Abstract
The objective of this study was to characterize the oncogenic actions of a recently identified cancer‐associated gene YWHAZ (also named as 14‐3‐3 ζ/δ) in urothelial carcinomas of the urinary bladder (UCUB). A genome‐wide study revealed YWHAZ to be involved in the amplicon at 8q22.3, and its genetic amplification was detected predominantly in muscle‐invasive bladder cancer (MIBC). Immunohistochemical staining confirmed the association of YWHAZ overexpression with higher tumor stages, lymph node/vascular invasion, and mitotic activity. Univariate and multivariate analyses further indicated the prognostic potential of YWHAZ for more aggressive cancer types. Both gene set enrichment analysis and STRING network studies suggested involvement of YWHAZ in regulating caspase‐mediated apoptosis. Ectopic expression of YWHAZ in bladder cells with low endogenous YWHAZ levels boosted cell resistance to doxorubicin and cisplatin, as well as to ionizing radiation. Conversely, YWHAZ‐knockdown using specific shRNA in cells with high endogenous YWHAZ levels diminished survival activity, suppressing cell growth and increasing cell death. Our findings confirm the essential role played by YWHAZ in sustaining cell proliferation during chemo/radiotherapy. Treatments based on anti‐YWHAZ strategies may thus be beneficial for UCUB patients overexpressing YWHAZ. © 2019 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: urothelial carcinomas of the urinary bladder (UCUB), YWHAZ (14‐3‐3 ζ/δ), caspase, apoptosis, chemo‐resistance, radio‐resistance
Introduction
Bladder cancer is the most common tumor of the urinary collecting system, and transitional cell carcinomas (urothelial carcinoma of the urinary bladder; UCUB) account for more than 90% of malignant tumors of the bladder 1. Risk factors causally related to the development of bladder cancer include cigarette smoking, occupational exposure to chemicals, and certain viral infections. Approximately 80% of all new cases of UCUB are classified as nonmuscle invasive (NMIBC; stages Ta/T1), also called superficial bladder cancer, while the remaining 20% of cases are muscle invasive (MIBC; stages T2–4) 1. Currently, the prognoses and treatments for these two types of UCUB differ markedly due to their distinct clinical features and outcomes. Recent studies using integrative cancer genomics approaches have advanced our understanding of the genomic landscapes of these two types of UCUB 2, 3, 4. However, their respective origins remain controversial 5, 6, 7, 8. Thus, molecules involved in the interplay between these two UCUB types are interesting targets to explore.
YWHAZ is a member of the 14‐3‐3 protein family and it functions as a central hub protein for many signal transduction pathways 9. Previous studies have shown that proteins in the 14‐3‐3 family are highly conserved and ubiquitously expressed in all eukaryotic organisms 9, 10. Although not functioning as enzymes, 14‐3‐3 proteins can form homo/heterodimers and bind to phosphorylated serine/threonine motifs on their target proteins, thereby altering the activity of their targets through post‐translational regulation 11. Upregulation of 14‐3‐3 isoforms was recently implicated in a variety of human cancers; however, understanding their specific clinical and biological significance awaits further investigation.
In recent years, YWHAZ has gained attention because its elevated expression associates with a variety of cancers, which indicates it may function as an oncoprotein 12, 13, 14, 15, 16, 17, 18, 19, 20. Clinicopathologic studies further support the relevance of YWHAZ in cancer malignancy and lymph node metastasis 18, 21. In particular, YWHAZ interacts with many apoptotic proteins, including NOXA, BAD, BAX, Raf kinases and caspase‐2, suggesting it plays a critical role in regulating apoptosis and allowing cellular adaptation to environmental stresses 22, 23, 24, 25. In addition, YWHAZ has been found to act as a critical regulator that switches TGF‐β's function from tumor suppressor to metastasis promoter through contextual changes of Smad's partner from p53 to Gli2 26. This evidence supports the idea that activities associated with aberrant YWHAZ expression contribute to its oncogenic function in UCUB by enhancing survival activity.
In the present study, we identified YWHAZ as a possible cancer driver that its amplification/overexpression associates with UCUB progression and poor clinical outcomes. Downstream network analyses and in vitro functional studies confirmed that UCUBs overexpressing YWHAZ gain the capacity to tolerate environmental stresses, including chemo/radiotherapy. Therefore, targeting YWHAZ may be an effective strategy for treating UCUBs exhibiting drug and/or radiation resistance.
Materials and methods
Cell lines and reagents
The BFTC905, HT1376, T24, 5637, TSGH8301, and RT4 human bladder cancer cell lines were purchased from the Bioresource Collection and Research Center (BCRC), Taiwan. These cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mixture (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) under a humidified atmosphere containing 5% CO2 at 37 °C. Cisplatin and doxorubicin were purchased from Merck (Darmstadt, Germany). For Western blotting, anti‐PARP‐1 (HPA045168) Ab was purchased from Sigma–Aldrich Corp. (St. Louis, MO, USA); anti‐YWHAZ (NBP1‐61188), anti‐BAk1 (NBP2‐67460), and anti‐BAX (NBP1‐28566) Abs were from Novus Biologicals LLC. (Littleton, CO, USA); anti‐CASP3 (ab13585), and anti‐CASP10 (ab177475) Abs were from Abcam (Cambridge, UK); and anti‐CASP7 (MAB6243) Abs were from Abnova Corp. (Taipei, Taiwan).
Human samples and tissue microarray preparation
Ten samples of normal urothelium and 60 UCUB samples (20 at stage Ta‐T1, 10 at stage T2, 17 at stage T3, and 13 at stage T4) were freshly collected from clinics and/or BioBank at the Kaohsiung Veterans General Hospital (KVGH) and Chi‐Mei Medical Center (CMMC) with the informed consent of each participant. The cancer tissues were initially stored in liquid nitrogen and then verified to contain >85% tumor cells by staining the frozen sections with hematoxylin. For tissue microarray preparation, 295 formalin‐fixed, paraffin‐embedded bladder cancer samples and 20 urothelium samples (three representative cores from each sample) were arranged onto tissue microarrays. The histological grading of UCUB was done according to the WHO classification. The tumor stages were determined based on American Joint Committee on Cancer (AJCC) TNM system in which primary tumor status and nodal metastasis were designated as T and N statuses, respectively. This study was reviewed and approved by the institutional review boards of KVGH and CMMC (VGHKS16‐CT11‐17 and 10302‐015, respectively).
DNA isolation for copy number alteration analysis
Genomic DNA was purified from frozen tissue samples using a DNeasy kit (Qiagen, Chatsworth, CA, USA) and subjected to genotyping using 250 K SNP arrays (Affymetrix, Santa Clara, CA, USA) according to the procedures provided by the manufacturers. Additional details of the method can be found in our earlier study 27. DNA samples from two cell lines, BFTC905 and TSGH8301, were similarly studied. The dChip program was used to normalize chip data using a model‐based (PM/MM) method, and copy number alterations in each sample were analyzed using the Hidden Markov Model with the assumption of diploidy for normal samples. Based on our earlier study 27, an amplified region with a cutoff of >2.6 copies in more than three consecutive SNPs was defined as an amplicon or genetic gain.
Dual‐color fluorescence in situ hybridization (FISH)
To confirm YWHAZ genetic gain/amplification in UCUBs, bacterial artificial chromosome clones RP11‐102K7 (red signal for 8q22.2 amplicon) and RP11‐327O12 (green signal for control) were purchased from Thermo/Invitrogen (Carlsbad, CA, USA). Based on our SNP array data, the RP11‐327O12 clone covers a region on chromosome 8 with no obvious copy number change in the UCUBs tested. The method for dual‐color FISH on tissue microarrays was described previously 28. Approximately 100 tumor cells per sample were examined under a fluorescence microscope (IX83, Olympus Corp., Shinjuku, Japan) and genetic amplification/gain was defined as a red‐to‐green signal ratio exceeding 2.5.
Immunohistochemistry
Mouse anti‐YWHAZ monoclonal antibodies (NBP1‐61188) were utilized for IHC studies using standard protocols with an Ab dilution of 1:100. Chromogen color was developed using an EnVision+System peroxidase kit (DAKO, Carpinteria, CA, USA) and the immunointensity of YWHAZ staining was scored independently by two pathologists and labeled as negative (0), weakly positive (+1), moderately positive (+2), strongly positive (+3), or intensely positive (+4). In cases where there was not consensus, a third investigator was invited to score the sample, and the final score was determined by the majority scores.
Data mining and functional interaction networks
The SNP array data from Dana‐Farber Cancer Institute (GSE39282) 29 were collected from the GEO data bank (https://www.ncbi.nlm.nih.gov/gds/) to validate our SNP array data. To know the biological relevance of genetic statuses, mRNA expression and protein levels of YWHAZ in UCUBs, data of the TCGA cohort 29, 30 were obtained from The Cancer Genome Atlas (TCGA) database via cBioportal (www.cbioportal.org). To map the key downstream pathways, normalized gene expression data (n = 404) were collected from the TCGA database, and gene set enrichment analysis (GSEA) was performed using GSEA Java (version 2.2.3) from Broad Institute 31, using which the analyses were based on the rank of mRNA expression levels in correlation with the YWHAZ expression. Genes up‐ or downregulated along with YWHAZ amplification/overexpression, with P values <1.00E−05 and false discovery rates (q values) < 1.00E−03, were also filtered out. The functional interactome among the up‐/downregulated genes were analyzed using the STRING protein–protein interaction database (http://string‐db.org/).
Cell‐based functional studies
YWHAZ cDNA was amplified using nested PCR and then cloned into the vector pcDNA6B/V5 (Clontech Lab., Mountain View, CA, USA). The resultant YWHAZ expression vector was transfected into RT4 cells using Lipofectamine 3000 (Thermo/Invitrogen), and the transfected cells were further enriched by incubation in the presence of 7 μg/ml blasticidin (Thermo/Invitrogen). Cells transfected with empty vector served as controls. For gene knockdown, specific shRNAs against YWHAZ and the scrambled control were purchased from the miRNA core at Academic Sinica, Taiwan. Growth of transfectants expressing different constructs was monitored daily using Alamar Blue assays (Thermo Fisher Scientific, Hampton, NH, USA). To assess cell viability, cells were stained with annexin V and propidium iodide (BD Biosciences, Franklin Lakes, NJ, USA) 48 h after drug or radiation treatments. To assay radiosensitivity, cells were exposed to gamma radiation generated using a Clinac‐6Ex electron linear accelerator (Varian Medical Systems, Inc., Palo Alto, CA, USA) with an electron beam energy of 6 MeV. Data were expressed as means ± SD from five replicates in each experimental group.
Quantitative reverse transcription polymerase chain reaction
RNA samples were collected from cells with different treatments using an RNA isolation kit (Qiagen), and the cDNA were prepared by using M‐MuLV reverse transcriptase enzyme according to the procedure provided by the manufacture (NEB Inc., Ipswich, MA, USA). Changes in the mRNA levels of YWHAZ and the downstream effectors were detected by SYBR Green‐based real‐time qPCR (Thermo Fisher Scientific) with the primers as listed in supplementary material, Table S1. Levels of GAPDH served as the internal control for qPCR data normalization. Data were expressed as means ± SD from triplicates in each experimental group.
Statistical analyses
SPSS V.14.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism (GraphPad Software Inc., San Diego, CA, USA) were utilized for statistical analyses. Statistical differences between two groups were evaluated using a t‐test. One‐way ANOVA was used to compare differences among multiple groups. Survival curves were sketched by the Kaplan–Meier method, and a log‐rank test was performed to assess prognostic significances. Parameters with univariate P values less than 0.05 were enrolled in multivariate tests conducted using a Cox proportional hazards model. Values of p < 0.05 were considered statistically significant.
Results
YWHAZ genetic gain/amplification is more frequent in advanced muscle‐invasive bladder cancer
A total of 72 urothelium samples, including 10 samples of normal bladder tissue; 20 UCUBs at stage Ta–T1, 10 at stage T2, 17 at stage T3, and 13 at stage T4; and 2 bladder cancer cell lines, were collected for genomic alteration analysis using a genome‐wide approach. Among the regions exhibiting genetic aberrations, we discovered a unique amplicon at chromosome 8q22.3, which was frequently detected in cancers at more advanced stages (T2–T4, muscle‐invasive bladder cancer [MIBC]) but rarely found in early stage cancers (T0/T1, nonmuscle‐invasive bladder cancer [NMIBC]) (Figure 1A,D, p = 0.043). Through amplicon mapping, we identified the smallest amplified region to be 97.5–103.3 Mb on chromosome 8 containing 28 annotated genes (Figure 1B; see supplementary material, Table S2). In addition to some RNA‐processing genes, YWHAZ was revealed to be the most cancer‐related gene in the 8q22.3 amplicon. Dual‐color FISH confirmed the existence of YWHAZ gain or amplification in UCUBs (Figure 1C), especially advanced‐stage cancers (T2–T4) (p < 0.001) with lymph node invasion (p = 0.027) and higher mitotic activity (HPF ≥ 10) (p = 0.003) (Table 1). SNP array data of the study cohort from Dana‐Faber Cancer Institute (GSE39282) containing 16 NMIBCs and 98 MIBCs were utilized to validate our data, and it confirmed a tendency toward YWHAZ genetic gain/amplification in MIBCs (Figure 1D, p = 0.047). Our data suggest an association between YWHAZ amplification/overexpression and more advanced UCUBs.
Figure 1.
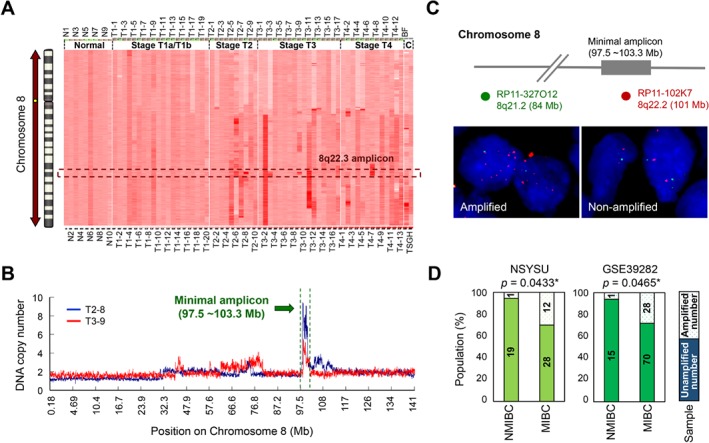
YWHAZ amplification/genetic gain is more frequently detected in MIBC. (A) Genetic alterations in 60 UCUB samples (20 at stage Ta‐T1, 10 at stage T2, 17 at stage T3, and 13 at stage T4) and 2 cell lines were determined using 250K SNP arrays. The SNP values on chromosome 8 were normalized to the average data from 10 samples of normal urothelium and are shown in a heatmap alone with genetic loci on the chromosome. (B) The minimal 8q22.3 amplicon was mapped to 97.5–103.3 Mb by overlapping amplified regions (with copy numbers >2.5) in UCUBs. (C) Dual‐color FISH was performed on UCUB tissue blocks. Red signals (RP11‐102K7) indicate the 8q22.3 amplicon region, while green signals (RP11‐327O12) indicate the control region on chromosome 8, with genetic copy numbers near 2.0. (D) Statistical analysis of 8q22.3 amplicon occurrence between NMIBC (stages Ta to T1) and MIBC (stages T2–T4) using a proportional t‐test. Two study cohorts were analyzed: one from our group (NSYSU; n = 60) and another from Dana–Farber Cancer Institute (GSE39282; n = 114). *p < 0.05, **p < 0.01, ***p < 0.001.
Table 1.
Correlation between YWHAZ amplification/protein overexpression and various clinicopathological factors
| 14‐3‐3 ζ/δ (n = 295) | YWHAZ gene (n = 289) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameters | Category | No. of case | Low (0 to ∼2+) | High (3+ to ∼4+) | P value | No. of cases | No amp. | Amp. | P value |
| Gender | Male | 216 | 161 | 55 | 0.282 | 213 | 122 | 91 | 0.416 |
| Female | 79 | 64 | 15 | 76 | 48 | 28 | |||
| Age (years) | <60 years | 81 | 66 | 15 | 0.222 | 80 | 48 | 32 | 0.894 |
| ≧60 years | 214 | 159 | 55 | 209 | 122 | 87 | |||
| Primary tumor (T) | Ta | 84 | 74 | 10 | <0.001*** | 83 | 59 | 24 | <0.001*** |
| T1 | 88 | 75 | 13 | 85 | 56 | 29 | |||
| T2–T4 | 123 | 76 | 47 | 121 | 55 | 66 | |||
| Nodal status (N) | N0 | 266 | 210 | 56 | 0.002** | 260 | 109 | 101 | 0.027* |
| N1–N2 | 29 | 15 | 14 | 29 | 11 | 18 | |||
| Histological grade | Low grade | 56 | 48 | 8 | 0.080 | 55 | 38 | 17 | 0.095 |
| High grade | 239 | 177 | 62 | 234 | 132 | 102 | |||
| Vascular invasion | Absent | 246 | 195 | 51 | 0.010* | 240 | 144 | 96 | 0.426 |
| Present | 49 | 30 | 19 | 49 | 26 | 23 | |||
| Perineurial invasion | Absent | 275 | 213 | 62 | 0.100 | 269 | 160 | 109 | 0.482 |
| Present | 20 | 12 | 8 | 20 | 10 | 10 | |||
| Tumor necrosis | Absent | 191 | 148 | 43 | 0.567 | 186 | 112 | 74 | 0.535 |
| Present | 101 | 77 | 27 | 103 | 58 | 45 | |||
| Mitotic activity (10 HPF) | <10 | 139 | 115 | 24 | 0.019* | 135 | 92 | 43 | 0.003** |
| ≥10 | 156 | 110 | 46 | 154 | 78 | 76 | |||
Amp, amplification. Statistically significant:
p < 0.05
p < 0.01
p < 0.001.
YWHAZ overexpression defines poor clinical outcomes
Based on the dataset from TCGA 30, samples with YWHAZ genetic gain/amplification tended to also express higher mRNA (by RNA seq) and protein (by RPPA; reverse phase protein arrays) levels in their cancer tissues (p = 0.013) (Figure 2A). This suggests YWHAZ could potentially serve as a cancer driver during UCUB development. The clinical impact of YWHAZ expression was assessed using IHC with bladder cancers at different clinical stages. Our data indicate a correlation between YWHAZ upregulation and tumor staging (Figure 2B). Using our criteria for IHC scoring (see Materials and methods), cancer samples were stratified into Low (with scores of 0 to +2) and High YWHAZ expression (scores of +3 or +4) groups. Consistent with the FISH results, cancers with high YWHAZ levels associated with muscle‐invasive cancer type (p < 0.001) and higher mitotic activity (HPF ≥ 10) (p = 0.019) (Table 1). Notably, cancers with high YWHAZ levels showed a higher tendency to invade the lymphatic and vascular systems (Table 1; p = 0.002 and p = 0.010, respectively). IHC staining also revealed highly YWHAZ‐positive cells with invasions into lymphatic and vascular systems (Figure 2C). Univariate and multivariate analyses revealed that both YWHAZ genetic gain/amplification and protein overexpression correlated with shorter disease‐specific survival (DSS) and metastasis‐free survival (MeFS) times (Table 2). Kaplan–Meier survival analyses confirmed patients with IHC scores of 0 to +2 had a better median DSS and MeFS times than those with scores of +3 to +4 (Figures 2D,E). These data suggest that YWHAZ overexpression is an indicator of aggressive phenotypes and poor prognosis for UCUB patients as other key clinical features such as tumor stage or mitotic activity.
Figure 2.
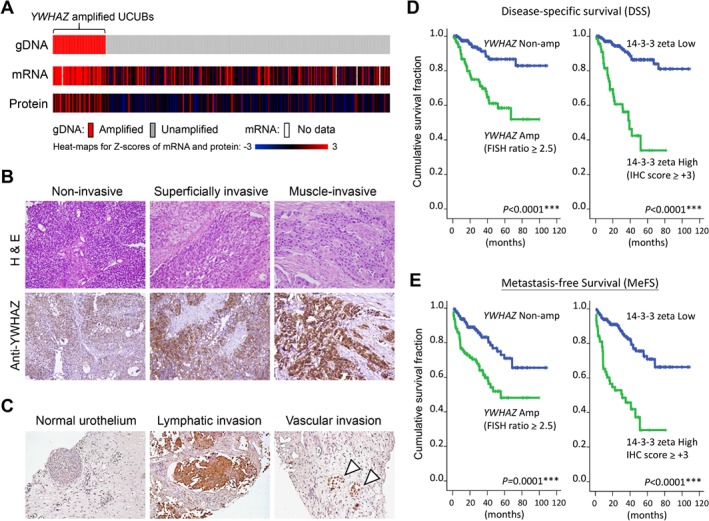
YWHAZ amplification/overexpression correlates with poor clinical outcome. (A) Data of copy number variations, mRNA expression (RNA‐seq) and protein expression (RPPA) in UCUBs from the TCGA group were collected using cBioportal (www.cbioportal.org) and aligned according to their genetic statuses. (B) Immunohistochemistry was performed with tissue microarrays containing UCUBs at different histopathological stages. Representative images of staining for cancers at noninvasive (Ta), superficially invasive (T1) and muscle‐invasive (T2–T4) statuses are shown together with the corresponding images of H&E staining. (C) Representative IHC images of bladder cancer cells undergoing lymphatic (middle) and vascular (right) invasion. Staining of normal urothelium (left) was used as the control. Kaplan–Meier survival analyses of UCUB patients were performed to compare median (D) DSS and (E) MeFS times based on YWHAZ genetic contents by dual‐color FISH and protein expression levels detected by IHC.
Table 2.
Univariate and multivariate log‐rank analyses for DSS and MeFS
| Univariate analysis | Multivariate analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSS | MeFS | DSS | MeFS | |||||||||
| Parameters | Category | Case no. | Event | P value | Event | P value | HR | 95% CI | P value | HR | 95% CI | P value |
| Gender | Male | 216 | 39 | 0.640 | 59 | 0.400 | ||||||
| Female | 79 | 11 | 16 | |||||||||
| Age (years) | <60 | 81 | 10 | 0.218 | 19 | 0.498 | ||||||
| ≧60 | 214 | 40 | 56 | |||||||||
| Primary tumor (T) | Ta | 84 | 1 | <0.001*** | 4 | <0.001*** | 1 | ‐ | <0.001*** | 1 | ‐ | 0.004** |
| T1 | 88 | 9 | 23 | 3.00 | 1.35–6.71 | 6.22 | 1.74–22.27 | |||||
| T2–4 | 123 | 40 | 48 | 27.78 | 2.83–250.00 | 7.77 | 2.16–27.98 | |||||
| Nodal status (N) | N0 | 266 | 41 | 0.002** | 61 | <0.001*** | 1 | ‐ | 0.702 | 1 | ‐ | 0.276 |
| N1–2 | 29 | 9 | 14 | 1.17 | 0.51–2.56 | 1.43 | 0.75–2.70 | |||||
| Histological grade | Low | 56 | 2 | 0.001** | 5 | <0.001*** | 1 | ‐ | 0.970 | 1 | ‐ | 0.572 |
| High | 239 | 48 | 70 | 1.03 | 0.19–4.91 | 1.36 | 0.47–3.99 | |||||
| Vascular invasion | Absent | 246 | 36 | 0.002** | 54 | <0.001*** | 1 | ‐ | 0.438 | 1 | ‐ | 0.920 |
| Present | 49 | 14 | 21 | 1.33 | 0.37–1.54 | 1.10 | 0.53–1.76 | |||||
| Perineurial invasion | Absent | 275 | 44 | 0.005** | 65 | <0.001*** | 1 | ‐ | 0.145 | 1 | ‐ | 0.133 |
| Present | 20 | 6 | 10 | 2.02 | 0.79–5.17 | 1.76 | 0.84–2.02 | |||||
| Tumor necrosis | Absent | 191 | 30 | 0.336 | 48 | 0.728 | ||||||
| Present | 104 | 20 | 27 | |||||||||
| Mitotic activity (HPF) | <10 | 139 | 12 | <0.001*** | 22 | <0.001*** | 1 | ‐ | 0.025* | 1 | ‐ | 0.020* |
| ≥10 | 156 | 38 | 53 | 2.18 | 1.10–4.29 | 1.87 | 1.11–3.15 | |||||
| YWHAZ protein | Low (0 to ∼2+) | 225 | 20 | <0.001*** | 39 | <0.001*** | 1 | ‐ | <0.001*** | 1 | ‐ | <0.001*** |
| High (3+ to ∼4+) | 70 | 30 | 36 | 4.31 | 2.33–7.97 | 3.28 | 2.01–5.37 | |||||
| YWHAZ gene | No amp. | 170 | 14 | <0.001*** | 30 | <0.001*** | 1 | ‐ | 0.005** | 1 | ‐ | <0.001*** |
| Amp. | 119 | 36 | 45 | 5.54 | 1.78–9.12 | 3.47 | 1.27–8.11 | |||||
Amp, amplification. Statistically significant:
p < 0.05
p < 0.01
p < 0.001.
YWHAZ overexpression downregulates signaling for endopeptidase‐mediated cell death
An earlier study suggested the key role played by YWHAZ as a central hub involved in various signal transduction pathways, especially those contributing to apoptosis regulation 9. To assess the functional role of YWHAZ in bladder cancer, we performed GSEA using the TCGA cohort to define possible pathways regulated by YWHAZ amplification/overexpression in cancer tissues. Although the concurrently upregulated genes were very diverse in terms of their cellular functions (see supplementary material, Table S3), the major pathways regulated by the concurrently downregulated genes were apoptotic cleavage of cellular proteins (p = 0.001) and innate immune system (p = 0.001) (Figure 3A). Other related pathways with P values less than 0.01 are summarized in supplementary material, Figure S1 and Table S4. STRING network analysis of the mutually exclusive genes further confirmed the downregulation of pathways involved in endopeptidase‐mediated cell death in YWHAZ‐overexpressing UCUBs (Figure 3B). Expression levels of the 10 major genes involved in the endopeptidase‐mediated cell death (shown in Figure 3B) were found significantly lower in YWHAZ amplified UCUBs as compared to the levels in UCUBs without (all the P values were less than 0.001) (Figure 3C).
Figure 3.
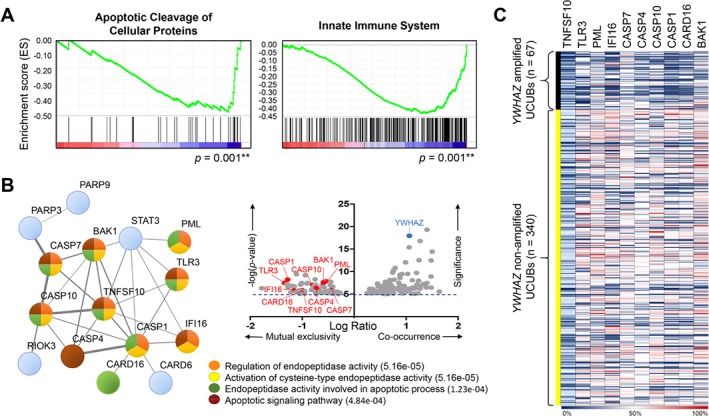
YWHAZ amplification/overexpression negatively regulates signaling pathways in caspase‐mediated apoptosis. (A) Gene set enrichment analysis was performed using mRNA expression data from the TCGA group. Two pathways, apoptotic cleavage of cellular proteins and innate immune system, were found associated with YWHAZ amplification/overexpression in UCUBs. (B) Scatter plot shows genes significantly (−log(p value) > 5.0) up‐ (log ratio > 0.3) or downregulated (log ratio < −0.3) along with YWHAZ amplification/overexpression (right upper panel). The functional protein interaction network of YWHAZ‐associated genes was analyzed using the STRING database (http://string‐db.org/) (left). Four major pathways were found to be involved in YWHAZ amplification/overexpression in UCUBs: regulation of endopeptidase activity, activation of cysteine‐type endopeptidase activity, endopeptidase activity involved in apoptotic process, and apoptotic signaling pathway with false discovery rates (q values) < 1.00 E‐03. (C) Gene expression heat map of 10 key genes involved in endopeptidase‐mediated cell death in UCUBs with or without YWHAZ amplification. Genetic and mRNA expression data were from the TCGA group.
The above findings suggest bladder cancer cells expressing higher levels of YWHAZ are less susceptible to environmental stress‐induced apoptosis than cells expressing lower levels of YWHAZ. To further test that idea, we overexpressed YWHAZ in RT4 cells, which endogenously express lower levels of YWHAZ than do other bladder cancer cell lines (Figures 4A,B). Forty‐eight hours after gene transfection, the expression levels of genes involved in endopeptidase‐mediated apoptosis were downregulated as compared to vector‐treated cells, including the DNA break sensor PARP1, pro‐apoptosis BCL‐2 proteins (BAK1 and BAX), and several caspase enzymes (CASP3, CASP7, CASP10) involved in apoptosis initiation and activation (Figure 4C). When RT4 cells were treated with clinical chemotherapy drugs, cells expressing YWHAZ showed higher tolerance/survival activity than cells transfected with empty vector (IC50 for doxorubicin: 602.6 versus 79.4 nm; IC50 for cisplatin: 20.9 versus 5.2 μm) (Figure 4D). Similarly, YWHAZ overexpression slowed the process of cell death triggered by 6 Gy irradiation in RT4 cells as compared to vector‐treated cells (Figure 4E). Apoptosis analyses using annexin‐V staining also confirmed survival advantages of YWHAZ overexpression when cells were treated with chemotherapy drugs or ionizing radiation (Figure 4F).
Figure 4.
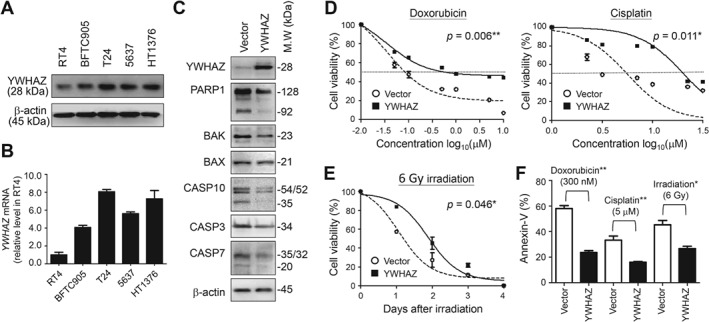
YWHAZ overexpression enhances survival activity to overcome environmental stresses induced by chemo−/radio‐therapy by downregulating caspase‐mediated apoptosis. (A) Western blotting was performed to determine endogenous YWHAZ levels in UCUB cell lines. Levels of β‐actin served as internal controls. (B) RT‐qPCR was performed to determine the relative expression levels of YWHAZ in UCUB cell lines using RT4 cells as the control. (C) Western blotting was performed to determine levels of the indicated effectors involved in caspase‐mediated apoptosis in RT4 cells 48 h after YWHAZ gene transfection. (D) RT4 cells overexpressing YWHAZ were treated with the indicated concentrations of doxorubicin and cisplatin. Cell viability was measured using Alamar Blue assays 4 days after drug treatments. (E) RT4 cells overexpressing YWHAZ were treated with 6 Gy irradiation, after which cell viability was measured every 24 h for 4 days. (F) Incidence rate of preapoptosis among treated cells 48 h after chemo−/radio‐therapy was determined by annexin‐V staining. Cells treated with empty vector served as controls in panels C to F. Data are expressed as means ± SD from five replicates in each experimental group. Paired t‐tests were performed to evaluate differences between vector‐ and YWHAZ‐treated cells. *p < 0.05, **p < 0.01, ***p < 0.001.
YWHAZ serves as a potential target for treating bladder cancer
Because our data suggest YWHAZ exerts a suppressive effect on apoptosis in bladder cancer, we next endeavored to generate several doxorubicin‐ or cisplatin‐resistant clones from RT4 cells. Notably, exposing the cells to different concentrations of drug induced changes in YWHAZ levels that were well correlated with the cells drug tolerance (Figure 5A). Twelve paired UCUBs before and after chemotherapy were collected for YWHAZ staining, and our data confirmed YWHAZ levels in UCUBs were increased during clonal evolution driven by drug treatments (Figure 5B). These findings suggest that YWHAZ plays an active role in mediating resistance to chemotherapeutic agents.
Figure 5.
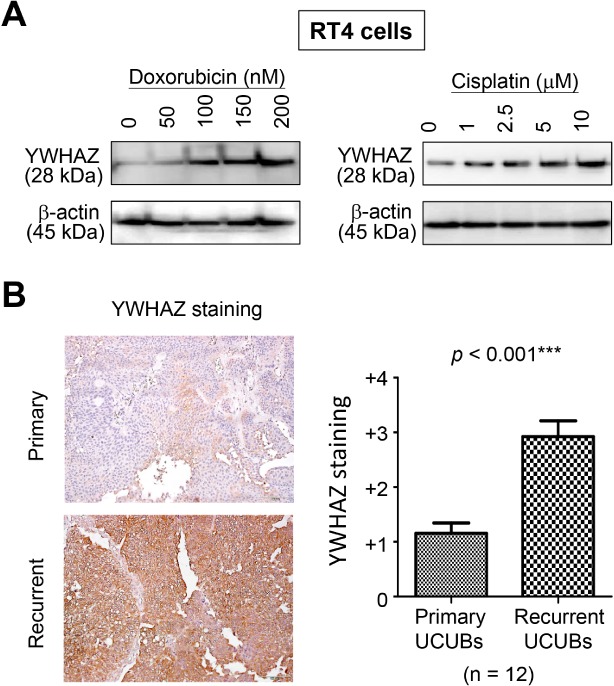
YWHAZ overexpression promotes chemo‐resistance. (A) Western blotting was performed to detect YWHAZ levels in RT4 cells showing drug resistance after exposure to various concentrations of doxorubicin or cisplatin. (B) Representative IHC images of paired UCUBs before (primary) and after (recurrent) chemotherapy (left). YWHAZ staining scores were shown in the bar chart using data from 12 paired UCUBs (right).
To test whether YWHAZ could serve as a potential target to reverse chemo/radioresistance, gene knockdown was performed using specific shRNAs in T24 cells, which were selected because of their relatively high endogenous YWHAZ levels (Figures 4A,B). The shRNA‐1 was chosen in our study because it showed highest gene reduction activity both by western blotting (Figure 6A) and RT‐qPCR (Figure 6B). Gene expression levels of the related pro‐apoptosis genes were found upregulated in T24 cells after YWHAZ knockdown (Figure 6C). When treated with chemotherapy drugs, the viability of shRNA‐1‐treated T24 cells was significantly lower than mock‐treated cells (IC50 for doxorubicin: 30.9 nm versus more than 10 μm; IC50 for cisplatin: 5.6 versus 125.9 μm) (Figure 6D,F). Moreover, reducing YWHAZ levels in T24 cells using shRNA‐1 also dramatically sensitized the cells to 6 Gy irradiation and markedly increased cell death. By contrast, the same treatment had no effect on mock‐treated cells (Figures 6E,F). To further assess the potential therapeutic benefits of inhibiting YWHAZ, we co‐cultured the cells with a low concentration of doxorubicin or cisplatin. When exposed to chemotherapeutic agents, the growth of shRNA‐1‐treated cells was significantly suppressed such that only a few colonies were detected, whereas the mock‐treated cells continued to grow and formed numerous colonies (Figures 6G,H). Our data support the point of view that YWHAZ overexpression is required for UCUB cells to adapt and survive under therapeutic stresses.
Figure 6.
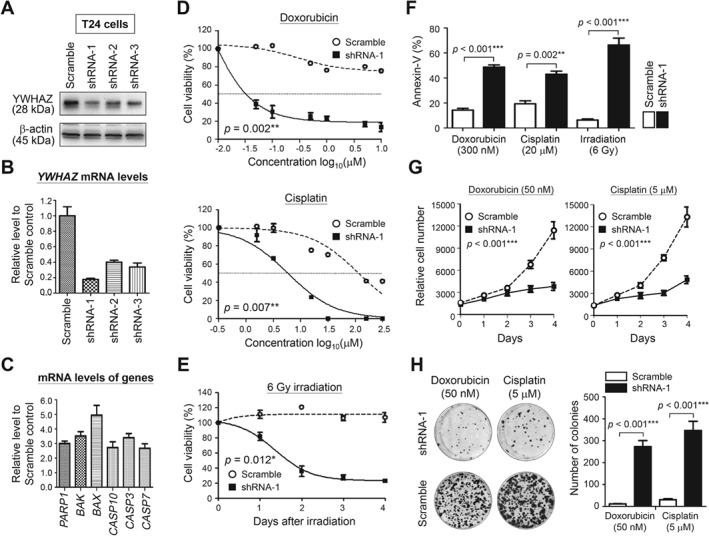
YWHAZ downregulation sensitizes cells to chemo/radiotherapy and suppresses cell growth. (A) Western blotting was performed to detect changes of YWHAZ levels in T24 cells treated with different shRNAs using β‐actin levels as the internal controls. RT‐qPCR was performed to detect mRNA levels of (B) YWHAZ in T24 cells treated with shRNAs and (C) the related apoptosis genes in shRNA‐1‐treated T24 cells. Cells treated with ‘scramble’ served as the control in panels A to C. (D) T24 cells transfected with shRNA‐1 were treated with the indicated concentrations of doxorubicin or cisplatin. Cell viability was measured by Alamar Blue assays 4 days after drug treatments. (E) T24 cells transfected with shRNA‐1 were treated with 6 Gy radiation, after which cell viability was assessed every 24 h for 4 days. (F) Incidence rate of preapoptosis among treated cells 48 h after chemo−/radio‐therapy was determined by annexin‐V staining. (G) Proliferation of shRNA‐1‐treated cells was monitored for 4 days under the treatment of 50 nm doxorubicin or 5 μm cisplatin. (H) Colony formation activity was studied on shRNA‐1‐treated T24 cells after 2 weeks treatment with 50 nm doxorubicin or 5 μm cisplatin (left). Colony numbers per 35‐mm dish are shown in bar charts (right). Cells treated with ‘scramble’ served as the controls in panels D to H. Data were expressed as means ± SD from five replicates in each experimental group. Paired t‐tests were performed to evaluate differences between ‘scramble’‐ and shRNA‐1‐treated cells. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
In this study, we confirmed YWHAZ amplification/overexpression in around 20% of UCUB samples; moreover, this genetic alteration was more frequently detected in MIBC than NMIBC (Figures 1 and 2). In addition, YWHAZ overexpression was found to be a potential prognostic marker indicative of poor clinical outcomes and shorter survival times (Figure 2 and Table 1). Mapping downstream signaling using GSEA and STRING revealed the key role played by YWHAZ in anti‐apoptosis pathways through downregulation of genes involved in caspase‐mediated cell death (Figure 3). A cell‐based functional study further confirmed that YWHAZ overexpression can drive bladder cancer toward resistance to chemo‐/radio‐therapy which can be proven by IHC staining on recurrent UCUBs after chemotherapy (Figures 4 and 5). Conversely, gene knockdown using a specific shRNA in YWHAZ‐overexpressing bladder cancer cells triggered marked increases in cell death after drug or radiation treatments (Figure 6). These data suggest YWHAZ a potent cancer driver that provides survival advantages against environmental stresses to UCUB cells. Consequently, targeting YWHAZ may be an effective strategy to increase the therapeutic efficacy of anticancer therapies.
YWHAZ amplification is also reported to be clinically significant in other cancer types, including breast 12, prostate 18, 20, lung 16, and oral 15, 17 cancers. Although YWHAZ has also been reported to act as a tumor suppressor during bladder cancer development 32, 33, our results consistently show that YWHAZ overexpression negatively regulates apoptosis, enabling cellular adaptation to the presence of chemotherapeutic drugs. In contrast to anti‐apoptotic effects, other proteins participating in diverse functions/pathways within cells were found to be upregulated along with YWHAZ overexpression (see supplementary material, Tables S3 and S4). Interestingly, some upregulated effectors implies altered cellular metabolism/homeostasis in cells overexpressing YWHAZ. For example, PYCR1 overexpression can alter amino acid metabolism via the glutamine‐proline regulatory axis 34, which promotes aggressiveness in human cancers 35, 36, 37, and is thus considered to be a useful therapeutic target 37, 38. Similarly, upregulation of SLC2A6 (A.K.A. GLUT6) was reported to promote glucose uptake 39, while POLR2K upregulation was found to increase nucleic acid metabolism 40 in aggressive cancers. Further study is therefore needed to investigate the impact of YWHAZ on bladder cancer metabolism.
In our study, we also found that YWHAZ amplification/overexpression associated with downregulation of innate immune responses, suggesting its involvement in tumor microenvironment remodeling (Figure 3A). In recent years, immune checkpoint blockage therapy has shown unprecedented anticancer activity against metastatic UCUBs 41. Nevertheless, more than half of patients using this approach receive no benefit, and some even have a worse outcome 42, 43, 44, indicating the need for a useful biomarker predictive of the therapeutic outcome of such treatments. It is known that innate immune system activation is critical for triggering spontaneous adaptive immune responses against cancer during immunotherapy 45, 46. Thus, combinational therapy to boost immune responses by inducing cytosolic DNA with irradiation or inflammatory tumor death using a bacterial adjuvant (e.g. Bacillus Calmette–Guérin treatment) were proposed as possible strategies to improve the therapeutic efficacy of immunotherapy for UCUB 41, 46, 47. In UCUBs with YWHAZ amplification/overexpression, genes involved in interferon signaling, TLR‐4 signaling, inflammasome network, antigen presentation/TCR recognition, and CD28 co‐stimulation were found significantly downregulated (see supplementary material, Table S4). More detailed studies are needed to determine whether YWHAZ could be used as a biomarker to define patients eligible for immune checkpoint blockage therapy and whether anti‐YWHAZ approach could be a useful strategy for improving the therapeutic efficacy of immunotherapy.
Although we defined the tendency to detect YWHAZ amplification/overexpression in UCUBs at more advanced stages, we are not yet able to confirm whether YWHAZ functions as a molecular link to drive malignant transition from NMIBC to MIBC. Using a 2D cell culture system, we found limited effects of YWHAZ levels on cell proliferation and invasion, either by protein overexpression in RT4 cells or gene knockdown in T24 cells (see supplementary material, Figure S2). But still, our GSEA analysis indicated the activation of RAP1 signaling pathway in YWHAZ‐overexpressing UCUBs (see supplementary material, Figure S1 and Table S4). Because RAP1 signaling pathway has been proven to be critical in controlling cell migration/invasion and adhesion dynamics via cadherin or integrin signaling 48, 49, 50, an environment that provides more cell–cell contact and cell–matrix adhesion might be necessary to study the impact of YWHAZ on cancer behaviors. In addition, recent studies indicate that YWHAZ acts in concert with the TGF‐β axis to control cancer‐stromal cell interaction and epithelial–mesenchymal transition 21, 26, 51, 52, which suggests other microenvironmental factors are involved in YWHAZ‐mediated carcinogenesis and cancer progression. Organoid cell cultures in a 3D condition or PDX models of tumors overexpressing YWHAZ may provide physiologically relevant contexts for us to better understand the potential roles of YWHAZ in determining UCUB invasiveness.
Since YWHAZ has been implicated in the carcinogenesis of many cancer types, the present study provides a more comprehensive investigation to address its functional roles in UCUB development. In addition to apoptosis regulation and TGF‐β signaling, we also revealed the potential involvement of YWHAZ in regulating critical cancer phenotypes, including cancer metabolism, innate immune responses, and T cell activation. Nevertheless, the association of YWHAZ amplification/overexpression to clinical outcomes and other relevant factors was evaluated retrospectively, our findings need to be confirmed with a prospective analysis on another cohort. With diverse cellular functions, YWHAZ could be a useful therapeutic target for treating advanced UCUB, one that enables achievement of better clinical outcomes when used in combination with other therapies.
Author contributions statement
SCL and JJCS designed the experiments and validated the data. CCY, CFL, IHC, MTL, GK, and CYC collected clinical samples, recorded clinical notes, and performed clinical association study. CFL, IHC, MTL, and CYC prepared the tissue microarray, carried out IHC staining, and then did sample scoring. ZJL, GK, CMC, and TH made the gene constructs, conducted cell‐based studies, and analyzed the data. IHC, PKK and GK performed data mining. CCY, CFL, SCL, and JJC confirmed the data and wrote the manuscript.
SUPPLEMENTARY MATERIAL ONLINE.
Figure S1. Reactomes/pathways associated with YWHAZ amplification/overexpression in UCUBs
Figure S2. Impacts of YWHAZ expression levels on cell proliferation and invasion in bladder cancer cells
Table S1. Sequences of oligonucleotide primers used in this study
Table S2. Genes involved in the chromosome 8q22.3 amplicon in urothelial carcinoma of urinary bladder (UCUB)
Table S3. Top 20 genes concurrently up‐regulated with YWHAZ amplification/overexpression in UCUBs
Table S4. Gene set enrichment analyses of genes concurrently up‐ or downregulated with YWHAZ amplification/overexpression in UCUBs
Supporting information
Figure S1. Reactomes/pathways associated with YWHAZ amplification/overexpression in UCUBs
Figure S2. Impacts of YWHAZ expression levels on cell proliferation and invasion in bladder cancer cells
Table S1 Sequences of oligonucleotide primers used in this study
Table S2 Genes involved in the chromosome 8q22.3 amplicon in urothelial carcinoma of urinary bladder (UCUB)
Table S3 Top 20 genes concurrently upregulated with YWHAZ amplification/overexpression in UCUBs
Table S4 Gene set enrichment analyses of genes concurrently up‐ or downregulated with YWHAZ amplification/ overexpression in UCUBs
Acknowledgements
The authors thank Mr. Stev Chao at China Medical University Hospital and Mr. Daniel Gracilla at National Sun Yatsen University for technical assistance. This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 105‐2622‐B‐110‐001‐CC2; 105‐2314‐B‐384‐003‐MY3; MOST 106‐2314‐B‐110‐001‐MY3), Ministry of Health and Welfare, Taiwan (MHW 10725), NSYSU‐KMU (107‐P012), National Health Research Institutes (NHRI‐107A1‐MG‐CO‐01‐1720‐2), and NSYSU‐KVGH (VGHNSU106‐003, VGHKS107‐125) joint research projects.
No conflicts of interest were declared.
Contributor Information
Shan‐Chih Lee, Email: sclee@csmu.edu.tw.
Jim J‐C Sheu, Email: jimsheu@mail.nsysu.edu.tw.
References
- 1. Antoni S, Ferlay J, Soerjomataram I, et al Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 2017; 71: 96–108. [DOI] [PubMed] [Google Scholar]
- 2. Glaser AP, Fantini D, Shilatifard A, et al The evolving genomic landscape of urothelial carcinoma. Nat Rev Urol 2017; 14: 215–229. [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pietzak EJ, Bagrodia A, Cha EK, et al Next‐generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017; 72: 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Erdmann J. Genetics: a clearer view. Nature 2017; 551: S44–S45. [DOI] [PubMed] [Google Scholar]
- 6. Bryan RT, Hussain SA, James ND, et al Molecular pathways in bladder cancer: part 2. BJU Int 2005; 95: 491–496. [DOI] [PubMed] [Google Scholar]
- 7. Bryan RT, Hussain SA, James ND, et al Molecular pathways in bladder cancer: part 1. BJU Int 2005; 95: 485–490. [DOI] [PubMed] [Google Scholar]
- 8. Mitra AP, Datar RH, Cote RJ. Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J Clin Oncol 2006; 24: 5552–5564. [DOI] [PubMed] [Google Scholar]
- 9. Aitken A. Post‐translational modification of 14‐3‐3 isoforms and regulation of cellular function. Semin Cell Dev Biol 2011; 22: 673–680. [DOI] [PubMed] [Google Scholar]
- 10. Aitken A. 14‐3‐3 proteins on the MAP. Trends Biochem Sci 1995; 20: 95–97. [DOI] [PubMed] [Google Scholar]
- 11. Muslin AJ, Tanner JW, Allen PM, et al Interaction of 14‐3‐3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 1996; 84: 889–897. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Zou L, Li Q, et al Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 2010; 16: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neal CL, Yu D. 14‐3‐3zeta as a prognostic marker and therapeutic target for cancer. Expert Opin Ther Targets 2010; 14: 1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi JE, Hur W, Jung CK, et al Silencing of 14‐3‐3zeta over‐expression in hepatocellular carcinoma inhibits tumor growth and enhances chemosensitivity to cis‐diammined dichloridoplatium. Cancer Lett 2011; 303: 99–107. [DOI] [PubMed] [Google Scholar]
- 15. Matta A, Bahadur S, Duggal R, et al Over‐expression of 14‐3‐3zeta is an early event in oral cancer. BMC Cancer 2007; 7: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan T, Li R, Todd NW, et al Up‐regulation of 14‐3‐3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res 2007; 67: 7901–7906. [DOI] [PubMed] [Google Scholar]
- 17. Lin M, Morrison CD, Jones S, et al Copy number gain and oncogenic activity of YWHAZ/14‐3‐3zeta in head and neck squamous cell carcinoma. Int J Cancer 2009; 125: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murata T, Takayama K, Urano T, et al 14‐3‐3zeta, a novel androgen‐responsive gene, is upregulated in prostate cancer and promotes prostate cancer cell proliferation and survival. Clin Cancer Res 2012; 18: 5617–5627. [DOI] [PubMed] [Google Scholar]
- 19. Nishimura Y, Komatsu S, Ichikawa D, et al Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer 2013; 108: 1324–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Menon R, Deng M, Ruenauver K, et al Somatic copy number alterations by whole‐exome sequencing implicates YWHAZ and PTK2 in castration‐resistant prostate cancer. J Pathol 2013; 231: 505–516. [DOI] [PubMed] [Google Scholar]
- 21. Chen CH, Chuang SM, Yang MF, et al A novel function of YWHAZ/beta‐catenin axis in promoting epithelial–mesenchymal transition and lung cancer metastasis. Mol Cancer Res 2012; 10: 1319–1331. [DOI] [PubMed] [Google Scholar]
- 22. Weerasekara VK, Panek DJ, Broadbent DG, et al Metabolic‐stress‐induced rearrangement of the 14‐3‐3zeta interactome promotes autophagy via a ULK1‐ and AMPK‐regulated 14‐3‐3zeta interaction with phosphorylated Atg9. Mol Cell Biol 2014; 34: 4379–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao X, He Y, Gao LM, et al Ser9‐phosphorylated GSK3beta induced by 14‐3‐3zeta actively antagonizes cell apoptosis in a NF‐kappaB dependent manner. Biochem Cell Biol 2014; 92: 349–356. [DOI] [PubMed] [Google Scholar]
- 24. Lee YK, Hur W, Lee SW, et al Knockdown of 14‐3‐3zeta enhances radiosensitivity and radio‐induced apoptosis in CD133(+) liver cancer stem cells. Exp Mol Med 2014; 46: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nutt LK, Buchakjian MR, Gan E, et al Metabolic control of oocyte apoptosis mediated by 14‐3‐3zeta‐regulated dephosphorylation of caspase‐2. Dev Cell 2009; 16: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu J, Acharya S, Sahin O, et al 14‐3‐3zeta turns TGF‐beta's function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell 2015; 27: 177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sheu JJ, Hua CH, Wan L, et al Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res 2009; 69: 2568–2576. [DOI] [PubMed] [Google Scholar]
- 28. Lai MT, Hua CH, Tsai MH, et al Talin‐1 overexpression defines high risk for aggressive oral squamous cell carcinoma and promotes cancer metastasis. J Pathol 2011; 224: 367–376. [DOI] [PubMed] [Google Scholar]
- 29. Riester M, Werner L, Bellmunt J, et al Integrative analysis of 1q23.3 copy‐number gain in metastatic urothelial carcinoma. Clin Cancer Res 2014; 20: 1873–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robertson AG, Kim J, Al‐Ahmadie H, et al Comprehensive molecular characterization of muscle‐invasive bladder cancer. Cell 2017; 171: 540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subramanian A, Tamayo P, Mootha VK, et al Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S, Jiang H, Wen H, et al Knockdown of tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein zeta (YWHAZ) enhances tumorigenesis both in vivo and in vitro in bladder cancer. Oncol Rep 2018; 39: 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu S, Wu Y, Yang T, et al Coexistence of YWHAZ amplification predicts better prognosis in muscle‐invasive bladder cancer with CDKN2A or TP53 loss. Oncotarget 2016; 7: 34752–34758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Craze ML, Cheung H, Jewa N, et al MYC regulation of glutamine‐proline regulatory axis is key in luminal B breast cancer. Br J Cancer 2018; 118: 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai F, Miao Y, Liu C, et al Pyrroline‐5‐carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non‐small cell lung cancer. Oncol Lett 2018; 15: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding J, Kuo ML, Su L, et al Human mitochondrial pyrroline‐5‐carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017; 38: 519–531. [DOI] [PubMed] [Google Scholar]
- 37. Zeng T, Zhu L, Liao M, et al Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol 2017; 34: 27. [DOI] [PubMed] [Google Scholar]
- 38. Loayza‐Puch F, Rooijers K, Buil LC, et al Tumour‐specific proline vulnerability uncovered by differential ribosome codon reading. Nature 2016; 530: 490–494. [DOI] [PubMed] [Google Scholar]
- 39. Byrne FL, Poon IK, Modesitt SC, et al Metabolic vulnerabilities in endometrial cancer. Cancer Res 2014; 74: 5832–5845. [DOI] [PubMed] [Google Scholar]
- 40. Kelly RS, Sinnott JA, Rider JR, et al The role of tumor metabolism as a driver of prostate cancer progression and lethal disease: results from a nested case‐control study. Cancer Metab 2016; 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rouanne M, Roumiguie M, Houede N, et al Development of immunotherapy in bladder cancer: present and future on targeting PD(L)1 and CTLA‐4 pathways. World J Urol 2018; 36: 1727–1740. [DOI] [PubMed] [Google Scholar]
- 42. Champiat S, Dercle L, Ammari S, et al Hyperprogressive disease is a new pattern of progression in cancer patients treated by Anti‐PD‐1/PD‐L1. Clin Cancer Res 2017; 23: 1920–1928. [DOI] [PubMed] [Google Scholar]
- 43. Champiat S, Ferrara R, Massard C, et al Hyperprogressive disease: recognizing a novel pattern to improve patient management. Nat Rev Clin Oncol 2018; 15: 748–762. [DOI] [PubMed] [Google Scholar]
- 44. Kato S, Goodman A, Walavalkar V, et al Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res 2017; 23: 4242–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol 2015; 33: 445–474. [DOI] [PubMed] [Google Scholar]
- 46. Corrales L, Matson V, Flood B, et al Innate immune signaling and regulation in cancer immunotherapy. Cell Res 2017; 27: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Liu J, Yang X, et al Bacillus Calmette–Guerin and anti‐PD‐L1 combination therapy boosts immune response against bladder cancer. Onco Targets Ther 2018; 11: 2891–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell‐cell junction formation. J Cell Sci 2007; 120: 17–22. [DOI] [PubMed] [Google Scholar]
- 49. Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol 2009; 21: 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang YL, Wang RC, Cheng K, et al Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med 2017; 14: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang XY, Ke AW, Shi GM, et al alphaB‐crystallin complexes with 14‐3‐3zeta to induce epithelial–mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology 2013; 57: 2235–2247. [DOI] [PubMed] [Google Scholar]
- 52. Lu J, Guo H, Treekitkarnmongkol W, et al 14‐3‐3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial–mesenchymal transition. Cancer Cell 2009; 16: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Reactomes/pathways associated with YWHAZ amplification/overexpression in UCUBs
Figure S2. Impacts of YWHAZ expression levels on cell proliferation and invasion in bladder cancer cells
Table S1 Sequences of oligonucleotide primers used in this study
Table S2 Genes involved in the chromosome 8q22.3 amplicon in urothelial carcinoma of urinary bladder (UCUB)
Table S3 Top 20 genes concurrently upregulated with YWHAZ amplification/overexpression in UCUBs
Table S4 Gene set enrichment analyses of genes concurrently up‐ or downregulated with YWHAZ amplification/ overexpression in UCUBs


