Abstract
Background
Manual width measurements of the middle cerebellar peduncle on MRI were shown to improve the accuracy of an imaging‐guided diagnosis of multiple system atrophy (MSA). Recently, automated volume segmentation algorithms were able to reliably differentiate patients with Parkinson's disease (PD) and the parkinsonian variant of MSA. The objective of the current study was to integrate probabilistic information of the middle cerebellar peduncle into an existing MRI atlas for automated subcortical segmentation and to evaluate the diagnostic properties of the novel atlas for the differential diagnosis of MSA (parkinsonian and cerebellar variant) versus PD.
Methods
Three Tesla MRI scans of 48 healthy individuals were used to establish an automated whole‐brain segmentation procedure that includes the volumes of the putamen, cerebellar gray and white matter, and the middle cerebellar peduncles. Classification accuracy of segmented volumes were tested in early‐stage MSA patients (18 MSA‐parkinsonism, 13 MSA‐cerebellar) and 19 PD patients using a C4.5 classifier.
Results
Putaminal and infratentorial atrophy were present in 77.8% and 61.1% of MSA‐parkinsonian patients, respectively. Four of 18 MSA‐parkinsonian patients (22.2%) had infratentorial atrophy without evidence of putaminal atrophy. Infratentorial atrophy was present in all MSA‐cerebellar patients, with concomitant putaminal atrophy in 46.2% of these cases. The diagnostic algorithm using putaminal and infratentorial volumetric information correctly classified all PD patients and 96.8% of MSA patients.
Conclusions
The middle cerebellar peduncle was successfully integrated into a subcortical segmentation atlas, and its excellent diagnostic accuracy outperformed existing volumetric MRI processing strategies in differentiating MSA patients with variable atrophy patterns from PD patients. © 2019 The Authors. Movement Disorders published by Wiley Periodicals, Inc. on behalf of International Parkinson and Movement Disorder Society.
Keywords: atlas, FreeSurfer, MRI, multiple system atrophy, Parkinson's disease
Multiple system atrophy (MSA) is a sporadic, adult‐onset α‐synucleinopathy with a rapidly progressive disease course and widespread neurodegeneration.1 Clinicopathological studies identified 2 motor phenotypes—a parkinsonian variant (MSA‐P) and a cerebellar variant (MSA‐C) with predominant degeneration in striatonigral and olivopontocerebellar nuclei and their projections, respectively.2 The key pathological findings include demonstration of neuronal cell loss and the appearance of widespread oligodendroglial α‐synuclein positive inclusions.3 Neuropathological studies revealed considerable variability in the severity and regional distribution of neuropathological changes between individual patients. In 2 series, 42% to 49% of MSA cases showed equally severe striatonigral degeneration (SND) and olivopontocerebellar atrophy (OPCA), whereas predominant SND or OPCA was seen in 34% and 17% of cases, respectively.4 Those studies failed to find a correlation between the severity of clinical symptoms and the extent of neuronal degeneration or the intracellular load of glial cytoplasmatic inclusions. Based on these observations, MR volumetry will likely require measures of both SND and OPCA to adequately capture MSA pathology. Within the olivopontocerebellar system, the middle cerebellar peduncle (MCP) was shown to be one of the most prominently affected brain regions, and imaging studies in patients with MSA and PD have shown that MSA is associated with a greater pontine and MCP atrophy compared with PD.5, 6, 7, 8, 9, 10, 11, 12
So far, diagnostic imaging studies in MSA have focused on the validation of either 2‐dimensional distance measures, calculation of indices, or whole‐brain voxel‐based analysis to identify disease characteristic patterns of atrophy.13, 14 Although such approaches have the potential to reveal disease‐related signal alterations, efforts to rank patterns of atrophy by their severity at the single‐subject level and hence disentangle the dimensions’ “signal distribution” and “signal quantity” are sparse.15, 16 Advances in MRI postprocessing algorithms including automated compartmentalization of the brain into multiple anatomic regions, volume calculation, and z transformation have provided an opportunity to localize and grade disease‐specific brain atrophy patterns in relation to age‐ and sex‐matched healthy control cohorts.17, 18 Recently, such an approach was shown to reliably differentiate progressive supranuclear palsy Richardson type and MSA‐P from Parkinson's disease.19 However, there is still room for improvement because current atlases lack probabilistic information on infratentorial brain structures that are relevant to MSA neuropathology. In particular, cerebellar volume, the MCP width, increased apparent diffusion coefficient within the MCP as well as the cerebellar hemispheres, and pons atrophy were shown to improve diagnostic accuracy in the differential diagnosis of MSA.9, 20, 21, 22
In the present study, we sought to further refine the currently available FreeSurfer‐based subcortical segmentation atlas by adding probabilistic information on the location of the MCP. The morphometric profile was determined in patients with clinically defined MSA‐P or MSA‐C to evaluate the relative contribution of MCP measurements to diagnostic accuracy and to estimate the added diagnostic yield of MCP measurements for the differential diagnosis of MSA and PD.
Materials and Methods
Participants
Forty‐eight healthy individuals (HCs) without evidence of any neurological disorder on careful clinical examination underwent conventional 3 T MRI and were used for the development of the novel atlas. Twenty‐six of these 48 healthy individuals were selected by investigator‐independent case‐control matching (see below) as a sex‐ and age‐matched healthy control group in our cross‐sectional study. MSA and PD patients were identified from the MRI database of the Movement Disorders outpatient clinic of the Department of Neurology at the Medical University of Innsbruck. Inclusion criteria were defined as: (1) a clinical diagnosis of probable MSA (either parkinsonian or cerebellar variant) or PD at the last visit according to consensus operational criteria,23, 24 and (2) a clinical follow‐up of at least 24 months. Exclusion criteria were dementia according to the Diagnostic and Statistical Manual IV criteria, white‐matter lesions grades 2 and 3, vascular or space‐occupying lesions within the cerebrum, or motion artifacts on MRI.
The study was approved by the Ethics Committee of the Medical University of Innsbruck.
MR Sequence
All MRI measurements were performed on a 3.0 Tesla whole‐body MR scanner (Magnetom Verio, Siemens, Erlangen, Germany) equipped with a 12‐channel head coil. All participants underwent the same MRI protocol, including whole‐brain T1‐weighted, fluid‐attenuated inversion‐recovery, T2‐, and proton density‐weighted as well as diffusion tensor imaging. The MRI parameters for the coronal T1‐weighted 3‐dimensional (3‐D) magnetization‐prepared rapid gradient echo were TR, 1800 milliseconds; TE, 2.18 milliseconds; inversion time, 900 milliseconds; slice thickness, 1.2 mm; matrix, 256 × 204 pixels; number of excitations, 1; flip angle, 9°; field of view, 220 × 165 mm.
Segmentation of subcortical regions from T1‐weighted 3‐D structural MRI data and estimation of structure volumes were performed using the FreeSurfer tool kit (version 6.0, available at http://surfer.nmr.mgh.harvard.edu/). The procedure automatically segments and labels brain structures based on (1) the prior probabilities of anatomical classes throughout an atlas space, (2) the prior probability obtained from frequency histograms within the atlas space providing the likelihood that a given anatomical class occurs at a given atlas location, and (3) the modeling of local spatial relationships between labeled structures as anisotropic nonstationary Markov random fields.18 The processing of T1‐weighted 3‐D structural MRIs included correction of motion artifacts, removal of nonbrain tissue, transformation into the Talairach reference space, segmentation of the subcortical white matter and deep gray‐matter volumetric structures, intensity normalization, tessellation of the gray matter, white‐matter boundary, automated topology correction, and surface deformation following intensity gradients.17, 18, 25, 26, 27 The procedure generated the intracranial volume as well as the volumes of the putamen, caudate, globus pallidum, thalamus, hippocampus, amygdala, brain stem, cerebellar white and gray matter, MCP, and the third and fourth ventricles. The preprocessing steps were visually inspected to ensure that no misalignment of brain structures had occurred. All MRI scans were included in the study. All imaging data were processed on an HP DL360p server with a total of 48 CPUs. Time needed to perform the automated volume calculation was approximately 8 hours for each subject, and 48 subjects were processed in parallel.
Atlas Development
The MCP was identified in all subjects by exploiting a validated probabilistic 3‐D atlas of cerebellar white‐matter structure.28 The observer independently segmented brain volume of the MCP was transformed to the individual subject's space using the deformation parameters obtained from the normalization procedure into Montreal Neurological Institute space by the software package SPM12 in Matlab 9.2. A subject's MCP volumes were reviewed and manually corrected if required in native space. Subsequently, the individual MCP maps were integrated into the default subcortical segmentation atlas as described previously.18 Three‐dimensional models of automatically segmented MCP in PD and MSA‐C are presented in the online supplement (Supplementary Fig. 1)
Statistical Analysis
Data analysis was performed using SPSS 24.0 and STATA 14.2. Case‐control matching was performed through the SPSS extension FUZZY with a fuzz of 0 for gender and 1.5 years for age.
Demographic data are presented as frequencies, means ± standard deviations, or median (interquartile range) according to data distribution. Gaussian distribution was confirmed by visual analysis of the Q‐Q plots and the Kolmogorov‐Smirnov test. Group differences of normally distributed data were analyzed by parametric tests (analysis of variance [ANOVA] or Student t test as applicable), non‐Gaussian distributed variables by nonparametric tests (Kruskal‐Wallis 1‐way ANOVA by ranks or Mann‐Whitney U test as applicable). Distributional differences were determined by the Pearson's chi‐square test for independence. Bonferroni correction for multiple testing was applied for post hoc testing.
For decision tree analysis, all volumetric measures were normalized by each subject's intracranial volume. Data were further processed by z‐transformation using mean centering and unit‐variance scaling of the sex‐ and age‐adjusted healthy control cohorts. Z scores ≥ ‐1.96 were omitted to prevent inclusion of insignificant cutoff values in the decision tree. A MRI‐based decision tree attempting to classify clinically diagnosed PD and MSA patients was developed exploiting the C4.5 classifier algorithm29, 30 implemented by the Waikato Environment of Knowledge Analysis machine learning software (WEKA 3.8.1). The following brain regions with potential discriminative value were considered: cerebellar white matter, cerebellar cortex, superior and middle cerebellar peduncle, putamen, caudate nucleus, pons, and midbrain. A 20‐fold cross‐validation was employed to estimate the classification performance of the decision tree.
Results
Demographics and Basic Clinical Information
Twenty‐six age‐matched healthy individuals, 31 MSA patients (18 MSA‐P, 13 MSA‐C), and 19 PD patients were included in the validation study. There was no significant difference between the study groups with regard to sex distribution (P = 0.882), age (P = 0.118), and disease duration (P = 0.787). Motor impairment was greater in MSA patients than in PD patients, as measured by Hoehn & Yahr staging (P < 0.001) and the UPDRS (P < 0.001). Detailed information on demographics and basic clinical information are provided in Table 1.
Table 1.
Demographics and basic clinical information
| HC sample | MSA | MSA‐P | MSA‐C | PD | P (post hoc testsa) | |
|---|---|---|---|---|---|---|
| Sample size | 26 | 31 | 18 | 13 | 19 | ‐ |
| Age at scan (years), mean ± SD | 60.2 ± 5.9 | 61.8 ± 8.0 | 63.2 ± 7.0 | 59.7 ± 9.1 | 64.2 ± 5.3 | 0.118 |
| Disease duration to scan (years), mean ± SD | ‐ | 2.6 ± 1.7 | 2.4 ± 1.8 | 2.8 ± 1.5 | 2.7 ± 1.4 | 0.787 |
| Sex (female/male) | 11 / 15 | 14/17 | 9/9 | 5/8 | 7/12 | 0.882 |
| Hoehn & Yahr staging, median (IQR) | ‐ | 3 (1) | 3 (1) | 3 (1) | 2 (1) | <0.001 PD vs MSA‐P, <0.001 PD vs MSA‐C, 0.005 MSA‐C, 0.844 |
| UPDRS III, mean ± SD | ‐ | 38.8 ± 13.3 | 40.9 ± 7.0 | 35.8 ± 19 | 25.5 ± 6.7 | <0.001 PD vs MSA‐P, <0.001 PD vs MSA‐C, 0.041 MSA‐P vs MSA‐C, 0.659 |
| UPDRS sum score, mean ± SD | ‐ | 62.0 ± 20.1 | 65.2 ± 10.5 | 57.6 ± 28.6 | 36.2 ± 9.8 | <0.001 PD vs MSA‐P, <0.001 PD vs MSA‐C, 0.003 MSA‐P vs MSA‐C, 0.675 |
| MMSE, mean ± SD | ‐ | 27.9 ± 2.2 | 28.2 ± 2.0 | 27.5 ± 2.4 | 29.0 ± 1.4 | 0.104 |
P values derived from group comparison between healthy controls (if applicable) and MSA‐P, MSA‐C, and PD patients. For post hoc testing Bonferroni correction for multiple testing was applied.
Morphometric MR Profile of MSA Variants
A detailed presentation of group differences is given in Table 2, and the frequency of atrophy of selected brain regions is presented in Table 3. In clinically classified MSA‐P patients putaminal and infratentorial atrophy were present in 77.8% and 61.1%, respectively. Isolated putaminal atrophy was seen in 38.8% of MSA‐P patients, whereas 22.2% showed isolated infratentorial atrophy without evidence of putaminal atrophy (Table 3, Fig. 1). Infratentorial atrophy was present in all MSA‐C patients, and 46.2% had concomitant putaminal atrophy. One PD patient (5.3%) who was clinically classified as having PD after 2 years of clinical follow‐up showed brain atrophy of the cerebellar white matter and the cerebellar cortex with volume reductions of more than 2 standard deviations compared with the mean of the sex‐ and age‐matched HC population. Automated brain volumetry of candidate brain regions was correlated with measures of disease severity (ie, UPDRS scores and Hoehn & Yahr staging; Supplementary Table 1). No correlation was found between disease duration and brain volumetry in candidate brain regions.
Table 2.
Total intracranial volume‐corrected volumes of different brain regions
| HC | PD | MSA‐P | MSA‐C | P a | |
|---|---|---|---|---|---|
| Cerebellar white matter | 9.71 ± 1.09 | 9.68 ± 1.24 | 8.28 ± 2.18 | 5.51 ± 1.26 | <0.001 HC vs IPD, 1.000 HC vs MSA‐P, 0.015 HC vs MSA‐C, <0.001 PD vs MSA‐P, 0.032 PD vs MSA‐C, <0.001 MSA‐P vs MSA‐C, <0.001 |
| Cerebellar cortex | 36.78 ± 2.7 | 36.38 ± 2.78 | 33.38 ± 4.69 | 29.74 ± 2.38 | <0.001 HC vs PD, 1.000 HC vs MSA‐P, 0.006 HC vs MSA‐C, <0.001 PD vs MSA‐P, 0.039 PD vs MSA‐C, <0.001 MSA‐P vs. MSA‐C 0.018 |
| Thalamus | 4.41 ± 0.27 | 4.52 ± 0.3 | 4.41 ± 0.31 | 4.30 ± 0.35 | 0.257 |
| Caudate | 2.11 ± 0.24 | 2.14 ± 0.23 | 1.9 ± 0.32 | 2.11 ± 0.20 | 0.019 HC vs PD, 1.000 HC vs MSA‐P, 0.048 HC vs MSA‐C, 1.000 PD vs MSA‐P, 0.034 PD vs MSA‐C, 1.000 MSA‐P vs MSA‐C, 0.138 |
| Putamen | 3.1 ± 0.23 | 3.06 ± 0.15 | 2.39 ± 0.43 | 2.72 ± 0.38 | <0.001 HC vs PD, 1.000 HC vs MSA‐P, <0.001 HC vs MSA‐C, 0.003 PD vs MSA‐P, <0.001 PD vs MSA‐C, 0.015 MSA‐P vs MSA‐C, 0.025 |
| Hippocampus | 2.6 ± 0.29 | 2.69 ± 0.23 | 2.54 ± 0.32 | 2.62 ± 0.22 | 0.453 |
| Amygdala | 0.97 ± 0.11 | 0.99 ± 0.09 | 0.96 ± 0.12 | 1.03 ± 0.10 | 0.266 |
| Pons | 9.7 ± 0.8 | 9.85 ± 1.02 | 9.01 ± 1.65 | 6.23 ± 1.18 | <0.001 HC vs IPD, 1.000 HC vs MSA‐P, 0.345 HC vs MSA‐C, <0.001 PD vs MSA‐P, 0.182 PD vs MSA‐C, <0.001 MSA‐P vs MSA‐C, <0.001 |
| Midbrain | 3.94 ± 0.29 | 3.97 ± 0.29 | 3.86 ± 0.35 | 3.47 ± 0.23 | <0.001 HC vs IPD, 1.000 HC vs MSA‐P, 1.000 HC vs MSA‐C, <0.001 PD vs HC, 1.000 PD vs MSA‐P, 1.000 PD vs MSA‐C, <0.001 MSA‐P vs MSA‐C, 0.003 |
| Superior cerebellar peduncle | 0.84 ± 0.08 | 0.95 ± 0.14 | 0.87 ± 0.17 | 0.74 ± 0.14 | <0.001 HC vs PD, 0.035 HC vs MSA‐P, 1.000 HC vs MSA‐C, 0.189 PD vs MSA‐P, 0.346 PD vs MSA‐C, <0.001 MSA‐P vs MSA‐C, 0.058 |
| Middle cerebellar peduncle | 6.56 ± 0.69 | 6.78 ± 0.86 | 5.75 ± 1.29 | 4.36 ± 0.70 | <0.001 HC vs IPD, 1.000 HC vs MSA‐P, 0.027 HC vs MSA‐C, <0.001 PD vs MSA‐P, 0.006 PD vs MSA‐C, <0.001 MSA‐P vs MSA‐C, <0.001 |
Values represent the ratio between the volume of the region divided by the subject‐specific total intracranial volume. In case of the bihemispheric presence of the region, the more affected side is presented. All numerical data are written as m × 10‐3.
P values derived from group comparison between healthy controls and MSA‐P, MSA‐C, and PD patients.
Table 3.
Frequency of z scores exceeding the 95% confidence interval of age‐ and sex‐matched healthy controls (ie, z score below −1.96) in selected regions
| Region | MSA, overall | MSA‐P | MSA‐C | PD |
|---|---|---|---|---|
| Cerebellar white matter | 21 (67.7%) | 8 (44.4%) | 13 (100%) | 1 (5.3%) |
| Cerebellar cortex | 19 (61.3%) | 9 (50.0%) | 10 (76.9%) | 1 (5.3%) |
| Middle cerebellar peduncle | 18 (58.1%) | 6 (33.3%) | 12 (92.3%) | 0 (0.0%) |
| Putamen | 20 (64.5%) | 14 (77.8%) | 6 (46.2%) | 0 (0%) |
| Pons | 13 (41.9%) | 2 (11.1%) | 11 (84.6%) | 0 (0.0%) |
| Midbrain | 1 (3.2%) | 0 (0.0%) | 1 (7.7%) | 0 (0.0%) |
Figure 1.
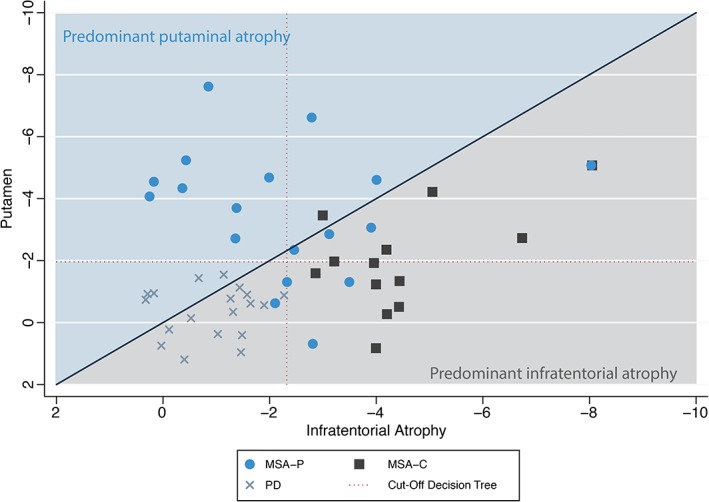
Scatterplot of brain atrophy patterns in MSA‐P compared with MSA‐C and PD patients. Atrophy of the putamen and the most affected region among the olivo‐ponto‐cerebellar trajectory (ie, middle cerebellar peduncle, cerebellar white matter, cerebellar cortex, pons, labeled “infratentorial atrophy”) are scattered to illustrate the predominant atrophy pattern in any individual patient. [Color figure can be viewed at wileyonlinelibrary.com]
Implications for the Differential Diagnosis of MSA Versus PD
Based on a C4.5 decision tree algorithm, atrophy measures of the putamen and the MCP were selected as the most useful parameters in differentiating MSA from PD. All PD patients and 93.5% of MSA patients (ie, sensitivity and specificity of 93.5% and 100%, respectively) were classified correctly (Fig. 2a). Cross‐validation of this model yields a weighted average F measure and a Cohen's kappa of 0.94 and 0.87, respectively. When the obtained cutoff values were exploited for the differential diagnosis of MSA‐P versus PD, a slightly lower sensitivity of 88.9% was observed while the specificity remained at 100% (overall correct classification, 94.6%). The algorithm unambiguously discriminated MSA‐C patients from PD patients with corresponding sensitivity and specificity rates of 100% each. The omission of the MCP volume resulted in a reduced weighted average F measure on cross‐validation of 0.90. In contrast, pooling of infratentorial atrophy measures (ie, selecting the most severely affected Z score of the MCP, cerebellar white matter, and cerebellar cortex) resulted in improved classification with 1 additional MSA patient being classified correctly (96.8% of MSA patients and all PD patients were classified correctly; Fig. 2b). Cross‐validation of this model yielded a weighted average F measure and Cohen's kappa of 0.94 and 0.87, respectively.
Figure 2.
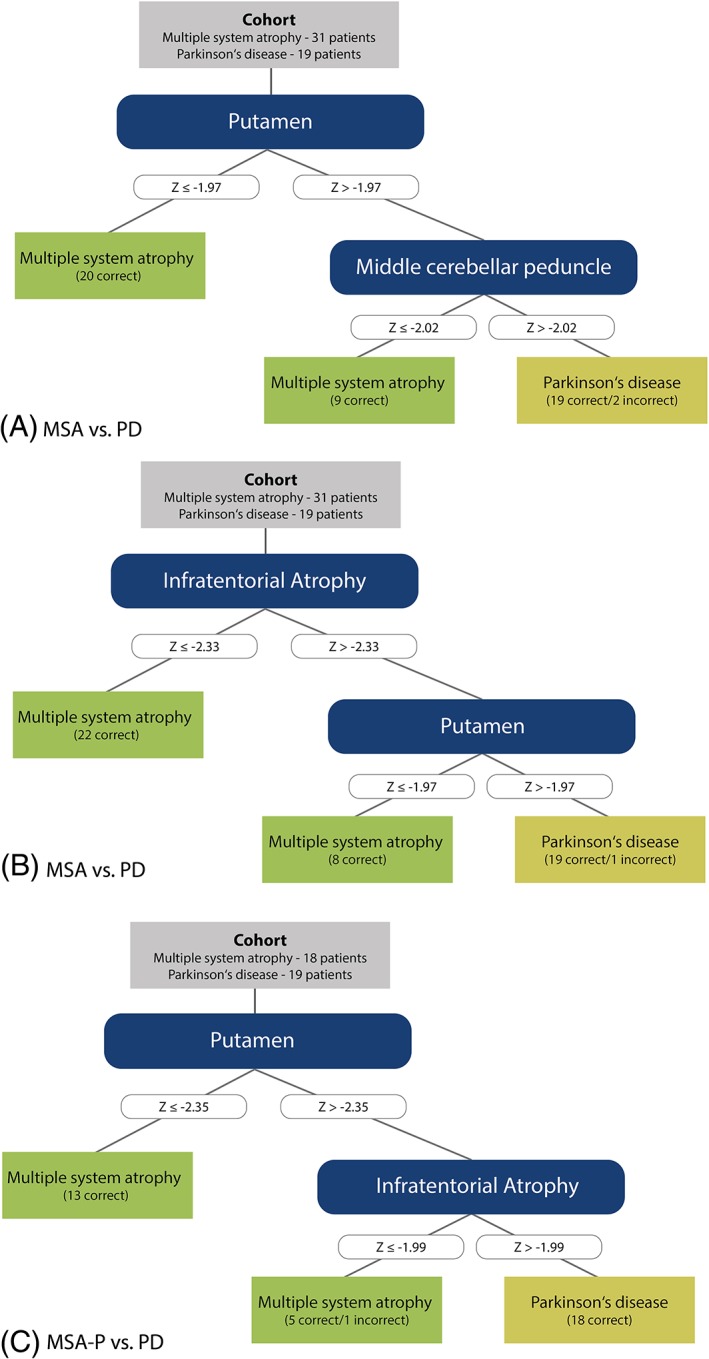
Diagnostic algorithm. (a) Decision tree classifying MSA and PD based on single‐region measurements. (b, c) Decision tree classifying MSA and PD (lit. b) as well as MSA‐P and PD (lit. c) based on single‐region measurements and pooled infratentorial atrophy score (minimum z score of middle cerebellar peduncle, cerebellar white matter, and cerebellar cortex). [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
In the present study, probabilistic information on the relative location of the MCP was integrated into the currently available standard atlas of FreeSurfer‐based subcortical segmentation. Automated volumetry in MSA patients showed that both putaminal and infratentorial atrophy is present in both MSA variants (ie, MSA‐P and MSA‐C). Intriguingly, 22.2% of MSA‐P patients revealed isolated infratentorial atrophy without evidence of putaminal atrophy. Based on these findings, we were able to develop an automated MRI decision support algorithm that includes volumetric information of infratentorial (ie, volumetry of the MCP, cerebellar gray and white matter) and putaminal atrophy, and this approach proved to be superior compared with existing volumetric MRI processing strategies in differentiating patients with MSA from PD on an individual subject level.
Gross anatomy examinations commonly distinguish 2 major patterns of neurodegeneration in MSA—OPCA and SND. Clinicopathological studies found that 42% to 49% of MSA cases showed equally severe striatonigral degeneration and olivopontocerebellar atrophy, whereas atrophy patterns with either predominant SND or OPCA were evident in 34% and 17% of cases, respectively.4 In the present study, infratentorial atrophy was present in all MSA‐C patients and 61.1% of MSA‐P patients, whereas putaminal atrophy was found in 77.8% of MSA‐P patients and 46.2% of MSA‐C patients. In addition, 4 of 18 MSA‐P patients showed isolated infratentorial atrophy. Although the observed frequency of cerebellar white‐matter atrophy was in the range of that obtained by the MCP volumetry in the entire MSA cohort, among MSA patients with normal putaminal volume, MCP atrophy yielded the largest area under the receiver operating characteristics curve when comparing MCP, cerebellar white matter, and cerebellar cortex volume (Supplementary Fig. 2).
These observations suggest that both SND and OPCA atrophy patterns need to be scrutinized by including the MCP to the cerebellar and putaminal assessment to obtain a reliable imaging‐supported diagnosis of MSA.
Early quantitative MRI studies primarily focused on planimetric measurements of infratentorial brain regions, and these studies suggested that MSA is associated with relatively greater pontine and MCP atrophy compared with PSP and PD patients.14 The latter observations led to the development of 2‐dimensional planimetric measurements including the MCP width and the MR Parkinson index that were shown to support the differential diagnosis of neurodegenerative parkinsonian disorders.10, 12, 31, 32, 33, 34 Attempts to reduce visual inspection biases of manual measurement methods and the requirement to establish 3‐dimensional volume measurements gave rise to the development of automated, atlas‐based analysis tools. Such 3‐dimensional and automated imaging efforts improved diagnostic accuracy rates for the differential diagnosis of degenerative parkinsonian disorders in recent years. By applying a support vector machine to classify regional volumetric MRI data, a multicenter study achieved sensitivity ranging from 79% to 87% and specificity from 87% to 96% for the diagnosis of PD, PSP, and MSA (combining MSA‐P and MSA‐C into 1 group).16 Another approach used mean values of gray‐mater density, diffusion tensor, and r2* signal within voxel‐clusters obtained from previous group analysis of patients with PD and MSA (including patients with MSA‐P and MSA‐C) and reported >95% discrimination accuracy of fractional anisotropy clusters of the cerebellum, brain stem, and white‐matter bilateral superior corona radiate combined with another mean diffusivity cluster of the right superior frontal gyrus.15 In the present study, monomodal volumetric measurements were harmonized by z transformation to rank affected brain areas according to their volume loss in individual patients. Furthermore, in an attempt to avoid the inclusion of volumes from brain areas within the normal range into the diagnostic algorithm, only measurements that showed a reduction of more than 2 standard deviations from the mean of the healthy control sample were considered. Finally, the 3‐dimensionally measured volume of the MCP, a brain region known to be frequently affected in patients with MSA, was added to the list of assessed brain volumes. The diagnostic algorithm was then generated by means of a C4.5 decision tree algorithm. This hierarchical stepwise classifier identified the volume of the putamen and the MCP as the most discriminative parameters to separate PD and MSA. Diagnostic accuracy was further improved by the introduction of a pooled infratentorial atrophy measure that represents the most severely affected z score of the middle cerebellar peduncle, cerebellar white matter, and cerebellar cortex. Only 1 patient with a diagnosis of MSA was incorrectly assigned to the PD group. This patient already had a substantial infratentorial atrophy (z score, ‐2.1) at a very early disease stage (symptom duration, 1.25 years), which is highly suspicious for a diagnosis of MSA; however, in this proof‐of‐concept design, the classifier C4.5 selected a more stringent cutoff to improve specificity at the expense of sensitivity. In addition, the pooled infratentorial atrophy measure was superior to single parameters to classify patients with MSA‐P from PD. Cross‐validation suggested a good fit of the model to an independent data set. The current model also extends previous work following a similar approach by introducing more accurate characterization of infratentorial atrophy and by applying the automated morphometric analysis on MSA‐C patients.19
The lack of postmortem confirmation is a potential limitation of the present study. However, we stringently applied validated clinical criteria, with consensus on the diagnosis from 2 experts in movement disorders being required. The final clinical classification was anchored on the last visit after an extended follow‐up period of at least 24 months. In addition, ancillary investigations including radiotracer imaging confirming nigrostriatal dopaminergic deficit and structural imaging excluding secondary causes were supportive of the degenerative nature of the movement disorder. The cross‐sectional design of the present study impedes a detailed characterization of the temporal evolution of MSA atrophy patterns; hence, future longitudinal MRI studies exploiting MRI volumetry should be performed in cases with early MSA to better characterize progression of brain atrophy over the course of the disease in MSA patients more precisely.
Despite the known benefits of decision tree algorithms like simplicity in constructing, robustness, and interpretability for clinicians, some potential limitations must also be considered. Decision trees often exhibit low diagnostic precision because of the strictly horizontal and vertical decision boundaries created by decision trees. Readers should also be aware that the decision trees, like any other statistical model, may change in structure if important variables are added or removed. Therefore, the decision trees should be viewed as descriptive explorative analysis explaining the data, but they are not confirming predictors.35, 36
In summary, the inclusion of the MCP in the automatized morphometric analysis corroborates previous neuropathological studies that showed that most MSA patients have a mixed SND and OPCA pathology and that brain atrophy patterns do not necessarily correspond to the clinical impression of predominant motor deficits. The combination of automated subcortical and supra‐ and infratentorial segmentation that included the volumetric measure of the MCP further improved the existing volumetric MRI decision support strategies on the diagnostic accuracy in separating MSA from PD. This approach will improve appropriate patient counseling and recruitment of homogenous patient cohorts into clinical studies. In addition, a more detailed automated brain segmentation based on anatomical MR sequences will also facilitate future longitudinal as well as multimodal imaging approaches.
Competing interests (for the past 12 months)
F.K. reports a grant from the MSA Coalition. K.S. reports grants from Oesterreichische Nationalbank, FWF Austrian Science Fund, Michael J. Fox Foundation, and International Parkinson and Movement Disorder Society and personal fees from International Parkinson and Movement Disorder Society, Boehringer Ingelheim, UCB, Lundbeck, Teva, AbbVie, and AOP Orphan Pharmaceuticals AG. S.B. reports personal fees from AbbVie, Gruenenthal, Allergan, Reata, and Ipsen and research grants from the European Union. G.K.W. reports receiving consulting and/or lecture fees from Affiris, Astra Zeneca, Boehringer Ingelheim, Ever Pharma, Lundbeck, Neuropore, Orion, and UCB as well as grant support from Medical University Innsbruck, Oesterreichische Nationalbank, FWF Austrian Science Fund, US MSA Coalition, Affiris, Astra Zeneca, and Boehringer Ingelheim. V.A. and S.P. are employed by Teva Pharmaceutical Industries, Ltd. E.R.G. reports lectures fees from Bracco and Bayer. W.P. has received consultancy and lecture fees in relation to clinical drug programs for PD from AbbVie, AstraZeneca, BIAL, Biogen, Britannia, Grünenthal, Intec, Ipsen, Lundbeck, Novartis, Neuroderm, Orion Pharma, Prexton, Roche, Sunovion, Sun Pharma, Takeda, Teva, UCB, and Zambon and royalties from Thieme, Wiley Blackwell, Oxford University Press and Cambridge University Press. G.G., R.S., I.Z., and C.S. have nothing to disclose.
Documentation of Author Roles
1. Research project: A. Conception, B. Organization, C. Execution;
2. Statistical Analysis: A. Design, B. Execution, C. Review and Critique;
3. Manuscript Preparation: A. Writing of the first draft, B. Review and Critique;
F.K.: 1A, 1B, 1C, 2A, 2B, 3A.
K.S.: 1A, 1B, 1C, 2C, 3B.
G.G.: 1A, 2A, 2C, 3B.
R.S.: 1C, 2C, 3B.
I.Z.: 1C, 2C, 3B.
S.B.: 1B, 1C, 2C, 3B.
E.R.G.: 1C, 2C, 3B.
G.K.W.: 1C, 2C, 3B.
W.P.: 1A, 1C, 2C, 3B.
C.S.: 1A, 1B, 1C, 2A, 2B, 3A.
Supporting information
SUPPLEMENTARY FIG. 1 Three‐dimensional models of the middle cerebellar peduncles in a PD patient (a) and a MSA‐C patient (b).
SUPPLEMENTARY FIG. 2 Receiver operating characteristic curve analysis of relevant brain regions in MSA patients without putaminal atrophy.
SUPPLEMENTARY TABLE 1 Correlation between clinical scores and volumetric MR measures in MSA and PD patients
Acknowledgments
The newly developed FreeSurfer‐compatible atlas for automated subcortical segmentation will be shared with interested parties on formal request to the authors.
Funding agency: This study was supported by the MSA Coalition.
Relevant conflicts of interests/financial disclosures: The authors have nothing to report in relation to this study.
The copyright line for this article was changed on July 29, 2019, after original print and online publication.
References
- 1. Krismer F, Wenning GK. Multiple system atrophy: insights into a rare and debilitating movement disorder. Nat Rev Neurol 2017;13(4):232–243. [DOI] [PubMed] [Google Scholar]
- 2. Ahmed Z, Asi YT, Sailer A, et al. The neuropathology, pathophysiology and genetics of multiple system atrophy. Neuropathol Appl Neurobiol 2012;38(1):4–24. [DOI] [PubMed] [Google Scholar]
- 3. Trojanowski JQ, Revesz T, Neuropathology Working Group on MSA . Proposed neuropathological criteria for the post mortem diagnosis of multiple system atrophy. Neuropathol Appl Neurobiol 2007;33(6):615–620. [DOI] [PubMed] [Google Scholar]
- 4. Ozawa T, Paviour D, Quinn NP, et al. The spectrum of pathological involvement of the striatonigral and olivopontocerebellar systems in multiple system atrophy: clinicopathological correlations. Brain 2004;127(Pt 12):2657‐2671. [DOI] [PubMed] [Google Scholar]
- 5. Oba H, Yagishita A, Terada H, et al. New and reliable MRI diagnosis for progressive supranuclear palsy. Neurology 2005;64(12):2050‐2055. [DOI] [PubMed] [Google Scholar]
- 6. Righini A, Antonini A, De Notaris R, et al. MR imaging of the superior profile of the midbrain: differential diagnosis between progressive supranuclear palsy and Parkinson disease. AJNR Am J Neuroradiol 2004;25(6):927–932. [PMC free article] [PubMed] [Google Scholar]
- 7. Warmuth‐Metz M, Naumann M, Csoti I, Solymosi L. Measurement of the midbrain diameter on routine magnetic resonance imaging: a simple and accurate method of differentiating between Parkinson disease and progressive supranuclear palsy. Arch Neurol 2001;58(7):1076–1079. [DOI] [PubMed] [Google Scholar]
- 8. Paviour DC, Price SL, Stevens JM, Lees AJ, Fox NC. Quantitative MRI measurement of superior cerebellar peduncle in progressive supranuclear palsy. Neurology 2005;64(4):675–679. [DOI] [PubMed] [Google Scholar]
- 9. Nicoletti G, Fera F, Condino F, et al. MR imaging of middle cerebellar peduncle width: differentiation of multiple system atrophy from Parkinson disease. Radiology 2006;239(3):825–830. [DOI] [PubMed] [Google Scholar]
- 10. Quattrone A, Nicoletti G, Messina D, et al. MR imaging index for differentiation of progressive supranuclear palsy from Parkinson disease and the Parkinson variant of multiple system atrophy. Radiology 2008;246(1):214–221. [DOI] [PubMed] [Google Scholar]
- 11. Hotter A, Esterhammer R, Schocke MF, Seppi K. Potential of advanced MR imaging techniques in the differential diagnosis of parkinsonism. Mov Disord 2009;24(Suppl 2):S711–S720. [DOI] [PubMed] [Google Scholar]
- 12. Mangesius S, Hussl A, Krismer F, et al. MR planimetry in neurodegenerative parkinsonism yields high diagnostic accuracy for PSP. Parkinsonism Relat Disord 2018;46:47–55. [DOI] [PubMed] [Google Scholar]
- 13. Lehericy S, Vaillancourt DE, Seppi K, et al. The role of high‐field magnetic resonance imaging in parkinsonian disorders: Pushing the boundaries forward. Mov Disord 2017;32(4):510–525. [DOI] [PubMed] [Google Scholar]
- 14. Heim B, Krismer F, De Marzi R, Seppi K. Magnetic resonance imaging for the diagnosis of Parkinson's disease. J Neural Transm (Vienna) 2017;124(8):915–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peran P, Barbagallo G, Nemmi F, et al. MRI supervised and unsupervised classification of Parkinson's disease and multiple system atrophy. Mov Disord 2018;33(4):600–608. [DOI] [PubMed] [Google Scholar]
- 16. Huppertz HJ, Moller L, Sudmeyer M, et al. Differentiation of neurodegenerative parkinsonian syndromes by volumetric magnetic resonance imaging analysis and support vector machine classification. Mov Disord 2016;31(10):1506–1517. [DOI] [PubMed] [Google Scholar]
- 17. Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 2000;97(20):11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002;33(3):341–355. [DOI] [PubMed] [Google Scholar]
- 19. Scherfler C, Gobel G, Muller C, et al. Diagnostic potential of automated subcortical volume segmentation in atypical parkinsonism. Neurology 2016;86(13):1242–1249. [DOI] [PubMed] [Google Scholar]
- 20. Messina D, Cerasa A, Condino F, et al. Patterns of brain atrophy in Parkinson's disease, progressive supranuclear palsy and multiple system atrophy. Parkinsonism Relat Disord 2011;17(3):172–176. [DOI] [PubMed] [Google Scholar]
- 21. Morisi R, Manners DN, Gnecco G, et al. Multi‐class parkinsonian disorders classification with quantitative MR markers and graph‐based features using support vector machines. Parkinsonism Relat Disord 2018;47:64–70. [DOI] [PubMed] [Google Scholar]
- 22. Nicoletti G, Lodi R, Condino F, et al. Apparent diffusion coefficient measurements of the middle cerebellar peduncle differentiate the Parkinson variant of MSA from Parkinson's disease and progressive supranuclear palsy. Brain 2006;129(Pt 10):2679–2687. [DOI] [PubMed] [Google Scholar]
- 23. Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J Neurol Neurosurg Psychiatry 1988;51(6):745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilman S, Wenning GK, Low PA, et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology 2008;71(9):670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- 26. Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex 2004;14(1):11–22. [DOI] [PubMed] [Google Scholar]
- 27. Segonne F, Dale AM, Busa E, et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage 2004;22(3):1060–1075. [DOI] [PubMed] [Google Scholar]
- 28. van Baarsen KM, Kleinnijenhuis M, Jbabdi S, Sotiropoulos SN, Grotenhuis JA, van Cappellen van Walsum AM . A probabilistic atlas of the cerebellar white matter. Neuroimage 2016;124(Pt A):724–732. [DOI] [PubMed] [Google Scholar]
- 29. Mudali D, Teune LK, Renken RJ, Leenders KL, Roerdink JB. Classification of Parkinsonian syndromes from FDG‐PET brain data using decision trees with SSM/PCA features. Comput Math Methods Med 2015;2015:136921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan JR. C4.5: programs for machine learning. Morgan Kaufmann Publishers Inc; 1993. [Google Scholar]
- 31. Gama RL, Tavora DG, Bomfim RC, Silva CE, de Bruin VM, de Bruin PF. Sleep disturbances and brain MRI morphometry in Parkinson's disease, multiple system atrophy and progressive supranuclear palsy ‐ a comparative study. Parkinsonism Relat Disord 2010;16(4):275–279. [DOI] [PubMed] [Google Scholar]
- 32. Hussl A, Mahlknecht P, Scherfler C, et al. Diagnostic accuracy of the magnetic resonance Parkinsonism index and the midbrain‐to‐pontine area ratio to differentiate progressive supranuclear palsy from Parkinson's disease and the Parkinson variant of multiple system atrophy. Mov Disord 2010;25(14):2444–2449. [DOI] [PubMed] [Google Scholar]
- 33. Morelli M, Arabia G, Salsone M, et al. Accuracy of magnetic resonance parkinsonism index for differentiation of progressive supranuclear palsy from probable or possible Parkinson disease. Mov Disord 2011;26(3):527–533. [DOI] [PubMed] [Google Scholar]
- 34. Nigro S, Arabia G, Antonini A, et al. Magnetic Resonance Parkinsonism Index: diagnostic accuracy of a fully automated algorithm in comparison with the manual measurement in a large Italian multicentre study in patients with progressive supranuclear palsy. Eur Radiol 2017;27(6):2665–2675. [DOI] [PubMed] [Google Scholar]
- 35. Bae J‐M. The clinical decision analysis using decision tree. Epidemiol Health 2014;36(0):e2014025–2014020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolpert DH. The Supervised Learning No‐Free‐Lunch Theorems In: Roy R, Köppen M, Ovaska S, Furuhashi T, Hoffmann F, eds. Soft Computing and Industry: Recent Applications. London: Springer London; 2002:25–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIG. 1 Three‐dimensional models of the middle cerebellar peduncles in a PD patient (a) and a MSA‐C patient (b).
SUPPLEMENTARY FIG. 2 Receiver operating characteristic curve analysis of relevant brain regions in MSA patients without putaminal atrophy.
SUPPLEMENTARY TABLE 1 Correlation between clinical scores and volumetric MR measures in MSA and PD patients


