Summary
The leaf outer epidermal cell wall acts as a barrier against pathogen attack and desiccation, and as such is covered by a cuticle, composed of waxes and the polymer cutin. Cutin monomers are formed by the transfer of fatty acids to glycerol by glycerol‐3‐phosphate acyltransferases, which facilitate their transport to the surface.
The extent to which cutin monomers affect leaf cell wall architecture and barrier properties is not known. We report a dual functionality of pathogen‐inducible GLYCEROL‐3‐PHOSPHATE ACYLTRANSFERASE 6 (GPAT6) in controlling pathogen entry and cell wall properties affecting dehydration in leaves.
Silencing of Nicotiana benthamiana NbGPAT6a increased leaf susceptibility to infection by the oomycetes Phytophthora infestans and Phytophthora palmivora, whereas overexpression of NbGPAT6a‐GFP rendered leaves more resistant. A loss‐of‐function mutation in tomato SlGPAT6 similarly resulted in increased susceptibility of leaves to Phytophthora infection, concomitant with changes in haustoria morphology. Modulation of GPAT6 expression altered the outer wall diameter of leaf epidermal cells. Moreover, we observed that tomato gpat6‐a mutants had an impaired cell wall–cuticle continuum and fewer stomata, but showed increased water loss.
This study highlights a hitherto unknown role for GPAT6‐generated cutin monomers in influencing epidermal cell properties that are integral to leaf–microbe interactions and in limiting dehydration.
Keywords: cell wall, cuticle, haustoria, Nicotiana benthamiana, oomycetes, Phytophthora, Solanum lycopersicum, stomata
Introduction
Most epidermal cells of the aerial parts of vascular plants are covered by a hydrophobic extracellular lipid barrier, known as the cuticle, which is composed of polymeric cutin and waxes (Yeats & Rose, 2013). The cutin matrix is a highly viscoelastic polymer with low tensile strength (Fich et al., 2016) that functions as a transpiration barrier (Schonherr, 1976) and also contributes mechanical strength to the underlying cell wall (Kolattukudy, 1980). Celluloses, hemicelluloses and pectins from the cell wall can be incorporated into the cutin matrix, thereby influencing its elasticity and stiffness (López‐Casado et al., 2007) and facilitating expansion during growth and development and in response to environmental cues (Bargel & Neinhuis, 2005; Underwood, 2012).
Cutin biosynthesis involves the esterification of oxygenated 16‐ or 18‐carbon fatty acids to glycerol (Beisson et al., 2012) through the action of glycerol‐3‐phosphate acyltransferases (GPAT4, GPAT6 and GPAT8). These enzymes have specificity for the second carbon of the glycerol (sn‐2 position) (Yang et al., 2012) and exhibit phosphatase activity that removes the phosphate group from glycerol‐3‐phosphate (Yang et al., 2010). Accordingly, mutants of the Arabidopsis thaliana GPAT4, GPAT6 and GPAT8 genes display reduced amounts of C16 and C18 fatty acid cutin monomers (Li et al., 2007; Mazurek et al., 2016). GPAT4 orthologues in Brassica napus are highly expressed in the seed coat, periderm and endodermis of roots (Chen et al., 2011b) and function in the development of reproductive organs (Chen et al., 2014). GPAT6 is involved in cutin synthesis in A. thaliana petals (Li‐Beisson et al., 2009) and tomato (Solanum lycopersicum) fruit (Petit et al., 2016) and was found to have multiple functions in stamen development and fertility (Li et al., 2012). Analysis of A. thaliana gpat6 knockout lines demonstrated that GPAT6 is essential for the accumulation of C16 cutin monomers (Li‐Beisson et al., 2009) and that the enzyme has a higher affinity for C16 and C18 ω‐oxidized acyl‐CoA substrates. A glossy fruit mutant of the tomato cv Micro‐Tom with increased total wax load, but lower amounts of total cutin in fruit cuticles, and a much thinner cuticle (Petit et al., 2014), were discovered to be a result of a point mutation in the GPAT6 gene that abolished enzymatic activity (Petit et al., 2016). Additionally, the Micro‐Tom gpat6‐a has perturbed pollen formation but is not male sterile (Petit et al., 2016).
The cuticle not only controls solute and gas exchange (Kerstiens, 1996a; Riederer & Schreiber, 2001) but also provides protection against pathogen invasion (Kerstiens, 1996a,b). Accordingly, to gain entry into host tissues, pathogens secrete hydrolytic enzymes, including cutinases, esterases, lipases and glycanases, which destroy the integrity of the cuticle–cell wall continuum (Belbahri et al., 2008; Blackman et al., 2014). For example, the oomycete Phytophthora infestans secretes cell wall‐ and cuticle‐degrading enzymes and forms surface appressoria that support tissue invasion. P. infestans is an economically important leaf pathogen of potato (Solanum tuberosum) and tomato (Haverkort et al., 2008) and can also infect wild tobacco species, including Nicotiana benthamiana (Becktell et al., 2006). During the early infection stages, P. infestans lives as a biotroph, proliferates an extensive intercellular hyphal network within the leaf mesophyll and projects short digit‐like haustoria into mesophyll cells to suppress immunity and support infection. In the later stages of infection, P. infestans switches to a necrotrophic lifestyle and kills the host tissue, resulting in necrotic lesions. Other Phytophthora species with similar lifestyles are not restricted to infecting aerial tissues. For example, the tropical pathogen P. palmivora can infect roots and shoots of many vascular and nonvascular host plants (Torres et al., 2016).
Cutin monomers and cell wall oligosaccharides released during a pathogen attack can serve as damage‐associated molecular patterns, allowing the plant cell to mount mitigating defence and wall repair responses, but conversely may also stimulate pathogen colonization by triggering the formation of appressoria (Gilbert et al., 1996). Phytophthora palmivora forms appressoria when exposed to cutin monomers in vitro (Wang et al., 2012) and cutin components have also been reported to trigger spore germination and cutinase expression in the fungus Botrytis cinerea (Leroch et al., 2013). The ectopic application of cutin monomers during root inoculation with P. palmivora was reported to restore full susceptibility to the Medicago truncatula GPAT mutant ram2 (Wang et al., 2012), suggesting that the oomycete in part relies on the presence of cutin‐derived signals to enhance its pathogenicity, as reducing GPAT activity enhances resistance to Phytophthora infection.
Here we document the importance of GPAT6 in leaf infections by oomycete and fungal pathogens, as well as its contribution to cell wall properties. We found that GPAT6 transcript abundance increases in response to Phytophthora infection, and that overexpression of GPAT6 results in increased resistance to oomycete infection. Furthermore, although gpat6 mutants are more susceptible to Phytophthora leaf infection, they display increased leaf resistance to B. cinerea, suggesting pathogen lifestyle‐specific differences. Changes in pathogen susceptibility are associated with altered thickness of the leaf cell wall plus cuticle, as well as altered transpiration and numbers of stomata. This is reflected in elevated transcript abundance of the immunity‐ and stomata development‐associated receptor‐like kinases SERK3/BAK1 and ERECTA in gpat6‐a leaves. Cuticle‐associated genes are consistently altered in leaves and fruits of gpat6‐a plants, whereas more variation exists in genes related to the cell wall and secondary metabolites. Although GPAT6‐like genes have been implicated in flower, fruit and seed development, our work uncovers a function in leaves of N. benthamiana and tomato where GPAT6 genes influence cell wall and cuticular properties associated with pathogen infection and water regulation.
Materials and Methods
Statistical analysis
Levene's tests were applied to check for heteroscedasticity between treatment groups. Following this, the appropriate two‐sample t‐test was applied, accounting for equal or unequal variances, to assess whether the means of two different treatment groups were significantly different, based on α = 0.5. Figures are labelled with asterisks to indicate P‐value range (i.e. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Microbial strains and cultivation
Phytophthora infestans strain 88069, previously described in van West et al. (1998), was grown at 18°C in the dark on rye sucrose agar plates. Zoospores were harvested from 14‐d‐old plates by adding 6 ml cold sterile H2O, incubating in the dark at 4°C for 45–60 min, then in the light at room temperature for 30 min and extracting the liquid by pipetting. Approximately 30 spores μl−1 sterile H2O were then used immediately for infection assays.
Phytophthora palmivora strain P16830‐YKDEL was previously described (Rey et al., 2013). P. palmivora was grown in a Conviron (Winnipeg, MB, Canada) A1000 Reach‐In Plant Growth Chamber at 25°C and 700 μmol intensity. For subculturing, rye sucrose agar plates were used with the addition of 50 μg ml−1 G418 (geneticin) to select for transformants. For production of zoospores, agar plates containing 10% unclarified V8 vegetable juice were used with the addition of 50 μg ml−1 G418 (geneticin). Harvesting of zoospores was performed as for P. infestans described earlier
Botrytis cinerea R190/11/3, isolated from Geranium by Robert Saville in 2011 (NIAB‐EMR, East Malling, UK) was grown on potato dextrose agar plates in a Conviron A1000 Reach‐In Plant Growth Chamber at 25°C and 700 μmol intensity and subcultured by excising an agar plug containing conidiophores and inverting it onto a fresh plate. Conidia for infection assays were harvested from 7‐d‐old potato dextrose agar plates but adding 6 ml cold sterile H2O, incubating in the light at room temperature for 1 h then gently agitating the conidiophores with a spatula to release the conidia. The concentration was adjusted to c. 30 conidia μl−1 sterile H2O.
Leaf infection assays
Droplets (10 μl) of identical Phytophthora zoospore (c. 1000 zoospores) or Botrytis conidia count (c. 5000 spores) were placed onto the abaxial side of all leaves of the experiment between their veins. Inoculated leaves were then incubated in a humidified chamber under illumination at 24°C for the stated time before imaging. Disease symptom images of P. infestans or P. palmivora lesions were obtained by placing the infected leaves on a light table emitting near‐UV light in the 380–500 nm range and covering them with a yellow filter. This created high contrast between uninfected zones that appear red/orange as a result of autofluorescence and infected zones that appear black as a result of cell death‐associated loss of autofluorescence. Leaves were imaged through the filter using a digital camera on a tripod with long exposure settings and lesion area was quantified using imagej. Quantification involved conversion to greyscale, inversion of the colours then application of a threshold (same threshold for all images) to isolate the nonautofluorescent necrotic lesions from autofluorescent background. Nonautofluorescent pixels were counted by making a selection around all of the lesions on a single leaf and then using the ‘Analyze Particles’ function to calculate the total necrotic area per leaf.
Phylogenetic analysis
Protein sequences from N. benthamiana, M. truncatula and S. lycopersicum homologous to A. thaliana GPATs were identified by search of the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using AtGPATs 1–9 as queries (sequence accession numbers are listed in Supporting information Fig. S1). Obtained sequences were then aligned using muscle (http://www.ebi.ac.uk/Tools/msa/muscle/) and a phylogenetic tree constructed using phyml (http://atgc.lirmm.fr/phyml/) with the Phylogeny.fr web tool (http://www.phylogeny.fr/). Branches with < 50% bootstrap support (100 iterations) were collapsed. The tree presented in Fig. S1 was rendered using treedyn (http://www.treedyn.org/) and annotated using gimp (https://www.gimp.org/).
Confocal microscopy
Confocal microscopy was performed using a Leica SP8 (Wetzlar, Germany) equipped with a white light laser (main laser power 70%, time gating) and a 63× water immersion objective and the following settings: pinhole, 1.00 AU; scan speed, 200 min–1; line averaging factor, 4; green fluorescent protein (GFP) excitation, 489 nm; emission window, 500–552 nm; mCherry excitation, 587 nm; emission window, 596–643 nm; plastid autofluorescence excitation, 489 nm; emission window, 650–700. Samples were mounted in water. Images of subcellular localiZation of NbGPAT6a‐GFP protein fusion were taken at 48 h postinoculation upon transient constitutive expression in N. benthamiana leaf.
Quantitative reverse transcription polymerase chain reaction (qRT‐PCR)
Total RNA was extracted from plant material using Qiagen RNeasy Plant Mini Kit (Qiagen), including 1% (v/v) β‐mercaptoethanol in the extraction buffer. RNA was then reverse‐transcribed to cDNA using the Roche Transcriptor First Strand cDNA Synthesis Kit. qPCR was performed in 384 well plates using a Roche LightCycler 480 SYBR Green I Master Mix in a Roche LightCycler 480 II machine. Three technical replicates were performed for each sample. Normalization of crossing point‐PCR‐cycle (Cp) values to an internal control was performed against NbEF1a, NbF‐BOX or NbL23 (Liu et al., 2012) for quantification of N. benthamiana transcripts and against PiWS21 (Yan & Liou, 2006) for P. infestans transcripts.
Expression analysis
Leaves of 6‐wk‐old tomato cv ‘Micro‐Tom’ or gpat6‐a mutant plants were subjected to a detached leaf infection assay (see earlier) and either zoospore suspension or water were applied to the lower epidermis. Leaf discs were harvested 72 h postinoculation (hpi). Three biological replicates per sample were obtained and subjected to RNA extraction and poly(A) selection. cDNA library preparation was performed with the TruSeq® RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer's protocol. cDNA sequencing of the 12 samples was performed with Illumina NextSeq 2500 in 100 paired‐end mode (Genewiz, Leipzig, Germany). Raw reads were subjected to quality control with fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then aligned back to the S. lycopersicum reference genome ITAG3.1 (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG3.1_release/) using star (v.2.5.2b) aligner. Raw counts were obtained with featurecounts (Liao et al. 2014), and only uniquely mapped and properly paired reads were considered further. Differentially expressed genes were identified with the deseq2 bioconductor package (Love et al. 2014) following four pairwise comparisons. Differentially expressed genes (absolute log fold‐change (LFC) ≥ 1.5 and adjusted P‐value ≤ 10−3) were used to generate volcano plots and upset plots using upsetr package (Conway et al., 2017).
Cryoscanning electron microscopy
Cryoscanning electron microscopy (cryo‐SEM) was performed on 6‐wk‐old N. benthamiana and tomato leaves using a Zeiss EVO HD15 (Oberkochen, Germany) with a Quorum cryo‐prep deck and cryo‐stage (Lewes, UK). Leaf sections were mounted, frozen, positioned inside the cryo‐prep deck and then fractured using a blade to allow for cross‐sectional imaging. Sublimation of samples for 3 min was used to remove surface ice and a 5 nm platinum coating was applied before imaging via secondary electron detection.
Cell wall porosity analysis
Discs (5 mm diameter) were excised from leaves using a cork borer and incubated for 1 h at room temperature with tetramethylrhodamine isothiocyanate (TRITC) : Dextran (0.1 mg ml−1, 150k mw; TDB Consultancy AB, Uppsala, Sweden) and Auramine O (0.01% w/v; Sigma). Images were acquired using a Leica SP8 equipped with white light laser (TRITC: excitation, 561 nm; emission window, 609–631 nm; AuramineO: excitation, 458 nm; emission window, 485–532 nm).
Water loss/dehydration analysis
Whole leaves were harvested, placed in a ventilated oven (30°C; MAXQ‐6000, Thermo Fisher Scientific, Waltham, MA, USA) and weighed over a time course.
Results
GPAT6 is induced during Phytophthora leaf infections
GPAT enzymes function in root interactions with symbiotic arbuscular mycorrhiza fungi and pathogenic Phytophthora oomycetes (Wang et al., 2012), but their roles in leaf interactions with pathogens have not been well characterized. To this end, we first identified all GPATs encoded in the tomato and N. benthamiana genomes and grouped them based on their phylogenetic relationship to the better characterized A. thaliana homologues (Fig. S1; Methods S1). This revealed the presence of three N. benthamiana genes grouping with AtGPAT1/2/3 that probably contribute to storage lipid biosynthesis (Zheng et al., 2003), and three genes associated with AtGPAT5/7 that may be involved in suberin biosynthesis (Beisson et al., 2007). The two clades associated with AtGPAT4/8 and AtGPAT6, implicated in A. thaliana cutin biosynthesis (Li et al., 2007; Li‐Beisson et al., 2009), contain two N. benthamiana genes each (Fig. S1). An additional group of six N. benthamiana genes form a clade together with MtRAM2 that is distinct from any A. thaliana GPAT genes, and were termed NbRAM2A‐F.
Previously published data on N. benthamiana root infection by P. palmivora reported that NbGPAT6a, but none of the other members of the GPAT4/6/8 clade, showed a consistent and significant transcriptional induction during all stages of infection (Fig. 1; Table S1) (Evangelisti et al., 2017).
Figure 1.
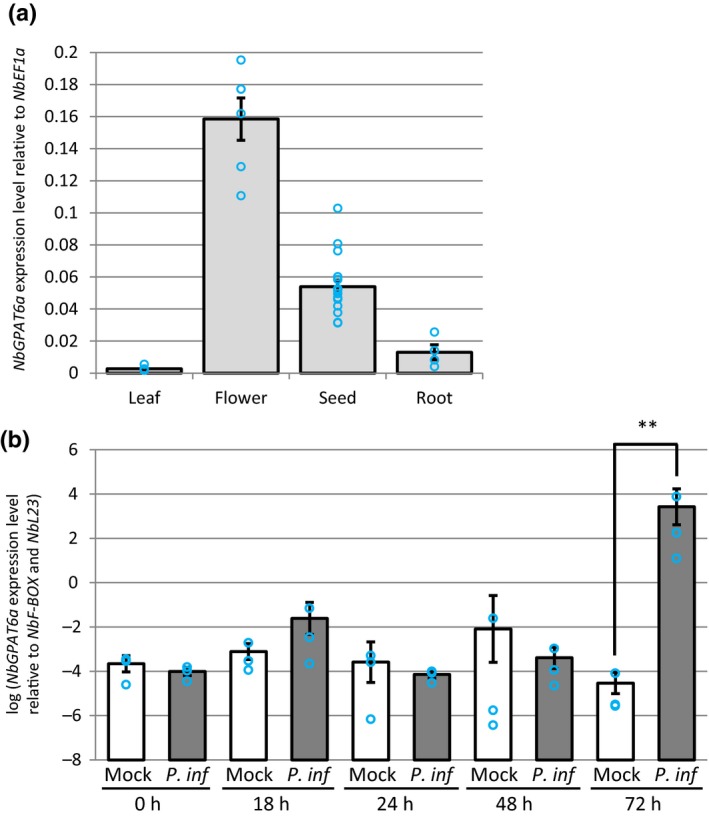
NbGPAT6a transcript abundance varies between Nicotiana benthamiana plant organs and is upregulated during Phytophthora infestans (P. inf) leaf colonization. (a) Expression level of NbGPAT6a in leaf, flower, seed and root tissues. Error bars represent ± SE of the mean of at least three biological replicates. Blue circles represent the average of three technical replicates. (b) Expression level of NbGPAT6a in leaf tissues is significantly increased at 72 h after inoculation with P. infestans. Error bars represent ± SE of the mean of three biological replicates (**, P < 0.01). Blue circles represent the average of three technical replicates.
Using qRT‐PCR, we detected NbGPAT6a expression in leaves, flowers, seeds and roots, with the highest steady‐state levels in flowers (Fig. 1). This is in agreement with GPAT6 expression patterns in other species (Li‐Beisson et al., 2009; Li et al., 2012; Petit et al., 2016).
To test whether NbGPAT6a expression levels increase during infection, we infected N. benthamiana leaves with P. infestans strain 88069 (van West et al., 1999) zoospore droplets and measured changes in NbGPAT6a expression over time. NbGPAT6a was highly induced in leaf tissues at 72 hpi with P. infestans (Fig. 1). We therefore conclude that NbGPAT6a expression is upregulated in roots and leaves infected with Phytophthora and that expression levels in leaves are elevated late during infection and so are not part of early, inducible, defence responses.
Constitutive expression of NbGPAT6a renders leaves resistant to Phytophthora infection
To address whether higher GPAT6a transcript abundances influence Phytophthora infection, we generated constitutive overexpression constructs by creating a translational fusion of the genomic NbGPAT6a open reading frame to the GFP reporter gene under control of the 35S promoter. We first investigated the subcellular distribution of the fusion protein upon transient expression in N. benthamiana leaves. GPAT6 is a predicted endoplasmic reticulum (ER)‐resident enzyme (Chen et al., 2011a) with two transmembrane domains (Fig. S2b) and we observed NbGPAT6‐GFP signals in the ER of leaf epidermal cells matching the subcellular distribution of ER‐targeted red fluorescent protein (RFP) (Fig. S2a). We then generated several independent N. benthamiana lines that stably and constitutively expressed NbGPAT6a‐GFP (Fig. 2).
Figure 2.
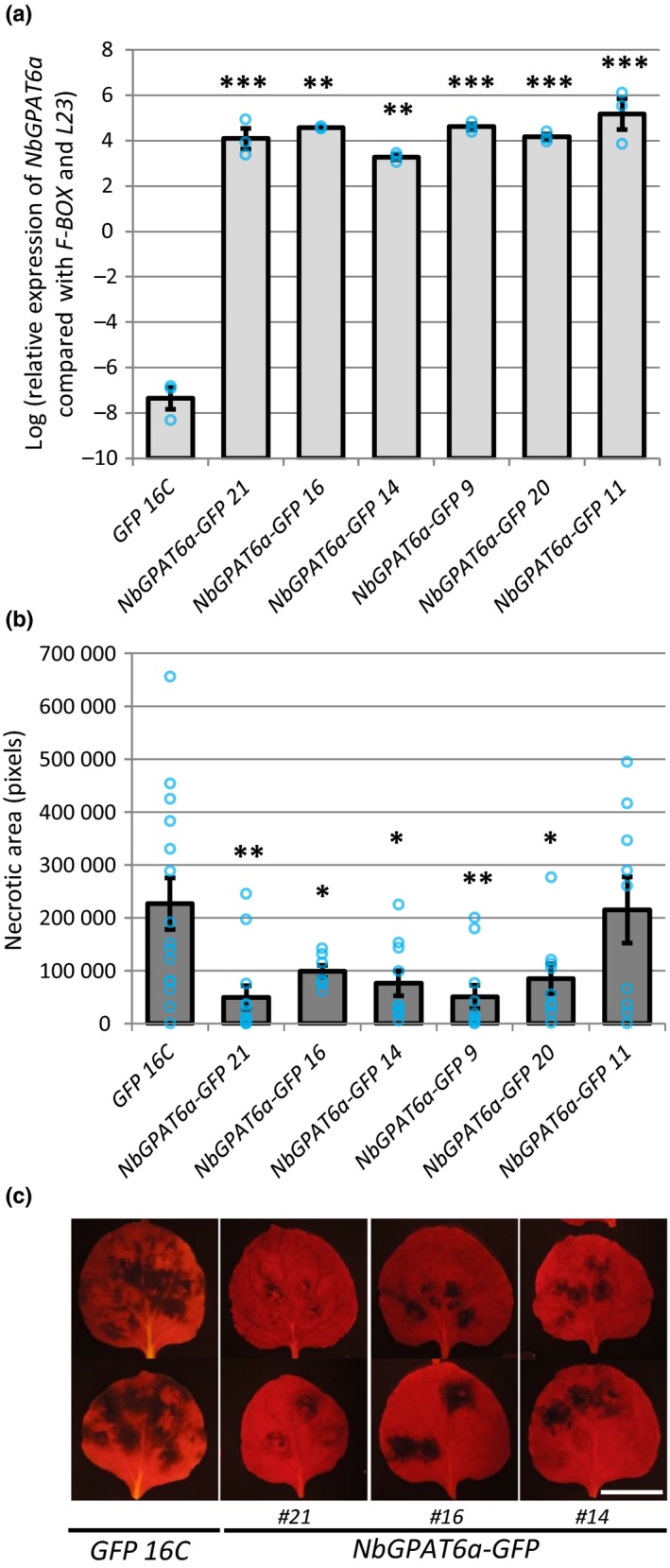
Constitutive overexpression of NbGPAT6a renders Nicotiana benthamiana leaves more resistant to infection by Phytophthora infestans. (a) Confirmation of NbGPAT6a‐GFP overexpression in transgenic N. benthamiana leaves, compared with control plants constitutively expressing GFP alone, quantified by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). Error bars represent ± SE of the mean of three biological replicates. Significant differences are indicated as: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Blue circles represent the average of three technical replicates. (b) Quantification of necrotic area present on leaves constitutively expressing NbGPAT6a‐GFP compared with the control expressing GFP alone following inoculation with P. infestans. Error bars represent ± SE of the mean of at least seven leaves from each transgenic line. Blue circles represent necrotic area on each leaf. (c) Representative images of leaves inoculated with P. infestans and imaged under blue light illumination using a yellow filter to record plastid red autofluorescence and to quantify necrotic area. Scale bar, 30 mm.
Overexpression of NbGPAT6a‐GFP resulted in a 73% increase in total leaf cutin, which was almost entirely a result of elevated concentrations of ω‐hydroxyl (OH) fatty acid (‐FA) cutin monomers (Fig. S3a). In particular, concentrations of hexadecane‐dioic acid, ω‐hydroxy hexadecanoic acid, ω‐hydroxy heptadecanoic acid, ω‐hydroxy‐octadecanoic acid and 10,16‐dihydroxy hexadecanoic acid showed significant increases relative to GFP16C (control) leaves (Fig. S3b).
When we tested NbGPAT6a‐GFP transgenic plants for their resistance to P. infestans leaf infections, we found that five of the six lines displayed smaller necrotic areas than the control lines (Fig. 2) without affecting overall morphology of P. infestans hyphae or haustoria within leaf epidermal cells of two independent NbGPATa‐GFP transgenic lines (Fig. S4). Notably, transient expression of NbGPAT6a‐GFP in fully expanded leaves followed 24 h later by P. infestans infection did not alter the extent of disease‐associated leaf necrosis (Fig. S5), suggesting that NbGPAT6‐mediated resistance is associated with longer‐term leaf development processes. Variation in these leaf development processes may contribute to the variation in resistance phenotype we observed across the six independent overexpressing lines.
Knockdown or knockout of GPAT6 renders leaves more susceptible to Phytophthora infection but more resistant to B. cinerea
To test whether reduced GPAT6 levels cause the opposite phenotype to increased levels, we established a NbGPAT6a‐specific virus‐induced gene silencing (VIGS) construct and demonstrated that it attenuated transcript abundances of NbGPAT6a, but not those of homologous transcripts (Figs 3, S6). We found that siNbGPAT6a‐mediated VIGS resulted in stronger leaf necrosis upon P. infestans infection (Fig. 3), suggesting a higher degree of susceptibility.
Figure 3.
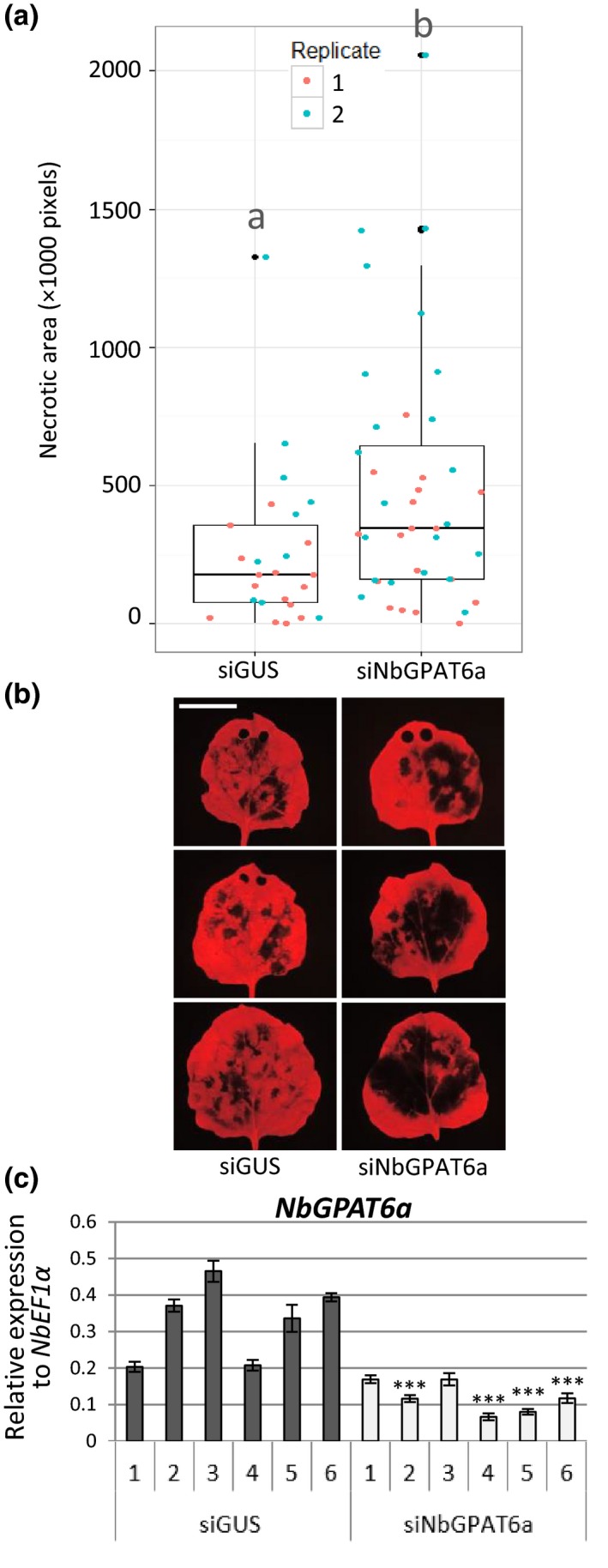
siNbGPAT6a‐mediated virus‐induced gene silencing (VIGS) results in stronger Nicotiana benthamiana leaf necrosis upon Phytophthora infestans infection. (a) Leaf necrotic area 5 d postinoculation (dpi) with P. infestans, as quantified by lack of red Chl fluorescence, two replicates combined (P = 0.0127). Data points for each replicate are denoted by different colours. Horizontal lines represent median and upper and lower quartiles. Whiskers extend to data points that are < 1.5 × interquartile range away from upper/lower quartile. Lower case letters ‘a’ and ‘b’ indicate significant differences between the means as compared using Student's t‐test (P < 0.05). (b) Representative UV images of leaf necrosis quantified in (a). Scale bar, 30 mm. Holes at the leaf tip in some images are the result of tissue samples taken for quantification of transcript abundance (these areas were excluded from necrotic area quantification). (c) Relative expression of NbGPAT6a (to NbEF1α) following VIGS using siNbGPAT6a or siGUS (control). Error bars represent ± SE of the mean of three biological replicates. Student's t‐test was used to compare mean relative expression in siNbGPAT6a samples with the lowest of control samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next tested whether GPAT6 contributes to Phytophthora infection in tomato using a gpat6‐a mutant in the ‘Micro‐Tom’ background (Petit et al., 2016). Detached leaf infection assays showed that the tomato gpat6‐a mutant was more susceptible to P. infestans (Fig. 4a,b) and P. palmivora infection (Fig. 4c), as evident by larger lesion sizes and higher expression levels of P. infestans sporulation marker transcript abundances at 48 hpi. When investigating infection structures, we found that P. infestans formed normal, digit‐like haustoria (55%) but also singly branched haustoria (45%) in epidermal cells of gpat6‐a mutant tomato leaves (65 haustoria counted; Fig. S7a). By contrast, almost exclusively digit‐like haustoria (92%, 61 haustoria counted) were formed in wild‐type (WT) leaves (Fig. S7b). Importantly, gpat6‐a mutants displayed less severe disease symptoms upon infection with the fungal pathogen B. cinerea (Fig. 4d). Taken together these data demonstrate that attenuating or knocking out the expression of GPAT6 genes has the opposite effect to GPAT6 gene overexpression, further supporting an important role for GPAT6 in ensuring full resistance to Phytophthora infections.
Figure 4.
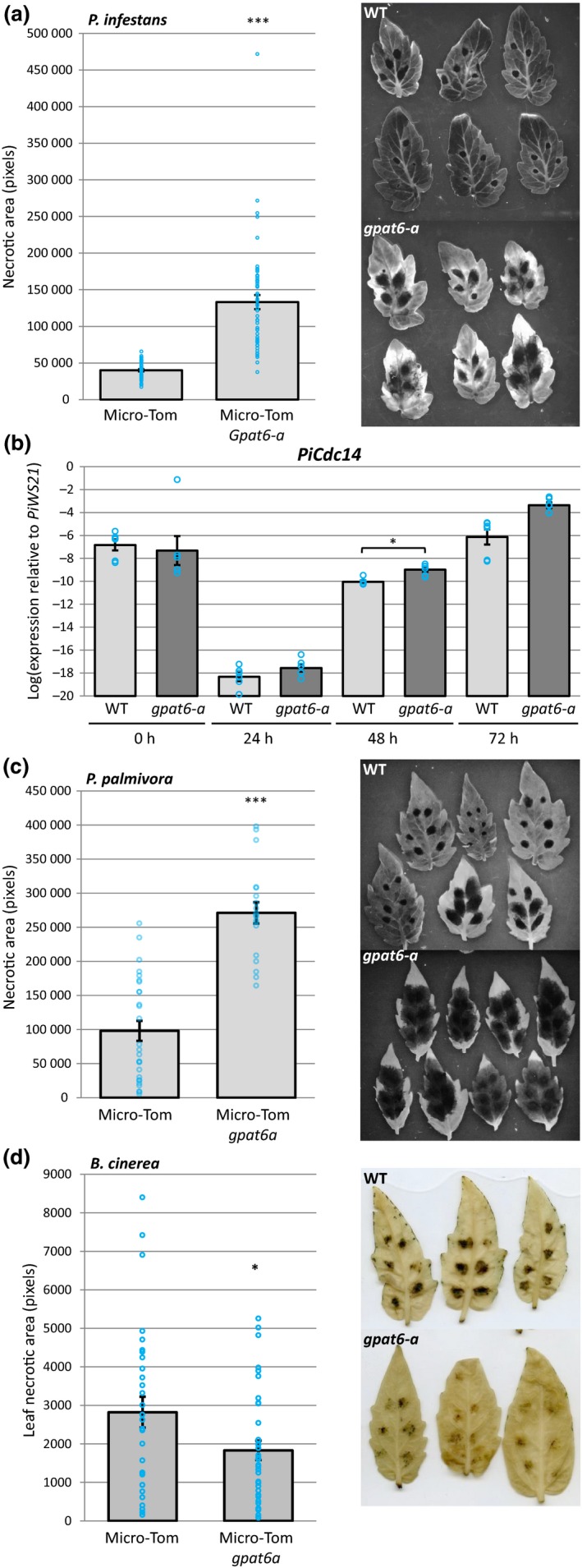
Tomato gpat6a mutants are more susceptible to Phytophthora infestans and Phytophthora palmivora infection but more resistant to Botrytis cinerea infection. (a) Quantification of Micro‐Tom leaf necrotic area at 72 h postinoculation with P. infestans for both wild‐type and gpat6‐a genotypes. Error bars represent ± SE of the mean. Each blue circle indicates the sum of necrotic area for one leaf, four inoculation sites. Representative UV images of wild‐type (WT) and gpat6‐a leaves used for necrotic area quantification are also shown. (b) Expression level of the sporulation‐specific PiCdc14 (sporulation marker) normalized to PiWS21 (40S ribosomal protein S3A). Error bars represent ± SE of the mean of three biological replicates (six technical replicates indicated by blue circles). *, P < 0.05. (c) Quantification of Micro‐Tom leaf necrotic area at 72 h postinoculation with P. palmivora for both WT and gpat6‐a genotypes. Errors bars represent ± SE of the mean (n = 27 for WT, n = 19 for gpat6‐a). Each blue circle indicates the sum of necrotic area for one leaf, six inoculation sites. ***, P < 0.001. (d) Quantification of leaf necrotic area at 6 d postinoculation with B. cinerea. Blue circles represent the total necrotic area for each leaf (six droplets of B. cinerea spore solution). Error bars represent ± SE of the mean (n = 30 for WT, n = 35 for gpat6‐a). Representative images of leaves used to quantify leaf necrotic area are also shown. This experiment was repeated twice with similar results.
Modulating GPAT6 expression alters the thickness of the outer cell walls of the leaf epidermis
The tomato gpat6‐a mutant was reported to have an altered fruit cuticle structure (Petit et al., 2016), which we hypothesized might be associated with the altered Phytophthora infection phenotypes described earlier. We imaged the cell wall and cuticle of NbGPAT6‐GFP N. benthamiana leaves that displayed different degrees of resistance to P. infestans infection, as well as tomato gpat6‐a mutant leaves, using cryo‐SEM. We observed that the outer epidermal cell wall was thinner in NbGPAT6a‐GFP lines compared with those expressing GFP alone (Fig. 5a,b), particularly in the line showing the highest P. infestans resistance (NbGPAT6a‐GFP #21).
Figure 5.
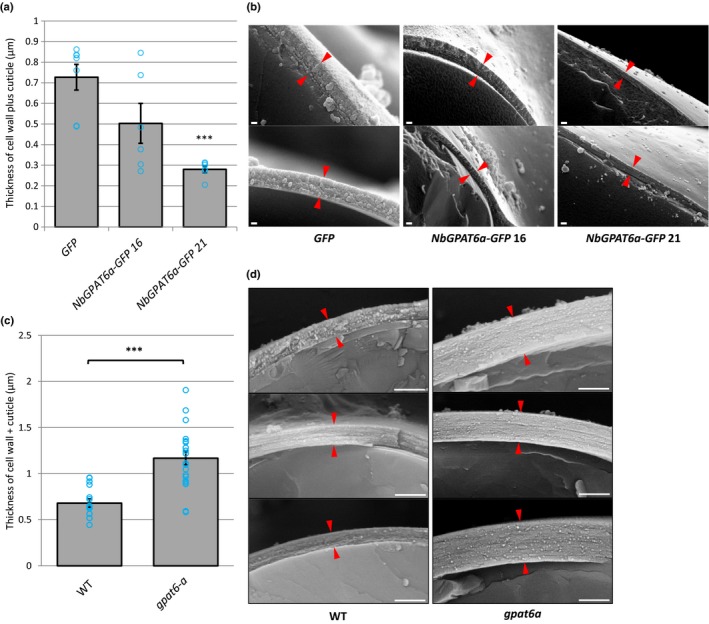
Outer epidermal cell wall thickness is correlated to GPAT6 expression levels. (a) Quantification of cell wall plus cuticle thickness in Nicotiana benthamiana leaves constitutively expressing NbGPAT6a‐GFP. Blue circles represent the mean of three measurements per image (n = 7 for GFP, n = 6 for NbGPAT6a‐GFP 16 and NbGPAT6a‐GFP 21). Error bars represent ± SE of the mean. ***, P < 0.001. (b) Representative cryoscanning electron microscopy (cryo‐SEM) images of transverse fractures used to quantify thickness. Red arrowheads indicate the boundary of cell wall plus cuticle. Scale bars, 200 nm. (c) Quantification of cell wall plus cuticle thickness in leaves of tomato gpat6‐a mutants. Blue circles represent the mean of three measurements per image (n = 15 for wild‐type (WT), n = 20 for gpat6‐a). Error bars represent ± SE of the mean. ***, P < 0.001. (d) Representative cryo‐SEM images of transverse fractures used to quantify thickness. Red arrowheads indicate the boundary of cell wall plus cuticle. Scale bars, 1 μm. This experiment was repeated twice with similar results.
Conversely, gpat6‐a leaf epidermal cells possessed a thicker cell wall (Figs 5c,d, S8a–e). This change in thickness was most prominent in the outer, but not the inner, periclinal wall of both the abaxial and adaxial leaf epidermis (Fig. S8e). Thus, the cell wall thickness inversely correlated with the level of GPAT6 expression.
GPAT6 enzymes are known to be involved in cutin biosynthesis (Li‐Beisson et al., 2009) and gpat6‐a tomato fruits have increased cuticle permeability to the dye toluidine blue. We found that gpat6‐a leaves did not show significantly altered permeability when toluidine blue was placed on the upper or lower epidermis, whereras abrasive treatment with bentonite/cellite resulted in full permeability, confirming the suitability of our staining procedure (Fig. S9a,b). To test for changes in wall porosity, we applied Dextran‐150 kDa‐TRITC to WT and gpat6‐a epidermis. Subsequent fluorescent imaging showed that TRITC‐labelled Dextran was incorporated to a greater extent in the gpat6‐a mutant, suggesting a larger porosity of the wall (Fig. 6).
Figure 6.
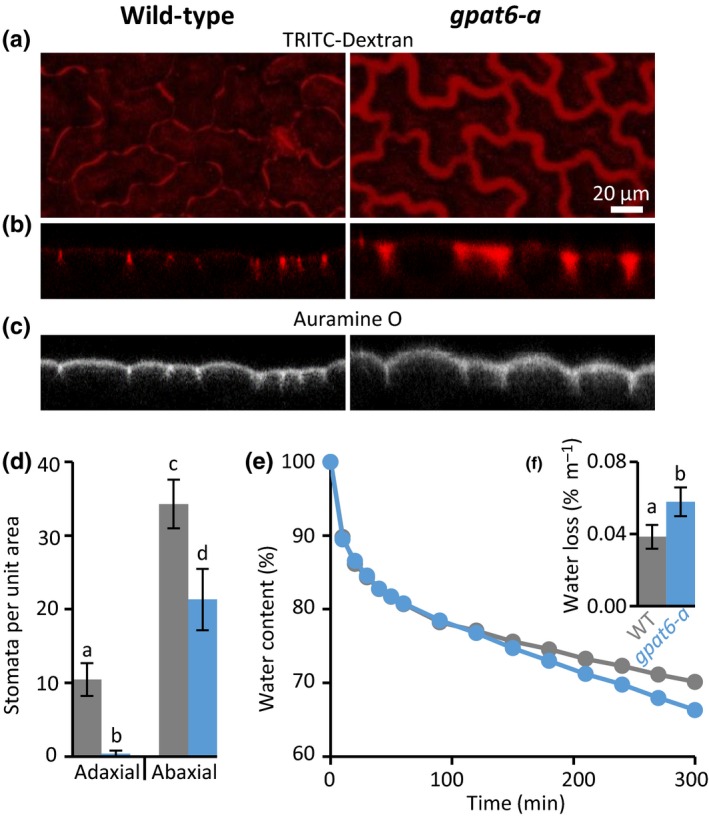
Leaf outer epidermal walls of tomato gpat6‐a mutant display altered porosity, reduced numbers of stomata and increased susceptibility to desiccation. Panels (a)–(c) are of identical magnification represented by the scale bar in (a). TRITC‐Dextran (150 kDa) distribution imaged from above (a) and confocal transects (b) are extended in gpat6‐a mutant epidermal walls. Auramine O was used to label cutin (c). (d) Stomatal numbers per unit leaf area; (e, f) water loss over time and total water loss. Error bars represent ± SD of six biological replicates. Different lowercase letters above error bars indicate significant difference between means.
Concomitantly, gpat6‐a tomato leaves showed an increased rate of water loss compared with the WT (Fig. 6e,f). This was significant in both the total amount of water loss and the relative water loss over time. We also observed that the gpat6‐a mutant leaves had fewer stomata than the WT (Figs 6d, S10a), but that the numbers increased to a value similar to the WT when gpat6‐a plants were grown under high humidity conditions (Fig. S10b). We did not observe changes in stomata numbers (Fig. S11a) or water loss over time in overexpressing N. benthamiana GPAT6a‐GFP lines with thinner walls (Fig. S11b,c), suggesting that the cuticle permeability was not altered, even though it was thinner. Furthermore, our analysis of the composition and overall architecture of the bulk leaf cell wall using cell wall antibodies did not reveal any significant alterations in GPAT6‐GFP‐overexpressing plants or the gpat6‐a mutant (Fig. S12).
The gpat6‐a leaf transcriptome reflects changes in cuticle and cell wall processes and stomatal patterning
To determine the impact of the gpat6‐a mutation on the tomato transcriptome, we carried out expression analysis of gpat6‐a and WT leaves from both P. infestans‐infected and control plants (Fig. 7), and compared our findings with previous expression data derived from the tomato fruit exocarp (Petit et al., 2016).
Figure 7.
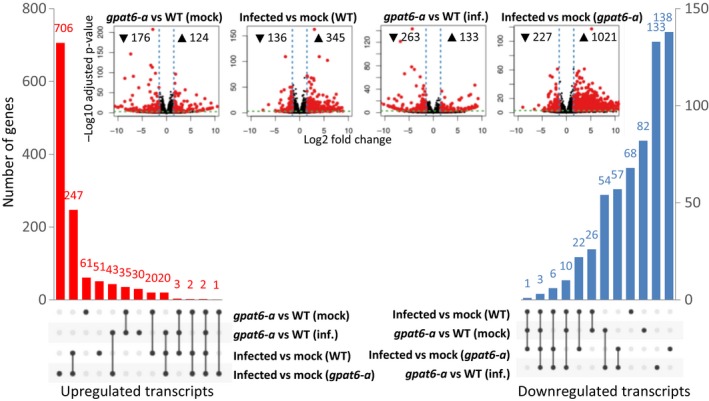
Transcriptome differences in gpat6‐a vs wild‐type (WT) tomato cv ‘Micro‐Tom’ leaves under control and infection conditions. Water or Phytophthora infestans 88069 zoospores was applied to detached leaves and harvested at 72 h postinoculation. Differentially upregulated (red) and downregulated (blue) transcripts (absolute log fold‐change ≥ 1.5 and adjusted P‐value ≤ 10−3) and pairwise comparison volcano plots (inset) are shown. This figure is based on Supporting Information Dataset S1.
Considering a LFC ≥ 1.5 with an adjusted P‐value of significance < 10E–3, we found the expression of 124 genes to be upregulated and 176 to be downregulated compared with uninfected leaves of gpat6‐a and WT plants (Fig. 7, Dataset S1). Infection of gpat6‐a plants resulted in a much higher number of induced genes (1021) compared with infection in WT leaves (345), congruent with gpat6‐a leaves being much more susceptible to P. infestans infection. Notably, there was a higher proportion of genes associated with immune responses (Pombo et al., 2014) induced in gpat6‐a leaves (Dataset S1). The differences in downregulated genes were more moderate, with 227 in gpat6‐a compared with 136 in WT‐infected plants.
Petit et al. (2016) highlighted 42 genes associated with lipid, secondary metabolite, and cell wall biosynthesis as differentially expressed in the gpat6‐a fruit exocarp. We found only 29 of these genes to be altered in the same direction (Table S1). Genes belonging to the ‘cuticle’ category responded similarly in leaves and fruit exocarp, with 18 of 21 genes similarly differentially expressed in both organs. However, genes in the ‘cell wall’ gene category responded differently and although we found seven out of 10 also repressed in leaves, none of the five reported by Petit et al. (2016) was induced. Two genes, annotated as pectin methyl esterase inhibitor and cellulose synthase‐like, showed opposite expression dynamics between leaves and fruit. Petit et al. (2016) assigned six differentially expressed genes to the secondary metabolism category – three repressed and three induced. Of these, two were also repressed and induced in our dataset, and two were also induced. In summary, the expression levels of cuticle‐associated genes were consistently altered in leaves and fruits of gpat6‐a tomato plants. More variation was observed in the cell wall and secondary metabolite categories. As alterations in the cell wall were seen to be most extensive in the outer wall of the epidermis, this represents an interesting target for future studies of expression patterns. However, P. infestans infection rapidly disrupts epidermal integrity, which represents a major technical challenge.
Even in uninfected leaves, genes associated with defence and immunity, such as disease resistance genes, protease inhibitors, pathogenicity‐related genes (PR‐3 and PR‐5), and chitinases were noted in the group of gpat6‐a repressed genes and absent from the group of induced genes (Dataset S1). Furthermore, seven genes encoding glutaredoxins were repressed in gpat6‐a, suggesting that this would increase susceptibility to redox stress. Together these data suggest that the increase in susceptibility of gpat6‐a can, in part, be attributed to constitutively lower levels of defence gene expression in uninfected gpat6‐a plants.
Six leucine‐rich repeat receptor like kinases (RLKs) were induced in gpat6‐a compared with WT plants (Dataset S1), including SERK3/BAK1 (Solyc01g056655), which showed the strongest transcriptional upregulation of all genes differentially expressed between gpat6‐a and WT leaves, although defence‐associated genes were more frequently repressed in gpat6‐a mutants. A homologue of the ERECTA homologue (Solyc01g057680), which is associated with negative regulation of stomata density in A. thaliana, was also induced. The observed induction of both SERK3/BAK1 and ERECTA may be related to the reduced number of stomata in gpat6‐a leaves. A. thaliana homologues of the other induced RLK encoding genes have been associated with pollen tube guidance (Solyc05g025780/LePRK3) (Gui et al., 2014), cell wall integrity sensing and resistance to Fusarium oxysporum root infections (Solyc01g014147, Solyc01g009930) (Van der Does et al., 2017) and regulating cell expansion through the transport of cell wall material (Solyc05g041750/PERK10) (Humphrey et al., 2014) but their roles in tomato have yet to be reported.
Discussion
Previous studies have implicated GPAT6 in the development of flowers and fruit, but its function in leaves has not been characterized. For example, A. thaliana GPAT6 is highly expressed in flowers (more than two‐fold higher in petals and sepals than in other GPAT genes) and is known to function in stamen development and fertility (Li et al., 2012), while its homologue in tomato has an additional function in fruit cutin biosynthesis (Petit et al., 2016). We demonstrate that despite its low steady‐state expression levels, GPAT6 fulfils an important role in leaves associated with epidermal outer cell wall properties that confer protection against dehydration, as well as infection by Phytophthora species.
Late transcriptional upregulation of NbGPAT6a during P. infestans infection is a mitigation response
Arabidopsis thaliana GPAT6 is a sn2‐acyltransferase that is involved in cutin biosynthesis. Our analysis shows that NbGPAT6a overexpression results in increased amounts of cutin monomers (Fig. S3), indicative of a conserved function. The late transcriptional upregulation of NbGPAT6 during P. infestans infection (Fig. 1) can be interpreted either as a pathogen‐controlled lipid harvesting strategy or, alternatively, as a mitigation response by the plant to tissue damage caused by pathogen colonization. It was recently hypothesized that obligate biotrophic fungal pathogens may exert a lipid parasitism, where the microbe benefits from plant fatty acid production (Jiang et al., 2017; Keymer & Gutjahr, 2018). In this context it was interesting to note a high frequency of aberrantly shaped haustoria in infected gpat6‐a tomato (Fig. S4). P. infestans haustoria are intracellular structures and characteristically digit‐shaped, although infrequently branched haustoria can also be observed (Blackwell, 1953). Whether an altered lipid metabolism in gpat6‐a tomato leaf epidermal cells might affect their development would require additional investigation using other lipid biosynthesis mutants. Nevertheless, gpat6‐a tomato mutants were more susceptible to both P. infestans and P. palmivora (Fig. 4), suggesting that the observed alteration in haustorium morphology does not impair infection and that the oomycete does not extensively rely on cutin monomers provided through GPAT6. It might also be informative to infect gpat6‐a tomato with obligate fungal pathogens, such as Oidium neolycopersici, to determine whether lipid parasitism is linked to an obligate biotrophic lifestyle. We conclude that Phytophthora‐controlled lipid harvesting is unlikely because overproduction of cutin monomers through overexpression of NbGPAT6a‐GFP did not result in increased pathogen‐caused symptoms; indeed, the leaves were more resistant to P. infestans infection (Fig. 2). Furthermore, Phytophthora‐derived cuticle and cell wall‐degrading enzymes probably release cutin monomers into the apoplast to enable sufficient uptake by the oomycete for continued infection in WT. We therefore propose that late transcriptional upregulation of NbGPAT6a during P. infestans infection is a mitigation response to tissue damage. Alternatively, it may be part of a delayed defence response, as various hydroxy fatty acid compounds have been implicated in pathogen resistance (Schweizer et al., 1996; Hou & Forman Iii, 2000; Wang et al., 2000), including against P. infestans.
Differences in oomycete and fungal leaf infections may be attributable to their lifestyles
A range of mutations have been reported to increase both leaf cuticle permeability and pathogen susceptibility (Tang et al., 2007). However, these mutants are all more resistant to B. cinerea (Ziv et al., 2018). Explanations for this apparent paradox include an increased release of disease resistance activators, antifungal diffusible components and improved uptake of elicitors in the mutants (Tang et al., 2007; Ziv et al., 2018). We observed that constitutively expressing NbGPAT6a lines did not display altered leaf permeability and increased water loss (Fig. S11), suggesting that other processes contribute to their increased resistance. Conversely, gpat6‐a mutants showed reduced expression levels of defence‐ and stress‐associated genes (Dataset S1), which may influence the infection outcome. The ability of pathogens to infect these mutants may depend on the pathogen infection biology or lifestyle. Phytophthora pathogens are hemibiotrophs, which initially require living host cells for infection (Fawke et al., 2015), whereas B. cinerea is considered a necrotroph that immediately kills the tissue (Van Kan et al., 2017). This may explain why gpat6‐a tomato leaves exhibit resistance to B. cinerea. Whereas a reliance on plant lipid biosynthesis was recently demonstrated for an obligate biotrophic fungal pathogen (Jiang et al., 2017), this remains to be reported for oomycetes.
The leaf cuticle layer may impose a physical restraint upon the outer facing epidermal cell wall
Our results suggest that GPAT6 influences various properties of the cell wall–cuticle superstructure. We show that loss of GPAT6 increased the thickness of the wall (Figs 5c,d, S8), and overexpression of NbGPAT6a‐GFP led to higher amounts of cutin monomers and reduced wall thickness (Fig. 5a,b). Interestingly, this effect was mainly observed in the outer face of the epidermal walls, which are also the only walls associated with a cuticle (Fig. S8e), while the overall composition of the leaf bulk cell wall was not altered (Fig. S12). This suggests that the outer epidermal cell wall may respond to its ‘cutin status’ and adapt its thickness, possibly through mechanical or biochemical sensing. An instantaneous and reversible increase in thickness of the cell wall has previously been observed upon abrasion of the cuticle (Xia et al., 2009). This suggests that physical properties of the wall allow it to flexibly expand in diameter and that cutin monomers contribute to preventing excessive expansion. Similar increases in wall thickness were reported when a tomato cutin synthase was mutated, leading to a substantially thinner fruit cuticle (Yeats et al., 2012). Although we observed that gpat6‐a mutant leaves have a thicker cell wall and cuticle, whereas those of leaves constitutively expressing NbGPAT6a are thinner, there was not such a clear contrast in terms of the rate of water loss or stomata numbers. Specifically, gpat6‐a leaves lose more water over time than those of WT (Fig. 6e,f), whereas leaves constitutively expressing NbGPAT6a lose the same amount of water as the WT (Fig. S11). This suggests that a deficiency in cutin monomers during development has a significant impact on permeability, whereas an excess of cutin monomers can be tolerated or compensated for by the plant and has no effect on permeability.
The increased porosity of gpat6‐a mutants with thicker walls may result from the looser packing of wall components, or from the low amounts of cutin monomers in the wall. This, in turn, may cause the observed increased rate of water loss, which is compensated for by altering the number of stomata.
Author contributions
SS, TAT, AG, SF and JKCR designed the research; SF, TAT, AG, EAF, IS, TY and SS performed research; SF, TAT, AG, EAF, IS, JKCR and SS analysed data; and SF, JKCR and SS wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Dataset S1 RNA‐seq of Solanum lycopersicum Micro‐tom WT and gpat6a leaves, infected with Phytophthora infestans.
Fig. S1 Phylogenetic relationship of N. benthamiana GPAT‐related proteins to homologues in M. truncatula, A. thaliana and S. lycopersicum.
Fig. S2 NbGPAT6a localizes to the endoplasmic reticulum and is predicted to have two transmembrane domains.
Fig. S3 Cutin content of NbGPAT6a‐GFP‐expressing leaves is dramatically increased relative to GFP 16C (control) leaves.
Fig. S4 Phytophthora infestans haustoria formed in wild‐type and NbGPAT6a‐GFP‐overexpressing Nicotiana benthamiana leaves are similar in structure.
Fig. S5 Transient expression of NbGPAT6a does not alter leaf necrosis triggered by P. infestans infection.
Fig. S6 Assessment of off‐target gene silencing following VIGS using siNbGPAT6a or siGUS (control).
Fig. S7 Phytophthora infestans forms digit‐like and branched haustoria in tomato gpat6‐a mutants.
Fig. S8 Thickness of outer cell wall plus cuticle is greater in gpat6‐a leaves than in the wild‐type.
Fig. S9 Leaves of gpat6‐a tomato mutants are largely impermeable to Toluidine Blue.
Fig. S10 Tomato gpat6‐a plants grown in water‐saturated atmosphere are not altered in their stomata numbers per leaf area.
Fig. S11 Stomata numbers and water loss over time remain unaffected in Nicotiana benthamiana overexpressing NbGPAT6‐GFP.
Fig. S12 Composition and overall architecture of the bulk leaf cell wall are not significantly altered in GPAT6‐GFP‐overexpressing plants or in the gpat6‐a mutant.
Methods S1 Description of experimental materials and methods used to generate supplementary data.
Table S1 Comparison of differentially regulated genes between WT and gpat6a Micro‐tom leaves identified by RNASeq in this study and that of Petit et al. (2016).
Acknowledgements
The authors would like to thank Clement Quan, Edouard Evangelisti, Liron Shenav, Firas Bou Daher and Ray Wightman (all SLCU, Cambridge) for technical and training support, Robert Saville (NIAB‐EMR, East Malling) for providing B. cinerea, Sam Brockington (Plant Sciences, Cambridge) for advice on phylogenetic analysis, Paul Knox (University of Leeds) for providing monoclonal antibodies, and Christophe Rothan (INRA, Paris) for providing tomato seeds. This research was funded by the Royal Society (RG120398, UF110073, UF160413) and the Gatsby Charitable Foundation (GAT3395/GLD). JR was supported by grants from the Plant Genome Research Program of the US National Science Foundation (IOS‐1339287) and the Agriculture and Food Research Initiative of the US Department of Agriculture (2016‐67013‐24732).
Data availability
The raw fastq data from the expression analysis are accessible at http://www.ncbi.nlm.nih.gov/sra/with accession number SRP158564. These data form the basis of Fig. 7 and Dataset S1. There are no restrictions on data availability.
References
- Bargel H, Neinhuis C. 2005. Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. Journal of Experimental Botany 56: 1049–1060. [DOI] [PubMed] [Google Scholar]
- Becktell M, Smart C, Haney C, Fry W. 2006. Host‐pathogen interactions between Phytophthora infestans and the Solanaceous Hosts Calibrachoa × hybridus, Petunia × hybrida, and Nicotiana benthamiana . Plant Disease 90: 24–32. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis . Plant Cell 19: 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li‐Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15: 329–337. [DOI] [PubMed] [Google Scholar]
- Belbahri L, Calmin G, Mauch F, Andersson JO. 2008. Evolution of the cutinase gene family: evidence for lateral gene transfer of a candidate Phytophthora virulence factor. Gene 408: 1–8. [DOI] [PubMed] [Google Scholar]
- Blackman LM, Cullerne DP, Hardham AR. 2014. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics 15: 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell EM. 1953. Haustoria of Phytophthora infestans and some other species. Transactions of the British Mycological Society 36: 138‐IN135. [Google Scholar]
- Chen X, Chen G, Truksa M, Snyder CL, Shah S, Weselake RJ. 2014. Glycerol‐3‐phosphate acyltransferase 4 is essential for the normal development of reproductive organs and the embryo in Brassica napus . Journal of Experimental Botany 65: 4201–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Snyder CL, Truksa M, Shah S, Weselake RJ. 2011a. sn‐Glycerol‐3‐phosphate acyltransferases in plants. Plant Signaling & Behavior 6: 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Truksa M, Snyder CL, El‐Mezawy A, Shah S, Weselake RJ. 2011b. Three homologous genes encoding sn‐glycerol‐3‐phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus . Plant Physiology 155: 851–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Gogleva A, Hainaux T, Doumane M, Tulin F, Quan C, Yunusov T, Floch K, Schornack S. 2017. Time‐resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection‐promoting Phytophthora palmivora effectors. BMC Biology 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawke S, Doumane M, Schornack S. 2015. Oomycete interactions with plants: infection strategies and resistance principles. Microbiology and Molecular Biology Reviews 79: 263–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fich EA, Segerson NA, Rose JK. 2016. The plant polyester cutin: biosynthesis, structure, and biological roles. Annual Review of Plant Biology 67: 207–233. [DOI] [PubMed] [Google Scholar]
- Gilbert R, Johnson A, Dean R. 1996. Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea . Physiological and Molecular Plant Pathology 48: 335–346. [Google Scholar]
- Gui C‐P, Dong X, Liu H‐K, Huang W‐J, Zhang D, Wang S‐J, Barberini ML, Gao X‐Y, Muschietti J, McCormick S. 2014. Overexpression of the tomato pollen receptor kinase LePRK1 rewires pollen tube growth to a blebbing mode. Plant Cell 26: 3538–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkort A, Boonekamp P, Hutten R, Jacobsen E, Lotz L, Kessel G, Visser R, van der Vossen E. 2008. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Research 51: 47–57. [Google Scholar]
- Hou C, Forman Iii R. 2000. Growth inhibition of plant pathogenic fungi by hydroxy fatty acids. Journal of Industrial Microbiology and Biotechnology 24: 275–276. [Google Scholar]
- Humphrey TV, Haasen KE, Aldea‐Brydges MG, Sun H, Zayed Y, Indriolo E, Goring DR. 2014. PERK–KIPK–KCBP signalling negatively regulates root growth in Arabidopsis thaliana . Journal of Experimental Botany 66: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D. 2017. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Kerstiens G. 1996a. Cuticular water permeability and its physiological significance. Journal of Experimental Botany 47: 1813–1832. [Google Scholar]
- Kerstiens G. 1996b. Plant cuticle: an integrated functional approach. Oxford, UK: BIOS Scientific Publishers. [Google Scholar]
- Keymer A, Gutjahr C. 2018. Cross‐kingdom lipid transfer in arbuscular mycorrhiza symbiosis and beyond. Current Opinion in Plant Biology 44: 137–144. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. 1980. Biopolyester membranes of plants: cutin and suberin. Science 208: 990–1000. [DOI] [PubMed] [Google Scholar]
- Leroch M, Kleber A, Silva E, Coenen T, Koppenhofer D, Shmaryahu A, Valenzuela PD, Hahn M. 2013. Transcriptome profiling of Botrytis cinerea conidial germination reveals upregulation of infection‐related genes during the prepenetration stage. Eukaryotic Cell 12: 614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J. 2007. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin‐like monomers. Proceedings of the National Academy of Sciences, USA 104: 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30: 923‐30. [DOI] [PubMed] [Google Scholar]
- Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, Yang ZN. 2012. Glycerol‐3‐phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Molecular Plant 5: 131–142. [DOI] [PubMed] [Google Scholar]
- Li‐Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. 2009. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proceedings of the National Academy of Sciences, USA 106: 22008–22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi L, Han C, Yu J, Li D, Zhang Y. 2012. Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS ONE 7: e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Casado G, Matas AJ, Domínguez E, Cuartero J, Heredia A. 2007. Biomechanics of isolated tomato (Solanum lycopersicum L.) fruit cuticles: the role of the cutin matrix and polysaccharides. Journal of Experimental Botany 58: 3875–3883. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Garroum I, Daraspe J, De Bellis D, Olsson V, Mucciolo A, Butenko MA, Humbel BM, Nawrath C. 2016. Connecting cutin composition to cuticle ultrastructure and physical properties of Arabidopsis petals. Plant Physiology 173: 1146–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Bres C, Just D, Garcia V, Mauxion J, Marion D, Bakan B, Joubès J, Domergue F, Rothan C. 2014. Analyses of tomato fruit brightness mutants uncover both cutin‐deficient and cutin‐abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiology 164: 888–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J, Bres C, Mauxion JP, Tai FW, Martin LB, Fich EA, Joubès J, Rose JK, Domergue F, Rothan C. 2016. The glycerol‐3‐phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiology 171: 894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fernandez‐Pozo N, Dunham DM, Fei Z, Martin GB. 2014. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector‐triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biology 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey T, Nars A, Bonhomme M, Bottin A, Huguet S, Balzergue S, Jardinaud MF, Bono JJ, Cullimore J, Dumas B et al 2013. NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytologist 198: 875–886. [DOI] [PubMed] [Google Scholar]
- Riederer M, Schreiber L. 2001. Protecting against water loss: analysis of the barrier properties of plant cuticles. Journal of Experimental Botany 52: 2023–2032. [DOI] [PubMed] [Google Scholar]
- Schonherr J. 1976. Water permeability of isolated cuticular membranes: the effect of pH and cations on diffusion, hydrodynamic permeability and size of polar pores in the cutin matrix. Planta 128: 113–126. [DOI] [PubMed] [Google Scholar]
- Schweizer P, Jeanguenat A, Whitacre D, Métraux J‐P, Mösinge E. 1996. Induction of resistance in barley against Erysiphe graminis f. sp. hordei by free cutin monomers. Physiological and Molecular Plant Pathology 49: 103–120. [Google Scholar]
- Tang D, Simonich MT, Innes RW. 2007. Mutations in LACS2, a long‐chain acyl‐coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis . Plant Physiology 144: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GA, Sarria GA, Martinez G, Varon F, Drenth A, Guest DI. 2016. Bud rot caused by Phytophthora palmivora: a destructive emerging disease of oil palm. Phytopathology 106: 320–329. [DOI] [PubMed] [Google Scholar]
- Underwood W. 2012. The plant cell wall: a dynamic barrier against pathogen invasion. Frontiers in Plant Science 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Boutrot F, Engelsdorf T, Rhodes J, McKenna JF, Vernhettes S, Koevoets I, Tintor N, Veerabagu M, Miedes E. 2017. The Arabidopsis leucine‐rich repeat receptor kinase MIK2/LRR‐KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genetics 13: e1006832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kan JA, Stassen JH, Mosbach A, Van Der Lee TA, Faino L, Farmer AD, Papasotiriou DG, Zhou S, Seidl MF, Cottam E et al 2017. A gapless genome sequence of the fungus Botrytis cinerea . Molecular Plant Pathology 18: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chin C‐K, Gianfagna T. 2000. Relationship between cutin monomers and tomato resistance to powdery mildew infection. Physiological and Molecular Plant Pathology 57: 55–61. [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE. 2012. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Current Biology 22: 2242–2246. [DOI] [PubMed] [Google Scholar]
- van West P, de Jong AJ, Judelson HS, Emons AM, Govers F. 1998. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genetics and Biology 23: 126–138. [DOI] [PubMed] [Google Scholar]
- van West P, Kamoun S, van‘t Klooster JW, Govers F. 1999. Internuclear gene silencing in Phytophthora infestans . Molecular Cell 3: 339–348. [DOI] [PubMed] [Google Scholar]
- Xia Y, Gao QM, Yu K, Lapchyk L, Navarre D, Hildebrand D, Kachroo A, Kachroo P. 2009. An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host & Microbe 5: 151–165. [DOI] [PubMed] [Google Scholar]
- Yan HZ, Liou RF. 2006. Selection of internal control genes for real‐time quantitative RT‐PCR assays in the oomycete plant pathogen Phytophthora parasitica . Fungal Genetics and Biology 43: 430–438. [DOI] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li‐Beisson Y, Beisson F, Feig M, Ohlrogge J. 2010. A distinct type of glycerol‐3‐phosphate acyltransferase with sn‐2 preference and phosphatase activity producing 2‐monoacylglycerol. Proceedings of the National Academy of Sciences, USA 107: 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li‐Beisson Y, Beisson F, Pollard M, Ohlrogge JB. 2012. A land‐plant‐specific glycerol‐3‐phosphate acyltransferase family in Arabidopsis: substrate specificity, sn‐2 preference, and evolution. Plant Physiology 160: 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LB, Viart HM, Isaacson T, He Y, Zhao L, Matas AJ, Buda GJ, Domozych DS, Clausen MH et al 2012. The identification of cutin synthase: formation of the plant polyester cutin. Nature Chemical Biology 8: 609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. 2013. The formation and function of plant cuticles. Plant Physiology 163: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. 2003. Arabidopsis AtGPAT1, a member of the membrane‐bound glycerol‐3‐phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv C, Zhao Z, Gao YG, Xia Y. 2018. Multifunctional roles of plant cuticle during plant‐pathogen interactions. Frontiers in Plant Science 9: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Dataset S1 RNA‐seq of Solanum lycopersicum Micro‐tom WT and gpat6a leaves, infected with Phytophthora infestans.
Fig. S1 Phylogenetic relationship of N. benthamiana GPAT‐related proteins to homologues in M. truncatula, A. thaliana and S. lycopersicum.
Fig. S2 NbGPAT6a localizes to the endoplasmic reticulum and is predicted to have two transmembrane domains.
Fig. S3 Cutin content of NbGPAT6a‐GFP‐expressing leaves is dramatically increased relative to GFP 16C (control) leaves.
Fig. S4 Phytophthora infestans haustoria formed in wild‐type and NbGPAT6a‐GFP‐overexpressing Nicotiana benthamiana leaves are similar in structure.
Fig. S5 Transient expression of NbGPAT6a does not alter leaf necrosis triggered by P. infestans infection.
Fig. S6 Assessment of off‐target gene silencing following VIGS using siNbGPAT6a or siGUS (control).
Fig. S7 Phytophthora infestans forms digit‐like and branched haustoria in tomato gpat6‐a mutants.
Fig. S8 Thickness of outer cell wall plus cuticle is greater in gpat6‐a leaves than in the wild‐type.
Fig. S9 Leaves of gpat6‐a tomato mutants are largely impermeable to Toluidine Blue.
Fig. S10 Tomato gpat6‐a plants grown in water‐saturated atmosphere are not altered in their stomata numbers per leaf area.
Fig. S11 Stomata numbers and water loss over time remain unaffected in Nicotiana benthamiana overexpressing NbGPAT6‐GFP.
Fig. S12 Composition and overall architecture of the bulk leaf cell wall are not significantly altered in GPAT6‐GFP‐overexpressing plants or in the gpat6‐a mutant.
Methods S1 Description of experimental materials and methods used to generate supplementary data.
Table S1 Comparison of differentially regulated genes between WT and gpat6a Micro‐tom leaves identified by RNASeq in this study and that of Petit et al. (2016).
Data Availability Statement
The raw fastq data from the expression analysis are accessible at http://www.ncbi.nlm.nih.gov/sra/with accession number SRP158564. These data form the basis of Fig. 7 and Dataset S1. There are no restrictions on data availability.


