Abstract
Background
Several studies have reported the benefits of olfactory training (OT) in the olfactory nervous system of mouse models. Therefore, in this study we performed next‐generation sequencing to evaluate the effects of OT on mRNA sequencing in the olfactory area.
Methods
Mice in each group were administered 300 mg of 3‐methylindole per kilogram of mouse weight. The olfactory function was evaluated by a food‐finding test once a week. The olfactory neuroepithelium was harvested for histologic examination and protein analysis. Subsequently, data analysis, gene ontology and pathway analysis, quantitative real‐time polymerase chain reaction of mRNA, and Western blot analysis were conducted.
Results
Mice were divided into 4 groups according to treatment. Control, anosmia, training, and steroid group mice resumed food finding. Olfactory Maker Protein, olfr1507, ADCY3, and GNAL mRNA expression was higher in the olfactory neuroepithelium of OT than anosmia group mice. In total, 26,364 mRNAs were analyzed. Comparison of the results of OT vs anosmia revealed that ADCY8,10, GFAP, NGF, NGFR, GFAP, and BDNF mRNAs were upregulated in the gene ontology.
Conclusion
OT improved olfactory function, as indicated by the food‐finding test. OT improved the olfactory recovery time to stimulate olfactory nerve regeneration. OT may initially stimulate the olfactory receptor, followed by neurogenesis. Steroid therapy and OT operated under completely different mechanisms in the upregulated gene study. These results indicate that OT may be one of the future modalities for treating olfactory impairment.
Keywords: olfactory disorders, olfaction, quality of life
Approximately 5% of the general population is considered anosmic, and an additional 15% is considered hyposmic.1 Despite numerous studies that have shown a regenerative ability for olfactory receptor neurons, few treatment options have been proven effective for dysosmia.2, 3 Numerous therapeutic strategies have been proposed, such as the use of strychnine, zinc, theophylline, lipoic acid, and caroverine.4, 5, 6, 7, 8, 9, 10, 11, 12 Although the effectiveness of most of these regimens is unclear, the usefulness of corticosteroids in sensorineural dysfunction‐related olfactory loss is established. This is supported by the observation that systemic steroids are helpful in sensorineural dysfunction‐related olfactory loss. However, some patients cannot use oral steroids because of systemic complications. In particular, the long‐term use of steroids can result in more severe complications, such as stroke, heart disease, and diabetes.13 Recently, olfactory training (OT) has been shown to improve olfactory function in humans.14, 15, 16 A recent systematic review and meta‐analysis suggested that OT may be an effective intervention for patients with olfactory dysfunction.17 Most other studies have reported positive outcomes of OT with regard to olfaction, without significant adverse effects. The olfactory system exhibits extraordinary plasticity because of mechanisms that have been investigated extensively at the cognitive and cellular levels.18, 19, 20 However, mechanisms underlying the neural plasticity of the olfactory system are still under investigation.
Therefore, in this study, we hypothesized that the OT would have positive effects in an anosmia model and aimed to determine the mechanism of olfactory disturbance recovery.
Materials and methods
Subjects
Thirty healthy female C57BL6 mice (age 9‐10 weeks, weight 18‐20 g) were randomly allocated to the following 4 groups: control, n = 6; anosmia, n = 8; training, n = 8; and steroid, n = 8. All animals received care according to our institutional animal care and use committee guidelines. 3‐Methylindol (3MI) was given intraperitoneally (IP) at a dose of 300 mg/kg of mouse weight, except for the control group. Dexamethasone 1 mg/kg IP was given to the steroid group.
Olfactory training
OT was performed over a period of 3 weeks. The mice were exposed 3 times daily to 4 odorants: pine, cinnamon, lemon, and peppermint. The lemon and cinnamon odorants were chosen as representatives of the 4 odor categories included in the Korean version of the Sniffin’ Sticks (KVSS) test.21 The animals were placed in a plastic case with a cover (total volume: 200 mm wide × 260 mm diameter × 130 mm height) with 1 of the 4 odorants in each. Each mouse was exposed to the odorants for 1 minute each, with a rest period of 2 minutes between each odorant to prevent olfactory fatigue.15
Behavioral tests
Induction and recovery from anosmia were assessed with a food‐finding test (FFT). The mice were deprived of food for 1 day and then released for 3 minutes into a T maze, in which a food pellet was buried beneath sawdust at the end of 1 of 2 horizontal arms.22 The food pellet (cheese, 1 × 2 cm) was randomly placed in 1 arm of the maze. Each test mouse started at the base of the T, and then walked forward and searched for the hidden food while the elapsed time was recorded. Sawdust was changed after each test. The subjects were considered to “pass” the test if they found the food within 3 minutes. Five repeated trials were completed for each test and the animals were considered not to be “anosmic” if they passed the test more than 3 times within 3 min. The FFT was performed at 5 different times for each mouse. The first FFT was performed before the 3MI injection to determine whether the olfactory functions of the subjects were normal. The second test was performed 3 days after the 3MI injection to evaluate whether the induction of anosmia was complete. The third FFT was performed 1 week after OT to evaluate recovery from anosmia. The fourth FFT was performed 2 weeks after OT. Finally, the fifth FFT was performed 3 weeks after OT.
Identification and harvest of whole olfactory neuroepithelium
The mice were euthanized at 2 and 3 weeks after OT. After the head was bisected, the olfactory neuroepithelium was used for immunohistologic examination, Western immunoblotting, and real‐time polymerase chain reaction (PCR). All the dissecting procedures were performed under an operating microscope. The posterior half of the nasal cavity contains convoluted structures formed by a bone core, which is called the ethmoturbinate, and is lined with olfactory epithelium, which contains olfactory receptor neurons. Under the microscope, the entire ethmoturbinate was harvested with microscissors.
Total RNA extraction
Total RNA was isolated using TRIzol reagent (Invitrogen, USA). RNA quantity was determined by spectrophotometer (Nanodrop One C; Thermo Fisher Scientific, USA) with a ratio of absorbance of >1.7 at 260 and 280 nm.
Library preparation and sequencing
For control and test RNAs, the construction of library was performed using QuantSeq 3' mRNA‐Seq Library Prep Kit (Lexogen, Inc, Austria) according to the manufacturer's instructions. In brief, each 500 ng total RNA were prepared and an oligo‐dT primer containing an Illumina‐compatible sequence at its 5' end was hybridized to the RNA and reverse transcription was performed. After degradation of the RNA template, second‐strand synthesis was initiated by a random primer containing an Illumina‐compatible linker sequence at its 5' end. The double‐stranded library was purified by using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The finished library is purified from PCR components. High‐throughput sequencing was performed as single‐end 75 sequencing using the NextSeq 500 (Illumina, Inc, USA).
Data analysis
The QuantSeq 3' mRNA‐Seq reads were aligned using Bowtie2 (Langmead and Salzberg, 2012). Bowtie2 indices were either generated from genome assembly sequence or the representative transcript sequences for aligning to the genome and transcriptome. The alignment file was used for assembling transcripts, estimating their abundances and detecting differential expression of genes. The differentially expressed gene was determined based on counts from unique and multiple alignments using coverage in Bedtools (Quinlan, 2010). The read count (RC) data were processed based on quantile normalization method using EdgeR within R (R Development Core Team, 2016) using Bioconductor (Gentleman et al, 2004). Gene classification was based on searches done by DAVID (http://david.abcc.ncifcrf.gov/) and Medline (http://www.ncbi.nlm.nih.gov/) databases.
Quantification by real‐time PCR
RNA was obtained using a Trizol prep method and cDNA synthesis was done using DNase I (Thermo Fisher, USA) with a cDNA synthesis kit (Promega, USA) following the manufacturer's method. Real‐time PCR was performed using GoTaq® qPCR Master Mix (Promega, USA) as a CFX96 real‐time system (Bio‐Rad, USA). Real‐time PCR reactions were optimized to 95°C for 2 minutes, 38 amplification cycles at 95°C for 15 seconds, 65°C for 1 minute. Primer sets were designed as follows: OMP, F: 5'‐CGACCTCACCAACCTCATGA‐3', R: 5'‐CATGACCTTGCGGATCTTGG‐3'; GNAL, F: 5'‐GACTACACACCCACAGACCA‐3', R: 5'‐GCCACGTAAATGATCGCAGT‐3'; Olfr1507, F: 5'‐GAAAGCCTTGTCCACCTGTG‐3', R: 5'‐ GGGTTCAGCAGAGGGGTTAT‐3'; Adcy3, F: 5'‐ GGACACGCTCACAAACATC‐3', R: 5'‐ GCCACATTGACCGTATTGC‐3'; β‐actin, F: 5'‐ CATCCGTAAAGACCTCTATGCCAAC‐3', R: 5'‐ATGGAGCCACCGATCCACA‐3'; GAPDH, F:5'‐TGTGTCCGTCGTGGATCTGA‐3', R: 5'‐TTGCTGTTGAAGTCGCAGGAG‐3'. Used as a reference to calculate fold change in target gene expression. The cycle threshold (Ct) values were estimated for the β‐actin and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as housekeeping gene and target genes.
Western blot analysis
For Western blot analysis, samples were obtained from experimental mice and dipped in liquid nitrogen and stored at ‐70°C. The sample was homogenized in 500 µL of protein extraction solution (Pro‐Prep; Intron Biotechnology, Korea) for 1 minute. The homogenate was lysed by incubation for 10 minutes on ice and then centrifuged at 14,000 rpm for 20 minutes at 4°C. Protein quantity was measured by bicinchoninic acid assay. Thirty micrograms of protein was mixed with a sample buffer and heated at 100°C for 10 minutes. The proteins were loaded per lane, resolved by 12% sodium dodecylsulfate‒polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to a polymer of nitrocellurose membrane (Amersham, Sweden). Completed transferred membranes were rinsed in Tris‐buffered saline (TBS) with 0.1% Tween‐20 and incubated for 1 hour in TBS containing 5% skim milk. Primary antibodies were diluted 1:1000 (rabbit anti‐OMP [Abcam, UK], rabbit anti‐ADCY3 [Novus, USA], and rabbit anti‐GAPDH [Cell Signaling, USA]), and 1:2000 (rabbit anti‐olfr1507 [Thermo Fisher Scientific, USA] and rabbit anti‐GNAL [Novus, USA]). After incubation with primary antibodies, the blots were incubated with goat anti‐rabbit secondary antibody conjugated to horseradish peroxidase (Bio‐Rad, USA). Immunoreactivity was visualized with Western Blotting ECL Reagent (Santa Cruz Biotechnology, USA). The signal intensity of the immunoreactive bands was quantified using a CS Analyzer 3.0 (ATTO Corp, Japan) and normalized to the corresponding signal for GAPDH.
Statistical analysis
Statistical analyses were performed using statistical software (SPSS version 19.0, SPSS, Inc). Differences among groups were analyzed using the Kruskal‐Wallis test. In cases of statistical significance, the ranked parameters were compared by one‐way analysis of variance and Bonferroni multiple comparison tests (PASW Statistics 18; SPSS, Inc). In all analyses, p < 0.05 was considered significant.
Results
Behavioral tests
The first FFT was administered to each mouse before a 3MI injection to determine whether the olfactory functions of the subjects were normal at the start of the experiment. All mice found the food within 30 seconds and passed the baseline test. None of the subjects were able to find the food within 3 minutes in any of the second tests. The anosmic mice were observed occasionally wandering around the maze and did not recognize the buried cheese when it was accidently exposed during a food‐finding trial. The third FFT was administered 1 week after OT.
All 8 anosmic mice and 8 OT mice and steroid‐treated mice failed to recognize the buried cheese in all trials at FFT 1. All 8 OT mice were observed to actively search and sniff in the sawdust and were able to find the food within 3 minutes on 4 occasions over 5 trials at FFT 3. OT mice found food significantly faster than the anosmia and steroid groups at FFT 3. Almost all mice showed olfaction recovery at FFT 4 based on the FFT (Fig. 1).
Figure 1.
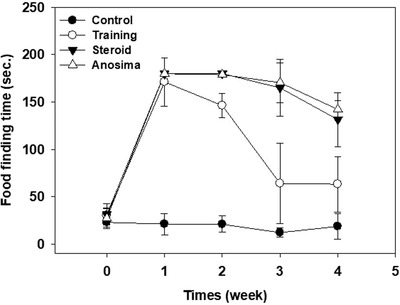
Olfactory‐trained mice found food significantly faster than the anosmia and steroid groups in FFT 3. Almost all mice showed recovery of olfaction at FFT 4. * p < 0.05. FFT = food‐finding test.
Real‐time PCR and Western blot analyses
OMP, Olfr1507, ADCY3, and GNAL mRNA levels in the olfactory neuroepithelium were evaluated using real‐time PCR. OMP, Olfr1507, ADCY3, and GNAL mRNA levels were significantly higher in the 2‐week OT group than in the anosmia group (control vs training in OMP, p < 0.01; anosmia vs training in Olfr1507, p < 0.05; anosmia vs training in ADCY3, p < 0.05; control vs training, p < 0.05; anosmia vs training in GNAL, p = 0.015; Fig. 2). However, at 3 weeks after OT, ADCY3 and GNAL mRNA levels were higher in the OT group than in the anosmia group (control vs training in ADCY3, p < 0.01; control vs training in GNAL, p < 0.01; Fig. 3).
Figure 2.
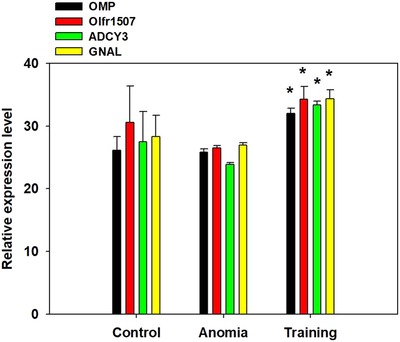
OMP, Olfr1507, ADCY3, and GNAL mRNA levels in olfactory neuroepithelium were evaluated using real‐time polymerase chain reaction. OMP, Olfr1507, ADCY3, and GNAL mRNA levels were significant higher in the 2‐week olfactory training group than the anosmia group (control vs training in OMP, p = 0.004; anosmia vs training in Olfr1507, p = 0.030; anosmia vs training in ADCY3, p = 0.023; control vs training, p = 0.024; anosmia vs training in GNAL, p = 0.015). * p < 0.05. ADCY3 = adenylyl cyclase 3; GNAL = guanine nucleotide‐binding protein G(olf) subunit alpha; Olfr1507 = olfactory receptor 1507; OMP = olfactory marker protein.
Figure 3.
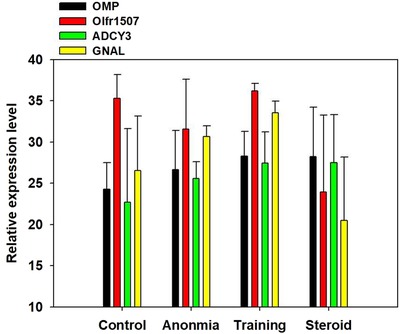
OMP, Olfr1507, ADCY3, and GNAL mRNA levels in the olfactory neuroepithelium were evaluated using real‐time polymerase chain reaction. ADCY3 and GNAL mRNA levels were higher in the olfactory training group than in the anosmia group at 3 weeks after olfactory training (control vs training in ADCY3, p = 0.009; control vs training in GNAL, p = 0.002). * p < 0.05. ADCY3 = adenylyl cyclase 3; GNAL = guanine nucleotide‐binding protein G(olf) subunit alpha; Olfr1507 = olfactory receptor 1507; OMP = olfactory marker protein.
Gene ontology
Scatterplots displaying differentially expressed mRNAs are presented in Figure 4. The gene ontology analysis results of the differentially expressed mRNAs are shown in Figures 5 and 6. We focused especially on neurogenesis, inflammatory response, extracellular matrix, and olfaction (fold change of 1.5, log2 normalized read count of 4 to minimize false counts). Comparison of the anosmia vs control groups revealed that 3656 mRNAs were upregulated and 19,626 were downregulated at 2 weeks. Further, 6910 mRNAs were upregulated and 16,371 were downregulated at 3 weeks. Comparison of the OT vs control groups revealed that 4505 mRNAs were upregulated and 18,777 were downregulated at 2 weeks. Moreover, 10,644 mRNAs were upregulated and 12,637 were downregulated at 3 weeks. Comparison of the steroid‐treated vs control groups revealed that 5074 mRNAs were upregulated and 4795 were downregulated at 2 weeks. In addition, 6839 mRNAs were upregulated and 16,442 were downregulated at 3 weeks. Comparison of the anosmia, steroid‐treated and OT groups revealed that olfactory receptor‐regulated gene and neurogenesis‐related genes were changed in a stepwise manner at 2 and 3 weeks progressively (Table 1). For gene ontology in our study, ADCY8 and ADCY10 genes were upregulated in the OT group after 2 weeks OT. Further, NGFR and NGFRAP1 genes were upregulated after 3‐week OT, not after 2‐week OT. Anosmia group showed increases in ADCY1 and 8 after 3‐week OT (Table 1).
Figure 4.
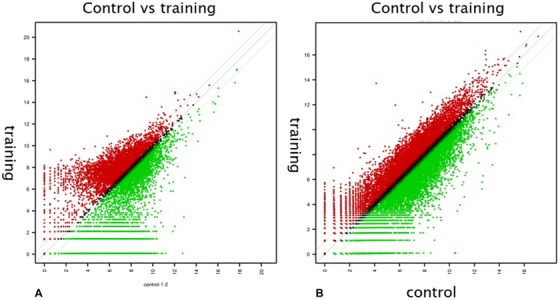
Scatterplots showing differentially expressed mRNAs. (A) Olfactory training group at 2 weeks. (B) Olfactory training group at 3 weeks. Red: expression level of y value is higher than expression level of x value; green: expression level of y value is lower than expression level of x value.
Figure 5.
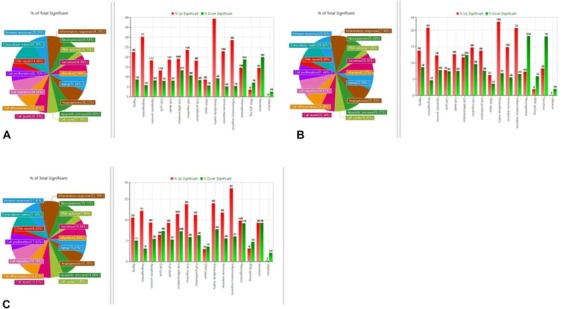
Gene ontology analysis and Venn diagram analysis of mRNA expression. (A) Anosmia group, (B) olfactory training group, and (C) steroid group after 2 weeks of olfactory training (fold change, 1.5; log2 normalized read counts of ≥4 were selected).
Figure 6.
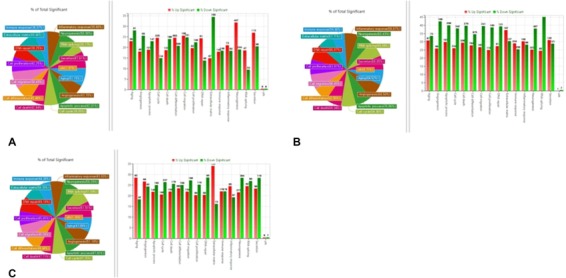
Gene ontology analysis and Venn diagram analysis of mRNA expression. (A) anosmia group, (B) olfactory training group, and (C) steroid group after 3 weeks of olfactory training (fold change, 1.5; log2 normalized read counts of ≥4 were selected).
Table 1.
Differential expression of mRNA related to recovery of olfactory disturbance*
| Gene symbol (2w) | Anosmia/control | OT/control | Gene symbol (3w) | Anosmia/control | OT/control |
|---|---|---|---|---|---|
| GDNF | 0.510 | 0.503 | GDNF | 1.912 | 0.256 |
| ADCY1 | 0.018 | 0.370 | ADCY1 | 7.607a | 2.564 |
| ADCY3 | 0.403 | 0.536 | ADCY3 | 0.635 | 0.382 |
| ADCY8 | 0.133 | 14.615a | ADCY8 | 6.355a | 0.604 |
| ADCY10 | 0.540 | 54.666a | ADCY10 | 1 | 1.005 |
| GFAP | 1.362 | 1.695a | GFAP | – | 1.798 |
| BDNF | 2.435 | 0.1159 | BDNF | 1.940 | 0.604 |
| NGFR | 0.104 | 0.102 | NGFR | 1.133 | 2.376a |
| NGF | 0.510 | 0.503 | NGF | 0.730 | 0.272 |
| NGFRAP | 0.0129 | 0.160a | NGFRAP | 1.346 | 2.175a |
*Fold change, 1.5; log2 normalized read counts of ≥4 were selected.
aSignificantly higher than control.
OT = olfactory training; 2w = after 2 weeks of OT; 3w = after 3 weeks of OT.
Western blot analysis
Western blot analysis was performed to detect protein expression of OMP and olfr1507 in the OT group compared with the anosmia group (Fig. 7). These results show that OT positively affected induction of OMP and Olfr1507 in the anosmia mouse model. Further, OT may accelerate OMP and Olfr1507 activation during olfactory recovery state.
Figure 7.
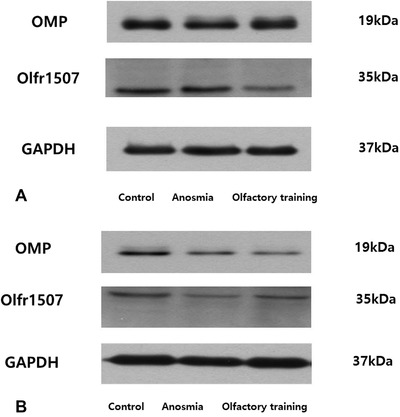
Western blot analysis was performed to detect protein expression of OMP and Olfr1507 in the olfactory training group compared with the anosmia group. Olfr1507 = olfactory receptor 1507; OMP = olfactory marker protein.
Discussion
Recent studies have shown that olfactory disorders occur at a much higher rate than previously assumed.21, 23 However, regardless of how long it may take for olfactory recovery, olfactory loss has been shown to have a severe impact on the quality of life in some patients.24 Despite the fact that numerous studies have shown a regenerative ability for olfactory receptor neurons, few treatment options have been proven effective for post‒upper respiratory infection dysosmia.2, 9 Repeated exposure of healthy individuals to odorants was shown to significantly increase olfactory sensitivity.25 Further, results of the OT metaanalysis were consistent with the reported results of individual studies, primarily with regard to improvement in threshold‐discrimination‐identification (TDI) score in patients with olfactory loss.17 Furthermore, in our study, we showed that OT effectively improved olfactory disturbance.26 We hypothesized that OT would have a positive effect in both human and animal models. As such, we suggested clarifying the mechanism of olfactory disturbance and recovery in this study using an anosmia mouse model.
There were several studies that established an animal model of olfactory dysfunction using 3MI or ZnSO4. A magnetic resonance imaging (MRI) technique identified mucosal damage to the nasal passages of mice resulting from exposure to respiratory toxicants in those studies27, 28, 29 3MI was chosen as a nasal toxicant because systemic administration of this compound in mice results in a well‐characterized necrotizing nasal lesion that is restricted to the olfactory mucosa.
OT improved olfaction significantly in the anomic mouse model in our study. The mRNA expression of OMP and Olfr1507 increased significantly in the OT group compared with the anosmia group after 2‐week OT. After 3 weeks of OT, mRNA expression of ADCY3 and GNAL increased significantly in the OT group relative to the anosmia group.
Most ADCY are integral membrane proteins involved in transducing extracellular signals into intracellular responses. GNAL are involved as modulators or transducers in various transmembrane signaling systems. In particular, G(olf) alpha mediates signal transduction within the olfactory neuroepithelium.30
Thus, the peak times of olfactory receptor stimulation and neurotrophic stimulation were faster in the OT group than in the anosmia group. Further, based on our findings, the order of stimulation was olfactory receptor stimulation followed by neurotrophic stimulation during olfaction recovery in the OT group.
As such, we surmised that the first mechanism of olfactory regeneration may be related to activation of olfactory receptor during OT, and the subsequent mechanism of olfactory recovery may be related to neurotrophic stimulation.
In the FFT, the OT group significantly recovered their olfaction after 2‐week OT. However, the steroid group recovered olfaction slowly, similar to the anosmia group, which recovered naturally.
Initially, we thought that the main effect of OT was to increase olfactory recovery in comparison to the anosmia and steroid groups. However, the final recovery results showed similar states in the 3 groups (anosmia, steroid‐treated, and OT groups) based on the results of our FFT.
We postulate that a fast recovery of olfaction can prevent embarrassment, despair, anxiety, and depression of olfactory disturbance and also increase the quality of life of patients. It is one of the clinical and scientific bases to conduct OT in the olfactory dysfunction state.
Regarding our gene ontology data, genes related with the extracellular matrix and inflammatory response were upregulated in the steroid group. In contrast, genes associated with the immune response and neurogenesis were increased simultaneously in the OT group. The results of our study show that the main olfactory recovery mechanism in the OT group was related to neurogenesis, which differed from the inflammatory effect in the steroid group.
We can hypothesize that OT initially stimulates the olfactory receptor directly during olfaction recovery. ADCY and GNAL enzymatic activity may be increased first in the OT group during the early recovery period. Subsequently, OT may stimulate the nervous system in a stepwise manner. As such, neurotrophic factors, such as GFAP and NGFR, may be increased in our gene ontology data. Further, these neurotrophic factors may stimulate neural plasticity of the central nervous system, which is associated with olfaction. In other studies, the olfactory system was shown to exhibit extraordinary plasticity due to mechanisms that have been extensively investigated at the cognition and cellular levels.18, 19 In addition, there is a decrease in olfactory function in many neurologic conditions.31 Neural plasticity with regard to olfactory loss may have widespread implications for brain function.19 It is known that sensory loss often entails functional and structural modifications in the central nervous system.32, 33 Therefore, the mechanisms of neural plasticity in the olfactory system are under study, because olfactory loss is among the first symptoms of neurodegenerative disorders, such as Alzheimer disease (AD) and Parkinson disease (PD).19, 31, 32, 33, 34, 35 In other studies, PD patients and controls underwent cognitive assessment with the neuropsychological battery and Mini‐Mental State Examination and olfactory assessment with Sniffin' Sticks screening. In their result, the cognitive domain correlated with olfactory loss in PD patients.36 Another study group investigated the utility and efficiency of the brief smell test in PD patients.37 In other studies, PD patients underwent an acute levodopa challenge for the clinical prediction of long‐term dopaminergic responses, and olfactory testing with the Sniffin' Sticks test was evaluated.38 The combination of responses to acute levodopa challenge with hyposmia according to the total olfactory score improved sensitivity for the early diagnosis of PD.38 The relationship between olfactory dysfunction, clinical symptoms, and the potential mechanisms of olfactory dysfunction in AD are currently under study.39 The mechanism of olfactory disturbance in AD involves neurotoxic effects, which include atrophy of hippocampal cells due to abnormally low concentrations of neurotrophic factors, including brain‐derived neurotrophic factor (BDNF).35
Understanding the extent and mechanisms of plasticity in the olfactory system may provide insight into the brain mechanisms underlying recovery and reorganization.19 These findings suggest that OT may induce extensive reorganizational processes in more than just olfactory areas; that is, OT may strengthen cognitive function beyond olfactory perception. In addition to the central effects of OT manifesting as network changes, there was a statistical improvement in the odor detection threshold in other studies.40, 41
When OT was conducted, BDNF was increased. Further, these neurotrophic factors can promote dopaminergic neuron differentiation. BDNF is a member of the neurotrophin family of growth factors,42 which are related to the canonical nerve growth factor. NGFR is one of the general nerve growth factors. As such, increasing NGFR might be evidence of nerve regeneration.43 NGFR serves as a low‐affinity common receptor subunit for multiple neurotrophins (NGF, BDNF, neurotrophin‐3 [NT‐3], and neurotrophin‐4/5 [NT‐4/5]).44 GFAP is the intermediate filament expressed in astrocytes. Increasing GFAP can stimulate astrocytes or astrocyte‐type olfactory ensheathing cells (OECs).45 In other words, OT can promote OEC production or activity. OECs are also viable candidates for cell therapy, which can improve neuropathic pain and motor function in patients with spinal cord injury through multiple mechanisms, including phagocytosis of axonal debris, migration toward glial scars, and secretion of neurotrophic factors.46
Olfactory neuronal plasticity showed increased activity in a functional MRI during OT in other studies.47 Functional connectivity analysis revealed different functional connectivity of the major olfactory areas before and after olfactory training. Before olfactory training, a widespread network encompassing largely non‐olfactory regions was observed. After olfactory training, these non‐olfactory network connections dispersed; only 1 significant connection with the major olfactory areas was retained.47
In summary, OT may stimulate significant olfactory recovery. The olfactory regeneration mechanism may be related to olfactory receptor stimulation first, followed by neurotrophic factor stimulation. OT may induce fast recovery of olfactory disturbance compared with spontaneous recovery and steroid treatment in our study. Further, the mechanism may be related to neuronal plasticity through neurotropic factors.
A limitation of our study was the lack of an ideal anosmia mouse model. Due to various causes of olfactory disturbance, trauma, post‒upper respiratory infection, nasal surgery, drug, etc, we will attempt to create a more suitable anosmia mouse model as the cause.
Conclusion
OT improved olfactory function and increased the olfactory recovery time. The olfactory regeneration mechanism may be related to olfactory receptor and neurotrophic factor stimulation. Our results indicate that olfactory training may be a useful modality for treating olfactory impairments in the future.
How to Cite this Article: Kim B‐Y, Park JY, Kim EJ, Kim BG, Kim SW, Kim SW. The neuroplastic effect of olfactory training to the recovery of olfactory system in mouse model. Int Forum Allergy Rhinol. 2019;9:715–723.
Funding sources for the study: Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (2017R1D1A1B04030364).
Potential conflict of interest: None provided.
References
- 1. Damm M, Pikart LK, Reimann H, et al. Olfactory training is helpful in postinfectious olfactory loss: a randomized controlled multicenter study. Laryngoscope. 2014;124:826‐831. [DOI] [PubMed] [Google Scholar]
- 2. Landis B, Giger R, Morabia A, et al. Olfaction: an epidemiological study on 1,046 subjects. Chem Senses. 2003;28:E33. [Google Scholar]
- 3. Schwob JE, Youngentob SL, Ring G, Iwema CL, Mezza RC. Reinnervation of the rat olfactory bulb after methyl bromide‐induced lesion: timing and extent of reinnervation. J Comp Neurol. 1999;412:439‐457. [DOI] [PubMed] [Google Scholar]
- 4. Deems DA, Doty RL, Settle RG, et al. Smell and taste disorders: a study of 750 patients from the University of Pennsylvania Smell and Taste Center. Arch Otorhinolaryngol Head Neck Surg. 1991;117:519‐528. [DOI] [PubMed] [Google Scholar]
- 5. Seiden AM, Duncan HJ. The diagnosis of a conductive olfactory loss. Laryngoscope. 2001;111:9‐14. [DOI] [PubMed] [Google Scholar]
- 6. Stoll AL, Oepen G. Zinc salts for the treatment of olfactory and gustatory symptoms in psychiatric patients: a case series. J Clin Psychiatry. 1994;55:309‐311. [PubMed] [Google Scholar]
- 7. Stevens MH. Steroid‐dependent anosmia. Laryngoscope. 2001;111:200‐203. [DOI] [PubMed] [Google Scholar]
- 8. Jafek BW, Moran DT, Eller PM, et al. Steroid‐dependent anosmia. Arch Otolaryngol Head Neck Surg. 1987;113:547‐549. [DOI] [PubMed] [Google Scholar]
- 9. Hirsch A, Dougherty D, Aranda J, et al. Medications for olfactory loss: pilot studies. J Neurol Orthop Med Surg. 1996;17:109‐114. [Google Scholar]
- 10. Levy LM, Henkin RI, Lin CS, Hutter A, Schellinger D. Increased brain activation in response to odors in patients with hyposmia after theophylline treatment demonstrated by fMRI. J Comput Assist Tomogr. 1998;22:760‐770. [DOI] [PubMed] [Google Scholar]
- 11. Hummel T, Heilmann S, Huttenbrink KB. Lipoic acid in the treatment of smell dysfunction following viral infection of the upper respiratory tract. Laryngoscope. 2002;112:2076‐2080. [DOI] [PubMed] [Google Scholar]
- 12. Quint C, Temmel AF, Hummel T, Ehrenberger K. The quinoxaline derivative caroverine in the treatment of sensorineural smell disorders: a proof‐of‐concept study. Acta Otolaryngol. 2002;122:877‐881. [PubMed] [Google Scholar]
- 13. Heilmann S, Huttenbrink KB, Hummel T. Local and systemic administration of corticosteroids in the treatment of olfactory loss. Am J Rhinol. 2004;18:29‐33. [PubMed] [Google Scholar]
- 14. Cain WS, Stevens JC, Nickou CM, Giles A, Johnston I, Garcia‐Medina MR. Life‐span development of odor identification, learning, and olfactory sensitivity. Perception. 1995;24:1457‐1472. [DOI] [PubMed] [Google Scholar]
- 15. Engen T, Bosack TN. Facilitation in olfactory detection. J Comp Physiol Psychol. 1969;68:320‐326. [DOI] [PubMed] [Google Scholar]
- 16. Livermore A, Laing DG. Influence of training and experience on the perception of multicomponent odor mixtures. J Exp Psychol Hum Percept Perform. 1996;22:267‐277. [DOI] [PubMed] [Google Scholar]
- 17. Pekala K, Chandra RK, Turner JH. Efficacy of olfactory training in patients with olfactory loss: a systematic review and meta‐analysis. Int Forum Allergy Rhinol. 2016;6:299‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson DA, Best AR, Sullivan RM. Plasticity in the olfactory system: lessons for the neurobiology of memory. Neuroscientist. 2004;10:513‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mainland JD, Bremner EA, Young N, et al. Olfactory plasticity: one nostril knows what the other learns. Nature. 2002;419:802. [DOI] [PubMed] [Google Scholar]
- 20. Qin W, Xuan Y, Liu Y, Jiang T, Yu C. Functional connectivity density in congenitally and late blind subjects. Cerebral Cortex. 2015;25:2507‐2516. [DOI] [PubMed] [Google Scholar]
- 21. Hong SC, Yoo YS, Kim ES, et al. Development of KVSS test (Korean version of Sniffin' sticks test). Korean J Otolaryngol Head Neck Surg. 1999;42:855‐860. [Google Scholar]
- 22. Kim HY, Kim JH, Dhong HJ, et al. Effects of statins on the recovery of olfactory function in a 3‐methylindole–induced anosmia mouse model. Am J Rhinol Allergy. 2012;26:e81‐e84. [DOI] [PubMed] [Google Scholar]
- 23. Hummel T, Rissom K, Hahner A, Reden J, Weidenbecher M, Huttenbrink KB. Effects of olfactory training in patients with olfactory loss. Laryngoscope. 2009;119:496‐499. [DOI] [PubMed] [Google Scholar]
- 24. Hummel T, Nordin S. Olfactory disorders and their consequences for quality of life—a review. Acta Otolaryngol. 2005;125:116‐121. [DOI] [PubMed] [Google Scholar]
- 25. Schriever VA, Lehmann S, Prange J, Hummel T. Preventing olfactory deterioration: olfactory training may be of help in older people. J Am Geriatr Soc. 2014;62:384‐386. [DOI] [PubMed] [Google Scholar]
- 26. Kim BG, Kim BY, Shin JH, Kim SW, Kim SW. The effect of olfactory training using korean version odorants: a preliminary study. Korean J Otorhinolaryngol Head Neck Surg. 2018;61:522‐527. [Google Scholar]
- 27. Kim JW, Hong SL, Lee CH, Jeon EH, Choi AR. Relationship between olfactory function and olfactory neuronal population in C57BL6 mice injected intraperitoneally with 3‐methylindole. Otolaryngol Head Neck Surg. 2010;143:837‐842. [DOI] [PubMed] [Google Scholar]
- 28. Cho HJ, Lee YH, Kim BR, et al. Newly developed method for mouse olfactory behavior tests using an automatic video tracking system. Auris Nasus Larynx. 2018;45:103‐110. [DOI] [PubMed] [Google Scholar]
- 29. Wiethoff AJ, Harkema JR, Koretsky AP, Brown WE. Identification of mucosal injury in the murine nasal airways by magnetic resonance imaging: site‐specific lesions induced by 3‐methylindole. Toxicol Appl Pharmacol. 2001;175:68‐75. [DOI] [PubMed] [Google Scholar]
- 30. Jones DT, Reed RR. Golf: an olfactory neuron specific G protein involved in odorant signal transduction. Science. 244:790‐795. [DOI] [PubMed] [Google Scholar]
- 31. Doty RL. The olfactory vector hypothesis of neurodegenerative disease: is it viable? Ann Neurol. 2008;63:7‐15. [DOI] [PubMed] [Google Scholar]
- 32. Qin W, Xuan Y, Liu Y, Jiang T, Yu C. Functional connectivity density in congenitally and late blind subjects. Cereb Cortex. 2015;25:2507‐2516. [DOI] [PubMed] [Google Scholar]
- 33. Bola M, Gall C, Moewes C, Fedorov A, Hinrichs H, Sabel BA. Brain functional connectivity network breakdown and restoration in blindness. Neurology. 2014;83:542‐551. [DOI] [PubMed] [Google Scholar]
- 34. Doty RL. Olfaction in Parkinson's disease and related disorders. Neurobiol Dis. 2012;46:527‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marine N, Boriana A. Olfactory markers of depression and Alzheimer's disease. Neurosci Biobehav Rev. 2014;45:262‐270. [DOI] [PubMed] [Google Scholar]
- 36. Camargo CHF, Jobbins VA, Serpa RA, Berbetz FA, Sabatini JS, Teive HAG. Association between olfactory loss and cognitive deficits in Parkinson's disease. Clin Neurol Neurosurg. 2018;173:120‐123. [DOI] [PubMed] [Google Scholar]
- 37. Cao M, Li Y, Gu Z, et al. Validation of the utility of the Brief Smell Identification Test in Chinese patients with Parkinson's disease. J Clin Neurosci. 2019;60:68‐72. [DOI] [PubMed] [Google Scholar]
- 38. Terroba Chambi C, Rossi M, Bril A, et al. Diagnostic value of combined acute levodopa challenge and olfactory testing to predict Parkinson's disease. Mov Disord Clin Pract. 2017;4:824‐828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuo LJ, Guo P, Liu L, et al. Analyses of clinical features and investigations on potential mechanisms in patients with Alzheimer's disease and olfactory dysfunction. J Alzheimers Dis. 2018;66:789‐799. [DOI] [PubMed] [Google Scholar]
- 40. Larsson M, Semb H, Winblad B, Amberla K, Wahlund LO, B¨ackman L. Odor identification in normal aging and early Alzheimer's disease: effects of retrieval support. Neuropsychology. 1999;13:47‐53. [DOI] [PubMed] [Google Scholar]
- 41. Zucco GM, Amodio P, Gatta A. Olfactory deficits in patients affected by minimal hepatic encephalopathy: a pilot study. Chem Senses. 2006;31:273‐278. [DOI] [PubMed] [Google Scholar]
- 42. Yan Q, Elliot J, Snider WD. Brain‐derived neurotrophic factor rescues spinal motor neurons from axotomy‐induced cell death. Nature. 1992;360:753‐755. [DOI] [PubMed] [Google Scholar]
- 43. Springer JE. Nerve growth factor receptors in the central nervous system. Exp Neurol. 1988;102:354‐365. [DOI] [PubMed] [Google Scholar]
- 44. Lindsay RM, Harmar AJ. Nerve growth factor regulates expression of neuropeptide genes in adult sensory neurons. Nature. 1989;337:362‐364. [DOI] [PubMed] [Google Scholar]
- 45. Lakatos A1, Barnett SC, Franklin RJ. Olfactory ensheathing cells induce less host astrocyte response and chondroitin sulphate proteoglycan expression than schwann cells following transplantation into adult CNS white matter. Exp Neurol. 2003;184:237‐246. [DOI] [PubMed] [Google Scholar]
- 46. Lu JI, Féron F, Mackay‐Sim A, Waite PM. Olfactory ensheathing cells promote locomotor recovery after delayed transplantation into transected spinal cord. Brain. 2002;125:14‐21. [DOI] [PubMed] [Google Scholar]
- 47. Kollndorfer K, Kowalczyk K, Hoche E, et al. Recovery of olfactory function induces neuroplasticity effects in patients with smell loss. Neural Plast. 2014;2014:140419. [DOI] [PMC free article] [PubMed] [Google Scholar]


