant Staphylococcus aureus (MRSA) activities. Furthermore, fasamycins A and B were found to inhibit the FabF enzyme of type II fatty acid (FASII) biosynthesis in bacteria [4], while naphthacemycins showed inhibition against several MRSA and methicillin-susceptible Staphylococcus aureus (MSSA) strains, in addition to an efficient circumvention of β-lactam resistance when administered in combination with imipenem [5,6,7]. As part of our ongoing screening program to search for unique scaffolds, we have identified a cluster of metabolites containing the rare phenylnaphthacene carbon framework from the soil bacterium, Streptomyces sp. MA37, which is a prolific producer of natural products including legonmycins [8], neocarazostatin A [9,10,11], legonoxamines [12], and a variety of organofluorines [13,14].
The current study establishes MS/MS molecular networks (MN) coupled with HPLC-UV-based techniques for the rapid chemical dereplication and structural feature identification of metabolites from complex crude microbial extracts of Streptomyces sp. MA37. The streamlined MN dereplication process was performed via the online Global Natural Product Social (GNPS) Molecular Networking platform [15,16]. Compounds with structurally-related motifs are clustered together in the network based on a similarity in the mass spectra, including fragmentation patterns, and when coupled with HPLC-UV, this provided an easy and rapid tool for the effective isolation of target scaffolds. This technique is very important, since dereplication and subsequent identifications of known motifs at an early stage in natural product research is a crucial step for the accelerated isolation and characterization of new chemical scaffolds, thereby saving enormous amounts of time, effort, and cost [17,18]. Subsequently, in the MA37 molecular network, a cluster of compounds was identified that showed characteristic UV patterns (226, 250, 286, 355, and 420 nm), suggesting a common chromophore [3,4,19]. Manual dereplication using AntiBase (2012) indicated that members of this cluster have not been previously reported.
The isolation was carried out and afforded accramycin A 1. This molecule was named as such due to the origin of the producer strain Streptomyces sp. MA37 from the Legon–Accra region in Ghana. Using LC-MS, UV, 1D, and 2D NMR techniques, the structure of 1 was determined to be an aromatic naphthacene-based natural product. When tested against Group B Streptococcus, accramycin A 1 showed a minimum inhibitory concentration (MIC) of 27 ug/mL. To our knowledge, this activity has never been reported in any naphthacene-type congeners, suggesting that the accramycin scaffold could be used as a basis for the development of potent drugs against Group B Streptococcus (GBS). Two further accramycin analogues, 2 and 3 (Figures S10–S14) were also identified in fraction S3; however, they were not isolable. Our results underline the enormous potential of the MS/MS molecular network coupled with UV-based approaches to identify and target unprecedented scaffolds.
2. Results and Discussion
A large fermentation (12 L) of Streptomyces sp. MA37 was performed at 28 °C at 180 rpm for 7 days. Subsequently, Diaion® HP-20 (3 g/50 mL) was added to the fermentation culture and left to shake overnight under the same incubation conditions. The mixture was filtered under vacuum, after which the resin and mycelia were extracted with 100% methanol. The methanol extract was concentrated under vacuum to give a total crude extract, followed by fractionation using Strata® C18-E solid phase extraction (SPE), which yielded six fractions (S1–6). All the fractions obtained were subjected to HPLC analysis with diode array detection to screen for metabolites bearing unique scaffolds. Fraction S3 showed the characteristic naphthacene UV spectra (226, 250, 286, 355, and 420 nm); hence, it was selected for further fractionation using semi-preparative reversed-phase HPLC, resulting in the isolation of accramycin A 1 (2.0 mg) (Figure 1).
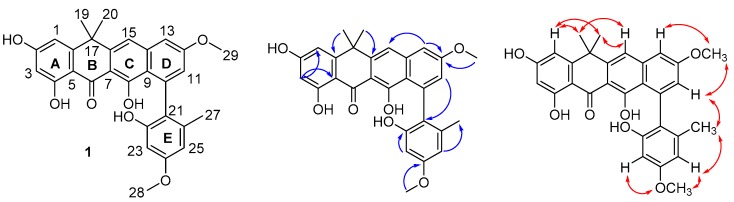
Figure 1.
Structure of accramycin A (1) with key HMBC (→) and NOESY (↔) correlations in DMSO-d6.
2.1. Structure Elucidation
Accramycin A 1 was obtained as a yellow powder. It showed UV absorption maxima at 226, 250, 286, 355, and 420 nm, which is distinctive of a fasamycin or naphthacemycin congener [5,19]. The molecular formula of 1 (Figure 1) was established as C29H26O7 by HR ESIMS (calculated [M + H]+ = 487.1751; observed [M + H]+ = 487.1741; ∆ = −2.1 ppm), indicating 17 degrees of unsaturation (Figures S1 and S2).
1H and 13C-NMR spectra and HSQC analysis revealed the presence of 29 carbons, including three methyl carbons at δC 20.6, 34.4, and 34.7; two methoxy signals at δC 55.3 and 55.6; seven sp2 methines at δC 99.2, 104.0, 106.6, 106.9, 112.1, 115.8, and 121.7; one sp3 quaternary carbon at δC 39.3; and sixteen sp2 quaternary carbons (δC 105.0–189.2) (Table 1, Figures S3–S9). Detailed analysis of the spectral data suggested that 1 was closely related to the previously described naphthacene, fasamycin C [19].
Table 1.
NMR data for accramycin A 1 (CD3OD, 600 MHz at 298 K).
| Position | 13C a | 1H, mult. (J, Hz) | Position | 13C a | 1H, mult. (J, Hz) |
|---|---|---|---|---|---|
| 1 | 112.1, CH | 6.49, d (2.4) | 16 | 147.4, C | - |
| 2 | 167.0, C b | - | 17 | 39.3, C | - |
| 3 | 104.0, CH | 5.99, d (2.4) | 18 | 154.5, C | - |
| 4 | 167.1, C | - | 19 | 34.4, CH3 | 1.70, s |
| 5 | 105.0, C | - | 20 | 34.7, CH3 | 1.69, s |
| 6 | 189.2, C | - | 21 | 125.3, C | - |
| 7 | 107.4, C | - | 22 | 155.2, C | - |
| 8 | 165.3, C | - | 23 | 99.2, CH | 6.32, d (2.4) |
| 9 | 118.7, C | - | 24 | 160.2, C | - |
| 10 | 141.0, C b | - | 25 | 106.9, CH | 6.37, d (2.4) |
| 11 | 121.7, CH | 6.71, d (2.4) | 26 | 138.2, C | - |
| 12 | 161.1, C | - | 27 | 20.6, CH3 | 1.91, s |
| 13 | 106.6, CH | 7.19, d (2.4) | 28 | 55.3, CH3 | 3.80, s |
| 14 | 141.8, C | - | 29 | 55.6, CH3 | 3.95, s |
| 15 | 115.8, CH | 7.47, s |
a Obtained from HSQC and HMBC, respectively; b estimated from DMSO-d6 spectrum.
The long-range couplings observed in the HMBC spectrum established the connectivity of rings A–D. The cross-peaks from H-1 (δH 6.49) to C-3 (δC 104.0) and C-5 (δC 105.0) revealed a 2,4-dihydroxy-substituted benzene substructure (ring A). The cross-peaks from H3-19 (δH 1.70) and H3-20 (δH 1.69) to C-16 (δC 147.4), C-17 (δC 39.3), and C-18 (δC 154.5) indicated a dimethyl motif attached to carbon 17 (ring B), and connected to ring A via C-5 and C-18. Carbon 6 resonated at δC 189.2, which is consistent with a ketone moiety conjugated to two aromatic rings in the structure. The cross-peaks from H-15 (δH 7.47) to C-7 (δC 107.4), C-9 (δC 118.7), and C-17 (δC 39.3) established the nature of ring C, which was condensed to ring B via C-7 and C-16. H-11 (δH 6.71) and H-13 (δH 7.19) in ring D were found to be arranged meta to one another based on their coupling constant (2.4 Hz). The HMBC correlations from H3-29 (δH 3.95) to C-12 (δC 161.1), and H3-28 (δH 3.80) to C-24 (δC 160.2) located the two methoxyl functions in rings D and E, respectively. Furthermore, ring E displayed two meta-coupled protons, H-23 (δH 6.32) and H-25 (δH 6.37, (J = 2.4 Hz), and an aromatic methyl group (δH 1.91, H3-27). The arrangement of the different substituents within this aromatic ring was confirmed by HMBC correlations from H3-27 to C-21 (δC 125.3), C-25 (δC 106.9), and C-26 (δC 138.2), and H-23 to C-21, C-22 (δC 155.2), and C-24 (δC 160.2), while the attachment of ring E to the rest of the molecule was established by the diagnostic correlation between H-11 and C-21.
The structure of 1 was further corroborated by analysis of the NOESY spectrum acquired in DMSO-d6 (Figure 1) and by comparison with the MS and NMR data of fasamycins and naphthacemycins in the literature [5,19], from which 1 differs with respect to the presence of a methoxy moiety (δH 3.95) attached to C-12 in ring D, thus establishing 1 as a new fasamycin congener, for which the name accramycin A is proposed.
Two further accramycin derivatives, 2 and 3, were tentatively identified in the molecular network (Figure S10) based on their UV spectra, but could not be isolated due to their minute quantities in the extract. The typical chlorine isotope pattern observed for compound 2 (m/z 521.1319 ([M + H]+: 523.1282, 3:1, both consistent with the molecular formula C29H25ClO7)) in the MN cluster indicated that it was a monochlorinated derivative of 1 (Figures S11 and S12). It is worth noting that chlorination is a feature which is commonly observed in the fasamycin [19] and naphthacemycin [5] series. Compound 3 (m/z 535.1478 ([M + H]+: 537.1447, 3:1, both consistent with the molecular formula C30H27ClO7)) (m/z 535.1500 ([M + H]+, C30H28ClO7+)) was identified as a homologue of compound 2 (Figure S13–S14).
2.2. Proposed acc Biosynthetic Gene Cluster and Pathway
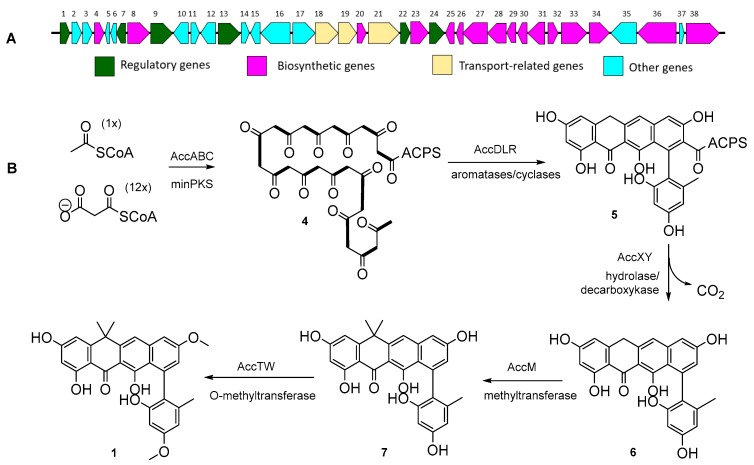
Given the structural similarity of accramycin A 1 with fasamycins [19], we predicted that 1 is originated from polyketide Type II synthase. In silico analysis of the annotated MA37 genome using antiSMASH 3.0 [20] revealed three putative type 2 PKS biosynthetic gene clusters. One biosynthetic gene cluster (BGC) is likely to be involved in the biosynthesis of accramycins (acc) showing high homology to the reported formicamycin BGC (KX859301) [19], and the other two share very high sequence homology to FD-594 (AB469194.1) [21] or hexaricin (KT713752.1) [22] biosynthesis. A pairwise comparison of the annotated proteins using BLAST [23] between the acc cluster with the formicamycin BGC showed high amino acid identity, strongly supporting that the acc BGC most likely encodes the biosynthesis of accramycins in MA37 (Table S1, Figure S15).
The acc gene cluster spans about 33 kb of genomic DNA and contains sixteen biosynthetic genes with catalytic functions, three putative transport-related genes, and six possible regulatory genes with deduced functions that were designated based on homology analysis (Figure 2A, Table 2). Thirteen open reading frames (orf 1–13) in the BGC appear to have no catalytic functions in the biosynthesis of 1. It is likely that the minimal PKS containing KSα (AccA), KSβ (AccB), and ACP (AccC) catalyze the decarboxylative condensation of one acetyl-CoA and twelve malonyl-CoAs to generate a tridecaketide intermediate 4 (Figure 2B), followed by cyclization catalyzed by three cylase/aromatases (AccD, AccL, and AccR) to yield 5. Further tailoring reactions occur through the action of additional putative enzymes, including PKS hydrolase (AccX) and decarboxylase (AccY) to transform 5 to 6.
Figure 2.
(A) Accramycin (acc) biosynthetic gene cluster in Streptomyces sp. MA37. (B) Proposed biosynthetic pathway of 1.
Table 2.
Deduced functions of open reading frames (ORFs) in accramycin (acc) biosynthetic gene cluster.
| No. | Gene | AA | Deduced Function | No. | Gene | AA | Deduced Function |
|---|---|---|---|---|---|---|---|
| 1 | accE | 242 | DNA binding response regulator | 20 | accL | 119 | Type II PKS cyclase |
| 2 | orf1 | 226 | Synthase | 21 | accK | 509 | Na+/H+ exchanger |
| 3 | orf2 | 210 | Glycosyl transferase | 22 | accJ | 145 | MarR family transcriptional regulator |
| 4 | accM | 220 | SAM-dependent methyl transferase | 23 | accG | 359 | Sensor histidine kinase |
| 5 | orf3 | 60 | Hypothetical protein | 24 | accF | 227 | LuxR family response regulator |
| 6 | orf4 | 38 | Unknown | 25 | accD | 152 | Polyketide cyclase |
| 7 | accN | 112 | Transcriptional regulator | 26 | accC | 97 | Acyl carrier protein |
| 8 | accO | 305 | NAD(P)-dependent oxidoreductase | 27 | accB | 415 | Beta-ketoacyl synthase |
| 9 | accP | 275 | MerR family transcriptional regulator | 28 | accA | 426 | Beta-ketoacyl synthase |
| 10 | orf5 | 193 | Nucleoside phosphorylase | 29 | accR | 131 | Cupin (cyclase/monooxygenase) |
| 11 | orf6 | 146 | VOC family protein | 30 | accS | 113 | Antibiotic biosynthesis monooxygenase |
| 12 | orf7 | 202 | Isomerase | 31 | accT | 350 | O-methyl transferase |
| 13 | accI | 303 | LysR family transcriptional regulator | 32 | accU | 113 | Antibiotic biosynthesis monooxygenase |
| 14 | orf8 | 108 | Hypothetical protein | 33 | accV | 430 | Halogenase |
| 15 | orf9 | 140 | Hypothetical protein | 34 | accW | 348 | O-methyl transferase |
| 16 | orf10 | 386 | Molybdopterin-binding protein | 35 | orf12 | 480 | Phenylalanine specific permease |
| 17 | orf11 | 480 | domain containing proteins | 36 | accX | 777 | hydrolase |
| 18 | accH | 491 | ABC transporter | 37 | orf13 | 43 | DUF-1232 domain containing protein |
| 19 | accI | 319 | ABC transporter | 38 | accY | 517 | decarboxylase |
Accramycin A 1 bears two methyl groups at position C-17 in the structure. This gem-dimethyl moiety can be found in benastatins [24], resistomysin [25], napthacemycins [5], fasamycins [4], tetarimycin [26], and L-755,805 [27]. Three putative methyltransferases (AccM, AccT, AccW) were identified in the acc gene cluster. AccM is closely related to BenF (42%/53%; CAM58795.1) [24,28] which catalyzes the geminal bis-methylation step in benastatin biosynthesis, and is likely to involve a similar PKS tailoring reaction in accramycin biosynthesis. The other two methyltransferases, AccT and AccW, showed the highest homology to ForT and ForW (76%/83%, WP_098245764.1; 50%/60%, WP_098245767.1) in the formicamycin pathway, respectively, which are likely to encode the O-methylation step of 1–3 at C-12 and C-24.
A further post-PKS transformation involved the halogenation of 1 to produce the minor metabolites 2 and 3. Analysis of the MA37 genome identified the putative FAD-dependent oxidoreductase AccV, which shows high homology (68%/78%, WP_098245766.1) to ForV halogenase in formicamycin BGC [19] Figure S14, Table S1), and is likely to catalyze the halogenation of 1, yielding 2 and 3.
2.3. Antibacterial Assay
Accramycin A 1 showed no activity against Gram-negative bacteria (Table 3). However, it showed moderate activity (MIC 27 µg/mL) against Group B Streptococcus (GBS; Streptococcus agalactiae) (Figure S16), and this activity has never been reported in any naphthacene-based antibiotics.
Table 3.
Minimum inhibitory concentration (MIC) of accramycin A 1 against a panel of bacterial pathogens.
| Pathogen | MIC (µg/mL) |
|---|---|
| Streptococcus B. ATCC 12386 | 27 |
| Escherichia coli ATCC 25922 | >50 |
| Pseudomonas aeruginosa ATCC 27853 | >50 |
GBS is the leading cause of life-threatening neonatal infections since 1970, and it also affects pregnant women, the elderly, the immunocompromised, and those with underlying chronic medical conditions [29,30]. The case fatality rate among adults has been reported to be markedly higher than among neonates, and continues to climb over the last two decades [29,30,31,32,33].
GBS remains susceptible to penicillin; however, increasing MICs (4-fold to 8-fold higher) are now described [33,34]. Reports have shown rising GBS resistance toward erythromycin and clindamycin, ranging between 25–52% and 12–41%, respectively [29,33,34,35,36]. The first two cases of invasive vancomycin-resistant GBS infection were documented in 2014 [37]. Continued investigation is necessary to monitor this emerging threat. The growing incidence of antibiotic resistance reinforces the need to develop alternative drugs.
The current study opens an avenue for possible structural modifications of the accramycin pharmacophore for the development of potent drugs against GBS. Structure–activity relationship studies on the accramycin scaffold would be of interest in the search for potential effective clinical candidates.
3. Materials and Methods
3.1. Fermentation
Streptomyces sp. MA37 was isolated from the rhizosphere of a Moracear Bark Cloth tree (Antiaris toxicaria), growing in the Legon Botanical Gardens, Accra, Ghana, Africa [13]. The seed culture of the MA37 strain was prepared by inoculating a single colony of the organism to 50 mL of ISP2 liquid medium (Oxoid glucose 2.0 g, Oxoid yeast extract 2.0 g, VWR malt extract 5.0 g in 500 mL milliQ H2O), and incubating for 7 days at 28 °C with shaking at 180 rpm (Incu-shake FL16-2). The seed culture was used to inoculate 12.0 L of ISP2 broth (1:100) in 24 baffled flasks (CorningTM polycarbonate flasks), each containing already autoclaved 500 mL of ISP2, and plugged with Fisherbrand™ polyurethane foam stoppers (Fisher Scientific, UK). The cultures were incubated for 7 days maintained at 28 °C with continuous agitation at 180 rpm. Subsequently, Diaion® HP-20 (3 g/50 mL) was added under sterile conditions to the fermentation culture and kept at the same temperature and shaking conditions for 18–24 h. The solution was filtered under vacuum, and the HP-20 resin was rinsed with milliQ water, and extracted thrice with 100% methanol (Fisher Chemical HPLC grade). All the methanol extracts were combined, concentrated under reduced pressure (Buchi Rotavapor R200, UK), and subjected to LC high-resolution electrospray ionization mass spectrometry (LC HRESIMS, UK) analysis.
3.2. Extraction and Isolation
The crude methanol extract was fractionated on Strata® C18-E solid phase extraction (SPE) (55 µm, 70 Å, 20 g/60 mL) cartridges, and yielded 6 fractions (S1–6). Fractionation was performed using 10 column volumes of milliQ H2O (S1), 25% MeOH (S2), by 50% MeOH (S3), followed by 75% MeOH (S4), and then 100% MeOH (S5). Finally, the SPE column was flushed with 100% MeOH with 0.05% of trifluoroacetic acid (S6) (Acros Organics). All the eluents were concentrated under reduced pressure (Buchi Rotavapor R200). Each fraction was subjected to LC-high-resolution electrospray ionization mass spectrometry (HRESIMS) analysis.
HPLC-UV analysis was carried out in all the fractions (S1–6) to screen and target unique scaffolds using an Agilent HPLC system (UK) (1260 Infinity) equipped with a diode array detector (DAD) (C-18 ACE 10 µM 10 × 250 mm column) with spectral scanning between 200–550 nm. The compounds of interest with the characteristic naphthacene UV maxima (226, 250, 286, 355, and 420 nm) were observed in fraction S3.
Analysis of the HPLC/HRESIMS data of fraction S3 revealed three resolved peaks with [M + H]+ at m/z 487.1741, 521.1318, and 535.1500, respectively. Dereplication of these masses was achieved using AntiBase (2012) software, indicating that all three molecules were previously unreported. Thus, fraction S3 was selected for further fractionation by reversed-phase semi-prep (C18 ACE 10 µM 10 × 250 mm column) HPLC (Agilent 1260 Infinity). The purification was carried out using a linear gradient from 20% H2O:MeOH:TFA (95:5:0.1) to 100% MeOH for 45 min with a solvent flow rate of 1.5 mL/min, to yield accramycin A 1 (2.0 mg). Two further accramycin derivatives, 2 and 3 were identified in fraction S3; however, they were not subjected to isolation due to their minute quantities in the extract.
Accramycin A 1: deep yellow powder. UV (CH3OH): 226, 250, 286, 355, and 420 nm; IR (neat) νmax 3320 cm−1, 2945 cm−1, 2832 cm−1, 1650 cm−1, 1403 cm−1, 1111 cm−1, 1021 cm−1; 1H, 13C NMR data, see Table 1; HRESIMS (positive mode) m/z calculated [M + H]+ = 487.1751; observed [M + H]+ = 487.1741; ∆ = −2.1 ppm.
3.3. MS/MS Molecular Networking
The MS/MS data of the crude extract and fractions (S1–6) were converted from .RAW to .mzXML file format using the ProteoWizard MSConvert software (version 3.0) ([38] for metabolite profiling and the identification of new chemical entities in Streptomyces sp. MA37 extracts. The .mzXML files were uploaded on the GNPS web platform to create the molecular network [15,39,40]. The GNPS settings used to obtain the consensus spectra were: Precursor ion mass tolerance of 0.02 Da and fragment ion tolerance of 0.02 Da. Edges were filtered by setting the default GNPS minimum cosine score above 0.7, minimum cluster size 2, network TopK 10, and more than 6 matched peaks. The spectral library matching was performed with a similar cosine threshold and minimum matched peaks. The remaining parameters were set as default, as suggested by the GNPS platform. The molecular network was generated using the GNPS “Classic” molecular networking workflow (METABOLOMICS-SNETS-V2 version 1.2.5,), and the GraphML data obtained were visualized using the circular layout in Cytoscape 3.6.1 [41].
3.4. Spectroscopic Analysis
High Resolution Electro-spray Ionization Mass Spectrometry (HR-ESIMS) was obtained using a LC MS Thermo Scientific MS system (LTQ Orbitrap) coupled to a Thermo Instrument HPLC system (Accela PDA detector, Accela PDA autosampler and Accela Pump) on a positive ESI mode (30.000), MS/MS resolution 7500, C18 (Sunfire 150 × 46 mm column) Reversed phase separation using 0.1% formic acid in water and 0.1% formic acid in acetonitrile gradient. The following conditions were used: Capillary voltage 45 V, auxiliary gas flow rate 10–20 arbitrary units, capillary temperature 200 °C, sheath gas flow rate 5 arbitrary units, spray voltage 4.5 kV, mass range 150–2000 amu (maximum resolution 30,000×). 1D and 2D NMR data were obtained on a Bruker AVANCE III HD 600 MHz (AscendTM14.1 Tesla, UK) with Prodigy TCITM cryoprobe at 298 K in DMSO-d6 and CD3OD. Trimethylsilane (TMS) was used as an internal standard. Deuterated solvents were obtained from Goss Scientific. The infrared spectrum was acquired using a PerkinElmer Spectrum version 10.4.00 Fourier transform infrared (FTIR) spectrometer (2013) (UK) equipped with an Attenuated Total Reflection (ATR) diamond cell.
3.5. Antibacterial Assay
Minimum inhibitory concentrations (MIC) were determined following the antibacterial assay protocols described in Lauritano (2016) [42] against the Gram-negative bacteria Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), and the Gram-positive bacterium Streptococcus B (ATCC 12386). All bacteria were cultured in Mueller–Hinton broth. The assays were performed in serial dilutions (in triplicates) in 96-well plates (Nunc, Thermo Fisher Scientific, United States), wherein a 50-µL suspension (log phase) of bacteria was incubated overnight at 37 °C and then supplemented with 50 µL of the test extract. The positive control consisted of the bacteria plus Milli-Q water (no test compound), while the negative control comprised the growth media and Milli-Q water. The absorbance was recorded after 24 h (OD600) in a Victor3 multilabel plate reader. The growth medium appeared clear in wells where the test compound prevented the growth or killed the bacteria; otherwise, it was cloudy. Activity threshold was set below 0.05 (OD600). The MIC was defined as the lowest concentration of a drug that inhibited visible bacterial growth.
4. Conclusions
With the rising GBS incidence and antibiotic resistance, we have yet to find more base scaffolds for the development of alternate drugs and drug leads. Herein, we report the isolation of Accramycin A 1 from the soil bacterium Streptomyces sp. MA37 by HPLC-UV aided by molecular networking. The structure of 1 was deduced by analysis of the LC MS, UV, 1D, and 2D NMR, and identified as a new naphthacene-type natural product. Preliminary studies on the antibacterial properties of 1 using GBS showed an MIC of 27 µg/mL. This is the first report of GBS activity amongst the naphthacene-based congeners, and suggests possible scaffold manipulation for the development of potent drugs against GBS based on the accramycin framework. Two accramycin analogues, 2 and 3, were tentatively identified in the MA37 fermentation broth by HPLC/HRESIMS analysis and molecular network. A putative accramycin biosynthetic pathway was proposed.
Supplementary Materials
The supplementary materials are available online.
Author Contributions
F.M., V.L., Q.F., R.E., and S.S., formal analysis and investigation. H.D., K.K., and F.M., funding acquisition and methodology. H.D., and F.M., writing original draft. K.K. and H.D. review and editing. H.D. supervision and project administration.
Funding
FM thanks the University of the Philippines for the Faculty, Reps and Staff Development Program (FRAS DP) for the PhD grant fellowship. HD and KK thank the financial supports of Leverhulme Trust-Royal Society Africa award (AA090088) and MRC African Research Leaders Award (MR/S00520X/1). R.E. and S.S. are grateful to British Council/Newton Fund for financial support through the Institutional Links scheme (Project No. 261781172). QF is grateful to the University of Aberdeen Elphinstone Scholarship and Scottish Funding Council/ScotCHEM for financial support through the PEER/PERCE Funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Martins A., Vieira H., Gaspar H., Santos S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs. 2014;12:1066–1101. doi: 10.3390/md12021066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welsch M.E., Snyder S.A., Stockwell B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010;14:347–361. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Z., Kallifidas D., Brady S.F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl. Acad. Sci. 2011;108:12629–12634. doi: 10.1073/pnas.1103921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Z., Chakraborty D., Dewell S.B., Reddy B.V.B., Brady S.F. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J. Am. Chem. Soc. 2012;134:2981–2987. doi: 10.1021/ja207662w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukumoto A., Kim Y.P., Iwatsuki M., Hirose T., Sunazuka T., Hanaki H., Omura S., Shiomi K. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. II. Structure elucidation. J. Antibiot. (Tokyo) 2017;70:568–573. doi: 10.1038/ja.2017.29. [DOI] [PubMed] [Google Scholar]
- 6.Fukumoto A., Kim Y.P., Matsumoto A., Takahashi Y., Suzuki M., Onodera H., Tomoda H., Matsui H., Hanaki H., Iwatsuki M., et al. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. I. The taxonomy of the producing strain, and the fermentation, isolation and antibacterial activities. J. Antibiot. (Tokyo) 2017;70:562–567. doi: 10.1038/ja.2017.28. [DOI] [PubMed] [Google Scholar]
- 7.Hirose T., Kojima Y., Matsui H., Hanaki H., Iwatsuki M., Shiomi K., Omura S., Sunazuka T. Total synthesis of (±)-naphthacemycin A 9, possessing both antibacterial activity against methicillin-resistant Staphylococcus aureus and circumventing effect of β-lactam resistance. J. Antibiot. (Tokyo) 2017;70:574–581. doi: 10.1038/ja.2016.141. [DOI] [PubMed] [Google Scholar]
- 8.Huang S., Tabudravu J., Elsayed S.S., Travert J., Peace D., Tong M.H., Kyeremeh K., Kelly S.M., Trembleau L., Ebel R., et al. Discovery of a single monooxygenase that catalyzes carbamate formation and ring contraction in the biosynthesis of the legonmycins. Angew. Chemie - Int. Ed. 2015;54:12697–12701. doi: 10.1002/anie.201502902. [DOI] [PubMed] [Google Scholar]
- 9.Su L., Lv M., Kyeremeh K., Deng Z., Deng H., Yu Y. A ThDP-dependent enzymatic carboligation reaction involved in Neocarazostatin A tricyclic carbazole formation. Org. Biomol. Chem. 2016;14:8679–8684. doi: 10.1039/C6OB01651K. [DOI] [PubMed] [Google Scholar]
- 10.Su L., Zhang R., Kyeremeh K., Deng Z., Deng H., Yu Y. Dissection of the neocarazostatin: a C 4 alkyl side chain biosynthesis by in vitro reconstitution. Org. Biomol. Chem. 2017:3843–3848. doi: 10.1039/C7OB00617A. [DOI] [PubMed] [Google Scholar]
- 11.Huang S., Elsayed S.S., Lv M., Tabudravu J., Rateb M.E., Gyampoh R., Kyeremeh K., Ebel R., Jaspars M., Deng Z., et al. Biosynthesis of Neocarazostatin A Reveals the Sequential Carbazole Prenylation and Hydroxylation in the Tailoring Steps. Chem. Biol. 2015;22:1633–1642. doi: 10.1016/j.chembiol.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Maglangit F., Tong M.H., Jaspars M., Kyeremeh K., Deng H. Legonoxamines A-B, two new hydroxamate siderophores from the soil bacterium, Streptomyces sp. MA37. Tetrahedron Lett. 2019;60:75–79. doi: 10.1016/j.tetlet.2018.11.063. [DOI] [Google Scholar]
- 13.Deng H., Ma L., Bandaranayaka N., Qin Z., Mann G., Kyeremeh K., Yu Y., Shepherd T., Naismith J.H., O’Hagan D. Identification of Fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by Genome Mining. ChemBioChem. 2014;15:364–368. doi: 10.1002/cbic.201300732. [DOI] [PubMed] [Google Scholar]
- 14.Ma L., Bartholome A., Tong M.H., Qin Z., Yu Y., Shepherd T., Kyeremeh K., Deng H., O’Hagan D., Su L., et al. Identification of a fluorometabolite from Streptomyces sp. MA37: (2R3S4S)-5-fluoro-2,3,4-trihydroxypentanoic acid. Chem. Sci. 2017;14:8679–8684. doi: 10.1039/C4SC03540B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox Ramos A.E., Evanno L., Poupon E., Champy P., Beniddir M.A. Natural products targeting strategies involving molecular networking: different manners, one goal. Nat. Prod. Rep. 2019 doi: 10.1039/C9NP00006B. [DOI] [PubMed] [Google Scholar]
- 17.Tabudravu J.N., Pellissier L., Smith A.J., Subko K., Autréau C., Feussner K., Hardy D., Butler D., Kidd R., Milton E.J., et al. LC-HRMS-Database screening metrics for rapid prioritization of samples to accelerate the discovery of structurally new natural products. J. Nat. Prod. 2019;82:211–220. doi: 10.1021/acs.jnatprod.8b00575. [DOI] [PubMed] [Google Scholar]
- 18.Chervin J., Stierhof M., Tong M.H., Peace D., Hansen K.Ø., Urgast D.S., Andersen J.H., Yu Y., Ebel R., Kyeremeh K., et al. Targeted Dereplication of Microbial Natural Products by High-Resolution MS and Predicted LC Retention Time. J. Nat. Prod. 2017;80:1370–1377. doi: 10.1021/acs.jnatprod.6b01035. [DOI] [PubMed] [Google Scholar]
- 19.Qin Z., Munnoch J.T., Devine R., Holmes N.A., Seipke R.F., Wilkinson K.A., Wilkinson B., Hutchings M.I. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. 2017;8:3218–3227. doi: 10.1039/C6SC04265A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., et al. AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo F., Yonezawa T., Komatsubara A., Mizoue K., Eguchi T. Cloning of the biosynthetic gene cluster for naphthoxanthene antibiotic FD-594 from Streptomyces sp. TA-0256. J. Antibiot. (Tokyo) 2011;64:123–132. doi: 10.1038/ja.2010.145. [DOI] [PubMed] [Google Scholar]
- 22.Tian J., Chen H., Guo Z., Liu N., Li J., Huang Y., Xiang W., Chen Y. Discovery of pentangular polyphenols hexaricins A–C from marine Streptosporangium sp. CGMCC 4.7309 by genome mining. Appl. Microbiol. Biotechnol. 2016;100:4189–4199. doi: 10.1007/s00253-015-7248-z. [DOI] [PubMed] [Google Scholar]
- 23.Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K., Connor R., Fiorini N., Funk K., Hefferon T., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019;47:D23–D28. doi: 10.1093/nar/gky1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoyama T., Naganawa H., Muraoka Y., Aoyagi T., Takeuchi T. Biosynthesis of benastatin A. J. Antibiot. (Tokyo) 1992;45:1767–1772. doi: 10.7164/antibiotics.45.1767. [DOI] [PubMed] [Google Scholar]
- 25.Jakobi K., Hertweck C. A Gene Cluster Encoding Resistomycin Biosynthesis in Streptomyces resistomycificus; Exploring Polyketide Cyclization beyond Linear and Angucyclic Patterns. J. Am. Chem. Soc. 2004;126:2298–2299. doi: 10.1021/ja0390698. [DOI] [PubMed] [Google Scholar]
- 26.Wyche T.P., Piotrowski J.S., Hou Y., Braun D., Deshpande R., McIlwain S., Ong I.M., Myers C.L., Guzei I.A., Westler W.M., et al. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J. Antibiot. (Tokyo) 2017;134:1415–1427. [Google Scholar]
- 27.Lam Y.K.T., Hensens O., Helms G., Williams D., Nallin M., Smith J., Gartner S., Rodrigueztt L.H., Stevens-miles S. L-755,805, A New Polyketide Endothelin Binding Inhibitor from an Actinomycete. Tetrahedron Lett. 1995;36:2013–2016. doi: 10.1016/0040-4039(95)00215-X. [DOI] [Google Scholar]
- 28.Xu Z., Schenk A., Hertweck C. Molecular analysis of the benastatin biosynthetic pathway and genetic engineering of altered fatty acid-polyketide hybrids. J. Am. Chem. Soc. 2007;129:6022–6030. doi: 10.1021/ja069045b. [DOI] [PubMed] [Google Scholar]
- 29.Nanduri S.A., Petit S., Smelser C., Apostol M., Alden N.B., Harrison L.H., Lynfield R., Vagnone P.S., Burzlaff K., Spina N.L., et al. Epidemiology of Invasive Early-Onset and Late-Onset Group B Streptococcal Disease in the United States, 2006 to 2015. JAMA Pediatr. 2008;299:2056–2065. doi: 10.1001/jamapediatrics.2018.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoff T.H., Farley M.M., Petit S., Craig A.S., Schaffner W., Gershman K., Harrison L.H., Lynfield R., Mohle-Boetani J., Zansky S., et al. Increasing Burden of Invasive Group B Streptococcal Disease in Nonpregnant Adults, 1990–2007. Clin. Infect. Dis. 2009;49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 31.Schrag S.J., Zywicki S., Farley M.M., Reingold A.L., Harrison L.H., Harrison L.H., Lefkowitz L., Hadler J.L., Danila R., Cieslak P., et al. Group B Streptococcal Disease in the WEra of Intrapartum Antibiotic Prophylaxis. N. Engl. J. Med. 2000;342:15–20. doi: 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 32.Blancas D., Santin M., Olmo M., Alcaide F., Carratala J., Gudiol F. Group B streptococcal disease in nonpregnant adults: Incidence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2004;23:168–173. doi: 10.1007/s10096-003-1098-9. [DOI] [PubMed] [Google Scholar]
- 33.Farley M.M. Group B Streptococcal Disease in Nonpregnant Adults. Clin. Infect. Dis. 2002;33:556–561. doi: 10.1086/322696. [DOI] [PubMed] [Google Scholar]
- 34.Berg B.R., Houseman J.L., TerSteeg Z.E., LeBar W.D., Newton D.W. Antimicrobial susceptibilities of group B streptococcus isolates from prenatal screening samples. J. Clin. Microbiol. 2014;52:3499–3501. doi: 10.1128/JCM.01781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamagni T.L., Keshishian C., Efstratiou A., Guy R., Henderson K.L., Broughton K., Sheridan E. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clin. Infect. Dis. 2013;57:682–688. doi: 10.1093/cid/cit337. [DOI] [PubMed] [Google Scholar]
- 36.Heelan J.S., Hasenbein M.E., McAdam A.J. Resistance of Group B Streptococcus to Selected Antibiotics, Including Erythromycin and Clindamycin. J. Clin. Microbiol. 2004;42:1263–1264. doi: 10.1128/JCM.42.3.1263-1264.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park C., Nichols M., Schrag S.J. Two Cases of Invasive Vancomycin-Resistant Group B Streptococcus Infection. N. Engl. J. Med. 2014;370:885–886. doi: 10.1056/NEJMc1308504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holman J.D., Tabb D.L., Mallick P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr Protoc Bioinforma. 2014;46:1–7. doi: 10.1002/0471250953.bi1324s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J.Y., Sanchez L.M., Rath C.M., Liu X., Boudreau P.D., Bruns N., Glukhov E., Wodtke A., De Felicio R., Fenner A., et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crüsemann M., O’Neill E.C., Larson C.B., Melnik A.V., Floros D.J., da Silva R.R., Jensen P.R., Dorrestein P.C., Moore B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017;80:588–597. doi: 10.1021/acs.jnatprod.6b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauritano C., Andersen J.H., Hansen E., Albrigtsen M., Escalera L., Esposito F., Helland K., Hanssen K.Ø., Romano G., Ianora A., et al. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016;28:1–12. doi: 10.3389/fmars.2016.00068. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.