Abstract
Alzheimer’s disease (AD) is a widespread dynamic neurodegenerative malady. Its etiology is still not clear. One of the foremost pathological features is the extracellular deposits of Amyloid-beta (Aβ) peptides in senile plaques. The interaction of Aβ and the receptor for advanced glycation end products at the blood-brain barrier is also observed in AD, which not only causes the neurovascular anxiety and articulation of proinflammatory cytokines, but also directs reduction of cerebral bloodstream by upgrading the emission of endothelin-1 to induce vasoconstriction. In this process, RAGE is deemed responsible for the influx of Aβ into the brain through BBB. In the current study, we predicted the interaction potential of the natural compounds vincamine, ajmalicine and emetine with the Aβ peptide concerned in the treatment of AD against the standard control, curcumin, to validate the Aβ peptide–compounds results. Protein-protein interaction studies have also been carried out to see their potential to inhibit the binding process of Aβ and RAGE. Moreover, the current study verifies that ligands are more capable inhibitors of a selected target compared to positive control with reference to ΔG values. The inhibition of Aβ and its interaction with RAGE may be valuable in proposing the next round of lead compounds for effective Alzheimer’s disease treatment.
Keywords: Alzheimer’s disease, natural compounds, binding energy, docking, Z-dock
1. Introduction
Alzheimer’s disease (AD) is a common progressive neurodegenerative disease affecting more than 46 million people worldwide. Figures in the USA for the year 2015 reported AD to affect over 5.3 million people, whereby in 2050, it is expected to grow to nearly 1 million new AD cases per year, with an estimated frequency to range from 11 million to 16 million [1]. Unfortunately, the etiology of AD is still not clear. One of the major pathological characteristics of AD tends to be extracellular deposition of Amyloid-beta (Aβ) peptides in senile plaques. The Aβ cascade-inflammatory hypothesis has been found to be the most probable therapeutic checkpoint for the treatment of AD [2]. Concerning structural perspectives, amyloid oligomers are spherical [3], surface-active entities [4] prone to form pore-like assemblies in the plasma membrane of brain cells [5].The circulating Aβ toxins are transported by the receptor for advanced glycation end products (RAGE), a multiligand receptor running transversely through the blood-brain barrier (BBB) into the brain. A RAGE-Aβ toxin interaction at the BBB directs the induction of oxidative stress, inflammatory responses and decreases the cerebral blood flow. RAGE specifically binds to the β-sheet fibrillar domain of Aβ toxins. Thereby, the regulation of RAGE action at the BBB, owing to its significance in disease progression, has been seen as a valuable treatment strategy for AD patients [6]. The deposition of amyloid in tissues changes its normal function, and it has been noticed that its elevated concentrations exert nonspecific toxic effects on cells by disturbing the integrity of membranes. Similarly, deposition of amyloid in tissues has been directly correlated with enhanced appearance of RAGE. In the brain of AD patients, the appearance of RAGE increases in glia and neurons. The penalty of Aβ ligation of RAGE has been shown to be relatively dissimilar for neurons as compared to microglia, where microglia become activated as a result of Aβ-RAGE interaction, as reflected by amplified motility and the expression of cytokines [7,8,9,10,11].
Curcumin, the very active constituent of turmeric, has various valuable properties, including anti-inflammatory, antioxidant and antitumor effects. Different studies so far have suggested that curcumin decreases the level of amyloid. Oxidized proteins avoid memory deficits and are thus helpful to patients with AD [12]. Inhibitors of Aβ aggregation can work by providing an adverse surface in amyloid oligomeric interaction [13]. Curcumin typically binds the target protein/peptide in the amyloidogenic pathway which makes it effiecient as an anti-aggregation agent [14,15]. This fact has been confirmed by staining studies of the amyloid deposits in in vivo systems [16,17]. Curcumin acts as an efficient β-sheet breaker in interactions with the Aβ peptide [18,19]. Following the above hypothesis in the present study, we have analyzed the Aβ aggregation inhibition potential of ajmalicine, emetine and vincamine compounds against curcumin as a standard control for their anti-Alzheimer’s potential.
2. Results and Discussion
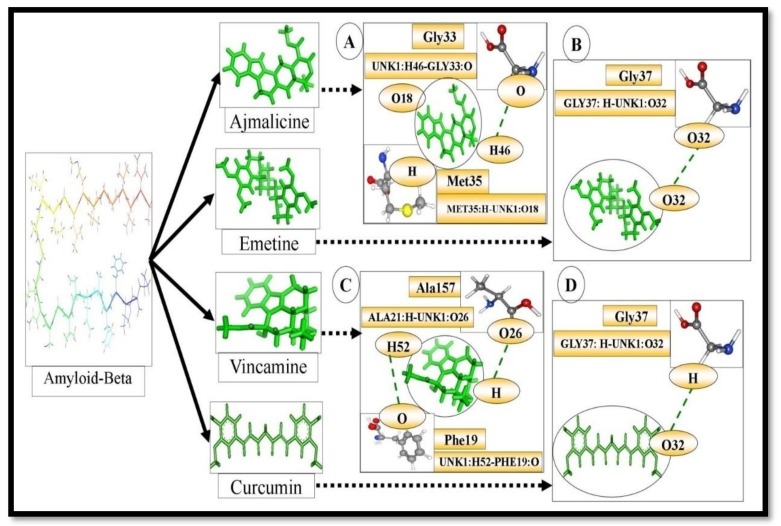
There is corpus evidence of AD being linked with oligomerization of β-Amyloid peptides [20]. Thereby, one of the procedures to adapt to AD is to discover compounds that can promote Aβ anti-aggregation and clearance [21]. The natural products or extracts reported in various preclinical and certain clinical studies provide valuable input to AD therapy [22,23]. Among them, curcumin is shown to inhibit Aβ aggregation and act as an antidote to Aβ-induced toxicity [14]. In the present investigation, Aβ was found to associate with Vnc through the amino acids Phe19, Phe20, Ala21, Asp23, Ile32, Gly33, Leu34, Met35 and Val36 (Figure 1C, Table 1); with Ajm through the amino acid residues Phe19, Phe20, Ala21, Gly22, Asp23, Ile32, Gly33,Leu34, Met35,and Val36 (Figure 1A, Table 1); and with Eme through the amino acid residues Ala21, Gl22, Asp23, Gly33, Leu34, Met35 and Val36 (Figure 1B, Table 1); as compared to standard curcumin, which was found to associate through the amino acid residues Ala21, Glu22, Asp23, Val24, Gly25, Leu34, Met35, Val36 and Gly37 (Figure 1D, Table 1). The amino acid residues Ala21, Asp23, Met35, and Val36 were seen in common interaction with selected ligand as well as curcumin in Aβ.
Figure 1.
(A) The complex shows interacting amino acid residues and hydrogen bonds formed between compound ajmalicine (the ligand, ajmalicine, has been shown in green ‘stick’ representation) and β-Amyloid (B) The complex interacting amino acid residues and hydrogen bonds formed between compound emetine (the ligand, emetine, has been shown in green ‘stick’ representation) and β-Amyloid (C) The complex interacting amino acid residues and hydrogen bonds formed between compound vincamine (the ligand, vincamine, has been shown in green ‘stick’ representation) and β-Amyloid. (D) The complex interacting amino acid residues and hydrogen bonds formed between compound curcumin (the ligand, curcumin, has been shown in green ‘stick’ representation) and β-Amyloid.
Table 1.
Interacting amino acid residues and the H-bond distance between β-Amyloid and natural compound.
| S.N | Target | Ligands Name | Interaction Amino Acid | H-bond Distance (Å) | H-bond |
|---|---|---|---|---|---|
| 1. | Β-Amyloid- | Vincamine | Phe19, Phe20, Ala21, Asp23, Ile32, Gly33, Leu34, Met35, and Val36 | 1.87477 2.11324 |
ALA21:H-UNK1: O26 UNK1: H52-PHE19:O |
| 2. | Ajmalicine | Phe19, Phe20, Ala21, Gly22, Asp23, Ile32, Gly33,Leu34, Met35, and Val36 | 1.81314 1.87937 |
MET35:H-UNK1: O18 UNK1: H46-GLY33:O |
|
| 3. | Emetine | Ala21, Gl22, Asp23, Gly33, Leu34, Met35, Val36, and Gly37 | 2.41182 | GLY37:H-UNK1: O32 | |
| 4. | Curcumin | Ala21, Glu22, Asp23, Val24, Gly25, Leu34, Met35, Val36, and Gly37. | 2.13938 | UNK1: H43-GLY37:O |
The free energy of binding for the interaction complexes ‘Vnc-Aβ’, ‘Ajm-Aβ’ and ‘Eme-Aβ’ were found to be −5.45, −6.66 and −6.99 kcal/mol respectively along with with their evaluated inhibition constants 249.96, 13.23 and 7.5 μM, respectively. While the free energy of binding and estimated inhibition constant for the ‘Cur-Aβ interaction’ was determined to be −3.61 kcal/mol and 136.2 µM (Table 2). Chloramphenicol (PubChem ID: 5959) was used as a negative control. The free energy of binding for the chloramphenicol with Aβ was found to be +1.16 kcal/mol. One H-bond UNK1:H43-GLY37: O was present in the ‘Cur-Aβ interaction’. The H-bonds distances were 2.13938 Å. Eight carbon atoms of Cur, namely C5, C4, C16, C10, C13, C8, C6, and C7 were found to be in hydrophobic interactions with amino acid residues Ala21, Leu34 and Val36 of the Aβ protein. C4 and C5 were found to be interacting with Ala21. C10, C13, and C16 were found to be interacting with Leu34 and C4, C5, C6, C7, and C8 were found to interact with Val36. In these interactions there are not any polar, pi-pi and cation-pi bonds present. ‘Van der Waals’, ‘hydrogen bond’ and ‘desolvation’ energy components for ‘Cur-Aβ interaction’ were −5.56 kcal/mol while the ‘electrostatic’ energy component was found to be −0.03 kcal/mol. The internal molecular energy component was found to be −5.59kcal/mol (Table 2). Similarly, on the other hand, two H-bonds ALA21:H-UNK1:O26 and UNK1:H52-PHE19:O were present in the ‘Vnc-Aβ interaction’. The H-bonds distances were 1.87477 and 2.11324 Å, respectively. Two carbon atoms of Vnc, namely C2 and C15 were found to be involved in hydrophobic interactions with amino acid residues Ala21 and the Aβ protein. One of the nitrogen atoms N1 and one H12 atom of Vnc were observed to make polar bonds with one amino acid residue Asp23, but no pi-pi and cation-pi bonds were present. ‘Van der Waals’, ‘hydrogen bond’ and ‘desolvation’ energy components for ‘Vnc-Aβ interaction’ were −5.33 kcal/mol while the ‘electrostatic’ energy component was found to be -0.78 kcal/mol. The internal molecular energy component was found to be −6.11kcal/mol (Table 2). In continuation, two H-bonds MET35:H-UNK1:O18 and UNK1:H46-GLY33:O were present in the ‘Ajm-Aβ interaction’. The H-bonds distances were 1.81314 and 1.87937 Å, respectively. Four carbon atoms of Ajm, namely C15, C18, C19, and C13 were found to be involved in hydrophobic interactions with amino acid residues Ala21, Val24 of the Aβ protein, in which C15 had interacted with Ala21; C18 and C19 were found to interact with Val24 and C13 interacted with Leu34. One of the hydrogen atoms H8 of Vnc was observed to make polar bonds with one amino acid residue Asp23. In this interaction, there were no pi-pi and cation-pi bonds. ‘Van der Waals’, ‘hydrogen bond’ and ‘desolvation’ energy components for ‘Ajm-Aβ interaction’ were −6.27 kcal/mol while the ‘electrostatic’ energy component was found to be -0.98 kcal/mol. The internal molecular energy component was found to be −7.25 kcal/mol (Table 2). Similarly, one H-bond GLY37:H-UNK1:O32 was present in the ‘Eme-Aβ interaction’. The H-bond distance was 2.41182 Å. Six carbon atoms of Eme, namely C5, C6, C7, C28, C27, C19 were found to be involved in hydrophobic interactions with amino acid residues Ala21, Leu34 and Val36 of the Aβ protein, in which C5, C6, C7 interacted with Ala21; C28, C27, C19 were found to interact with Leu34 and C5 and C6 interacted with Val36. Two hydrogen atoms H29 and H30 of Eme were observed to make polar bonds with one amino acid residue, Asp23. In this interaction, there were no pi-pi and cation-pi bonds present. ‘Van der Waals’, ‘hydrogen bond’ and ‘desolvation’ energy components for ‘Eme-Aβ interaction’ were −8.2 kcal/mol while the ‘electrostatic’ energy component was found to be −0.88 kcal/mol. The internal molecular energy component was found to be −9.08 kcal/mol (Table 2). It is appropriate to mention that ΔG value obtained through computational study can only propose the efficiency of binding for a ligand-enzyme pair [24].
Table 2.
Interaction energies of Aβ with ligands (Kcal/mol) obtained from molecular docking analysis.
| S. No |
Target | Ligand Name | Binding Energy (Kcal/mol) | Inhibition Constant (Ki) | vdw+hb+ Desolvation Energy (Kcal/mol) | Internal Molecular Energy (Kcal/mol) | Electrostatic Energy (Kcal/mol) |
|---|---|---|---|---|---|---|---|
| 1. | β-Amyloid | Vincamine | −5.45 | 249.96 µM | −5.33 | −6.11 | −0.78 |
| 2. | Ajmalicine | −6.66 | 13.23 µM | −6.27 | −7.25 | −0.98 | |
| 3. | Emetine | −6.99 | 7.5 µM | −8.2 | −9.08 | −0.88 | |
| 4. | Curcumin | −3.61 | 136.2 µM | −5.56 | −5.59 | −0.03 |
2.1. Protein-Protein Interaction Study
An earlier study reported the suitable interface of RAGE and Aβ at the luminal membrane of the BBB, proposing that RAGE acts as a transporter protein for circulating Aβ across the BBB [25]. This fact is supported in another research where RAGE mediates the entry of Aβ1-40 and Aβ1-42 into the hippocampus and cortex across the BBB [26]. The Aβ-RAGE connection at the BBB does not just result in neurovascular stress and articulation of proinflammatory cytokines (TNF-𝛼 and IL-6), in addition, it prompts diminished cerebral bloodstream by improving the emission of endothelin-1 to instigate vasoconstriction [27].
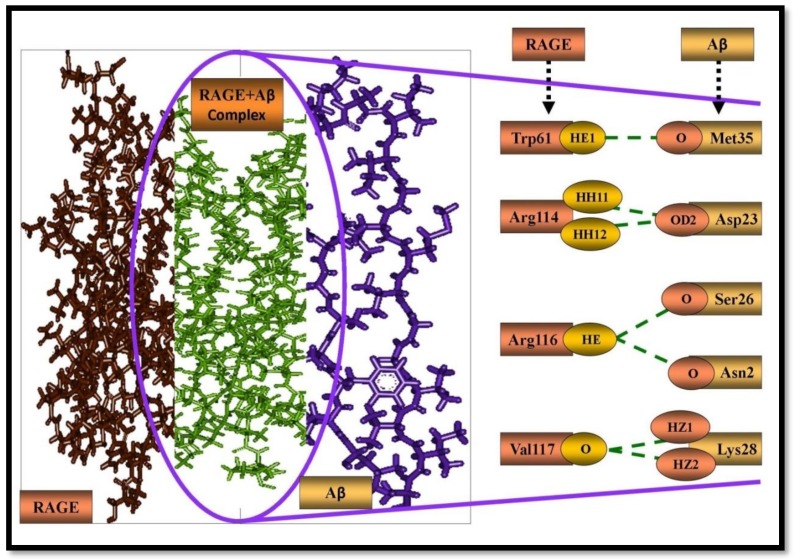
We therefore further deciphered the interaction impact of Aβ on RAGE in terms of binding efficiency and interacting amino acid residues. For this reason, we connected the Z-dock technique by figuring out the Z-dock score of protein-protein association between Aβ and RAGE and compared it with docked protein complexes of (Aβ+Ajm, Aβ+Eme, and Aβ+Vnc) with RAGE. The Z-dock score for Aβ and RAGE interaction was found to be 1269.55. Met35, Asp23, Ser26, Asn27 and Lys28 of the Aβ and Trp61, Arg114, Arg116, Val117 amino acid residues of RAGE were found to participate in the interaction (Figure 2).
Figure 2.
Protein-protein interaction. (A) Aβ, (B) RAGE, (C) the complex of Aβ and RAGE obtained by the protein-protein docking method. Purple and brown stick color representations are the amino acid residues of Aβ and RAGE, respectively, involved in H-bond formation.
In the complex of Aβ and RAGE, seven H-bonds were found in which Met35 interacts with Trp61. Two amino acid residues of Asp23 were found to interact with Arg114, Ser26 interacted with Arg116, Asn27 interacted with Arg116, and two amino acid residues of Lys28 interacted with two amino acid residues of Val117 (Table 3).
Table 3.
Acid residues involved in hydrogen bond formation in the protein-protein interaction.
| S S.No |
Amino Acid Residues of Aβ | Amino Acid Residues of RAGE | Donor Atom | Acceptor Atom | H-Bonds | Distance of H-Bond (Å) |
|---|---|---|---|---|---|---|
| 1. | Met35 | Trp61 | TRP61:HE1 | MET35:O | TRP61:HE1 - MET35:O | 2.31028 |
| 2. | Asp23 | Arg114 | ARG114:HH11 | ASP23:OD2 | ARG114:HH11 -ASP23:OD2 | 1.40911 |
| 3. | Asp23 | Arg114 | ARG114:HH12 | ASP23:OD2 | ARG114:HH12 -ASP23:OD2 | 2.02749 |
| 4. | Ser26 | Arg116 | ARG116:HE | SER26:O | ARG116:HE - SER26:O | 2.48357 |
| 5. | Asn27 | Arg116 | ARG116: HE | ASN27:O | ARG116: HE - ASN27:O | 2.04783 |
| 6. | Lys28 | Val117 | LYS28:HZ1 | VAL117:O | LYS28:HZ1 - VAL117:O | 2.15927 |
| 7. | Lys28 | Val117 | LYS28:HZ2 | VAL117:O | LYS28:HZ2 - AVAL117:O | 1.10912 |
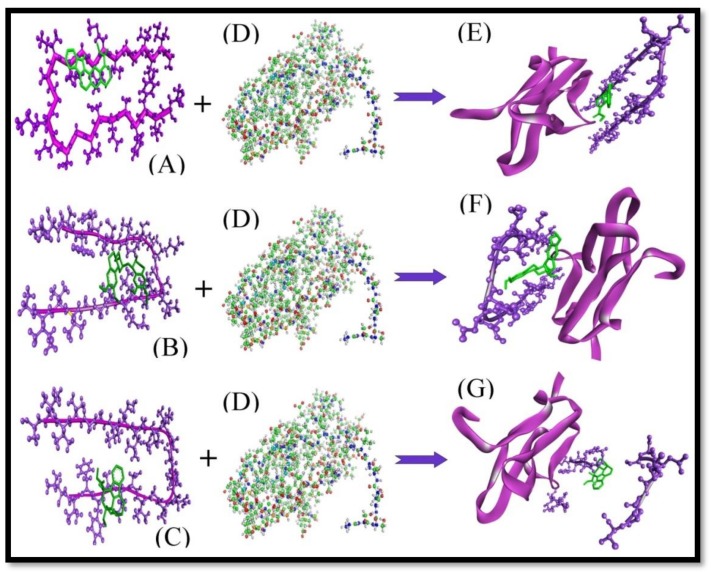
The Z-dock score of the complexes (Aβ+Ajm, Eme+Aβ, and Aβ+Vnc) with RAGE was found to be 911.83, 940.69 and 907.98, respectively. In these complexes, no H-bond formation had taken place. The interaction complexes are shown in (Figure 3).
Figure 3.
The complex-protein interaction. (A) The complex of Aβ+Ajm, (B) the complex of Aβ+Eme, (C) the complex of Aβ+Vnc. (D) Structure of RAGE protein. (E) The interacting complex structure of (Aβ+Ajm) with RAGE obtained by the protein-protein docking method. (F) The interacting complex structure of (Aβ+Eme) with RAGE obtained by the protein-protein docking method. (G) The interacting complex structure of (Aβ+Vnc) with RAGE obtained by the protein-protein docking method.
Results clearly reflected a significant decrease in the Z-dock score of Aβ and RAGE from 1269.55 to 911.83, 940.69 and 907.98 for (Aβ+Ajm, Eme+Aβ, and Aβ+Vnc) with RAGE (Table 4) along with loss of H-Bond formation.
Table 4.
Score of protein-protein and complex-protein interactions.
| S.No. | Protein-Protein interaction | Z-dock Score |
|---|---|---|
| 1. | Aβ+RAGE | 1269.55 |
| Complex-Protein Interaction | Z-dock Score | |
| 2. | (Aβ+Ajm)+ RAGE | 911.83 |
| 3. | (Aβ+Eme)+ RAGE | 940.69 |
| 4. | (Aβ+Vnc)+ RAGE | 907.98 |
With the help of the above results, one could say that the ligands Ajm, Eme and Vnc play a very important role in avoiding the interaction between Aβ and RAGE.
2.2. Comparison of Vnc, Ajm, and Eme With Positive Control Curcumin
Curcumin has a BBB of 0.913545 which is higher than that of Vnc (0.871568) and Eme (0.875498), but lower than Ajm (1.9897). Human Intestinal Absorption (HIA) value for curcumin was 94.40% (Table S1) which is higher than that of Ajm (93.31%), but lower than Vnc (95.95%) and Eme (96.59%). Thus, Vnc and Eme may be better absorbed by the human body than curcumin. With the help of the PreADME server, we have analyzed the toxicity of Vnc, Ajm, Eme leads and curcumin. Curcumin, Vnc, and Eme are nonmutagenic while Ajm is mutagenic. Vnc, Ajm, and Eme are non-carcinogenic in the mouse-like curcumin. Curcumin and Eme are carcinogenic in rats while Vnc and Ajm are non-carcinogenic in rats.
Thereby, the values of Ki and ΔG, interacting amino acid residues, H-bond, polar and hydrophobic interactions and properties of Vnc, Ajm and Eme against curcumin obtained through computational studies probably suggest the efficacy of the proposed leads in AD therapy. The “computational” studies, with reference to the protein (Aβ) and ligands (Vnc, Ajm, and Eme) and protein-protein interactions, are expected to frame the premise of future treatment against a few neurological disarranges [28]. This article may be considered as an extension of our ongoing research work whereby we have reported inhibition of the related neuro-toxic Aβ by selected ligands and an inhibition study with the well-known ligand, curcumin.
3. Materials and Methods
3.1. Preparation of Receptor-Protein Structures
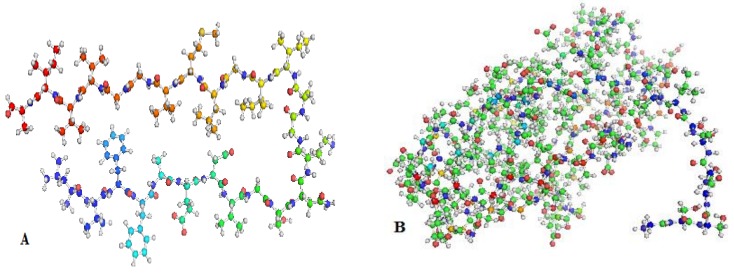
The three-dimensional structures of the β-amyloid (Aβ) (PDB ID: 2BEG) and RAGE (PDB ID: 2ENS) were obtained using RCSB-PDB (www.rcsb.org) (Figure 4). The PDB file was cleaned and the heteroatoms (HETATM) of the receptor were removed manually since these are non-standard residues of the protein [29].
Figure 4.
The three dimensional structures of the protein. (A) β-Amyloid (PDB ID: 2BEG). (B) Receptor for advanced glycation end products (RAGE) (PDB ID: 2ENS).
Chimera was used for energy minimization. The steric collision was detached for the steepest gradient minimization. The steepest descent steps and the size were 1000 and 0.02 Å, respectively. The consumer gradient steps were 1000 and the gradient step size was 0.02 Å [30,31].
3.2. Preparation of Ligand Structure
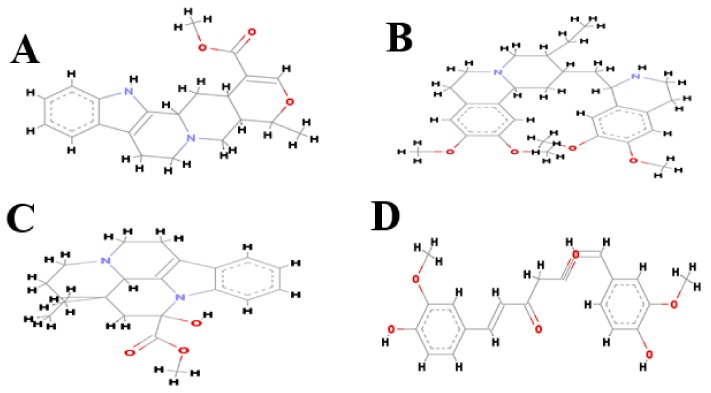
The simplified molecular input line entry specification notations of the inhibitors ajmalicine (Ajm), emetine (Eme) and vincamine (Vnc) were obtained from the PubChem database. The online display of CORINA (http://www.molecular-networks.com/ products/ Corina) was used to build the 2D structures of the ligands (Figure 5A–D). Before docking, these inhibitors were energy minimized by Chimera software [32] and saved in PDP format. Gasteiger charges [33] were applied to the ligands and they were further exposed to single-step minimization.
Figure 5.
2D chemical structure of the compounds: (A) ajmalicine, (B) emetine, (C) vincamine, (D) curcumin.
3.3. Molecular Interaction Study
Molecular interaction examinations were performed via the Autodock version 4.2 suite with the Cygwin interface instrument [34,35]. Molecular docking methods were used for target and ligand interaction to obtain the top conformations on the foundation of binding energy (kcal/mol). Before applying the docking algorithm, we marked all water molecules in proteins that had been expelled from the focused protein structure. The hydrogen molecules were included in the target molecules. After, Kollman united charges and solvation parameters were applied to proteins. Gasteiger partial charges were added to the ligands atom. A framework box was set to cover the greatest piece of chosen protein for ligand cooperation. The value was set to standard 60 × 60 × 60 Å in X, Y and Z as the organization of the network point with the default estimation of framework focuses separating at 0.375 Å. The Lamarckian genetic algorithm (LGA) was connected at the receptor protein and ligand for adaptable docking calculations [36,37]. The LGA parameters, as well as the populace measure, vitality assessments, transformation rate, hybrid rate, and step size, were set to: 150; 2,500,000; 27,000; 0.02; 0.8 and; 0.2 Å, respectively. The LGA runs were set to a standard of 10 runs. We watched all adaptations of protein with ligand complex. They were then examined for the connection orientations in terms of binding energies of the docked structure utilizing Discovery Studio Visualizer.
3.4. Protein-Protein Interaction Analysis
Z-dock was utilized for protein-protein interactions (Aβ+RAGE) [38] as well as for protein-ligand complexes and protein (Aβ+Ajm, Aβ+Eme, and Aβ+Vnc & RAGE) interactions. Z-dock is one of the most unbeaten suites that encompass large calculation facility in Critical Assessment of Predicted Interactions (CAPRI) [39]. Z-dock is an original phase rigid body molecular docking algorithm that utilizes a fast Fourier transform (FFT) calculation to calculate progress performance for translational searching [40].
4. Conclusions
The present study proposes that all the selected compounds can be absorbed by the human body, by passing through the BBB, and have high inhibition potential. With the help of docking, we have proposed the inhibition of Aβ and degradation of amyloid peptide aggregation. Our compounds have shown the higher efficiency to bind with Aβ (Vnc: −5.45 Kcal/Mol, Ajm: −6.66 Kcal/Mol, Eme: −6.99 Kcal/Mol) compared to standard Cur: −3.61 Kcal/Mol. Z-dock scores calculated from protein-protein and complex-protein connections further support that the selected compounds have potential for disaggregation.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no (RG-1440-009).
Abbreviations
| AD | Alzheimer’s disease |
| Aβ | Amyloid-beta |
| RAGE | Receptor for advanced glycation end products |
| BBB | Blood-brain barrier |
| Vnc | Vincamine |
| Ajm | Ajmalicine |
| Eme | Emetine |
Supplementary Materials
Table S1: Human Intestinal Absorption (HIA) value of compounds.
Author Contributions
S.S.A., S.M.D.R. and M.H.S. were involved in the conceptual design, methodology, research work, data interpretation and critically helped in writing the manuscript; H.K. and L.R. analyzed the data; S.A.A., R.U., and H.M.M. helped in language correction and drafting the manuscript.
Funding
Please add: This research received no external funding.
Conflicts of Interest
The authors confirm that this article content has no conflict of interest.
References
- 1.Alzheimer’s Association Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.McGeer P.L., McGeer E.G. The amyloid cascade-inflammatory hypothesis of Alzheimer disease: Implications for therapy. Acta Neuropathol. 2013;126:479–497. doi: 10.1007/s00401-013-1177-7. [DOI] [PubMed] [Google Scholar]
- 3.Glabe C.G. Structural Classification of Toxic Amyloid Oligomers. J. Boil. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kayed R., Sokolov Y., Edmonds B., McIntire T.M., Milton S.C., Hall J.E., Glabe C.G. Permeabilization of Lipid Bilayers Is a Common Conformation-dependent Activity of Soluble Amyloid Oligomers in Protein Misfolding Diseases. J. Boil. Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 5.Quist A., Doudevski I., Lin H., Azimova R., Ng D., Frangione B., Kagan B., Ghiso J., Lal R. Amyloid ion channels: A common structural link for the protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deane R.J. Is RAGE still a therapeutic target for Alzheimer’s disease? Future Med. Chem. 2012;4:915–925. doi: 10.4155/fmc.12.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan S.D., Chen X., Fu J., Chen M., Zhu H., Roher A., Slattery T., Zhao L., Nagashima M., Morser J., et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 8.Du Yan S., Zhu H., Zhu A., Golabek A., Du H., Roher A., Yu J., Soto C., Schmidt A.M., Stern D., et al. Receptor-dependent cell stress and amyloid accumulation in systemic amyloidosis. Nat. Med. 2000;6:643–651. doi: 10.1038/76216. [DOI] [PubMed] [Google Scholar]
- 9.Paresce D.M., Ghosh R.N., Maxfield F.R. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–565. doi: 10.1016/S0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang H.Y., Lee D.H., D’Andrea M.R., Peterson P.A., Shank R.P., Reitz A.B. Aβ (1–42) binds to α-7 nicotinic acetylcholine receptor with high affinity. J. Biol. Chem. 2000;275:5626–5632. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 11.Yaar M., Zhai S., Pilch P.F., Doyle S.M., Eisenhauer P.B., Fine R.E., Gilchrest B.A. Binding of beta-amyloid to the p75 neurotrophin receptor induces apoptosis. J. Clin. Investig. 1997;100:2333–2340. doi: 10.1172/JCI119772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Fang Y., Xu Y., Lian Y., Xie N., Wu T., Zhang H., Sun L., Zhang R., Wang Z. Curcumin Improves Amyloid β-Peptide (1-42) Induced Spatial Memory Deficits through BDNF-ERK Signaling Pathway. PLoS ONE. 2015;10:e0131525. doi: 10.1371/journal.pone.0131525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefani M., Rigacci S. Protein Folding and Aggregation into Amyloid: The Interference by Natural Phenolic Compounds. Int. J. Mol. Sci. 2013;14:12411–12457. doi: 10.3390/ijms140612411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bratkovic I.H., Gaspersic J., Smid L.M., Bresjanac M., Jerala R. Curcumin binds to the α-helical intermediate and to the amyloid form of prion protein—A new mechanism for the inhibition of PrPSc accumulation. J. Neurochem. 2008;104:1553–1564. doi: 10.1111/j.1471-4159.2007.05105.x. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa D., Taguchi H., Yamamoto A., Shirai N., Hirao K., Tooyama I. Curcuminoid Binds to Amyloid-β1-42 Oligomer and Fibril. J. Alzheimer’s Dis. 2011;24:33–42. doi: 10.3233/JAD-2011-102100. [DOI] [PubMed] [Google Scholar]
- 16.Yang F., Lim G.P., Begum A.N., Ubeda O.J., Simmons M.R., Ambegaokar S.S., Chen P.P., Kayed R., Glabe C.G., Frautschy S.A., et al. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 17.Pandey N., Strider J., Nolan W.C., Yan S.X., Galvin J.E. Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol. 2008;115:479–489. doi: 10.1007/s00401-007-0332-4. [DOI] [PubMed] [Google Scholar]
- 18.Na Zhao L., Chiu S.-W., Benoit J., Chew L.Y., Mu Y. The Effect of Curcumin on the Stability of Aβ Dimers. J. Phys. Chem. B. 2012;116:7428–7435. doi: 10.1021/jp3034209. [DOI] [PubMed] [Google Scholar]
- 19.Balasubramanian K. Molecular orbital basis for yellow curry spice curcumin’s prevention of Alzheimer’s disease. J. Agric. Food Chem. 2006;54:3512–3520. doi: 10.1021/jf0603533. [DOI] [PubMed] [Google Scholar]
- 20.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 21.Ngo S.T., Li M.S. Top-leads from natural products for the treatment of Alzheimer’s disease: Docking and molecular dynamics study. Mol. Simul. 2013;39:279–291. doi: 10.1080/08927022.2012.718769. [DOI] [Google Scholar]
- 22.Williams P., Sorribas A., Howes M.J.R. Natural products as a source of Alzheimer’s drug leads. Nat. Prod. Rep. 2011;28:48–77. doi: 10.1039/C0NP00027B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howes M.-J.R., Houghton P.J. Plants used in Chinese and Indian traditional medicine for improvement of memory and cognitive function. Pharm. Biochem. Behav. 2003;75:513–527. doi: 10.1016/S0091-3057(03)00128-X. [DOI] [PubMed] [Google Scholar]
- 24.Alam A., Shaikh S., Ahmad S., Ansari M., Shakil S., Rizvi S.M.D., Shakil S., Imran M., Haneef M., Abuzenadah A., et al. Molecular Interaction of Human Brain Acetylcholinesterase with a Natural Inhibitor Huperzine-B: An Enzoinformatics Approach. CNS Neurol. Disord. Drug Targets. 2014;13:487–490. doi: 10.2174/18715273113126660163. [DOI] [PubMed] [Google Scholar]
- 25.Mackic J.B., Bading J., Ghiso J., Walker L., Wisniewski T., Frangione B., Zlokovic B.V. Circulating amyloid-βpeptide crosses the blood-brain barrier in aged monkeys and contributes to Alzheimer’s disease lesions. Vascul. Pharmacol. 2002;38:303–313. doi: 10.1016/S1537-1891(02)00198-2. [DOI] [PubMed] [Google Scholar]
- 26.Martel C.L., Mackic J.B., McComb J.G., Ghiso J., Zlokovic B.V. Blood-brain barrier uptake of the 40 and 42 amino acid sequences of circulating Alzheimer’s amyloid βin guinea pigs. Neurosci. Lett. 1996;206:157–160. doi: 10.1016/S0304-3940(96)12462-9. [DOI] [PubMed] [Google Scholar]
- 27.Deane R., Du Yan S., Submamaryan R.K., LaRue B., Jovanovic S., Hogg E., Welch D., Manness L., Lin C., Yu J., et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat. Med. 2003;9:907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 28.Awasthi M., Singh S., Pandey V.P., Dwivedi U.N. Alzheimer’s disease: An overview of amyloid beta dependent pathogenesis and its therapeutic implications along with in silico approaches emphasizing the role of natural products. J. Neurol. Sci. 2016;361:256–271. doi: 10.1016/j.jns.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Kalim M., Khan A., Baig M.H., Arif J.M., Lohani M., Jamal F. Efficacy of natural inhibitors against pkc: An in silico approach to combat cancer. Recent Res. Sci. Technol. 2011;3:90–93. [Google Scholar]
- 30.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Gasteiger J., Marsili M. A new model for calculating atomic charges in molecules. Tetrahedron Lett. 1978;19:3181–3184. doi: 10.1016/S0040-4039(01)94977-9. [DOI] [Google Scholar]
- 34.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 35.Rarey M., Kramer B., Lengauer T., Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;26:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 36.Goodsell D.S., Morris G.M., Olson A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996;9:1–5. doi: 10.1002/(SICI)1099-1352(199601)9:1<1::AID-JMR241>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad S.S., Akhtar S., Danish Rizvi S.M., Kamal M.A., Sayeed U., Khan M.K.A., Siddiqui M.H., Arif J.M. Screening and Elucidation of Selected Natural Compounds for Anti-Alzheimer’s Potential Targeting BACE-1 Enzyme: A Case Computational Study. Curr. Comput. Aided Drug Des. 2017;13:311–318. doi: 10.2174/1573409913666170414123825. [DOI] [PubMed] [Google Scholar]
- 38.Chen R., Li L., Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins Struct. Funct. Bioinform. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 39.Hwang H., Vreven T., Pierce B.G., Hung J.-H., Weng Z. Performance of ZDOCK and ZRANK in CAPRI Rounds 13–19. Proteins Struct. Funct. Bioinform. 2010;78:3104–3110. doi: 10.1002/prot.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowdhury R., Rasheed M., Keidel D., Moussalem M., Olson A., Sanner M., Bajaj C. Protein-Protein Docking with F2Dock 2.0 and GB-Rerank. PLoS ONE. 2013;8:e51307. doi: 10.1371/journal.pone.0051307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.