Abstract
The male‐biased prevalence of certain neurodevelopmental disorders and the sex‐biased outcomes associated with stress exposure during gestation have been previously described. Here, we hypothesized that genes distinctively targeted by only one or both homologous proteins highly conserved across therian mammals, SOX3 and SRY, could induce sexual adaptive changes that result in a differential risk for neurodevelopmental disorders. ChIP‐seq/chip data showed that SOX3/SRY gene targets were expressed in different brain cell types in mice. We used orthologous human genes in rodent genomes to extend the number of SOX3/SRY set (1,721). These genes were later found to be enriched in five modules of coexpressed genes during the early and mid‐gestation periods (FDR < 0.05), independent of sexual hormones. Genes with differential expression (24, p < 0.0001) and methylation (40, p < 0.047) between sexes were overrepresented in this set. Exclusive SOX3 or SRY target genes were more associated with the late gestational and postnatal periods. Using autism as a model sex‐biased disorder, the SOX3/SRY set was enriched in autism gene databases (FDR ≤ 0.05), and there were more de novo variations from the male autism spectrum disorder (ASD) samples under the SRY peaks compared to the random peaks (p < 0.024). The comparison of coexpressed networks of SOX3/SRY target genes between male autism and control samples revealed low preservation in gene modules related to stress response (99 genes) and neurogenesis (78 genes). This study provides evidence that while SOX3 is a regulatory mechanism for both sexes, the male‐exclusive SRY also plays a role in gene regulation, suggesting a potential mechanism for sex bias in ASD.
Keywords: neurodevelopmental disorder, sex, SOX3, SRY, stress
ABBREVIATIONS
- ASD
autism spectrum disorder
- ADHD
attention deficit hyperactivity disorder
- CNV
copy number variation
- ChIP
chromatin immunoprecipitation
- CON
Control
- DIOPT
Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool
- DNV
de novo variations
- FDR
false discovery rate
- FPKM
fragments per kilobase million
- GWAS
genome‐wide association study
- HMG
high‐mobility‐group
- ID
intellectual disability
- MAF
minor allele frequency
- MSET
modular single‐set enrichment test
- NPCs
neural progenitor cells
- PCW
postconceptional week
- PGC
psychiatry genomics consortium
- PMI
post mortem interval
- RAS
renin angiotensin system
- RDNV
rare de novo variation
- RPKM
reads per kilobase million
- RVIS
residual variation intolerance score
- SHR
spontaneously hypertensive rat
- TF
transcription factor
- TSS
transcription start site
- WGCNA
weighted gene coexpression network analysis
1. BACKGROUND
Sex impacts the brain's structure and functional organization, as reflected by differences in neuropsychiatric disorders between the sexes (Joel & McCarthy, 2017). Accordingly, several brain regions that demonstrate sexual dimorphism have already been associated with psychiatric disorders, suggesting that sex is an underlying factor in their prevalence. For example, neurodevelopmental disorders, such as autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD) and schizophrenia, are more prevalent in men (Bale & Epperson, 2015).
This sexual dimorphism in brain gene expression reflects differences in the regulatory mechanisms that could be attributed to either hormonal regulation or chromosome complement. The use of “four core genotypes” mice models revealed that sexual dimorphism is not exclusively hormone‐dependent (Arnold & Chen, 2009). The gene dosage effect of the sex chromosomes is attributed to several mechanisms, such as Y‐linked genes, X‐linked genes, mosaicism, skewed X‐inactivation, X imprinting and heterochromatin sink (Ober, Loisel, & Gilad, 2008; Schaafsma & Pfaff, 2014).
The regulatory roles of the Y chromosome on autosomal/X‐linked genes have also been described (Bellott et al., 2014; Case et al., 2013; Lemos, Branco, & Hartl, 2010). In Drosophila melanogaster, for example, a class of autosomal/X genes has been reported to consistently change expression levels across multiple Y introgression experiments (Branco, Tao, Hartl, & Lemos, 2013; Lemos, Araripe, & Hartl, 2008; Sackton & Hartl, 2013). Moreover, a set of autosomal genes was sensitive to the sex chromosome complement in mice (Wijchers et al., 2010). Recent studies regarding brain sexual dimorphism noted several differences in gene expression regulatory mechanisms (e.g., genetic and epigenetic factors; Chen et al., 2016) that could contribute to sex‐biased prevalence in neuropsychiatric disorders (Ratnu, Emami, & Bredy, 2017).
In this context, two members of the SOX gene family, SOX3 and SRY, can contribute to these differences. The SOX3 gene, which is located at the X chromosome, plays a critical role in the central nervous system and hypothalamic development (Bergsland et al., 2011; Brunelli, Casey, Bell, Harland, & Lovell‐Badge, 2003; Cheah & Thomas, 2015; Topalovic et al., 2017) and undergoes dosage compensation in females (Graves, 2015). The SRY gene is located at the Y chromosome and evolved from SOX3 (Cortez et al., 2014; Foster & Graves, 1994; Skaletsky et al., 2003), sharing an high‐mobility‐group (HMG) box domain that is highly conserved among species (Bergstrom, Young, Albrecht, & Eicher, 2000).
The Sry locus is primarily known as the developmental switch responsible for testes determination. It is transcribed in males at specific periods, even in tissues that are not involved in gonad sexual differentiation (Mayer, Mosler, Just, Pilgrim, & Reisert, 2000; Turner, Ely, Prokop, & Milsted, 2011). For example, increased SRY expression was observed after induced differentiation into human dopaminergic neurons (Czech et al., 2012), and it also modulates dopamine biosynthesis through MAOA (Dewing et al., 2006) and Th regulation (Milsted et al., 2004). Few studies have explored its role in neurodevelopment, however, it seems to be crucial for brain development (Loke, Harley, & Lee, 2015).
The contributions of the Sry locus to the hypertensive phenotype are well established in the spontaneously hypertensive rat (SHR), which has also been used as a model for neurodevelopmental disorders more prevalent in males due the presence of symptoms related to ADHD (Sagvolden, Russell, Aase, Johansen, & Farshbaf, 2005) and ASD (Zhang‐James et al., 2014).
Differences between SRY and SOX3 are likely related to distinct domains outside the HMG box, as a chimeric SRY with SOX3‐HMG‐box domain rescued SRY protein regulation function (Bergstrom et al., 2000). Ectopic Sox3 expression in bipotential gonad cells (which can differentiate into either testis or ovary) caused XX male reversal in mice (Sutton et al., 2011), suggesting that SOX3 and SRY target a set of common genes. Sry expression in the kidney is also involved in sex‐specific regulation of the blood pressure by the renin‐angiotensin system (RAS), showing activation of the REN promoter, while SOX3 inhibits it (Araujo et al., 2015). Collectively, these data suggest that the regulation exerted by SRY plays a role in sexual differentiation that is not dependent on circulating gonadal hormones and that the association of both transcription factors (TFs) could act as a regulatory mechanism.
We hypothesized that SOX3, SRY or both could differentially regulate sets of genes, according to cell type, brain region and developmental time; these sets of genes act as regulators of the normal differences between the sexes during development. In that case, exclusive SRY and SOX3/SRY target genes could be potentially associated with sex‐biased neurodevelopmental disorders. Using an integrated bioinformatics approach, we identified the target genes of SOX3 and/or SRY. Next, we evaluated the expression level and regulation of these genes in brain samples. Finally, we used ASD gene databases to access whether SOX3 and/or SRY target genes were enriched for this sex‐biased neurodevelopmental disorder. By providing evidence of the differential regulation exerted by SOX3 and/or SRY targets during neurodevelopment, our findings provide insights into a mechanism through which sex factors may contribute to ASD male predisposition and provide a new perspective for studies of other sex‐biased neurodevelopmental disorders.
2. METHODS
2.1. ChIP‐seq/chip annotation and peak datasets
Using published datasets of SRY and SOX3 ChIP‐seq/chip, we identified putative targets of SRY and SOX3 regulation by annotating each peak to the nearest gene provided by RefSeq/UCSC genome annotation databases, according to the corresponding gene assembly using BEDTools (v.2.25.0; Quinlan & Hall, 2010). To explore SRY, we used three published ChIP‐chip datasets from human, mouse and rat organisms. The human SRY ChIP‐chip was performed using embryonic carcinoma cells (Ntera2) in NimbleGen Array Service onto 1.5‐kb human promoter arrays composed of 23,467 EntrezGenes. From these genes, 1,280 genes were identified among 1,343 SRY peaks (Jin, O'Geen, Iyengar, Green, & Farnham, 2007).
A second dataset from mouse SRY ChIP‐chip used embryonic (E11.5) gonadal cells in a 385K RefSeq Promoter Array of MM8 assembly composed of the closest 27,530 RefSeq genes, corresponding to 18,878 mouse EntrezGenes (Li, Zheng, & Lau, 2014). From these, 3,738 genes were identified among 3,083 SRY peaks. The third dataset from rat SRY ChIP‐chip used embryonic (E13) gonadal cells in Nimblegene's Rat ChIP‐chip 3x720K RefSeq Promoter Array, composed of the closest 16,168 RefSeqs genes, and 252 genes were identified in analyses of the 262 possible SRY peaks (Bhandari, Haque, & Skinner, 2012). Finally, two studies were used for SOX3 ChIP‐seq annotation, and both used neural progenitor cells (NPCs). We found 9,719 peaks corresponding to the closest 7,984 RefSeq genes and 5,307 EntrezGenes in the first study (Bergsland et al., 2011) and 8,067 peaks corresponding to closest 7,523 RefSeq genes and 5,150 EntrezGenes in the second study (McAninch & Thomas, 2014).
The Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (DIOPT; Hu et al., 2011) was used to identify orthologs between mice and human, applying the best match criterion. This tool uses ten orthology prediction algorithms across species, four based on phylogenetic models (Phylome, TreeFam, EnsemblCompara, OrthoDB), five based on sequence similarity (Homologene, OMA, Roundup, InParanoid, OrthoMCL) and one based on network protein–protein interactions (Isobase). For ortholog genes in rats, only Compara (BioMart) was used (Vilella et al., 2009).
To build the SRY and SOX3 peaks datasets, we used the LiftOver tool (Rosenbloom et al., 2014) to convert the genome coordinates of each species to human hg19 assembly, using default parameters. This process resulted in 4,486 SRY peaks (1,343 from humans, 2,956 from mice and 187 from rats) and 14,032 SOX3 peaks converted from a mouse model, 5,385 from Bergsland et al. (2011) and 8,647 from McAninch & Thomas (2014).
2.2. Random peak sets and enrichment analysis
Datasets from SRY and SOX3 peaks were used to build 1,000 random peak sets with the same chromosome distribution and length and number of peaks, using the shuffleBed function in BEDTools (Quinlan & Hall, 2010). The empirical p value was calculated as a proportion of the number of random sets that resulted in overlaps that were equal to or greater than the group compared.
2.3. Databases
A summary table concerning the datasets provided by individual studies used in this study is available in Supporting Information Table S1.
2.3.1. Gene expression datasets from mouse brains
Two expression datasets of mouse brain were used. The first dataset was composed of seven developmental stages (four embryonic stages [E11.5, E13.5, E15.5, E18.5] and three postnatal stages [P4, P14, and P28]) and four brain structures analyzed through in situ hybridization (midbrain, forebrain, hindbrain and spinal cord). The analysis revealed 1,721 genes, represented by 1,715 EntrezGenes, which were expressed in at least one stage (Thompson et al., 2014).
The second dataset was composed of the following: (a) a pool of five primary brain cell cultures from postnatal day 1 (P1), containing expression data of astrocytes and oligodendrocytes (divisions 1, 2.5 and 4), (b) a pool of three brain samples from embryos (16.5 postconceptional days) to explore cortical neurons, and (c) adult microglia obtained from P60, resulting in 19,501 identified transcripts (19,427 genes; Sharma et al., 2015). We selected transcripts with expression data available for at least one sample ≥1 RPKM (reads per kilobase per million) using 21 samples (triplicates of each cell type: microglia, astrocyte, three divisions of oligodendrocytes and two from cortical neurons), totaling 14,109 transcripts (14,102 genes). The combined datasets resulted in 14,526 genes expressed in the brain samples.
2.3.2. Tissue‐specific gene expression
Our analysis included data from a database composed of 842 Ensembl genes and their expression in 13 tissues obtained from 103 C57BL/6 mice samples and 348 human samples. These samples demonstrated similar tissue‐specific expression patterns across mouse and human genes (Lin et al., 2014). The genes were reannotated, resulting in 840 EntrezGenes across the following tissues: adrenal (2 genes), brain (162 genes), kidney (44 genes), liver (89 genes), lung (14 genes), mammary (2 genes), muscle (51 genes), ovary (2 genes), sigmoid (11 genes), spleen (4 genes), small bowel (15 genes), stomach (11 genes), and testis (430 genes).
2.3.3. Cell type specificity measurement
The cell‐type‐specific gene expression patterns were measured using an adapted Shannon's entropy calculation (Schug et al., 2005) in a published dataset containing gene expression data from the mouse brain samples (Sharma et al., 2015). Only transcripts that showed expression in at least two of the three replicates, in no less than one cell type, were considered for the analysis. We used the median expression value in each cell type. Only transcripts with H ≤ 2 and Q value ≤7 were considered for further analysis. Low H values indicated that the transcript is expressed in few cell types, and low Q values indicated that it was highly expressed in a specific cell type. This analysis resulted in 2,907 genes with a cell‐type‐specific pattern (lowest Q values) for the following cell types: adult microglia (821 genes), cortical neurons div 05 (283 genes) and division 10 (595 genes), oligodendrocytes div 01 (107 genes), division 2.5 (100 genes), division 04 (180 genes), and astrocytes (821 genes).
2.3.4. Gene expression datasets from human brains
The BrainSpan Atlas database was used to select genes that demonstrated at least 1 RPKM in any brain region, in at least one sample (Miller et al., 2014). It included 383 samples of 32 individuals (19 males and 13 females) from 8 postconceptional weeks (PCW) to 7 years old. Gene annotation was performed using BioMart (Guberman et al., 2011) and org.Hs.eg.db (Carlson, 2015), both in R (R Core Team, 2015). In total, 27,636 Ensembl transcripts (20,283 EntrezGenes) were considered for the analyses. To study the differences between sexes, we performed an enrichment analysis between the sex differentially expressed genes and SOX3/SRY target genes. These sex differentially expressed genes (FDR < 0.01, fold difference > 2; 122 with higher expression in males and 37 with higher expression in females) were identified by Kang et al. (2011) in an analysis of human brain samples from 16 brain regions collected from 31 males and 26 females between the ages of 5.7 PCW and 82 years.
To investigate the differences in DNA methylation during brain development, we used previously published methylation data from 100 males and 79 females brain samples. These samples aged from 23 to 184 postconceptional days, interrogating 8,059 CpG sites modulated during brain development (Spiers et al., 2015). There were 521 CpG sites located on autosomes (from 8,059 probes, Bonferroni‐corrected p value<1.22E−7), consisting of 389 CpG sites located next to unique genes and 118 in intergenic regions. All coordinates from the CpG sites were annotated using the human genome reference (hg19).
2.3.5. ASD risk genes and variants
For the ASD databases, we used the SFARI Gene database (Abrahams et al., 2013), with four different levels of stringency thresholds: scores 1 and 2 with genes associated with syndromes (n = 84), scores 1 and 2 without genes associated with syndromes (n = 56), scores 1, 2, 3 and 4 with genes associated with syndromes (n = 665) and scores 1, 2, 3 and 4 without genes associated with syndromes (n = 598). In addition, we included an exome dataset with 12,510 de novo mutations found in individuals with autism (9,293 mutations) and their unaffected sibling controls (3,217 mutations), as reported by Sanders and collaborators (Sanders et al., 2015). This dataset contained 4,620 samples from 3,152 affected individuals and 1,468 controls from the Simon Simplex Collection (SSC) and the Autism Sequencing Consortium (ASC). We also used a dataset from the PGC‐ASD consortium (Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium, 2015) containing GWAS data from 5,305 ASD‐diagnosed cases and 5,305 pseudo controls of European descent (PGC ASD Euro). To select the SNPs for our study, we used a previous study that calculated the polygenic risk for ASD (AUT8; Cross‐Disorder Group of the Psychiatric Genomics Consortium, 2013). We used only those SNPs that presented an association with a p value ≤10E−5, resulting in 20 genes and 10 genes. Additionally, we utilized an exome dataset of genes associated with developmental disorders and intellectual disability through de novo variant enrichment (n = 94; DDD; McRae et al., 2017). We used 140,556 de novo mutations from whole genome sequencing (Yuen et al., 2017) from simplex and multiplex families composed of 2,062 males (1,388 probands and 674 affected siblings) individuals and 558 females (353 probands and 205 affected siblings).
2.3.6. Residual variation intolerance score (RVIS)
To select genes intolerant to mutation, we used the RVIS applied to the ESP6500 population (Petrovski, Wang, Heinzen, Allen, & Goldstein, 2013). Genes within the 25th percentile, representing the most intolerant genes with three cut‐offs, according to MAF (Minor Allele Frequency; MAF ≤ 0.01, 0.01 < MAF ≤ 0.1, or 0.1 < MAF ≤ 1), were selected, resulting in 473, 350, and 332 genes, respectively.
2.4. Enrichment analysis
To perform the MSET analysis (Eisinger, Saul, Driessen, & Gammie, 2013), we selected the background genes from chip annotation (GPL18145‐6740). Genes were reannotated using RefSeq genes from UCSC (Rosenbloom et al., 2014). In total, chip annotation represented 27,530 RefSeq genes corresponding to 18,878 EntrezGenes in mouse UCSC mm9. For the ChIP‐seq analysis, we used the entire background of UCSC mm9, resulting in 24,467 EntrezGenes. Using a human model, genes from ChIP‐chip were annotated using RefSeq UCSC hg19, corresponding to 20,378 EntrezGenes. All analyses were performed with 10,000 permutations to calculate the p value using the MSET algorithm. To perform permutations, the algorithm resamples 10,000 times the same number of entries using the entire background, empirically calculating the p value, according to the number of random sets that reached the same or greater numbers of overlaps divided by the number of permutations (Eisinger et al., 2013). The empirical p value were adjusted for multiple testing using FDR in R (R Core Team, 2015). For functional enrichment analyses, we used WebGestalt (Wang, Duncan, Shi, & Zhang, 2013) with the whole genome as the background. We considered those categories with at least five genes and an adjP ≤ 0.05 (after the Benjamini–Hochberg correction) to be significant. We generated word clouds using https://www.wordclouds.com/.
2.5. STRING analysis and TF annotation
STRING (Szklarczyk et al., 2017) analyses were performed using either SRY or SOX3 proteins as seeds, with up to 100 neighbors. All types of evidence were included, with a minimum confidence level of 0.4. Transcription binding factor data were obtained using the TF checkpoint database (Chawla, Tripathi, Thommesen, Lægreid, & Kuiper, 2013), which includes nine different databases composed of 3,479 TFs. To assign the motif domain in TF enrichment analysis to the proper TF, we used the UniProt domain or the TF name and searched for it in the TRANSFAC v7.0 public database (Matys et al., 2006).
2.6. Coexpression network analysis
We used previously published male prefrontal cortex samples consisting of 12 ASD‐affected and 15 controls (CON) from GSE28521, containing the expression data of 10,769 probes (9,742 unique EntrezGenes; Voineagu et al., 2011). Only SRY/SOX3 related genes expressed in the brain dataset were selected, resulting in 1,262 probes. Probes representing the same genes were averaged (n = 1,097), and those expressed in at least 50% of the samples from each group were selected, resulting in 1,036 genes. The Weighted Gene Coexpression Network Analysis (WGCNA) was applied to the expression data to build an unsigned coexpression network to characterize the features of the coexpressed gene modules (Langfelder & Horvath, 2007, 2008; Langfelder, Luo, Oldham, & Horvath, 2011). Further methodological details are available in the Supporting Information .
3. RESULTS
3.1. Selecting putative SRY and/or SOX3 target genes in mouse dataset
Datasets from ChIP‐chip SRY (Li et al., 2014) and ChIP‐seq SOX3 (Bergsland et al., 2011; McAninch & Thomas, 2014) from mice resulted in 3,738 and 7,610 targeted genes, respectively, with an overlap of 1,253 genes. Thus, we analyzed three datasets: SRY‐exclusive (2,485 genes), SOX3‐exclusive (6,357 genes) and SRY/SOX3 dataset (1,253 genes; Table 1, Figure 1a). A detailed flowchart with all steps involved in these analyses is available in the Supporting Information (Figure S1).
Table 1.
Mouse genes target by SOX3 and SRY
| Protein | Method | Tissue or cell | Number of peaks | Closest RefSeq annoation | Entrez gene | Total genes | References |
|---|---|---|---|---|---|---|---|
| SRYa | ChIP‐chip | Gonad cells from embrionic 13.5 | 3,083 | 5,704 | 3,738 | 3,738 | Li et al. (2014) |
| SOX3b | ChIP‐seq | ES cells (E14.1) | 9,719 | 7,984 | 5,307 | 7,610 | (Bergsland et al. (2011) |
| NPCs cells from ES cell monolayer | 8,067 | 7,523 | 5,150 | McAninch and Thomas (2014) |
ChIP‐chip background (27,530 RefSeq and 18,878 entrez gene).
Mouse background UCSC mm9 (34,917 RefSeq and 24,467 entrezgene).
Figure 1.
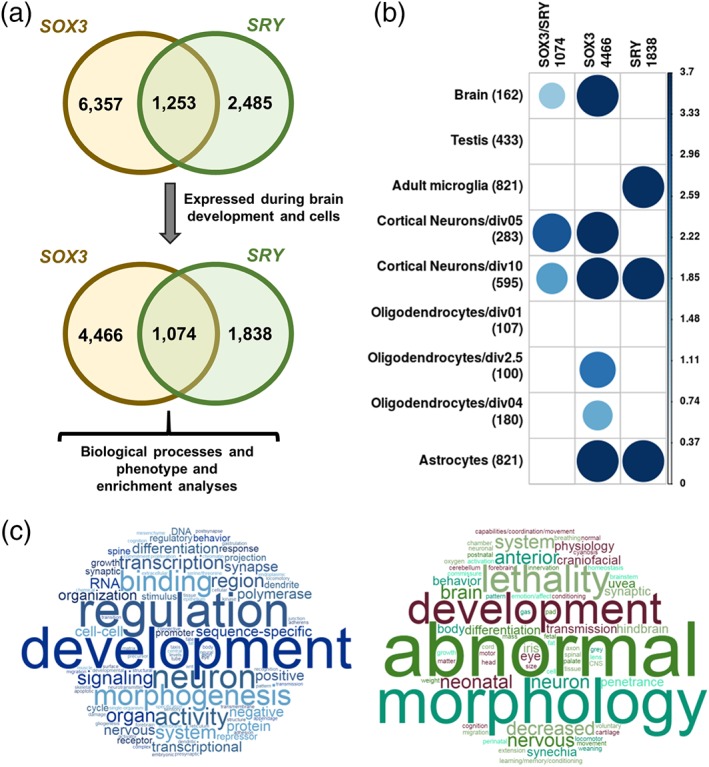
Overview of the analysis in mouse model. (a) Briefly, analysis from SOX3 ChIP‐seq (Bergsland et al., 2011; McAninch & Thomas, 2014) and SRY ChIP‐chip (Li et al., 2014) to identify target genes regulated by both protein. Genes were filtered out, according to brain tissue expression (Sharma et al., 2015; Thompson et al., 2014), resulting in 4,466 SOX3 target genes, 1,838 SRY target genes and 1,074 SOX3/SRY target genes. (b) To investigate whether these datasets related to brain, enrichment analysis was performed in several databases, with the respective number of genes shown in the rows using MSET (Eisinger et al., 2013), with 10,000 permutations and FDR correction. The color scale represents −log10(FDR). (c) GO analysis (left panel) and phenotype (right panel) analysis was performed using Webgestalt (Wang et al., 2013) and was considered to be relevant those categories with at least 10 genes and p value adjusted by the Benjamini–Hochberg p value <0.05, represented by a word cloud. This last analysis revealed several phenotypes relevant to ASD. The phenotypes related to abnormal nervous development, growth and size, craniofacial abnormalities, behavior and neurological phenotype, mortality during fetal or postnatal period, normal phenotype, vision/eye phenotype, and abnormal oxygen level [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Selecting putative SRY and/or SOX3 target genes during brain development
To maximize the probability of studying only those genes related to brain development, SRY and SOX3 target genes were selected from expression databases of prenatal and postnatal periods of brain development (Thompson et al., 2014) and specific cell types (microglia, astrocytes, oligodendrocytes and cortical neurons; Sharma et al., 2015). From 10,095 genes, 7,378 (73%) were expressed in at least one brain sample. From this number, 4,466 represented 70% of all targeted by SOX3, 1,838 (74%) by SRY, and 1,074 (86%) by both SOX3 and SRY (SOX3/SRY; Supporting Information Table S2). As the data from ChIP‐chip SRY (Li et al., 2014) came from embryonic gonadal cells and not NPCs as the SOX3, we also performed an enrichment analysis with tissue specific pattern of large scale RNA‐seq datasets from 13 tissues (Lin et al., 2014), using brain and testis genes. The analysis showed that for the SOX3/SRY set, only the brain tissue‐specific dataset was overrepresented, with 15 genes (9.26%, FDR [false discovery rate] = 3.70E−02, FE [Fold Enrichment] = 1.79); the testis tissue‐specific had an overlap of one gene (0.02%, FDR = 1, FE = 0.055). Similar results were observed for the SOX3 exclusive gene set with 54 genes (33.33%, FDR = 2.00E−04, FE = 1.83) for the brain and 26 genes (6%, FDR = 1, FE = 0.337) for the testes. No enrichment was observed for SRY exclusive gene sets (Supporting Information Table S3).
3.3. Were putative SRY and/or SOX3 targets enriched in the genes with nervous system cell‐specific expression patterns?
We evaluated the expression levels in brain cell types. Cell specific expression patterns were defined by Shannon entropy (Schug et al., 2005), using a dataset of 19,501 transcripts expressed in astrocytes, oligodendrocytes, adult microglia and cortical neurons (Sharma et al., 2015). Only transcripts that reported entropy (H) below or equal to two were selected. Each transcript was then assigned to the tissue that had the lowest Q value, identifying those transcripts preferentially expressed in a cell type but not exclusively expressed in a specific cell type (Supporting Information Figure S2).
Comparing the cell‐specific expression to the SOX3/SRY dataset, there were an overrepresentation in two groups of cortical neurons (division 05: FDR = 7.00E−04, FE = 2.69 and division 10: FDR = 8.80E−03, FE = 1.54). For the SRY exclusive target genes, adult microglia FDR = 2.00E−04, FE = 1.68), cortical neurons (division 10: FDR = 2.00E−04, FE = 1.59) and astrocytes (FDR = 2.00E−04, FE = 1.47) were overrepresented. For the SOX3 exclusive target genes, overrepresentation was found for the cortical neurons (division 05: FDR = 2.00E−04, FE = 1.99 and division 10: FDR = 1.00E−04, FE = 1.64), oligodendrocytes (division 2.5: FDR = 1.80E−03, FE = 1.7 and division 4: FDR = 1.37E−02, FE = 1.51) and astrocytes (FDR = 2.00E−04, FE = 1.47; Figure 1b, Supporting Information Table S4).
3.4. Were putative SOX3 and/or SRY targets enriched for biological processes related to neurodevelopment?
To investigate the biological function of the SOX3/SRY dataset, we performed an enrichment analysis within the biological processes from gene ontology, which revealed several categories related to brain development, including dendrite development (30 genes, R = 2.41, adjP = 1.42E−03), neuron migration (23 genes, R = 2.5, adjP = 2.86E−03), axon development (48 genes, R = 1.77, adjP = 4.10E−03) and synapse organization (29 genes, R = 1.89, adjP = 1.90E−02). The other categories were skeletal system development (52 genes, R = 1.79, adjP = 2.47E−03), cell–cell signaling by wnt (40 genes, R = 1.73, adjP = 1.55E−02) and embryonic organ development (46 genes, R = 1.58, adjP = 2.53E−02; Supporting Information Table S5). While enrichment analysis from SOX3 dataset was similar to the one from the SOX3/SRY dataset (Supporting Information Table S6), the SRY dataset showed only the cell‐substrate junction category (64 genes, R = 1.53, adjP = 4.49E−02) enriched.
Moreover, the SOX3/SRY dataset phenotype analysis showed enrichment for abnormal nervous system physiology (162 genes, R = 1.63E−05, adjP = 1.63E−05), abnormal brain development (70 genes, R = 1.77, adjP = 1.77E−03), abnormal synaptic transmission (72 genes, R = 1.79, adjP = 1.16E−03), abnormal axon extension (12 genes, R = 3.15, adjP = 3.69E−02), abnormal neuronal migration (16 genes, R = 2.62, adjP = 3.69E−02) and craniofacial phenotype (92 genes, R = 1.41, adjP = 3.69E−02). Phenotypes related to behavior were also identified in the abnormal cued conditioning behavior (19 genes, R = 3.01, adjP = 6.22E−03) and abnormal emotion/affect behavior (58 genes, R = 1.69, adjP = 1.15E−02) categories (Supporting Information Table S7, Figure 1c).
3.5. Identification of putative human SRY and/or SOX3 target genes
To investigate these findings in a human model, we identified orthologous genes between the mouse and human genome and improved our original dataset with additional datasets of SRY ChIP‐chip assays in humans and rats (Supporting Information Table S8). We verified that SRY and SOX3 are conserved among species by aligning the SOX3 HMG‐box domains from mice and humans, showing that both domains were identical. SRY was highly conserved among species, with 54 identical residues (78%) and four similar residues (6%; Figure 2a, Supporting Information Figure S3).
Figure 2.
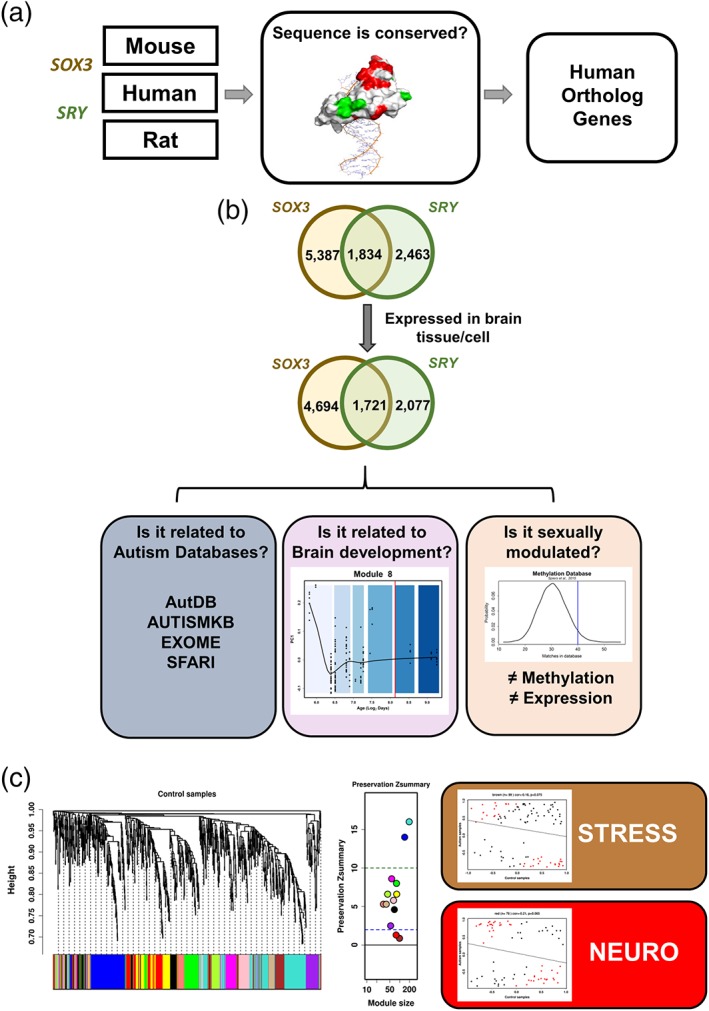
Summary of designed experiments in human model. (a) Chromatin immunoprecipitation of SOX3 and SRY from mouse, human, and rat models were selected in the literature. First, HMG‐box DNA binding motif sequence conservation was evaluated among the species; after confirming it, human orthologues genes were identified. (b) SOX3 and SRY targets (first Venn diagram) were filtered for brain expression (second Venn diagram), then three main experiments were performed: one regarding ASD databases (purple box), other brain development (lilac box) and, the last regarding differential sex modulation (pink box). (c) Preservation module coexpression analysis of 1,721 SOX3/SRY set comparing ASD and CON showed that least preserved modules brown and red were primarily related to the stress response and neurogenesis, respectively [Color figure can be viewed at wileyonlinelibrary.com]
The identification of orthologous genes between species showed 4,297 unique genes with SRY binding sites (Bhandari et al., 2012; Jin et al., 2007; Li et al., 2014) and 7,221 unique genes with SOX3 binding sites (Bergsland et al., 2011; McAninch & Thomas, 2014) for humans. From these genes, 3,798 (88%) SRY and 6,415 (89%) and SOX3 target genes, respectively, were expressed during brain development (Figure 2b, Supporting Information Table S9). There was an overlap of 1,721 genes between SOX3 and SRY (SOX3/SRY set), and 2,077 and 4,694 genes were targeted exclusively by SRY (SRY set) and SOX3 (SOX3 set), respectively. From the SOX3/SRY set, 19 genes are located in the X chromosome (1%); from the SRY set, 73 and 2 genes are located on the X (3.5%) and Y (0.1%) chromosomes; and from the SOX3 set, 122 (2.6%) and three (0.06%) genes are located on X and Y chromosomes, respectively.
As the DNA binding domains from both proteins were conserved, we searched for evidence of whether the three gene sets had the same regulatory modulation mechanism among the species. We used the poised H3K4me3/H3K27me3 bivalent epigenetic state in male germ cells from five mammalian species; this state is closely related to the gene regulatory programs that govern somatic development (Lesch, Silber, McCarrey, & Page, 2016). Poised genes are those that present H3K4me3 and H3K27me3 marks in a window of 4 kb from TSS (≥0.5 reads per million) and expression ≤5 FPKM. This definition identified 405 poised promoter genes among the species and 1,929 between the mice and humans (Lesch et al., 2016).
Thus, we sought to analyze the SOX3/SRY set independently of the species by performing an enrichment analysis to identify which genes from the SRY/SOX3 set maintain marks that are conserved across species. This analysis showed that the SOX3/SRY set was overrepresented in 78 (19%, FDR = 1.00E−04, FE = 2.31) genes conserved across five species and 310 (16%, FDR = 1.00E−04, FE = 1.93) genes conserved between mice and humans. Similar enrichment results were observed for the SOX3 set. However, only the poised promoters conserved in mouse and human were enriched for the SRY set (Supporting Information Table S10 and Figure S4).
Functional annotation analyses of the SOX3/SRY set identified overrepresentation for the immune system, metabolism, nervous system development, and gene expression and nervous system component regulation, such as synapses, axons and the neuronal spine (Table 2 and Supporting Information Tables S11 and S12). Similar results were observed for the SOX3 exclusive gene set (Supporting Information Table S13), and no enrichment was found for the SRY exclusive gene set. A summary of the designed experiments is shown in Figure 2, and a detailed flowchart of the analyses is available in the Supporting Information (Figure S5).
Table 2.
Functional categories enriched according to 1,721 SOX3/SRY target genes expressed in brain
| GO type | GO ID | GO category name | C | O | E | R | p |
|---|---|---|---|---|---|---|---|
| Gene ontology enrichment analysis | |||||||
| BP | GO:0007399 | Nervous system development | 1,724 | 298 | 174.17 | 1.71 | 1.19E−19 |
| BP | GO:0010468 | Regulation of gene expression | 3,344 | 492 | 337.83 | 1.46 | 1.79E−19 |
| BP | GO:0060255 | Regulation of macromolecule metabolic process | 4,151 | 581 | 419.36 | 1.39 | 2.33E−19 |
| BP | GO:0022008 | Neurogenesis | 1,140 | 218 | 115.17 | 1.89 | 2.33E−19 |
| BP | GO:0048699 | Generation of neurons | 1,073 | 209 | 108.4 | 1.93 | 2.33E−19 |
| BP | GO:0006351 | Transcription, DNA‐dependent | 3,220 | 470 | 325.3 | 1.44 | 2.95E−18 |
| BP | GO:0045595 | Regulation of cell differentiation | 1,012 | 196 | 102.24 | 1.92 | 2.95E−18 |
| BP | GO:0031326 | Regulation of cellular biosynthetic process | 3,325 | 482 | 335.91 | 1.43 | 2.95E−18 |
| BP | GO:0006355 | Regulation of transcription, DNA‐dependent | 2,876 | 430 | 290.55 | 1.48 | 2.95E−18 |
| BP | GO:2000112 | Regulation of cellular macromolecule biosynthetic process | 3,119 | 458 | 315.1 | 1.45 | 3.08E−18 |
| BP | GO:2001141 | Regulation of RNA biosynthetic process | 2,897 | 432 | 292.67 | 1.48 | 3.20E−18 |
| BP | GO:0048646 | Anatomical structure formation involved in morphogenesis | 1,594 | 273 | 161.03 | 1.7 | 3.29E−18 |
| BP | GO:0030182 | Neuron differentiation | 990 | 192 | 100.02 | 1.92 | 4.91E−18 |
| BP | GO:0048468 | Cell development | 1,461 | 255 | 147.6 | 1.73 | 5.24E−18 |
| MF | GO:0044212 | Transcription regulatory region DNA binding | 328 | 73 | 31.33 | 2.33 | 1.10E−10 |
| MF | GO:0001067 | Regulatory region nucleic acid binding | 336 | 74 | 32.1 | 2.31 | 1.10E−10 |
| MF | GO:0005488 | Binding | 11,955 | 1,239 | 1,142 | 1.08 | 2.68E−10 |
| CC | GO:0000785 | Chromatin | 296 | 62 | 27.48 | 2.26 | 1.80E−08 |
| CC | GO:0045202 | Synapse | 489 | 79 | 45.39 | 1.74 | 6.84E−06 |
| CC | GO:0030425 | Dendrite | 355 | 57 | 32.95 | 1.73 | 2.00E−04 |
| CC | GO:0031252 | Cell leading edge | 257 | 44 | 23.86 | 1.84 | 3.00E−04 |
| CC | GO:0030424 | Axon | 286 | 47 | 26.55 | 1.77 | 5.00E−04 |
| CC | GO:0044309 | Neuron spine | 159 | 30 | 14.76 | 2.03 | 6.00E−04 |
| CC | GO:0042995 | Cell projection | 1,230 | 152 | 114.18 | 1.33 | 6.00E−04 |
| CC | GO:0043197 | Dendritic spine | 159 | 30 | 14.76 | 2.03 | 6.00E−04 |
| CC | GO:0016607 | Nuclear speck | 148 | 28 | 13.74 | 2.04 | 1.20E−03 |
| CC | GO:0044456 | Synapse part | 370 | 54 | 34.35 | 1.57 | 3.40E−03 |
| CC | GO:0097060 | Synaptic membrane | 215 | 35 | 19.96 | 1.75 | 3.70E−03 |
| CC | GO:0030027 | Lamellipodium | 121 | 23 | 11.23 | 2.05 | 3.70E−03 |
| CC | GO:0014069 | Postsynaptic density | 116 | 22 | 10.77 | 2.04 | 4.60E−03 |
| CC | GO:0044327 | Dendritic spine head | 116 | 22 | 10.77 | 2.04 | 4.60E−03 |
| KEGG pathway analysis | |||||||
| KEGG | Systemic lupus erythematosus | 136 | 40 | 5.43 | 7.37 | 5.41E−22 | |
| KEGG | Pathways in cancer | 326 | 54 | 13.01 | 4.15 | 2.50E−17 | |
| KEGG | Wnt signaling pathway | 150 | 35 | 5.99 | 5.85 | 3.99E−16 | |
| KEGG | MAPK signaling pathway | 268 | 42 | 10.69 | 3.93 | 6.74E−13 | |
| KEGG | Focal adhesion | 200 | 32 | 7.98 | 4.01 | 3.23E−10 | |
| KEGG | Neurotrophin signaling pathway | 127 | 23 | 5.07 | 4.54 | 1.21E−08 | |
| KEGG | Calcium signaling pathway | 177 | 27 | 7.06 | 3.82 | 1.87E−08 | |
| KEGG | Regulation of actin cytoskeleton | 213 | 30 | 8.5 | 3.53 | 1.87E−08 | |
| KEGG | Axon guidance | 129 | 22 | 5.15 | 4.27 | 5.59E−08 | |
| KEGG | Metabolic pathways | 1,130 | 75 | 45.09 | 1.66 | 3.58E−05 | |
| KEGG | Leukocyte transendothelial migration | 116 | 16 | 4.63 | 3.46 | 3.74E−05 | |
| KEGG | Viral myocarditis | 70 | 12 | 2.79 | 4.3 | 4.27E−05 | |
Abbreviations. BP = biological process; C = number of reference genes in the category; CC = cellular component; E = expected number in the category; MC = molecular component; O = number of genes in the gene set and also in the category; p = p value adjusted by multiple test adjustment (FDR); R = ratio of enrichment.
3.6. Do putative SRY and/or SOX3 target genes have male‐biased expression, and were and were they specifically modulated during brain development?
Using MSIgDB/WebGestalt, we found that genes from the SOX3/SRY set were overrepresented in several TFs, including sex hormone receptors (Supporting Information Table S14). Thus, a set of genes were directly regulated by androgens (AR, 65 genes, 3%, p < 1E−05) and estrogen (ER, 112 genes, 6.5%, p < 1E−10); this set of genes totaled 167 genes that are directly targeted by AR and/or ER (i.e., 9.8%; Supporting Information Table S15).
To determine the involvement of the SRY/SOX3 set in neurodevelopment, we used modules of genes coexpressed during neocortical development (Parikshak et al., 2013). We identified nine enriched modules, one of which showed enrichment for ER targets (Table 3 and Figure 3). Thus, AR (n = 65) and ER (n = 112) target genes were tested as two independent datasets. Comparing both analyses, eight modules were enriched only in the SOX3/SRY set, four (M3, M5, M11 and M14) were all composed of genes that have their highest expression levels during the early (8–12 postconception week, PCW) period and one (M2) during the mid‐fetal period (13–17 PCW). These genes were involved with biological processes that are primarily related to zinc ion binding, nucleic acid metabolic process, translational elongation and viral transcription (Supporting Information Table S16). The other three modules (M1, M16 and M18) demonstrated higher expression levels in the postbirth period, with M1 and M18 showing expression in the early periods of gestation. Only the M8 module was enriched for the SOX3/SRY set and ER, and it was composed of genes that have their highest expression levels during the early fetal period (8–12 PCW; Supporting Information Figure S6).
Table 3.
Enrichment analysis of specific neurodevelopmental modules
| Modulesa | SOX3/SRY BRAIN | SOX3 | SRY | AR | ER | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # genes | FE | p value | FDR | # genes | FE | p value | FDR | # genes | FE | p value | FDR | # genes | FE | p value | FDR | # genes | FE | p value | FDR | |
| M1 | 75 | 1.47 | 8.00E−04 | 2.72E−03 | 217 | 1.58 | 1.00E−04 | 2.43E−04 | 56 | 0.952 | 6.77E−01 | 7.67E−01 | 5 | 2.59 | 4.61E−02 | 3.92E−01 | 2 | 6.00E−01 | 8.55E−01 | 1.00E+00 |
| M2 | 78 | 1.29 | 1.15E−02 | 2.79E−02 | 217 | 1.33 | 1.00E−04 | 2.43E−04 | 78 | 1.16 | 9.27E−02 | 1.58E−01 | 2 | 0.876 | 6.75E−01 | 1.00E+00 | 4 | 1.02E+00 | 5.56E−01 | 1.00E+00 |
| M3 | 115 | 1.66 | 1.00E−04 | 5.67E−04 | 274 | 1.41 | 1.00E−04 | 2.43E−04 | 104 | 1.3 | 3.10E−03 | 8.78E−03 | 1 | 0.383 | 9.27E−01 | 1.00E+00 | 4 | 8.90E−01 | 6.69E−01 | 1.00E+00 |
| M4 | 30 | 1.02 | 4.71E−01 | 6.16E−01 | 94 | 1.27 | 6.10E−03 | 9.43E−03 | 55 | 1.72 | 1.00E−04 | 5.67E−04 | 0 | 0 | 1.00E+00 | 1.00E+00 | 0 | 0.00E+00 | 1.00E+00 | 1.00E+00 |
| M5 | 80 | 1.33 | 4.90E−03 | 1.39E−02 | 239 | 1.49 | 1.00E−04 | 2.43E−04 | 80 | 1.15 | 1.05E−01 | 1.62E−01 | 5 | 2.2 | 7.38E−02 | 4.18E−01 | 2 | 5.11E−01 | 9.06E−01 | 1.00E+00 |
| M6 | 18 | 0.697 | 9.64E−01 | 1.00E+00 | 70 | 1.01 | 4.81E−01 | 6.29E−01 | 31 | 1.07 | 3.75E−01 | 4.55E−01 | 0 | 0 | 1.00E+00 | 1.00E+00 | 0 | 0.00E+00 | 1.00E+00 | 1.00E+00 |
| M8 | 33 | 2 | 1.00E−04 | 5.67E−04 | 66 | 1.46 | 3.00E−04 | 6.38E−04 | 29 | 1.49 | 1.98E−02 | 4.81E−02 | 0 | 0 | 1.00E+00 | 1.00E+00 | 6 | 5.60E+00 | 7.00E−04 | 1.19E−02 |
| M9 | 17 | 0.963 | 5.98E−01 | 7.26E−01 | 42 | 0.894 | 8.16E−01 | 9.79E−01 | 18 | 0.888 | 7.37E−01 | 7.83E−01 | 1 | 1.5 | 4.77E−01 | 1.00E+00 | 0 | 0.00E+00 | 1.00E+00 | 1.00E+00 |
| M10 | 2 | 0.789 | 7.26E−01 | 8.23E−01 | 4 | 0.617 | 9.21E−01 | 9.79E−01 | 1 | 0.35 | 9.48E−01 | 9.48E−01 | 0 | 0 | 1.00E+00 | 1.00E+00 | 0 | 0.00E+00 | 1.00E+00 | 1.00E+00 |
| M11 | 63 | 1.59 | 1.00E−04 | 5.67E−04 | 134 | 1.25 | 1.90E−03 | 3.23E−03 | 53 | 1.17 | 1.25E−01 | 1.77E−01 | 0 | 0 | 1.00E+00 | 1.00E+00 | 6 | 2.33E+00 | 4.91E−02 | 2.09E−01 |
| M12 | 11 | 0.366 | 1.00E+00 | 1.00E+00 | 46 | 0.57 | 1.00E+00 | 1.00E+00 | 57 | 1.78 | 1.00E−04 | 5.67E−04 | 1 | 0.881 | 6.79E−01 | 1.00E+00 | 0 | 0.00E+00 | 1.00E+00 | 1.00E+00 |
| M13 | 67 | 1.12 | 1.75E−01 | 2.98E−01 | 199 | 1.26 | 1.00E−04 | 2.43E−04 | 98 | 1.44 | 1.00E−04 | 5.67E−04 | 2 | 0.884 | 6.68E−01 | 1.00E+00 | 3 | 1.28E+00 | 3.54E−01 | 1.00E+00 |
| M14 | 53 | 1.34 | 2.01E−02 | 4.27E−02 | 106 | 0.912 | 8.73E−01 | 9.79E−01 | 73 | 1.63 | 1.00E−03 | 3.40E−03 | 1 | 0.64 | 7.77E−01 | 1.00E+00 | 1 | 3.89E−01 | 9.26E−01 | 1.00E+00 |
| M15 | 28 | 1.12 | 2.96E−01 | 4.19E−01 | 79 | 1.15 | 9.10E−02 | 1.29E−01 | 38 | 1.34 | 3.99E−02 | 8.48E−02 | 1 | 1.06 | 6.14E−01 | 1.00E+00 | 4 | 2.45E+00 | 8.61E−02 | 2.93E−01 |
| M16 | 50 | 1.32 | 2.28E−02 | 4.31E−02 | 137 | 1.34 | 1.00E−04 | 2.43E−04 | 53 | 1.21 | 8.14E−02 | 1.54E−01 | 5 | 3.51 | 1.30E−02 | 2.21E−01 | 6 | 2.44E+00 | 3.94E−02 | 2.09E−01 |
| M17 | 73 | 1.09 | 2.44E−01 | 3.77E−01 | 248 | 1.39 | 1.00E−04 | 2.43E−04 | 86 | 1.12 | 1.39E−01 | 1.82E−01 | 2 | 0.79 | 7.21E−01 | 1.00E+00 | 5 | 1.15E+00 | 4.41E−01 | 1.00E+00 |
| M18 | 89 | 1.43 | 4.00E−04 | 1.70E−03 | 202 | 1.2 | 1.50E−03 | 2.83E−03 | 102 | 1.38 | 6.00E−04 | 2.55E−03 | 1 | 0.424 | 9.13E−01 | 1.00E+00 | 9 | 2.22E+00 | 2.04E−02 | 1.73E−01 |
#: Number of overlapping genes. FE = fold enrichment.
Bold: p value (10,000 permutations) or FDR ≤ 0.05.
Results from Parikshak et al. (2013).
Figure 3.
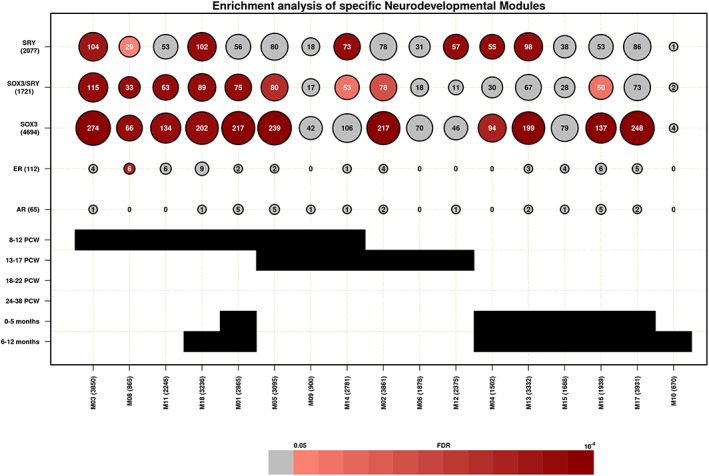
Enrichment analysis in neurodevelopment modules: Each line represents a gene set with the respective number of genes or a gestation period and each column is represented by neurodevelopmental gene modules with respective number of genes (Parikshak et al., 2013), totaling 17 modules. The circles are proportional with gene overlap in each dataset, and the numbers represent the number of overlapping genes. Colors are proportional to significant enrichment, those scaled in red were those that reached the threshold of FDR < 0.05 [Color figure can be viewed at wileyonlinelibrary.com]
In the SOX3 set, a total of 11 modules were enriched. Three new modules were enriched (M4, M13 and M17), while the other eight modules were also enriched for the SOX3/SRY set. The expression level of the M17 module increases throughout the gestational period, stabilizing after birth. The M4 and M13 modules were also shared with the SRY set, with both modules having their highest expression levels after birth. Regarding the SRY set, a total of seven modules were enriched. As stated above, two modules are shared with the SOX3 set, and four are also present in the SOX3/SRY gene set (M3, M8, M14 and M18), all showing the highest expression level in the first periods of gestation, with M18 also peaking after birth; one module (M12) is exclusive to this set. The highest expression level of the M12 module was during the mid‐fetal period, and its biological processes are primarily related to cilium, cell projection and axoneme. Most enriched modules that are exclusive to the SOX3 and SRY sets are characterized by having their highest expression levels after birth. These modules were related to synapses signaling, ion channel transmembrane and oxidative phosphorylation (Supporting Information Table S16).
We also compared the SOX3/SRY set to 37 females and 123 males genes that showed sex biased expression during brain development (Kang et al., 2011). There was no overlap between the female‐biased gene set and the SOX3/SRY set. However, the male‐biased gene set was enriched in the SOX3/SRY set (19.5%, 24/123, p < 1E−04, 10,000 pe, FE = 2.37; Figure 4a). The genes potentially regulated by AR and/or ER and that are part of the SOX3/SRY set were not enriched for either set, with only two overlapping genes (ZIC1 ‐ targeted by ER and ZNF423 targeted by AR). Using the SOX3 or SRY set, no enrichment was observed in the female or male biased expression.
Figure 4.
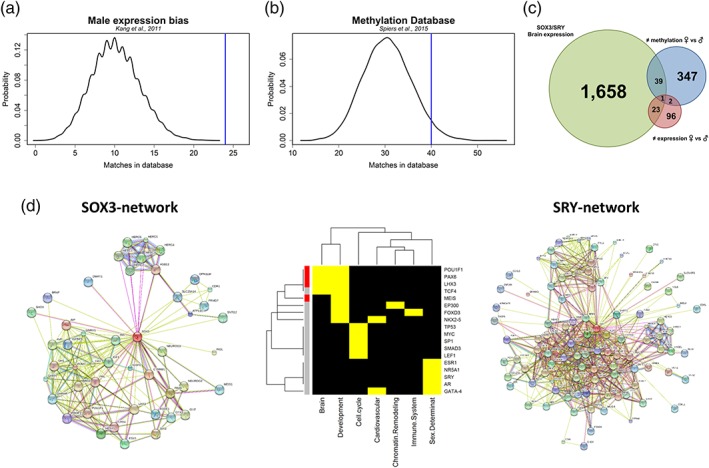
Enrichment analysis using MSET (Eisinger et al., 2013) (a) Dataset of 389 unique autosomal genes that were differentially methylated between 100 males and 79 females fetal brain samples, aged 23–184 days postconception (Spiers et al., 2015) (b) A dataset of 159 genes that were differentially expressed between 31 males and 26 females from brain samples aged 5.7 postconception weeks to 82 years (Kang et al., 2011). In both graphics, the x‐axis indicates the number of overlapping genes between datasets (Amethylation and B‐expression vs. 1,721 SOX3/SRY/brain target genes), and the y‐axis indicates the proportion of this overlap according to the number of permutations (n = 10,000). The blue line indicates the number of overlapping genes, with 1,721 target genes of SOX3/SRY, and the histogram shows the probability of density, according to the simulations. The methylation dataset had 40 genes (10%, p value = 0.0466), and the expression dataset had 24 genes (19.5%, p value <0.0001). (c) A Venn diagram of the SOX3/SRY target genes expressed in the brain (n = 1,721, green circle), showing the differentially methylated dataset (n = 389 autosomal genes, blue circle) and the male‐bias expression dataset (n = 123 genes, red circle). The size of the circle is proportional to the total number of genes. The gene in the overlapping region is COX7A2. (d) SRY and SOX3‐network. Networks of each protein SOX3 (left) and SRY (right) are shown. The heatmap shows 18 transcription factors (rows) found in the enrichment analysis of 1,721 SOX3/SRY and related functions in the columns. Yellow and black indicate the presence and absence, respectively. Four are in SOX3‐network related to brain development(red blocks), and 14 TFs are in SRYnetwork (gray blocks) and are related to sex determination, cell cycle, cardiovascular, and other function as histone regulation, brain development and immune system [Color figure can be viewed at wileyonlinelibrary.com]
3.7. Do putative SRY and/or SOX3 target genes show different methylation target sites between males and females during brain development?
We compared the SOX3/SRY, SOX3 and SRY sets to 389 unique autosomal genes that were differentially methylated between males and females during brain development (Spiers et al., 2015). No enrichment was observed in the SOX3 set, while the SRY set identified 52 genes (p = .0068, 10,000 pe, FE = 1.41) represented by 57 CpGs sites, with 32 sites more methylated in males and 25 in females. Using the SOX3/SRY set, we found an overlap of 40 genes (10%, 40/389, p = 4.66E−02, 10,000 pe, FE = 1.31) represented by 45 CpG sites, with 19 sites more methylated in males and 26 in females (Figure 4b, Supporting Information Table S17). Moreover, the genes regulated by AR and/or ER within the SOX3/SRY set were not enriched within the 389 genes, with only HOXC4, SGK1, SPTBN1, TOMM40, CHD6 and ZNF503 appearing in both lists. From the 45 differentially methylated CpG sites between sexes, twenty‐seven (60%, 26 genes) were located at precisely the same genomic coordinates of the SRY peaks, including all 19 CpGs more methylated in males. Only four CpG sites (4 genes, 9%) overlapped SOX3 peaks. We found that the SOX3 peaks (~ 328 bp) were smaller than the SRY peaks (~1,879 bp) and that they were distributed farther than 3 kb upstream or downstream from the methylated CpG sites (Supporting Information Figure S7 and Table S18).
COX7A2 is one of the genes in the overlap list (Figure 4c, Supporting Information Figure S8). It is hypomethylated and more expressed in males during brain development, with its CpG sites located within SRY and SOX3 peaks with histone marks (H3K4me3). These histone marks are regulatory marks of active promoter regions (Barski et al., 2007; Kimura, 2013), indicating that both proteins may be involved in the regulation of this region.
The different roles of SRY and SOX3 could be related to their protein partners. Using STRING (Szklarczyk et al., 2017), two networks were built using the same parameters. The SRY‐network comprised 101 genes with 649 edges (average [avg] node degree 12.9 and avg. local clustering coefficient of 0.736), whereas the SOX3‐network comprised 47 genes and 223 edges (avg node degree 9.49 and avg local clustering coefficient of 0.79). The two networks only share ten genes (i.e., CTNNB1, HERC5, HERC6, HERC1, HERC3, BRD2, HERC2, HERC4, DMRT1, SHOX). When we investigated only the TFs in each network, SRY and SOX3 showed 41 (41%) and 22 genes (47%), respectively, with five common TFs (CTNNB1, HERC1, BRD2, DMRT1, and SHOX). A previous enrichment analysis identified 18 TFs (Table 4, Supporting Information Table S14), with four in the SOX3‐network targeting 233 SOX3/SRY genes and 14 TFs in the SRY‐network targeting 821 SOX3/SRY genes. The four TFs from the SOX3‐network (MEIS1, PAX6, POU1F1, and LHX3) have known roles in neurodevelopment, particularly pituitary gland and eye development. The TFs inform that SRY‐network plays a broader role, such as sex determination, cell cycle and differentiation, immune system, and neural development (Figure 4d).
Table 4.
Transcription factor in SOX3 and SRY networks (STRING)
| Entrez gene | STRING ID | SOX3 | SRY | Unirprot_Human | Family | TF enrichment | Related function (NCBI, RefSeq) |
|---|---|---|---|---|---|---|---|
| 367 | AR | N | Y | F1D8N5,P10275 | Androgen receptor | hsa_V$AR_01, hsa_V$AR_Q2, hsa$AR_Q6 | Steroid‐hormone activated transcription factor, sex determination related gene |
| 1482 | NKX2‐5 | N | Y | P52952 | Homeobox | hsa_V$NKX25_01, hsa_V$NKX25_2 | Heart formation and development |
| 2033 | EP300 | N | Y | Q09472, Q7Z6C1 | NA | hsa_V$P300_01 | Chromatin remodeling, histone acetyltransferase |
| 2099 | ESR1 | N | Y | A8KAF4, G4XH65, P03372, Q9UBT1 | Oestrogen receptor | hsa_V$ER_Q6, hsa_V$ER_Q6_01, hsa_V$ER_Q6_02 | Hormone binding, sex development |
| 2516 | NR5A1 | N | Y | F1D8R8, Q13285 | Retinoic acid receptor | hsa_TGACCTTG_V$SF1_Q6, hsa_V$SF1_Q6 | Sex determination related gene |
| 2626 | GATA4 | N | Y | B3KUF4, P43694 | Zf‐GATA | hsa_V$GATA4_Q3 | Embryogenesis, testicular differentiation, cardiovascular |
| 4088 | SMAD3 | N | Y | P84022, Q9P0T0 | MH1 | hsa_V$SMAD3_Q6 | Cell cycle |
| 4211 | MEIS1 | Y | N | F5GYS8, O00470 | Homeobox | hsa_TGACAGNY_V$MEIS1_01, hsa_V$MEIS1_01, hsa_V$MEIS1AHOXA9_01 | Crucial for normal development |
| 4609 | MYC | N | Y | P01106 | bHLH | hsa_V$MYCMAX_01, hsa_V$MYCMAX_02, has_V$MYCMAX_03 | Cell cycle |
| 5080 | PAX6 | Y | N | F1T0F8, P26367, Q66SS1 | PAX | hsa_V$PAX6_01 | Neural development, particularly eye |
| 5449 | POU1F1 | Y | N | P28069 | POU | hsa_V$POU1F1_Q6 | Brain and pituitary development |
| 6667 | SP1 | N | Y | P08047 | zf‐C2H2 | hsa_GGGCGGR_V$SP1_Q6, hsa_V$SP1_Q6, hsa_V$SP1_Q6_01 | Cell cycle |
| 6736 | SRY | N | Y | A7WPU8, Q05066 | HMG | hsa_V$SRY_01, hsa_V$SRY_02 | Sex determination related gene |
| 6925 | TCF4 | N | Y | B3KVA4, H3BTP3, P15884 | bHLH | hsa_V$TAL1BETAITF2_01 | Nervous system development |
| 7157 | TP53 | N | Y | H2EHT1, K7PPA8, P04637, Q53GA5 | P53 | hsa_V$P53_02 | Cell cycle |
| 8022 | LHX3 | Y | N | F1T0D5, F1T0D7, F1T0D9, Q9UBR4 | Homeobox | hsa_YTAATTAA_V$LHX3_01, hsa_V$LHX3_01 | Pituitary development |
| 27022 | FOXD3 | N | Y | Q9UJU5 | Fork head | hsa_V$FOXD3_01 | Immune system |
| 51176 | LEF1 | N | Y | Q659G9, Q9UJU2 | HMG | hsa_CTTTGA_V$LEF1_Q2, hsa_CTTTGT_V$LEF1_Q2, hsa_V$LEF1_Q2, hsa_V$LEF1_Q6 | Cell differentiation, hair cell |
3.8. Were putative SRY and/or SOX3 targets overrepresented in the ASD genes?
To investigate whether our gene sets could be involved in a sex‐biased neurodevelopmental disorder, we used ASD as a model and searched for their enrichment in the SFARI databases (Abrahams et al., 2013). The SOX3/SRY and the SOX3 exclusive gene sets were enriched in the thresholds that we established for the SFARI Gene database (Table 5, Figure 5). We also compared these gene sets with whole exome sequencing datasets from families, using rare de novo variations (RDNVs; Sanders et al., 2015), GWAS data (Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium, 2015) and ASD transcriptome studies (Gupta et al., 2014; Voineagu et al., 2011; Supporting Information Table S19). These other databases used as SFARI combine both GWAS (common variants) and exome (rare variations), which have different sample power analyses. The analyses revealed that the SOX3/SRY set was enriched for all databases, except for GWAS (AUT8: 2 genes, FDR = 0.348, FE = 2.63, PGC ASD Euro: 1 gene, FDR = 0.776, FE = 0.697). Comparing all datasets to SRY or SOX3 set, only one GWAS database (PGC ASD Euro: 10 genes, FDR = 8.7E−03, FE = 2.4) was enriched for the SOX3 set, and no overrepresentation was observed in any database using SRY set. We reasoned that this result may be a consequence of the small size of these databases. However, assuming that the SOX3/SRY set represents highly conserved genes and is related to the differences between sexes in brain neurodevelopment, we expected that the data obtained from GWAS databases, which only have common variation, would not be enriched.
Table 5.
Autism related genes databases enrichment analysis
| SOX3/SRY | SOX3 | SRY | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Database | Number of genes | # genes | FE | p value | FDR | # genes | FE | p value | FDR | # genes | FE | p value | FDR |
| DDD (2017)a | 93 | 22 | 2.8 | 1.00E−04 | 1.50E−04 | 34 | 1.4 | 1.54E−02 | 1.54E−02 | 6 | 0.633 | 9.23E−01 | 9.30E−01 |
| Sanders et al. (2015)b | 5,413 | 536 | 1.19 | 1.00E−04 | 1.50E−04 | 1,593 | 1.17 | 1.00E−04 | 2.00E−04 | 573 | 1.05 | 7.87E−02 | 4.72E−01 |
| SFARI categories 1–4 (w/o syndromic genes) | 597 | 78 | 1.57 | 1.00E−04 | 1.50E−04 | 238 | 1.6 | 1.00E−04 | 2.00E−04 | 50 | 0.834 | 9.30E−01 | 9.30E−01 |
| SFARI categories 1–4 (w/ syndromic genes) | 665 | 92 | 1.66 | 1.00E−04 | 1.50E−04 | 259 | 1.56 | 1.00E−04 | 2.00E−04 | 57 | 0.853 | 9.24E−01 | 9.30E−01 |
| SFARI 1–2 (w/o syndromic genes) | 56 | 9 | 1.94 | 3.52E−02 | 3.52E−02 | 22 | 1.54 | 1.40E−02 | 1.54E−02 | 5 | 0.892 | 6.71E−01 | 9.30E−01 |
| SFARI 1–2 (w/ syndromic genes) | 84 | 14 | 2 | 9.50E−03 | 1.14E−02 | 31 | 1.43 | 1.39E−02 | 1.54E−02 | 8 | 0.946 | 6.13E−01 | 9.30E−01 |
Figure 5.
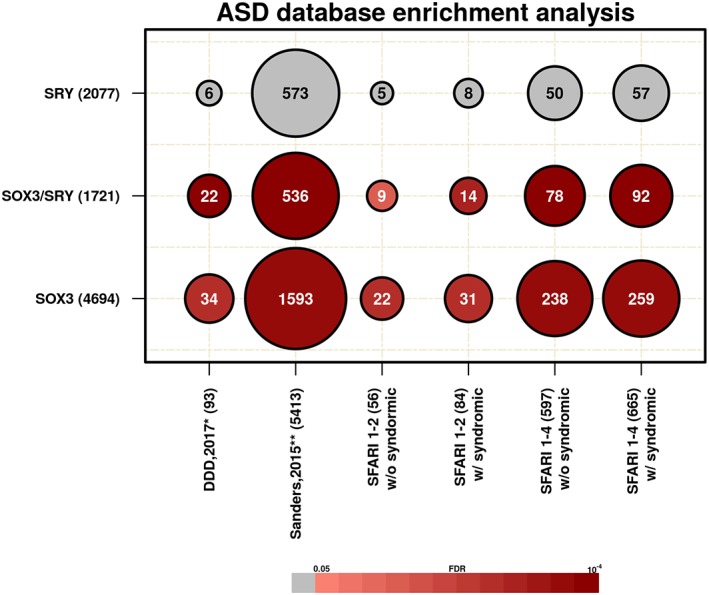
Enrichment analysis in ASD databases: Each line represents a gene set with the respective number of genes, and each column is represented by ASD databases with the respective number of genes. The circles are proportional with gene overlap in each dataset and the numbers represent the number of overlapping genes. Colors are proportional to significant enrichment, those scaled in red were those that reached the threshold of FDR <0.05. *Data from deciphering developmental disorders (DDD; McRae et al., 2017), **Data from exome study (Sanders et al., 2015) [Color figure can be viewed at wileyonlinelibrary.com]
Then, using the database of residual variation intolerance score (RVIS; Petrovski et al., 2013), we observed that all three gene sets (SOX3/SRY, SOX3 set, and SRY set) showed enrichment in genes that do not tolerate variations (only the SOX3/SRY set was enriched in the dataset of genes intolerant to mutations) and that are involved in early fetal brain development (15 genes, FDR = 3.20 E−03, FE = 2.47; Choi et al., 2016; Supporting Information Table S19).
Finally, we used the genomic coordinates from the de novo variations (DNVs) from whole genome sequencing of probands (1,392 males and 353 females ASD) and affected siblings (674 males and 206 females ASD; Yuen et al., 2017) and compared them with SRY and SOX3 peak coordinates. If the regulation of binding sites of one or both proteins is important in the context of disease, we should observe a larger overlap of DNVs under the peaks than randomly. We built 1,000 random sets of genomic coordinates from each dataset peak with the same number of peaks and chromosome distribution and compared the overlaps as a control. Using the SRY peaks, we identified an enrichment of DNVs in the male probands with 242 DNVs (p value = 0.024, Supporting Information Figure S9a) and 184 DNVs (p value = 0.017) in the male affected siblings. This result was not observed using SOX3 peaks, resulting in 111 DNVs (p value = 0.173, Supporting Information Figure S9b) using probands and 83 DNVs (p value = 0.202) in the affected siblings. In the female dataset using SRY peaks, we identified an enrichment of DNVs with 78 DNVs in probands (p value = 0.004, Supporting Information Figure S9c) and 42 DNVs in affected siblings (p value = 0.682). In contrast, for SOX3 peaks, 36 of the DNVs in the probands were observed (p value = 0.069, Supporting Information Figure S9d), and 14 DNVs were observed in the affected siblings (p value = 0.968), representing no difference. To confirm that the SRY overlapping was higher because we used human SRY peaks dataset, we investigated the origins of SRY peaks. Using a human model, the 242 DNVs from male probands were under 57 (23.55%) SRY peaks, with eight peaks also identified in the mouse model. Another 184 (76.03%) DNVs were found to intersect with the mouse SRY peaks, and one (0.42%) DNV intersected with SRY peaks from the rats. The 78 DNVs from female probands were under 17 (22%) SRY peaks from a human model, and 61 (78%) DNVs were found to intersect with SRY peaks from the mouse model. This result shows that for the male ASD dataset, the context of SRY or SRY‐complex binding is important.
Half of the DNVs identified under SRY peaks in male (124, 51%) and female (39, 50%) samples are upstream from a known transcription start site (TSS; Supporting Information Figure S10), and 23% of DNVs in male (n = 56) and female (n = 18) samples are downstream, up to 1 kb from TSS, suggesting that these variations could be in regulatory regions. Regarding DNVs under the SOX3 peaks, a smaller percentage is upstream from TSS in male (n = 30, 27%) and female (n = 6, 17%) samples, and only 9 (12%) and 5 (14%) DNVs are downstream, up to 1 kb, from TSS in male and female samples, respectively. Although SOX3 peaks are not overrepresented in DNVs in the ASD samples compared to the random sets, 72 (34%) of 214 genes, representing the 242 DNVs that are under SRY peaks, are also targets of the SOX3 protein.
3.9. Disruption of the coexpression module in male individuals with autism showed enrichment in the pathways related to stress and neurodevelopment
A coexpression network analysis was used for the SOX3/SRY set by comparing the prefrontal brain samples from 12 ASD and 15 control male individuals (Voineagu et al., 2011). Female samples were excluded from this analysis, as the samples were scarce. There were no significant between‐group differences in age or postmortem interval (PMI; Wilcoxon test, p value >0.05). We identified 13 modules in control samples, all of which were preserved with Z‐summaries ≥10, showing that their coexpressed network was robust. The 13 modules were evaluated in the ASD samples (Figure 6, Supporting Information Table S20), showing that the red and brown modules were disrupted. They were enriched in pathways related to ER‐nucleus signaling pathway, ER stress response, apoptosis, regulation of nervous system development and neuron differentiation (Table 6, Supporting Information Tables [Link], [Link], [Link], [Link], [Link], [Link], [Link]). This analysis is limited, as we applied the WGCNA on a subset of genes and substantial coexpression information may have been lost, biasing the modules identification.
Figure 6.
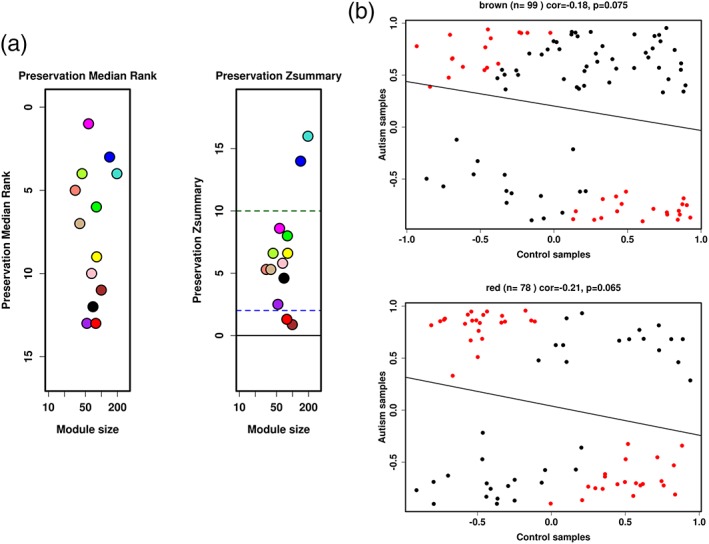
Module preservation analysis preservation analysis resulted in 13 gene modules, which are labeled with unique colors. These modules were generated using WGCNA with control samples. (a) Left panel shows the median preservation rank (y‐axis) in relation to the module size (x‐axis). Each circle represents a module labeled in different colors, with the lowest rank scores represented by brown (MedianRank = 11), black (MedianRank = 12), purple (MedianRank = 13), and red (MedianRank = 13) modules. The right panel shows the Z‐summary score (y‐axis) as a function of module size. The dashed lines represent thresholds 2 and 10: ≥10: High preservation; 2< Z‐summary <10: Moderate preservation; <2: Low preservation. The least preserved modules are represented by brown (Z‐summary = 0.87) and red (Z‐summary = 1.3). (b) Scatter plots of kME of control (x‐axis) and autism (y‐axis) samples. Each dot corresponds to kME, which is the correlation of each gene in the module with the module eigenmode (eigengene‐based connectivity). The red dots represent the genes that showed greater differences between the ASD and CON samples. Higher correlations indicate that the internal module coexpression structure is more preserved. Only the brown and red modules were represented because they demonstrated the lowest cor.kME. Pearson correlation (cor.kME) and the corresponding p value are represented above the respective graphic. The upper panel represents the brown module, with 99 genes (cor = −0.18, p = .075), consisting of genes enriched in categories related to the stress response. The lower panel represents the red module, with 78 genes (cor = − 0.21, p = .065), consisting of genes enriched in categories primarily related to neurogenesis [Color figure can be viewed at wileyonlinelibrary.com]
Table 6.
Functional enrichment analysis of least preserved modules between ASD and CON
| GO type | GO category name | GO ID | C | O | E | R | p |
|---|---|---|---|---|---|---|---|
| Brown module (n = 99 genes) | |||||||
| Biological process | ER‐nucleus signaling pathway | GO:0006984 | 97 | 5 | 0.56 | 8.87 | 1.67E−02 |
| Biological process | Response to endoplasmic reticulum stress | GO:0034976 | 113 | 5 | 0.66 | 7.62 | 3.34E−02 |
| Biological process | Negative regulation of apoptotic process | GO:0043066 | 572 | 10 | 3.32 | 3.01 | 3.87E−02 |
| Cellular component | Synapse part | GO:0044456 | 370 | 6 | 2 | 3.01 | 4.77E−02 |
| Disease | HIV | PA447230 | 755 | 10 | 1.73 | 5.77 | 5.87E−05 |
| Disease | Stress | PA445752 | 464 | 6 | 1.07 | 5.63 | 1.40E−03 |
| Disease | HIV infections | PA446213 | 382 | 5 | 0.88 | 5.7 | 2.90E−03 |
| Transcription factor | hsa_V$HIF1_Q5 | 2,120 | 242 | 7 | 0.56 | 12.6 | 1.29E−05 |
| Transcription factor | hsa_V$PPAR_DR1_Q2 | 2,248 | 257 | 7 | 0.59 | 11.87 | 1.63E−05 |
| Transcription factor | hsa_V$HIF1_Q3 | 2,273 | 225 | 5 | 0.52 | 9.68 | 4.00E−04 |
| hsa_TAATAAT,MIR‐126 | 876 | 220 | 7 | 0.51 | 13.86 | 2.97E−06 | |
| Red module (n = 78 genes) | |||||||
| Biological process | Regulation of cell development | GO:0060284 | 495 | 10 | 2.3 | 4.35 | 1.18E−02 |
| Biological process | Regulation of synaptic plasticity | GO:0048167 | 96 | 5 | 0.45 | 11.21 | 1.18E−02 |
| Biological process | Regulation of nervous system development | GO:0051960 | 440 | 8 | 2.05 | 3.91 | 3.30E−02 |
| Biological process | Regulation of neuron differentiation | GO:0045664 | 331 | 7 | 1.54 | 4.55 | 3.30E−02 |
| Biological process | Neuron differentiation | GO:0030182 | 990 | 12 | 4.6 | 2.61 | 3.81E−02 |
| Biological process | Generation of neurons | GO:0048699 | 1,073 | 12 | 4.99 | 2.41 | 4.75E−02 |
| Disease | Central nervous system diseases | PA443657 | 438 | 6 | 0.79 | 7.57 | 0.0003 |
| Disease | Mental disorders | PA447208 | 564 | 7 | 1.02 | 6.86 | 0.0003 |
| Transcription factor | hsa_GGGCGGR_V$SP1_Q6 | 2,452 | 2,891 | 24 | 5.23 | 4.59 | 4.06E−09 |
| Transcription factor | hsa_GCCATNTTG_V$YY1_Q6 | 2,459 | 419 | 5 | 0.76 | 6.6 | 1.20E−03 |
Abbreviations. C = number of reference genes in the category; E = expected number in the category; O = number of genes in the gene set and also in the category; p = p value adjusted by multiple test adjustment (FDR); R = ratio of enrichment.
Thus, we compared the SOX3/SRY set to the 12 modules built from a comprehensive dataset of 104 brain samples, containing 57 (40 unique) control individuals and 47 (32 unique) autistic individuals, from multiple cortical tissues (BA19, n = 62; BA10, n = 14; and BA44, n = 28; Gupta et al., 2014). This analysis revealed enrichment for six modules, all related to glial or neuronal function biological processes (Table 7). Three of six modules were already described as being related to ASD (mod1, mod5, and mod6; Gupta et al., 2014). Mod1 and mod6 showed enrichment to functional categories related to nervous system development, synapses and neurotransmitter, while mod5 identified enrichment in categories related to immune system (Table 8, Supporting Information Tables [Link], [Link], [Link], [Link]). Similar analyses were performed with 24 modules built from the cortex brain samples of 48 control and 49 ASD individuals (Parikshak et al., 2016). This analysis resulted in an enrichment of 10 modules, four of which were already associated with ASD, three of which were upregulated (CTX.M9, CTX.M19, and CTX.M20) and one downregulated (CTX.M16; Table 9). The CTX.M9 and CTX.M20 modules were enriched in genes related to metabolism and development; only CTX.M9 was enriched in response to oxidative stress. CTX.M16 was composed of genes related to cognition and the regulation of neurotransmitter signaling and neuron development. CTX.M19 showed enrichment in immune system stimulation (Supporting Information Tables [Link], [Link], [Link], [Link], [Link], [Link]).
Table 7.
Enrichment analysis of coexpressed ASD modules
| SOX3/SRY | Gupta modulesa | |||||||
|---|---|---|---|---|---|---|---|---|
| Gupta modulesa | Number of genes | # genes | FE | p value | FDR | Directiona | p valuea | GO functiona |
| mod1 | 1,587 | 172 | 1.35 | 1.00E−04 | 2.00E−04 | − | 8.29E−03 | Neuronal |
| mod2 | 1,290 | 166 | 1.56 | 1.00E−04 | 2.00E−04 | + | 8.72E−01 | Neuronal |
| mod3 | 1,046 | 134 | 1.56 | 1.00E−04 | 2.00E−04 | − | 7.61E−02 | Glial |
| mod4 | 758 | 65 | 1.12 | 1.89E−01 | 2.84E−01 | − | 7.64E−01 | Translational |
| mod5 | 747 | 94 | 1.54 | 1.00E−04 | 2.00E−04 | + | 9.64E−04 | Glial |
| mod6 | 644 | 88 | 1.67 | 1.00E−04 | 2.00E−04 | + | 6.39E−03 | Neuronal |
| mod7 | 590 | 79 | 1.63 | 1.00E−04 | 2.00E−04 | + | 1.08E−01 | Glial |
| mod8 | 446 | 38 | 1.07 | 3.46E−01 | 3.77E−01 | + | 3.64E−02 | Transcription |
| mod9 | 368 | 39 | 1.3 | 5.39E−02 | 9.24E−02 | + | 2.55E−01 | Transcription |
| mod10 | 267 | 23 | 1.11 | 3.35E−01 | 3.77E−01 | − | 6.47E−01 | Translational |
| mod11 | 237 | 22 | 1.11 | 3.38E−01 | 3.77E−01 | + | 5.84E−02 | Glial |
| mod12 | 83 | 2 | 0.564 | 8.79E−01 | 8.79E−01 | + | 1.43E−01 | Neuronal |
Direction: A positive (+) sign indicates upregulated gene expression in autism cases.
#: Number of overlapping genes. FE = fold enrichment.
Bold: p value calculated in 10,000 permutations.
Data provided by Gupta et al. (2014).
Table 8.
Functional enrichment analysis of SOX3/SRY set compared to Gupta modules related to ASD
| Ma | GO type | GO ID | GO category name | C | O | E | R | p |
|---|---|---|---|---|---|---|---|---|
| mod1 | BP | GO:0006813 | Potassium ion transport | 215 | 48 | 17.26 | 2.78 | 1.93E−08 |
| BP | GO:0006836 | Neurotransmitter transport | 193 | 40 | 15.50 | 2.58 | 1.65E−06 | |
| BP | GO:0050890 | Cognition | 251 | 43 | 20.15 | 2.13 | 8.10E−05 | |
| BP | GO:0007626 | Locomotory behavior | 188 | 33 | 15.10 | 2.19 | 4.81E−04 | |
| BP | GO:0007215 | Glutamate receptor signaling pathway | 71 | 17 | 5.70 | 2.98 | 1.01E−03 | |
| BP | GO:0002209 | Behavioral defense response | 33 | 10 | 2.65 | 3.77 | 4.13E−03 | |
| BP | GO:0006914 | Autophagy | 437 | 55 | 35.09 | 1.57 | 1.08E−02 | |
| CC | GO:0098793 | Presynapse | 317 | 77 | 27.45 | 2.81 | 0.00E+00 | |
| Phenotype | HP:0002167 | Neurological speech impairment | 697 | 92 | 50.22 | 1.83 | 4.89E−07 | |
| Phenotype | HP:0100851 | Abnormal emotion/affect behavior | 436 | 56 | 31.42 | 1.78 | 3.88E−04 | |
| Phenotype | HP:0000708 | Behavioral abnormality | 900 | 96 | 64.85 | 1.48 | 3.88E−04 | |
| Phenotype | HP:0001263 | Global developmental delay | 888 | 94 | 63.99 | 1.47 | 6.39E−04 | |
| Phenotype | HP:0002011 | Morphological abnormality of the central nervous system | 1,691 | 153 | 121.85 | 1.26 | 1.84E−03 | |
| Phenotype | HP:0012433 | Abnormal social behavior | 69 | 14 | 4.97 | 2.82 | 1.29E−02 | |
| Phenotype | HP:0001249 | Intellectual disability | 1,039 | 99 | 74.87 | 1.32 | 1.86E−02 | |
| Phenotype | HP:0100852 | Abnormal fear/anxiety‐related behavior | 103 | 17 | 7.42 | 2.29 | 3.13E−02 | |
| Phenotype | HP:0012447 | Abnormal myelination | 269 | 33 | 19.38 | 1.70 | 4.30E−02 | |
| mod5 | BP | GO:0034340 | Response to type I interferon | 79 | 31 | 3.38 | 9.18 | 0.00E+00 |
| BP | GO:0098542 | Defense response to other organism | 443 | 51 | 18.93 | 2.69 | 2.46E−08 | |
| BP | GO:0007249 | I‐kappaB kinase/NF‐kappaB signaling | 257 | 35 | 10.98 | 3.19 | 1.78E−07 | |
| BP | GO:0034341 | Response to interferon‐gamma | 155 | 23 | 6.62 | 3.47 | 2.23E−05 | |
| BP | GO:0045088 | Regulation of innate immune response | 349 | 36 | 14.91 | 2.41 | 9.98E−05 | |
| BP | GO:0032606 | Type I interferon production | 111 | 16 | 4.74 | 3.37 | 1.14E−03 | |
| BP | GO:0070741 | Response to interleukin‐6 | 29 | 8 | 1.24 | 6.46 | 1.14E−03 | |
| BP | GO:0002253 | Activation of immune response | 482 | 41 | 20.60 | 1.99 | 1.14E−03 | |
| BP | GO:0035456 | Response to interferon‐beta | 24 | 7 | 1.03 | 6.82 | 1.95E−03 | |
| BP | GO:0032612 | Interleukin‐1 production | 68 | 10 | 2.91 | 3.44 | 1.54E−02 | |
| mod6 | BP | GO:0099531 | Presynaptic process involved in chemical synaptic transmission | 154 | 18 | 4.94 | 3.64 | 8.43E−04 |
| BP | GO:0001505 | Regulation of neurotransmitter levels | 192 | 19 | 6.16 | 3.08 | 3.47E−03 | |
| BP | GO:0051962 | Positive regulation of nervous system development | 443 | 30 | 14.22 | 2.11 | 1.08E−02 | |
| BP | GO:0044708 | SinglE−organism behavior | 393 | 26 | 12.62 | 2.06 | 2.79E−02 | |
| CC | GO:0098793 | Presynapse | 317 | 29 | 10.70 | 2.71 | 1.37E−04 | |
| CC | GO:0030424 | Axon | 379 | 29 | 12.79 | 2.27 | 1.51E−03 | |
| CC | GO:0098984 | Neuron to neuron synapse | 193 | 16 | 6.51 | 2.46 | 3.01E−02 | |
| MF | GO:0016247 | Channel regulator activity | 128 | 15 | 4.28 | 3.50 | 3.20E−03 |
Abbreviations. BP = Biological Process, CC = Cellular Component, MC = Molecular Component, C = number of reference genes in the category, O = number of genes in the gene set and also in the category, E = expected number in the category, R = Ratio of enrichment; p = p value adjusted by multiple test adjustment (FDR).
Data provided by Gupta and collaborators (Gupta et al., 2014).
Table 9.
Enrichment analysis of coexpressed ASD modules
| SOX3/SRY | ||||||
|---|---|---|---|---|---|---|
| Database | Number of genes | # genes | FE | p value | FDR | ASDa |
| CTX.M1 | 321 | 28 | 1.08 | 3.61E−01 | 5.42E−01 | |
| CTX.M2 | 1,468 | 151 | 1.3 | 6.00E−04 | 3.60E−03 | |
| CTX.M3 | 1,211 | 132 | 1.31 | 8.00E−04 | 3.84E−03 | |
| CTX.M4 | 233 | 23 | 1.22 | 1.91E−01 | 3.27E−01 | [−] |
| CTX.M5 | 108 | 17 | 1.9 | 7.50E−03 | 2.00E−02 | |
| CTX.M6 | 69 | 0 | 0 | 1.00E+00 | 1.00E+00 | |
| CTX.M7 | 98 | 15 | 2.07 | 5.80E−03 | 2.00E−02 | |
| CTX.M8 | 86 | 8 | 1.14 | 4.06E−01 | 5.51E−01 | |
| CTX.M9 | 507 | 71 | 1.7 | 1.00E−04 | 8.00E−04 | [+] |
| CTX.M10 | 257 | 21 | 1.01 | 5.15E−01 | 6.18E−01 | [−] |
| CTX.M12 | 172 | 24 | 1.68 | 6.90E−03 | 2.00E−02 | |
| CTX.M13 | 171 | 12 | 1 | 5.48E−01 | 6.26E−01 | |
| CTX.M14 | 127 | 5 | 0.524 | 9.70E−01 | 1.00E+00 | |
| CTX.M15 | 130 | 12 | 1.1 | 4.13E−01 | 5.51E−01 | |
| CTX.M16 | 278 | 34 | 1.46 | 1.69E−02 | 4.06E−02 | [−] |
| CTX.M17 | 191 | 23 | 1.48 | 3.36E−02 | 7.33E−02 | |
| CTX.M18 | 300 | 31 | 1.26 | 1.13E−01 | 2.12E−01 | |
| CTX.M19 | 274 | 35 | 1.53 | 7.40E−03 | 2.00E−02 | [+] |
| CTX.M20 | 331 | 63 | 2.32 | 1.00E−04 | 8.00E−04 | [+] |
| CTX.M21 | 108 | 9 | 1.03 | 5.07E−01 | 6.18E−01 | |
| CTX.M22 | 221 | 22 | 1.32 | 1.15E−01 | 2.12E−01 | |
| CTX.M23 | 228 | 15 | 0.842 | 7.92E−01 | 8.64E−01 | |
| CTX.M24 | 1,234 | 92 | 1.06 | 2.79E−01 | 4.46E−01 | |
| CTX.M25 | 813 | 117 | 1.75 | 1.00E−04 | 8.00E−04 | |
#: Number of overlapping genes. FE = fold enrichment.
Bold: p value (10,000 permutations) or FDR ≤ 0.05.
Results from Parikshak et al. (2016).
4. DISCUSSION
There was evidence supporting the hypothesis that sexual dimorphism appears before gonadal formation in animal models, suggesting that sex chromosomes play a role in the differences between sexes (Arnold, 2004; Arnold & Burgoyne, 2004; Wolstenholme, Rissman, & Bekiranov, 2013). The regulatory capacities exerted by SRY (Y‐linked protein) and SOX3 (X‐linked protein) in the brain and their capacity to regulate autosomal genes are well established today (Collignon et al., 1996; Mayer et al., 2000; McAninch & Thomas, 2014; Reisert & Pilgrim, 1991). Therefore, we hypothesized that genes regulated by X and, Y factors in a differential manner could produce adaptive sex changes but also contribute to the differential risk for neurodevelopmental disorders. Accordingly, although the regulation exerted by SOX3 was not considered to be a sex‐specific factor, males have two different regulatory proteins, whereas females have one, suggesting the existence of a group of genes that are potentially associated with the sex‐biased prevalence in neurodevelopmental disorders.
We were limited by the available data in the literature, which provided only SRY and SOX3 target genes in different cell types and from distinct species. Ideally, to understand the role of SOX3 and SRY in neurodevelopment, the same cell type from the brains of a single species should be analyzed. To overcome this difficulty, we first analyzed datasets from only mice
In this analysis, we selected target genes from both proteins expressed during different brain periods. As the SRY data came from gonad cells, to increase the certainty that our selected datasets would be more associated with brain expression, we compared them with the patterns of brain and testis specific tissue expression. Although we have used data from adult brains and testes, the SOX3 and SOX3/SRY targets were enriched for only brain expression patterns, and even for exclusive SRY targets, no specific pattern of testis expression was observed.
SOX3 has been clearly involved in brain development, hence the overrepresentation of its targets in all neural cells and brain tissue was expected (Cheah & Thomas, 2015; McAninch & Thomas, 2014). Likewise, exclusive targets of SRY were enriched in most neural cell types. However, the SOX3/SRY target genes demonstrated biased expression in the brain, only for genes with specific expression in cortical neuron differentiation. As astrocytes and oligodendrocytes appear later during neurodevelopment (Budday, Steinmann, & Kuhl, 2015), we could speculate whether the action of SOX3 and SRY on the same target genes could be more restricted to earlier neurodevelopmental periods.
The SOX3/SRY set showed enrichment for functional and phenotypic categories related to neurodevelopment, neuron projection, craniofacial abnormalities, head size, behavior and motor coordination, features with clear sexual dimorphism (Giedd, Castellanos, Rajapakse, Vaituzis, & Rapoport, 1997; Ngun, Ghahramani, Sánchez, Bocklandt, & Vilain, 2011; Sacher, Neumann, Okon‐Singer, Gotowiec, & Villringer, 2013). Only exclusive targets of SRY or SOX3 were enriched in the astrocytes, and only exclusive SRY targets were enriched for microglia, suggesting that our approach was able to identify potentially different regulatory mechanisms in different cell types that are important for brain and sex differences (Lenz & McCarthy, 2015).
After confirming that SRY and SOX3 had highly conserved HMG‐box among species, according to protein alignment and literature evidence (Araujo et al., 2015; Collignon et al., 1996; Wood & Episkopou, 1999), we improved our datasets using mouse, rat and human data to identify the target genes and selected only those genes expressed during human brain development (up to 7 years of age). Moreover, using the data from Lesch et al. (2016), we showed that the SOX3/SRY target genes were under the same regulation programs, independently of the analyzed species. In SRY, this regulatory potential was demonstrated by the ectopic activation of human SRY in mice, and this activation was able to alter gene expression in the brain, heart, lung, and liver (Kido, Sun, & Lau, 2017).
The SOX3/SRY target genes were related to brain development, metabolism and DNA transcriptional regulation, suggesting that they could be important in determining the metabolic differences between sexes. Different brain expression patterns between sexes have been previously reported in the literature in both human and animal models, particularly during neurodevelopment (Ellegren & Parsch, 2007; Peper & Koolschijn, 2012; Reinius & Jazin, 2009; Trabzuni et al., 2013; Xu, Burgoyne, & Arnold, 2002; Ziats & Rennert, 2014; Ziats & Rennert, 2013).
Our SOX3/SRY set was overrepresented in eight neurodevelopmental coexpression modules; most demonstrated higher gene expression levels during the early‐gestation and mid‐gestation periods (Parikshak et al., 2013). The exclusive targets from SOX3 and SRY, except for the sets that overlap with the SOX3/SRY targets, were more associated with the post‐birth expression modules. Parikshak previously described that early gestational modules were more related to processes associated with cell division and differentiation (promoting organogenesis), whereas the process from mid gestational period to after birth was more associated with synaptic transmission and regulation of plasticity. To better understand the putative action of SOX3 and SRY on the same targets, we focused on the modules in the intrauterine period that were enriched in the SOX3/SRY set, independently of sex hormone modulation (Lombardo et al., 2012; McCarthy, Arnold, Ball, Blaustein, & De Vries, 2012; Neufang et al., 2009; Peper & Koolschijn, 2012).
Biological processes related to zinc ion binding, known to play important roles in brain development, were overrepresented in the modules (Levenson & Morris, 2011; McKenzie, Fosmire, & Sandstead, 1975; Sandstead, 2003; Sandstead, Fosmire, McKenzie, & Halas, 1975). Zinc binding deficiency during the prenatal period results in impaired brain growth, which also affects cognitive functions and behavior (Bhatnagar & Taneja, 2001; Hagmeyer, Haderspeck, & Grabrucker, 2014; Kirksey et al., 1991, 1994; Tyszka‐Czochara et al., 2014). Low serum zinc levels were also observed in neuropsychiatric disorders (Pfaender & Grabrucker, 2014), including ASD (Grabrucker, 2014). In rats, prenatal zinc deficiency yielded more aggressive female offspring compared to controls, a phenotype that was not observed in male offspring (Halas, Reynolds, & Sandstead, 1977). Zinc plays a role in synaptic plasticity (Grabrucker et al., 2011), which is different between the sexes (Kang et al., 2011; Mottron et al., 2015), suggesting that modulation of zinc‐related pathways might be sex‐specific. Using Kang data (Kang et al., 2011), the analysis of sex biased expression during brain development supported a possible role of the SOX3/SRY set, independently of hormones.
Moreover, zinc is involved with DNA methylation mechanisms (Hong, Keen, Mizuno, Johnston, & Tamura, 2000; Jing et al., 2015; Wallwork & Duerre, 1985). We demonstrated that SOX3/SRY and SRY sets were enriched for genes differentially methylated between sexes during neurodevelopment. Again, approximately 60% of these differentially methylated CpG sites are located precisely over SRY peaks; this finding is interesting because SRY plays an epigenetic role in several organisms (Clement, Bhandari, Sadler‐Riggleman, & Skinner, 2011; Dewing et al., 2006; Sekido, 2014; Wu, Chen, Li, Lau, & Shih, 2009).
We observed that the SRY peaks were larger than the SOX3 peaks, what could result in bias, as larger peaks have a higher chance to overlap with a CpG site. However, analyses of the distribution of the distance of the SOX3/SRY peaks, in relation to the CpG sites, revealed that most of the SOX3 peaks were located more than 3 kb upstream or downstream, indicating that even if the SOX3 peaks were as large as the SRY peaks (1,879 nucleotides), it would not overlap with the CpG sites. Thus, the SOX3 regulatory role is likely to be something other than the regulation provided by CpG site methylation, including in neurodevelopment (Bergsland et al., 2011; Brunelli et al., 2003; Bylund, Andersson, Novitch, & Muhr, 2003; McAninch & Thomas, 2014; Sutton et al., 2011).
The differential modulation exerted by SRY and SOX3 was reinforced by the network analyses. We investigated just a small fraction of each network and found a considerable difference in the TF distribution. While the SOX3 network showed TFs that were closely linked to neurodevelopment, the SRY network showed a larger network with a higher number of TF related to a wide range of biological processes, including cell cycle, chromatin remodeling, and neural development. For instance, differences in regulation were observed in the renin (REN) promoter, where SRY and SOX3 promote and repress activation, respectively (Araujo et al., 2015). This finding indicates that even if both proteins target the same gene, the regulation that each exerts can differ, particularly in the context of sex‐biased diseases.
To investigate these findings in the context of neuropsychiatry disorders, we selected ASD as a model, as it is a sex‐biased neurodevelopmental disorder with a ratio of 4:1 (boys:girls; Werling, 2016). Two groups of developmentally regulated expression modules have been associated with ASD: those most highly expressed during early pregnancy and those with higher expression during the stage from late pregnancy to after birth (Wang, Zhao, Lachman, & Zheng, 2018). Exclusive SRY or SOX3 gene sets are associated with postbirth modules, while the SOX3/SRY set is most related to the early periods of neurodevelopment. Genes from neurodevelopmental modules M2 and M3, which were also overrepresented in our SOX3/SRY set, were reported to be enriched within RDNV from ASD exome studies (Parikshak et al., 2013), suggesting that our SOX3/SRY set could be important in the context of ASD in the early period of gestation.
To reinforce our hypothesis, early differences in brain cell development between sexes were observed, even before hormonal production (Arnold & Burgoyne, 2004; Dewing, Shi, Horvath, & Vilain, 2003; Rinn & Snyder, 2005). The early and early‐mid periods (8–17 PCW) are important to neuronal proliferation and migration (Stiles & Jernigan, 2010; Tau & Peterson, 2010), and impairments in neuronal migration are a characteristic that was observed in ASD (Reiner, Karzbrun, Kshirsagar, & Kaibuchi, 2016). Moreover, we showed that although all three datasets were enriched for genes that do not tolerate variations, as previously demonstrated for ASD genes (Petrovski et al., 2013), only the SOX3/SRY target was enriched in the dataset of genes intolerant for mutations involved in the early fetal brain development, reinforcing the hypothesis that they are important for programing early male/female brain development.
One example of an ASD‐associated risk gene that has shown sex‐differential effects is MAOA, in which genetic variants in the promoter region of MAOA have been shown to interfere with an SRY binding site and to pose a higher risk to males (Dimas et al., 2012; Verma et al., 2014). We complemented our analyses using ASD databases to investigate the potential of this differential regulation. We found a higher proportion of DNVs over SRY peaks compared to random peaks in both male probands and affected sibling samples, suggesting that these variants can interfere with the binding sites of SRY or SRY‐complex in male individuals. This finding was observed in female datasets with large numbers of DNVs under SRY‐peaks, compared to random sets; however, females do not harbor the SRY gene. We cannot confirm whether these variations are located in functional sites, as we did not investigate the putative TF that could bind to these sites. Although this result was not observed while investigating the SOX3 peaks, 34% of the genes with DNVs under SRY peaks are also targets of the SOX3 protein, suggesting the involvement of SOX3/SRY in this dataset. Our results suggest that both proteins play differential regulatory roles between sexes during brain development, with their targets overrepresented in ASD datasets.
Moreover, we observed differences in the coexpression gene network of the SOX3/SRY dataset between adult ASD and controls. Indeed, two modules showed low preservation scores, consisting of categories that were primarily related to stress, synapses and neuronal development. Identification of the categories associated with the endoplasmic reticulum and apoptosis regulation indicated pathways that were also related to the stress response (Hicks & Machamer, 2005; Schröder, 2008), along with genes regulated by PPAR and HIF1 TFs. PPAR demonstrated increased expression levels during pregnancy after stress exposure in male but not female placentas (Holler, Westin, Jiricny, & Schaffner, 1988; Mueller & Bale, 2008), and HIF1 showed increased expression under oxidative stress (Pialoux et al., 2009; Zhu et al., 2003).
Several studies have demonstrated that impairments in neurodevelopment and synapse formation associated with ASD (Bourgeron, 2015; Codina‐Solà et al., 2015; De Rubeis et al., 2014; Devlin & Scherer, 2012; Sanders et al., 2015; Voineagu et al., 2011) were also associated with sex bias models of prenatal stress exposure (Bale & Epperson, 2015; Murmu et al., 2006; Xu et al., 2013; Zagron & Weinstock, 2006).
Implications of epigenetics mechanisms underlying sex models have already been demonstrated in healthy humans (Maschietto et al., 2017) and those under stressful conditions (Mueller & Bale, 2008; Pilsner et al., 2012; Tobi et al., 2009), with males frequently demonstrating worse outcomes (Bale & Epperson, 2015; Wainstock, Shoham‐Vardi, Glasser, Anteby, & Lerner‐Geva, 2015; Weinstock, 2011) In addition, Y chromosome regulation is affected by stress conditions, as exemplified by exposure to heavy metals, resulting in upregulation of SRY genes (Glahn et al., 2008) and activation of catecholamine synthesis, in a hormone‐independent manner (Lee & Harley, 2012). SP1 (TF) was identified in the SOX3/SRY set and SRY‐network. The SP1 binding site has a CpG, which can be affected by DNA methylation (Douet, Heller, & Le Saux, 2007; Holler et al., 1988; Zhu et al., 2003) and acts synergistically with SRY (Sekido, 2014; Wu et al., 2009). Evidence indicates that promoter regions DNMT3A and B provide binding sites for SRY (Yanagisawa, Ito, Yuasa, & Maruyama, 2002), suggesting that modulations in SRY expression could affect methylation patterns. EP300, a histone acetyltransferase, was identified in the SRY network, as a partner that modifies the chromatin state. Together this evidence suggests that SRY may recruit or regulate epigenetic factors under a stress response.
Contributions to the sex‐skewed prevalence of ASD seem to be related to naturally sexually dimorphic processes, which could modulate the impact of variants in a sex specific manner (Werling, 2016; Werling, Parikshak, & Geschwind, 2016). We provided support for this hypothesis and suggested that sex‐differential regulation exerted solely by SRY or in combination with SOX3 in different cells or gestation time could be a contributing factor associated with male stress exposure vulnerability (Wainstock et al., 2015).
We are aware that our work has limitations, such as combining datasets from different species, requiring a new annotation pipeline that does not cover all data. In addition, the SOX3 ChIP‐seq experiments were performed in neuronal cells, whereas the SRY experiments were performed using gonadal cells. Thus, we first evaluated SRY peaks in a murine model, which has clear evidence of differential regulation exerted by sex chromosome with the “four core genotype” model (Arnold & Chen, 2009). Some studies have modest sample sizes, which may interfere with the identification of differentially expressed or methylated genes during neurodevelopment. We only used ASD as a model, therefore, we cannot conclude whether our findings can be extended to other neurodevelopmental disorders.
5. CONCLUSIONS
While the hypothesis of the sex chromosome complement is not new, we provided a new perspective in which the sex chromosomes could contribute to the sex bias prevalence in ASD, in addition to genetic variation, skewed X inactivation and escaping X chromosome genes. The regulatory roles of SOX3 and SRY over autosomal genes could be attributed in part to their specific partners during brain development. Although we cannot predict how much the sex chromosomes contribute to ASD, we provide a mechanism that explores the effect of sex chromosome genes and their regulatory roles over autosomal genes that can contribute to male bias in ASD and possibly to other neurodevelopmental disorders.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Not applicable. All data from animal or humans used in this study are in the public domain collected from public databases or published articles.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All the data used in this study and their source are described in Supporting Information Table S1.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING INFORMATION
FAPESP grant number 2011/14658‐2 supported this work and provided scholarships to MM (2015/06281‐7), ACT (2014/00041‐1), BCGL (2014/00591‐1), VNSR (2011/04956‐6), and ASF (2014/10488‐3). CAPES (Brazilian Federal Agency for Support and Evaluation of Graduate Education) provided scholarships to VNSR (PROEX‐33002010073P7), VDG (PROEX‐1669479), and ARB (DS Program). ASF was supported by an Institutional Scholarship from UFABC (Universidade Federal do ABC) and CAPES (DS‐1750212).
AUTHOR'S CONTRIBUTIONS
ACT, MM, and HB: designed the study and wrote the manuscript. BCGL, VNSR, ACFS, and ACT: performed database and studies selection and statistical analysis. ARB, ASF, JGCP, VHCT, VDG, and ACT: performed bioinformatical analyses. All authors read and approved the manuscript.
Supporting information
Supplementary Figures
Table S1 Summary of datasets studies used in this study
Table S2 SRY and SOX3 target genes from ChIP‐chip/seq experiments
Table S3 MSET analyses using tissue‐specific pattern gene expression database (Lin et al., 2014)
Table S4 MSET analyses using tissue‐specific pattern gene expression database of brain cell types
Table S5 GO (noRedundant) enrichment analysis SOX3/SRY (n = 847 out of 1,074) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S6 GO (noRedundant) enrichment analysis of SOX3 (n = 3,181 out of 4,466) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S7 Phenotype enrichment analysis SOX3/SRY (n = 658 out of 1,074) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S8 Human orthologs genes of SOX3/SRY putative binding sites
Table S9 Table of target genes SOX3/SRY and Brain expression up 7 yrs
Table S10 MSET analyses using core promoters histone marks
Table S11 GO enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S12 KEGG enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S13 GO (noRedundant) enrichment analysis of SOX3 (n = 3,649 out of 4,694) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S14 Transcription Factor enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S15 Genes directly target of AR and/or ER according to Webgestalt output
Table S16 GO enrichment analysis (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10, Top10)
Table S17 40 autosomal genes differentially methylated among brain development male vs female samples
Table S18 The number of peaks obtained of SRY and SOX3 after conversion through genomic coordinates and median peak size.
Table S19 MSET analysis using several lists related to ASD and Early Brain
Table S20 Table of SOX3/SRY genes according to modules
Table S21 GO enrichment analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S22 Transcription Factor analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S23 Disease enrichment analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S24 microRNA analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S25 GO enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S26 Disease enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S27 TF enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S28 GO enrichment analysis (mod1, n = 1,587 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S29 Phenotype enrichment analysis (mod1, n = 1,587 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S30 GO enrichment analysis (mod5, n = 747 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S31 GO enrichment analysis (mod6, n = 644 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S32 GO enrichment analysis (CTX.M9, n = 507 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S33 GO enrichment analysis (CTX.M16, n = 278 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S34 HPO enrichment analysis (CTX.M16, n = 278 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S35 GO enrichment analysis (CTX.M19, n = 274 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S36 HPO enrichment analysis (CTX.M19, n = 274 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S37 GO enrichment analysis (CTX.M20, n = 331 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
ACKNOWLEDGMENTS
The State of São Paulo Research Foundation FAPESP and the National Council for Scientific and Technological Development (CNPq) supported this research. The authors thank Vanessa de Jesus Rodrigues de Paula and Kyle Wiley for reviewing the manuscript.
Tahira AC, Barbosa AR, Feltrin AS, et al. Putative contributions of the sex chromosome proteins SOX3 and SRY to neurodevelopmental disorders. Am J Med Genet Part B. 2019;180B:390–414. 10.1002/ajmg.b.32704
Funding information Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Grant/Award Number: DS ProgramDS‐1750212PROEX‐1 669479PROEX‐33002010073P7; Fundação de Amparo à Pesquisa do Estado de São Paulo, Grant/Award Number: 2011/04956‐62011/14658‐22014/00041‐12014/00591‐12014/10488‐32015/06281‐7; Universidade Federal do ABC, Grant/Award Number: Institutional Scholarship; UFABC; CAPES, Grant/Award Number: DS‐1750212; FAPESP, Grant/Award Numbers: 2014/10488‐3, 2011/04956‐6, 2014/00591‐1, 2014/00041‐1, 2015/06281‐7, 2011/14658‐2
REFERENCES
- Abrahams, B. S. , Arking, D. E. , Campbell, D. B. , Mefford, H. C. , Morrow, E. M. , Weiss, L. A. , … Packer, A. (2013). SFARI gene 2.0: A community‐driven knowledgebase for the autism spectrum disorders (ASDs). Molecular Autism, 4(1), 36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo, F. C. , Milsted, A. , Watanabe, I. K. M. , Del Puerto, H. L. , Santos, R. A. S. , Lazar, J. , … Prokop, J. W. (2015). Similarities and differences of X and Y chromosome homologous genes, SRY and SOX3, in regulating the renin‐angiotensin system promoters. Physiological Genomics, 47(5), 177–186. 10.1152/physiolgenomics.00138.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, A. P. (2004). Sex chromosomes and brain gender. Nature Reviews Neuroscience, 5(9), 701–708. 10.1038/nrn1494 [DOI] [PubMed] [Google Scholar]
- Arnold, A. P. , & Burgoyne, P. S. (2004). Are XX and XY brain cells intrinsically different? Trends in Endocrinology & Metabolism, 15(1), 6–11. 10.1016/j.tem.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Arnold, A. P. , & Chen, X. (2009). What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Frontiers in Neuroendocrinology, 30(1), 1–9. 10.1016/j.yfrne.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium . (2015). Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium. Retrieved from http://www.med.unc.edu/pgc/results-and-downloads
- Bale, T. L. , & Epperson, C. N. (2015). Sex differences and stress across the lifespan. Nature Neuroscience, 18(10), 1413–1420. 10.1038/nn.4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski, A. , Cuddapah, S. , Cui, K. , Roh, T.‐Y. , Schones, D. E. , Wang, Z. , … Zhao, K. (2007). High‐resolution profiling of histone methylations in the human genome. Cell, 129(4), 823–837. [DOI] [PubMed] [Google Scholar]
- Bellott, D. W. , Hughes, J. F. , Skaletsky, H. , Brown, L. G. , Pyntikova, T. , Cho, T.‐J. , … Page, D. C. (2014). Mammalian Y chromosomes retain widely expressed dosage‐sensitive regulators. Nature, 508(7497), 494–499. 10.1038/nature13206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland, M. , Ramsköld, D. , Zaouter, C. , Klum, S. , Sandberg, R. , & Muhr, J. (2011). Sequentially acting Sox transcription factors in neural lineage development. Genes and Development, 25(23), 2453–2464. 10.1101/gad.176008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom, D. E. , Young, M. , Albrecht, K. H. , & Eicher, E. M. (2000). Related function of mouse SOX3, SOX9, and SRY HMG domains assayed by male sex determination. Genesis, 28(3–4), 111–124. [PMC free article] [PubMed] [Google Scholar]
- Bhandari, R. K. , Haque, M. M. , & Skinner, M. K. (2012). Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS One, 7(9), e43380 10.1371/journal.pone.0043380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, S. , & Taneja, S. (2001). Zinc and cognitive development. The British Journal of Nutrition, 85(Suppl 2), S139–S145. [DOI] [PubMed] [Google Scholar]
- Bourgeron, T. (2015). From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature Reviews. Neuroscience, 16(9), 551–563. 10.1038/nrn3992 [DOI] [PubMed] [Google Scholar]
- Branco, A. T. , Tao, Y. , Hartl, D. L. , & Lemos, B. (2013). Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis‐specific gene expression in Drosophila simulans . Heredity, 111(1), 8–15. 10.1038/hdy.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli, S. , Casey, E. S. , Bell, D. , Harland, R. , & Lovell‐Badge, R. (2003). Expression of SOX3 throughout the developing central nervous system is dependent on the combined action of discrete, evolutionarily conserved regulatory elements. Genesis, 36(1), 12–24. 10.1002/gene.10193 [DOI] [PubMed] [Google Scholar]
- Budday, S. , Steinmann, P. , & Kuhl, E. (2015). Physical biology of human brain development. Frontiers in Cellular Neuroscience, 9, 257 10.3389/fncel.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund, M. , Andersson, E. , Novitch, B. G. , & Muhr, J. (2003). Vertebrate neurogenesis is counteracted by Sox1‐3 activity. Nature Neuroscience, 6(11), 1162–1168. 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- Carlson, M. (2015). org.Hs.eg.db: genome wide annotation for human. Retrieved from http://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html
- Case, L. K. , Wall, E. H. , Dragon, J. A. , Saligrama, N. , Krementsov, D. N. , Moussawi, M. , … Teuscher, C. (2013). The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Research, 23(9), 1474–1485. 10.1101/gr.156703.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, K. , Tripathi, S. , Thommesen, L. , Lægreid, A. , & Kuiper, M. (2013). TFcheckpoint: A curated compendium of specific DNA‐binding RNA polymerase II transcription factors. Bioinformatics, 29(19), 2519–2520. 10.1093/bioinformatics/btt432 [DOI] [PubMed] [Google Scholar]
- Cheah, P.‐S. , & Thomas, P. Q. (2015). SOX3 expression in the glial system of the developing and adult mouse cerebellum. Springerplus, 4, 400 10.1186/s40064-015-1194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.‐Y. , Lopes‐Ramos, C. M. , Kuijjer, M. L. , Paulson, J. N. , Sonawane, A. R. , Fagny, M. , … DeMeo, D. L. (2016). Sexual dimorphism in gene expression and regulatory networks across human tissues. bioRxiv, 82289 10.1101/082289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Shooshtari, P. , Samocha, K. E. , Daly, M. J. , Cotsapas, C. , & King, B. (2016). Network analysis of genome‐wide selective constraint reveals a gene network active in early fetal brain intolerant of mutation. PLoS Genetics, 12(6), e1006121 10.1371/journal.pgen.1006121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, T. M. , Bhandari, R. K. , Sadler‐Riggleman, I. , & Skinner, M. K. (2011). SRY directly regulates the neurotrophin 3 promoter during male sex determination and testis development in rats. Biology of Reproduction, 85(2), 277–284. 10.1095/biolreprod.110.090282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina‐Solà, M. , Rodríguez‐Santiago, B. , Homs, A. , Santoyo, J. , Rigau, M. , Aznar‐Laín, G. , … Cuscó, I. (2015). Integrated analysis of whole‐exome sequencing and transcriptome profiling in males with autism spectrum disorders. Molecular Autism, 6, 21 10.1186/s13229-015-0017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon, J. , Sockanathan, S. , Hacker, A. , Cohen‐Tannoudji, M. , Norris, D. , Rastan, S. , … Lovell‐Badge, R. (1996). A comparison of the properties of Sox‐3 with Sry and two related genes, Sox‐1 and Sox‐2. Development (Cambridge, England), 122(2), 509–520. [DOI] [PubMed] [Google Scholar]
- Cortez, D. , Marin, R. , Toledo‐Flores, D. , Froidevaux, L. , Liechti, A. , Waters, P. D. , … Kaessmann, H. (2014). Origins and functional evolution of Y chromosomes across mammals. Nature, 508(7497), 488–493. 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- Cross‐Disorder Group of the Psychiatric Genomics Consortium . (2013). Identification of risk loci with shared effects on five major psychiatric disorders: A genome‐wide analysis. Lancet (London, England), 381(9875), 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech, D. P. , Lee, J. , Sim, H. , Parish, C. L. , Vilain, E. , & Harley, V. R. (2012). The human testis‐determining factor SRY localizes in midbrain dopamine neurons and regulates multiple components of catecholamine synthesis and metabolism. Journal of Neurochemistry, 122(2), 260–271. 10.1111/j.1471-4159.2012.07782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis, S. , He, X. , Goldberg, A. P. , Poultney, C. S. , Samocha, K. , Ercument Cicek, A. , … Buxbaum, J. D. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 515(7526), 209–215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, B. , & Scherer, S. W. (2012). Genetic architecture in autism spectrum disorder. Current Opinion in Genetics & Development, 22(3), 229–237. 10.1016/j.gde.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Dewing, P. , Chiang, C. W. K. , Sinchak, K. , Sim, H. , Fernagut, P.‐O. , Kelly, S. , … Vilain, E. (2006). Direct regulation of adult brain function by the male‐specific factor SRY. Current Biology: CB, 16(4), 415–420. 10.1016/j.cub.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Dewing, P. , Shi, T. , Horvath, S. , & Vilain, E. (2003). Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Molecular Brain Research, 118(1–2), 82–90. 10.1016/S0169-328X(03)00339-5 [DOI] [PubMed] [Google Scholar]
- Dimas, A. S. , Nica, A. C. , Montgomery, S. B. , Stranger, B. E. , Raj, T. , Buil, A. , … Dermitzakis, E. T. (2012). Sex‐biased genetic effects on gene regulation in humans. Genome Research, 22(12), 2368–2375. 10.1101/gr.134981.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet, V. , Heller, M. B. , & Le Saux, O. (2007). DNA methylation and Sp1 binding determine the tissue‐specific transcriptional activity of the mouse Abcc6 promoter. Biochemical and Biophysical Research Communications, 354(1), 66–71. 10.1016/j.bbrc.2006.12.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger, B. E. , Saul, M. C. , Driessen, T. M. , & Gammie, S. C. (2013). Development of a versatile enrichment analysis tool reveals associations between the maternal brain and mental health disorders, including autism. BMC Neuroscience, 14(1), 147 10.1186/1471-2202-14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H. , & Parsch, J. (2007). The evolution of sex‐biased genes and sex‐biased gene expression. Nature Reviews. Genetics, 8(9), 689–698. 10.1038/nrg2167 [DOI] [PubMed] [Google Scholar]
- Foster, J. W. , & Graves, J. A. (1994). An SRY‐related sequence on the marsupial X chromosome: Implications for the evolution of the mammalian testis‐determining gene. Proceedings of the National Academy of Sciences of the United States of America, 91(5), 1927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd, J. N. , Castellanos, F. X. , Rajapakse, J. C. , Vaituzis, A. C. , & Rapoport, J. L. (1997). Sexual dimorphism of the developing human brain. Progress in Neuro‐Psychopharmacology and Biological Psychiatry, 21(8), 1185–1201. 10.1016/S0278-5846(97)00158-9 [DOI] [PubMed] [Google Scholar]
- Glahn, F. , Schmidt‐Heck, W. , Zellmer, S. , Guthke, R. , Wiese, J. , Golka, K. , … Foth, H. (2008). Cadmium, cobalt and lead cause stress response, cell cycle deregulation and increased steroid as well as xenobiotic metabolism in primary normal human bronchial epithelial cells which is coordinated by at least nine transcription factors. Archives of Toxicology, 82(8), 513–524. 10.1007/s00204-008-0331-9 [DOI] [PubMed] [Google Scholar]
- Grabrucker, A. M. (2014). A role for synaptic zinc in ProSAP/Shank PSD scaffold malformation in autism spectrum disorders. Developmental Neurobiology, 74(2), 136–146. 10.1002/dneu.22089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker, A. M. , Knight, M. J. , Proepper, C. , Bockmann, J. , Joubert, M. , Rowan, M. , … Boeckers, T. M. (2011). Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. The EMBO Journal, 30(3), 569–581. 10.1038/emboj.2010.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, J. A. M. (2015). Evolution of vertebrate sex chromosomes and dosage compensation. Nature Reviews Genetics, 17(1), 33–46. 10.1038/nrg.2015.2 [DOI] [PubMed] [Google Scholar]
- Guberman, J. M. , Ai, J. , Arnaiz, O. , Baran, J. , Blake, A. , Baldock, R. , … Kasprzyk, A. (2011). BioMart central portal: An open database network for the biological community. Database: The Journal of Biological Databases and Curation, 2011: bar041 10.1093/database/bar041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S. , Ellis, S. E. , Ashar, F. N. , Moes, A. , Bader, J. S. , Zhan, J. , … Arking, D. E. (2014). Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity‐dependent genes in autism. Nature Communications, 5, 5748 10.1038/ncomms6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmeyer, S. , Haderspeck, J. C. , & Grabrucker, A. M. (2014). Behavioral impairments in animal models for zinc deficiency. Frontiers in Behavioral Neuroscience, 8, 443 10.3389/fnbeh.2014.00443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halas, E. S. , Reynolds, G. M. , & Sandstead, H. H. (1977). Intra‐uterine nutrition and its effects on aggression. Physiology & Behavior, 19(5), 653–661. 10.1016/0031-9384(77)90040-3 [DOI] [PubMed] [Google Scholar]
- Hicks, S. W. , & Machamer, C. E. (2005). Golgi structure in stress sensing and apoptosis. Biochimica et Biophysica Acta, 1744(3), 406–414. 10.1016/j.bbamcr.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Holler, M. , Westin, G. , Jiricny, J. , & Schaffner, W. (1988). Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes & Development, 2(9), 1127–1135. 10.1101/gad.2.9.1127 [DOI] [PubMed] [Google Scholar]
- Hong, K. H. , Keen, C. L. , Mizuno, Y. , Johnston, K. E. , & Tamura, T. (2000). Effects of dietary zinc deficiency on homocysteine and folate metabolism in rats. The Journal of Nutritional Biochemistry, 11(3), 165–169. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Flockhart, I. , Vinayagam, A. , Bergwitz, C. , Berger, B. , Perrimon, N. , & Mohr, S. E. (2011). An integrative approach to ortholog prediction for disease‐focused and other functional studies. BMC Bioinformatics, 12(1), 357 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, V. X. , O'Geen, H. , Iyengar, S. , Green, R. , & Farnham, P. J. (2007). Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Research, 17, 807–817. 10.1101/gr.6006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, M. , Rech, L. , Wu, Y. , Goltz, D. , Taylor, C. G. , & House, J. D. (2015). Effects of zinc deficiency and zinc supplementation on homocysteine levels and related enzyme expression in rats. Journal of Trace Elements in Medicine and Biology: Organ of the Society for Minerals and Trace Elements (GMS), 30, 77–82. 10.1016/j.jtemb.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Joel, D. , & McCarthy, M. M. (2017). Incorporating sex as a biological variable in neuropsychiatric research: Where are we now and where should we be? Neuropsychopharmacology, 42(2), 379–385. 10.1038/npp.2016.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. J. , Kawasawa, Y. I. , Cheng, F. , Zhu, Y. , Xu, X. , Li, M. , … Sestan, N. (2011). Spatio‐temporal transcriptome of the human brain. Nature, 478(7370), 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido, T. , Sun, Z. , & Lau, Y.‐F. C. (2017). Aberrant activation of the human sex‐determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Scientific Reports, 7(1), 4113 10.1038/s41598-017-04117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H. (2013). Histone modifications for human epigenome analysis. Journal of Human Genetics, 58, 439–445. 10.1038/jhg.2013.66 [DOI] [PubMed] [Google Scholar]
- Kirksey, A. , Rahmanifar, A. , Wachs, T. D. , McCabe, G. P. , Bassily, N. S. , Bishry, Z. , … Jerome, N. W. (1991). Determinants of pregnancy outcome and newborn behavior of a semirural Egyptian population. The American Journal of Clinical Nutrition, 54(4), 657–667. [DOI] [PubMed] [Google Scholar]
- Kirksey, A. , Wachs, T. D. , Yunis, F. , Srinath, U. , Rahmanifar, A. , McCabe, G. P. , … Jerome, N. W. (1994). Relation of maternal zinc nutriture to pregnancy outcome and infant development in an Egyptian village. The American Journal of Clinical Nutrition, 60(5), 782–792. [DOI] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2007). Eigengene networks for studying the relationships between co‐expression modules. BMC Systems Biology, 1, 54 10.1186/1752-0509-1-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted gene co‐expression network analysis. BMC Bioinformatics, 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , Luo, R. , Oldham, M. C. , & Horvath, S. (2011). Is my network module preserved and reproducible? PLoS Computational Biology, 7(1), e1001057 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , & Harley, V. R. (2012). The male fight‐flight response: A result of SRY regulation of catecholamines? BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 34(6), 454–457. 10.1002/bies.201100159 [DOI] [PubMed] [Google Scholar]
- Lemos, B. , Araripe, L. O. , & Hartl, D. L. (2008). Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science (New York, N.Y.), 319, 91–93. 10.1126/science.1148861 [DOI] [PubMed] [Google Scholar]
- Lemos, B. , Branco, A. T. , & Hartl, D. L. (2010). Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proceedings of the National Academy of Sciences of the United States of America, 107, 15826–15831. 10.1073/pnas.1010383107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, K. M. , & McCarthy, M. M. (2015). A starring role for microglia in brain sex differences. The Neuroscientist, 21, 306–321. 10.1177/1073858414536468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch, B. J. , Silber, S. J. , McCarrey, J. R. , & Page, D. C. (2016). Parallel evolution of male germline epigenetic poising and somatic development in animals. Nature Genetics, 48(8), 888–894. 10.1038/ng.3591 [DOI] [PubMed] [Google Scholar]
- Levenson, C. W. , & Morris, D. (2011). Zinc and neurogenesis: Making new neurons from development to adulthood. Advances in Nutrition (Bethesda, MD), 2(2), 96–100. 10.3945/an.110.000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zheng, M. , & Lau, Y.‐F. (2014). The sex‐determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Reports, 8(3), 723–733. 10.1016/j.celrep.2014.06.055 [DOI] [PubMed] [Google Scholar]
- Lin, S. , Lin, Y. , Nery, J. R. , Urich, M. A. , Breschi, A. , Davis, C. A. , … Snyder, M. P. (2014). Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America, 111(48), 17224–17229. 10.1073/pnas.1413624111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke, H. , Harley, V. , & Lee, J. (2015). Biological factors underlying sex differences in neurological disorders. The International Journal of Biochemistry & Cell Biology, 65, 139–150. 10.1016/j.biocel.2015.05.024 [DOI] [PubMed] [Google Scholar]
- Lombardo, M. V. , Ashwin, E. , Auyeung, B. , Chakrabarti, B. , Taylor, K. , Hackett, G. , … Baron‐Cohen, S. (2012). Fetal testosterone influences sexually dimorphic gray matter in the human brain. The Journal of neuroscience : the official journal of the Society for Neuroscience, 32(2), 674–680. 10.1523/JNEUROSCI.4389-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschietto, M. , Bastos, L. C. , Tahira, A. C. , Bastos, E. P. , Euclydes, V. L. V. , Brentani, A. , … Brentani, H. (2017). Sex differences in DNA methylation of the cord blood are related to sex‐bias psychiatric diseases. Scientific Reports, 7, 44547 10.1038/srep44547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys, V. , Kel‐Margoulis, O. V. , Fricke, E. , Liebich, I. , Land, S. , Barre‐Dirrie, A. , … Wingender, E. (2006). TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Research, 34(Database issue), D108–D110. 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A. , Mosler, G. , Just, W. , Pilgrim, C. , & Reisert, I. (2000). Developmental profile of Sry transcripts in mouse brain. Neurogenetics, 3(1), 25–30. [DOI] [PubMed] [Google Scholar]
- McAninch, D. , & Thomas, P. (2014). Identification of highly conserved putative developmental enhancers bound by SOX3 in neural progenitors using ChIP‐Seq. PLoS One, 9(11), e113361 10.1371/journal.pone.0113361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, M. M. , Arnold, A. P. , Ball, G. F. , Blaustein, J. D. , & De Vries, G. J. (2012). Sex differences in the brain: The not so inconvenient truth. The Journal of Neuroscience, 32(7), 2241–2247. 10.1523/JNEUROSCI.5372-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, J. M. , Fosmire, G. J. , & Sandstead, H. H. (1975). Zinc deficiency during the latter third of pregnancy: Effects on fetal rat brain, liver, and placenta. The Journal of Nutrition, 105(11), 1466–1475. [DOI] [PubMed] [Google Scholar]
- McRae, J. F. , Clayton, S. , Fitzgerald, T. W. , Kaplanis, J. , Prigmore, E. , Rajan, D. , … Hurles, M. E. (2017). Prevalence and architecture of de novo mutations in developmental disorders. Nature, 542(7642), 433–438. 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. a. , Ding, S.‐L. , Sunkin, S. M. , Smith, K. a. , Ng, L. , Szafer, A. , … Lein, E. S. (2014). Transcriptional landscape of the prenatal human brain. Nature, 508(7495), 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milsted, A. , Serova, L. , Sabban, E. L. , Dunphy, G. , Turner, M. E. , & Ely, D. L. (2004). Regulation of tyrosine hydroxylase gene transcription by Sry. Neuroscience Letters, 369(3), 203–207. 10.1016/j.neulet.2004.07.052 [DOI] [PubMed] [Google Scholar]
- Mottron, L. , Duret, P. , Mueller, S. , Moore, R. D. , Forgeot d'Arc, B. , Jacquemont, S. , & Xiong, L. (2015). Sex differences in brain plasticity: A new hypothesis for sex ratio bias in autism. Molecular Autism, 6(1), 33 10.1186/s13229-015-0024-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, B. R. , & Bale, T. L. (2008). Sex‐specific programming of offspring emotionality following stress early in pregnancy. Journal of Neuroscience, 28(36), 9055–9065. 10.1523/JNEUROSCI.1424-08.2008.Sex-Specific [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu, M. S. , Salomon, S. , Biala, Y. , Weinstock, M. , Braun, K. , & Bock, J. (2006). Changes of spine density and dendritic complexity in the prefrontal cortex in offspring of mothers exposed to stress during pregnancy. The European Journal of Neuroscience, 24(5), 1477–1487. 10.1111/j.1460-9568.2006.05024.x [DOI] [PubMed] [Google Scholar]
- Neufang, S. , Specht, K. , Hausmann, M. , Güntürkün, O. , Herpertz‐Dahlmann, B. , Fink, G. R. , & Konrad, K. (2009). Sex differences and the impact of steroid hormones on the developing human brain. Cerebral Cortex, 19(2), 464–473. 10.1093/cercor/bhn100 [DOI] [PubMed] [Google Scholar]
- Ngun, T. C. , Ghahramani, N. , Sánchez, F. J. , Bocklandt, S. , & Vilain, E. (2011). The genetics of sex differences in brain and behavior. Frontiers in Neuroendocrinology, 32(2), 227–246. 10.1016/J.YFRNE.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober, C. , Loisel, D. A. , & Gilad, Y. (2008). Sex‐specific genetic architecture of human disease. Nature Reviews. Genetics, 9(12), 911–922. 10.1038/nrg2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak, N. N. , Luo, R. , Zhang, A. , Won, H. , Lowe, J. K. , Chandran, V. , … Geschwind, D. H. (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155(5), 1008–1021. 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak, N. N. , Swarup, V. , Belgard, T. G. , Irimia, M. , Ramaswami, G. , Gandal, M. J. , … Geschwind, D. H. (2016). Genome‐wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature, 540(7633), 423–427. 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peper, J. S. , & Koolschijn, P. C. M. P. (2012). Sex steroids and the organization of the human brain. The Journal of Neuroscience, 32(20), 6745–6746. 10.1523/JNEUROSCI.1012-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski, S. , Wang, Q. , Heinzen, E. L. , Allen, A. S. , & Goldstein, D. B. (2013). Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genetics, 9(8), e1003709 10.1371/journal.pgen.1003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaender, S. , & Grabrucker, A. M. (2014). Characterization of biometal profiles in neurological disorders. Metallomics: Integrated Biometal Science, 6(5), 960–977. 10.1039/c4mt00008k [DOI] [PubMed] [Google Scholar]
- Pialoux, V. , Mounier, R. , Brown, A. D. , Steinback, C. D. , Rawling, J. M. , & Poulin, M. J. (2009). Relationship between oxidative stress and HIF‐1 alpha mRNA during sustained hypoxia in humans. Free Radical Biology & Medicine, 46(2), 321–326. 10.1016/j.freeradbiomed.2008.10.047 [DOI] [PubMed] [Google Scholar]
- Pilsner, J. R. , Hall, M. N. , Liu, X. , Ilievski, V. , Slavkovich, V. , Levy, D. , … Gamble, M. V. (2012). Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS One, 7(5), e37147 10.1371/journal.pone.0037147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A. R. , & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2015). R: A language and environment for statistical computing. Retrieved from https://www.r-project.org/
- Ratnu, V. S. , Emami, M. R. , & Bredy, T. W. (2017). Genetic and epigenetic factors underlying sex differences in the regulation of gene expression in the brain. Journal of Neuroscience Research, 95, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, O. , Karzbrun, E. , Kshirsagar, A. , & Kaibuchi, K. (2016). Regulation of neuronal migration, an emerging topic in autism spectrum disorders. Journal of Neurochemistry, 136(3), 440–456. 10.1111/jnc.13403 [DOI] [PubMed] [Google Scholar]
- Reinius, B. , & Jazin, E. (2009). Prenatal sex differences in the human brain. Molecular Psychiatry, 14(11), 987–989. [DOI] [PubMed] [Google Scholar]
- Reisert, I. , & Pilgrim, C. (1991). Sexual differentiation of monoaminergic neurons––Genetic or epigenetic? Trends in Neurosciences, 14(10), 468–473. [DOI] [PubMed] [Google Scholar]
- Rinn, J. L. , & Snyder, M. (2005). Sexual dimorphism in mammalian gene expression. Trends in Genetics, 21, 298–305. 10.1016/j.tig.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Rosenbloom, K. R. , Armstrong, J. , Barber, G. P. , Casper, J. , Clawson, H. , Diekhans, M. , … Kent, W. J. (2014). The UCSC genome browser database: 2015 update. Nucleic Acids Research, 43(Database issue), D670–D681. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher, J. , Neumann, J. , Okon‐Singer, H. , Gotowiec, S. , & Villringer, A. (2013). Sexual dimorphism in the human brain: Evidence from neuroimaging. Magnetic Resonance Imaging, 31(3), 366–375. 10.1016/j.mri.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Sackton, T. B. , & Hartl, D. L. (2013). Meta‐analysis reveals that genes regulated by the Y chromosome in Drosophila melanogaster are preferentially localized to repressive chromatin. Genome Biology and Evolution, 5, 255–266. 10.1093/gbe/evt005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagvolden, T. , Russell, V. A. , Aase, H. , Johansen, E. B. , & Farshbaf, M. (2005). Rodent models of attention‐deficit/hyperactivity disorder. Biological Psychiatry, 57, 1239–1247. 10.1016/j.biopsych.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Sanders, S. J. , He, X. , Willsey, A. J. , Ercan‐Sencicek, A. G. , Samocha, K. E. , Cicek, A. E. , … State, M. W. (2015). Insights into autism Spectrum disorder genomic architecture and biology from 71 risk loci. Neuron, 87(6), 1215–1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstead, H. H. (2003). Zinc is essential for brain development and function. The Journal of Trace Elements in Experimental Medicine, 16(4), 165–173. 10.1002/jtra.10042 [DOI] [Google Scholar]
- Sandstead, H. H. , Fosmire, G. J. , McKenzie, J. M. , & Halas, E. S. (1975). Zinc deficiency and brain development in the rat. Federation Proceedings, 34(1), 86–88. [PubMed] [Google Scholar]
- Schaafsma, S. M. , & Pfaff, D. W. (2014). Etiologies underlying sex differences in autism Spectrum disorders. Frontiers in Neuroendocrinology, 35(3), 255–271. 10.1016/j.yfrne.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Schröder, M. (2008). Endoplasmic reticulum stress responses. Cellular and Molecular Life Sciences, 65(6), 862–894. 10.1007/s00018-007-7383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, J. , Schuller, W.‐P. , Kappen, C. , Salbaum, J. M. , Bucan, M. , & Stoeckert, C. J. (2005). Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biology, 6(4), R33 10.1186/gb-2005-6-4-r33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido, R. (2014). The potential role of SRY in epigenetic gene regulation during brain sexual differentiation in mammals. Advances in Genetics, 86, 135–165. 10.1016/B978-0-12-800222-3.00007-3 [DOI] [PubMed] [Google Scholar]
- Sharma, K. , Schmitt, S. , Bergner, C. G. , Tyanova, S. , Kannaiyan, N. , Manrique‐Hoyos, N. , … Simons, M. (2015). Cell type‐ and brain region‐resolved mouse brain proteome. Nature Neuroscience, 18(12), 1819–1831. 10.1038/nn.4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky, H. , Kuroda‐Kawaguchi, T. , Minx, P. J. , Cordum, H. S. , Hillier, L. , Brown, L. G. , … Page, D. C. (2003). The male‐specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature, 423(6942), 825–837. 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Spiers, H. , Hannon, E. , Schalkwyk, L. C. , Smith, R. , Wong, C. C. Y. , O'Donovan, M. C. , … Mill, J. (2015). Methylomic trajectories across human fetal brain development. Genome Research, 25(3), 338–352. 10.1101/gr.180273.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles, J. , & Jernigan, T. L. (2010). The basics of brain development. Neuropsychology Review, 20(4), 327–348. 10.1007/s11065-010-9148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, E. , Hughes, J. , White, S. , Sekido, R. , Tan, J. , Arboleda, V. , … Thomas, P. (2011). Identification of SOX3 as an XX male sex reversal gene in mice and humans. The Journal of Clinical Investigation, 121(1), 328–341. 10.1172/JCI42580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D. , Morris, J. H. , Cook, H. , Kuhn, M. , Wyder, S. , Simonovic, M. , … von Mering, C. (2017). The STRING database in 2017: Quality‐controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research, 45(D1), D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau, G. Z. , & Peterson, B. S. (2010). Normal development of brain circuits. Neuropsychopharmacology, 35(1), 147–168. 10.1038/npp.2009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C. L. , Ng, L. , Menon, V. , Martinez, S. , Lee, C. K. , Glattfelder, K. , … Jones, A. R. (2014). A high‐resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron, 83(2), 309–323. 10.1016/j.neuron.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi, E. W. , Lumey, L. H. , Talens, R. P. , Kremer, D. , Putter, H. , Stein, A. D. , … Heijmans, B. T. (2009). DNA methylation differences after exposure to prenatal famine are common and timing‐ and sex‐specific. Human Molecular Genetics, 18(21), 4046–4053. 10.1093/hmg/ddp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalovic, V. , Krstic, A. , Schwirtlich, M. , Dolfini, D. , Mantovani, R. , Stevanovic, M. , & Mojsin, M. (2017). Epigenetic regulation of human SOX3 gene expression during early phases of neural differentiation of NT2/D1 cells. PLoS One, 12(9), e0184099 10.1371/journal.pone.0184099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni, D. , Ramasamy, A. , Imran, S. , Walker, R. , Smith, C. , Weale, M. E. , … Ryten, M. (2013). Widespread sex differences in gene expression and splicing in the adult human brain. Nature Communications, 4, 2771 10.1038/ncomms3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, M. E. , Ely, D. , Prokop, J. , & Milsted, A. (2011). Sry, more than testis determination? American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 301(3), R561–R571. 10.1152/ajpregu.00645.2010 [DOI] [PubMed] [Google Scholar]
- Tyszka‐Czochara, M. , Grzywacz, A. , Gdula‐Argasińska, J. , Librowski, T. , Wiliński, B. , & Opoka, W. (2014). The role of zinc in the pathogenesis and treatment of central nervous system (CNS) diseases. Implications of zinc homeostasis for proper CNS function. Acta Poloniae Pharmaceutica, 71(3), 369–377. [PubMed] [Google Scholar]
- Verma, D. , Chakraborti, B. , Karmakar, A. , Bandyopadhyay, T. , Singh, A. S. , Sinha, S. , … Rajamma, U. (2014). Sexual dimorphic effect in the genetic association of monoamine oxidase a (MAOA) markers with autism spectrum disorder. Progress in Neuro‐Psychopharmacology & Biological Psychiatry, 50, 11–20. 10.1016/j.pnpbp.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Vilella, A. J. , Severin, J. , Ureta‐Vidal, A. , Heng, L. , Durbin, R. , & Birney, E. (2009). EnsemblCompara GeneTrees: Complete, duplication‐aware phylogenetic trees in vertebrates. Genome Research, 19(2), 327–335. 10.1101/gr.073585.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu, I. , Wang, X. , Johnston, P. , Lowe, J. K. , Tian, Y. , Horvath, S. , … Geschwind, D. H. (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature, 474(7351), 380–384. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainstock, T. , Shoham‐Vardi, I. , Glasser, S. , Anteby, E. , & Lerner‐Geva, L. (2015). Fetal sex modifies effects of prenatal stress exposure and adverse birth outcomes. Stress, 18(1), 49–56. 10.3109/10253890.2014.974153 [DOI] [PubMed] [Google Scholar]
- Wallwork, J. C. , & Duerre, J. A. (1985). Effect of zinc deficiency on methionine metabolism, methylation reactions and protein synthesis in isolated perfused rat liver. The Journal of Nutrition, 115(2), 252–262. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Duncan, D. , Shi, Z. , & Zhang, B. (2013). WEB‐based GEne SeT AnaLysis toolkit (WebGestalt): Update 2013. Nucleic Acids Research, 41(Web Server issue), W77–W83. 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Zhao, D. , Lachman, H. M. , & Zheng, D. (2018). Enriched expression of genes associated with autism spectrum disorders in human inhibitory neurons. Translational Psychiatry, 8(13). 10.1038/s41398-017-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock, M. (2011). Sex‐dependent changes induced by prenatal stress in cortical and hippocampal morphology and behaviour in rats: An update. Stress, 14(6), 604–613. 10.3109/10253890.2011.588294 [DOI] [PubMed] [Google Scholar]
- Werling, D. M. (2016). The role of sex‐differential biology in risk for autism spectrum disorder. Biology of Sex Differences, 7(1), 58 10.1186/s13293-016-0112-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling, D. M. , Parikshak, N. N. , & Geschwind, D. H. (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nature Communications, 7, 10717 10.1038/ncomms10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers, P. J. , Yandim, C. , Panousopoulou, E. , Ahmad, M. , Harker, N. , Saveliev, A. , … Festenstein, R. (2010). Sexual dimorphism in mammalian autosomal gene regulation is determined not only by sry but by sex chromosome complement as well. Developmental Cell, 19(3), 477–484. 10.1016/j.devcel.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Wolstenholme, J. T. , Rissman, E. F. , & Bekiranov, S. (2013). Sexual differentiation in the developing mouse brain: Contributions of sex chromosome genes. Genes, Brain, and Behavior, 12(2), 166–180. 10.1111/gbb.12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, H. B. , & Episkopou, V. (1999). Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre‐gastrulation to early somite stages. Mechanisms of Development, 86(1–2), 197–201. [DOI] [PubMed] [Google Scholar]
- Wu, J. B. , Chen, K. , Li, Y. , Lau, Y.‐F. C. , & Shih, J. C. (2009). Regulation of monoamine oxidase a by the SRY gene on the Y chromosome. FASEB Journal, 23(11), 4029–4038. 10.1096/fj.09-139097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Burgoyne, P. S. , & Arnold, A. P. (2002). Sex differences in sex chromosome gene expression in mouse brain. Human Molecular Genetics, 11(12), 1409–1419. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Yang, B. , Yan, C. , Hu, H. , Cai, S. , Liu, J. , … Shen, X. (2013). Effects of duration and timing of prenatal stress on hippocampal myelination and synaptophysin expression. Brain Research, 1527, 57–66. 10.1016/j.brainres.2013.06.025 [DOI] [PubMed] [Google Scholar]
- Yanagisawa, Y. , Ito, E. , Yuasa, Y. , & Maruyama, K. (2002). The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents. Biochimica et Biophysica Acta – Gene Structure and Expression, 1577(3), 457–465. 10.1016/S0167-4781(02)00482-7 [DOI] [PubMed] [Google Scholar]
- Yuen, R. K. C. , Merico, D. , Bookman, M. , Howe, J. L. , Thiruvahindrapuram, B. , Patel, R. V. , … Scherer, S. W. (2017). Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nature Neuroscience, 20(4), 602–611. 10.1038/nn.4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagron, G. , & Weinstock, M. (2006). Maternal adrenal hormone secretion mediates behavioural alterations induced by prenatal stress in male and female rats. Behavioural Brain Research, 175(2), 323–328. 10.1016/j.bbr.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Zhang‐James, Y. , Yang, L. , Middleton, F. A. , Yang, L. , Patak, J. , & Faraone, S. V. (2014). Autism‐related behavioral phenotypes in an inbred rat substrain. Behavioural Brain Research, 269, 103–114. 10.1016/j.bbr.2014.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W.‐G. , Srinivasan, K. , Dai, Z. , Duan, W. , Druhan, L. J. , Ding, H. , … Otterson, G. A. (2003). Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Molecular and Cellular Biology, 23(12), 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats, M. N. , & Rennert, O. M. (2013). Sex‐biased gene expression in the developing brain: Implications for autism spectrum disorders. Molecular Autism, 4(1), 10 10.1186/2040-2392-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats, M. N. , & Rennert, O. M. (2014). Identification of differentially expressed microRNAs across the developing human brain. Molecular Psychiatry, 19(7), 848–852. 10.1038/mp.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Table S1 Summary of datasets studies used in this study
Table S2 SRY and SOX3 target genes from ChIP‐chip/seq experiments
Table S3 MSET analyses using tissue‐specific pattern gene expression database (Lin et al., 2014)
Table S4 MSET analyses using tissue‐specific pattern gene expression database of brain cell types
Table S5 GO (noRedundant) enrichment analysis SOX3/SRY (n = 847 out of 1,074) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S6 GO (noRedundant) enrichment analysis of SOX3 (n = 3,181 out of 4,466) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S7 Phenotype enrichment analysis SOX3/SRY (n = 658 out of 1,074) (Mus musculus, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S8 Human orthologs genes of SOX3/SRY putative binding sites
Table S9 Table of target genes SOX3/SRY and Brain expression up 7 yrs
Table S10 MSET analyses using core promoters histone marks
Table S11 GO enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S12 KEGG enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S13 GO (noRedundant) enrichment analysis of SOX3 (n = 3,649 out of 4,694) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum of genes in the category = 5).
Table S14 Transcription Factor enrichment analysis (n = 1,721) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S15 Genes directly target of AR and/or ER according to Webgestalt output
Table S16 GO enrichment analysis (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10, Top10)
Table S17 40 autosomal genes differentially methylated among brain development male vs female samples
Table S18 The number of peaks obtained of SRY and SOX3 after conversion through genomic coordinates and median peak size.
Table S19 MSET analysis using several lists related to ASD and Early Brain
Table S20 Table of SOX3/SRY genes according to modules
Table S21 GO enrichment analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S22 Transcription Factor analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S23 Disease enrichment analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S24 microRNA analysis (brown module, n = 99 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S25 GO enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S26 Disease enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S27 TF enrichment analysis (red module, n = 78 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 5)
Table S28 GO enrichment analysis (mod1, n = 1,587 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S29 Phenotype enrichment analysis (mod1, n = 1,587 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S30 GO enrichment analysis (mod5, n = 747 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S31 GO enrichment analysis (mod6, n = 644 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S32 GO enrichment analysis (CTX.M9, n = 507 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S33 GO enrichment analysis (CTX.M16, n = 278 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S34 HPO enrichment analysis (CTX.M16, n = 278 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S35 GO enrichment analysis (CTX.M19, n = 274 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S36 HPO enrichment analysis (CTX.M19, n = 274 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Table S37 GO enrichment analysis (CTX.M20, n = 331 genes) (Homo sapiens, P‐value ≤0.05, multiple test correction BH, minimum = 10)
Data Availability Statement
All the data used in this study and their source are described in Supporting Information Table S1.


