Abstract
The ability, propensity and need to mount an immune response vary both among individuals and within a single individual over time.
A wide array of parameters has been found to influence immune state in carefully controlled experiments, but we understand much less about which of these parameters are important in determining immune state in wild populations.
Diet can influence immune responses, for example when nutrient availability is limited. We therefore predict that natural dietary variation will play a role in modulating immune state, but this has never been tested.
We measured carbon and nitrogen stable isotope ratios in an island population of house mice Mus musculus domesticus as an indication of dietary variation, and the expression of a range of immune‐related genes to represent immune state.
After accounting for potential confounding influences such as age, sex and helminth load, we found a significant association between carbon isotope ratio and levels of immune activity in the mesenteric lymph nodes, particularly in relation to the inflammatory response.
This association demonstrates the important interplay between diet and an animal's response to immune challenges, and therefore potentially its susceptibility to disease.
A plain language summary is available for this article.
Keywords: Diet, eco‐immunology, house mouse, immune response, stable isotope analysis
1. INTRODUCTION
At any given time, an animal's “immune state” can be considered as the numbers, concentrations and distribution of the various cells and molecules that make up the immune system (Abolins et al., 2018). This immune state is highly variable both within and among individuals and is ultimately determined by the many immune challenges (i.e. antigens) encountered throughout the animal's life, as well as the individual's ability (Watson et al., 2016) and propensity (Jackson et al., 2014) to respond to them. While we have a good mechanistic understanding of how, for example, certain genes or nutrients might influence the immune response, less is understood about the relative importance of such influences in determining variation in the wild (Pedersen & Babayan, 2011). Elucidating the key drivers of this variation will play an important role in understanding susceptibility to many forms of disease.
We know through laboratory studies and food supplementation experiments that an animal's diet can have a major influence on its immune state. Immune responses are energetically costly (Lochmiller & Deerenberg, 2003), and therefore, the amount of energy acquired through the diet can influence the amount that is allocated to the immune system (Forbes et al., 2016). Furthermore, certain diet‐derived nutrients play a particularly important role in the immune system, so the quantities consumed can be a limiting factor in the strength of an immune response (Saino, Ferrari, Romano, Martinelli, & Møller, 2003; Webb, Leslie, Lochmiller, & Masters, 2003). Diet may also influence the gut microbiota, with far‐reaching consequences for the host's immune system (Murphy, Velazquez, & Herbert, 2015; Rosshart et al., 2017).
While experimental manipulations demonstrate that diet can influence immune state, there is a lack of evidence (outside of humans; see Barbaresko, Koch, Schulze, & Nöthlings, 2013) for whether natural dietary variation does indeed explain a significant proportion of variation in immune state. A recent study on wild house mice Mus musculus domesticus by Abolins et al. (2018) took important steps towards identifying the relative contributions of a range of host and environmental variables to immune state. Their analysis suggests that intrinsic host factors such as age and condition are more important than parasitic infections in influencing a wild animal's immune state. In addition, distinct populations show particular immune phenotypes, in a way which is not directly related to the extent of genetic differentiation. Importantly though, the study by Abolins et al. (2018) does not include any measures of the diet of the mice in question. Mice show a highly flexible diet (Sage, 1981), and we predict an association between natural dietary variation and immune state, but this remains to be tested.
Stable isotope analysis (SIA) is a method by which the proportions of stable isotopes of elements (such as carbon 13C:12C and nitrogen 15N:14N) can be used to determine certain key ecological parameters, including dietary variation (Ben‐David & Flaherty, 2012; Kelly, 2000; Peterson & Fry, 1987). Food sources vary in their isotope ratios, and this variation is incorporated by consumers in a predictable manner (DeNiro & Epstein, 1978, 1981). For example, differences in carbon isotope ratios can identify whether the diet is derived from a marine or terrestrial source (Hobson, 1987; Peterson & Fry, 1987), and differences in nitrogen isotope ratios can be used to determine dietary source and relative trophic position (Minagawa & Wada, 1984; Schoeninger, DeNiro, & Tauber, 1983). Importantly, SIA is a less biased means of estimating diet compared to other approaches such as gut content or faecal analysis, which are strongly influenced by the digestibility of the food items (Stapp, 2002). In addition, depending on the type of tissue sampled, SIA provides an estimate of the average diet over a period of weeks or months rather than a single snapshot in time (Tieszen, Boutton, Tesdahl, & Slade, 1983). On the other hand, a limitation of SIA is that while it provides an index of dietary variation, it does not identify the particular food sources involved unless a range of potential sources is also sampled in depth (see Methods: Stable Isotope Analysis for further details).
We aimed to investigate whether dietary variation in the wild house mouse M. musculus domesticus is associated with changes in immunological state. We measured carbon and nitrogen isotope ratios from mouse muscle tissue at a number of different geographical locations within a single island population, as a proxy for dietary variation. We characterized immune state by measuring expression of a number of immune‐related genes in the spleen and mesenteric lymph nodes (MLNs) and the concentration of various cytokines within the blood. We also recorded some habitat indices, mouse biometrics and gut parasite burdens. We predicted that isotope values will vary by sampling site, but that in addition they will be associated with changes in the immune state.
2. MATERIALS AND METHODS
2.1. Sample collection
We conducted fieldwork on the Isle of May (56°11'N, 2°33'W), an island off the coast of Scotland, UK, covering an area of 45 ha (Figure 1). The island is largely treeless and is mostly covered by different types of bird‐modified maritime grassland (Wright, Wal, Wanless, & Bardgett, 2010). Wild house mice are present on all parts of the island and are feral and non‐commensal (Triggs, 1991).
Figure 1.
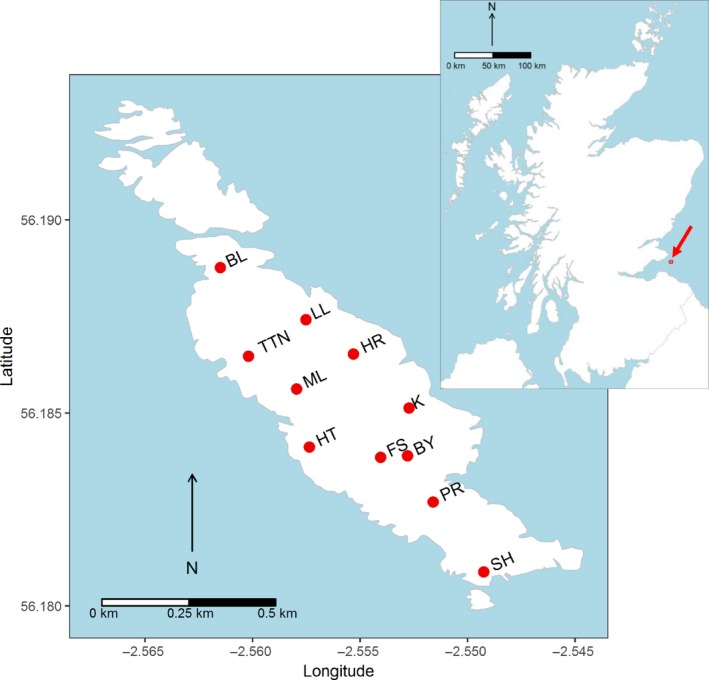
Approximate trapping locations on the Isle of May, along with the location of the isle within Scotland (inset). BL: Burnett's Leap; BY: Byres; FS: Fluke Street; HR: Holyman's Road; HT: High Tarn; K: Kettle; LL: Low Light; ML: Main Light; PR: Priory ruins; RO: Rona; SH: South Horn; TTN: Three Tarn Nick
We trapped house mice over the course of 4 days between 9 and 12 October 2015. At each of eleven locations on the island (Figure 1), we placed 16 traps in pairs approximately every 2 m along a transect. We used primarily Longworth traps (Longworth Scientific Instrument Co.), along with small numbers of Ugglan (Granhab) and home‐made “Jordan” traps (Perrow & Jowitt, 1995). Equal proportions of trap types were used at each of the trapping locations in case there were any differences in trapping efficiency. All traps were baited with a commercially available wild bird seed mix, and hay was provided as insulation.
We checked the traps twice daily, and any non‐pregnant mice captured were taken to be culled. Mice were euthanized by a rising concentration of CO2 with death confirmed by exsanguination.
2.2. Habitat variables
To avoid over‐fitting models with large numbers of explanatory variables, we chose two key variables to represent habitat variation among sampling sites. Firstly, we categorized habitat type according to the dominant plant species found at each site, as this influences not only the plant food sources available but also other sources such as invertebrates. These categories were based on a vegetation survey carried out in 2008 (R. van der Wal, unpublished data); no major vegetation changes since that date were evident. Each site was dominated either by the perennial grass Yorkshire fog Holcus lanatus or by sea campion Silene uniflora.
We also recorded the density of breeding puffins, since, in other island populations, mice have been recorded scavenging and preying on seabird eggs and juveniles (Angel, Wanless, & Cooper, 2009; Cuthbert & Hilton, 2004). Furthermore, seabirds represent a potential source of marine‐derived nutrients in the diet of the mice, and such nutrients have a distinctive isotopic signature (Hobson, 1987). Values for “puffin density” are based on counts of occupied Atlantic puffin Fratercula arctica burrows from a census carried out in April/May 2017 (Newell, Harris, Burthe, & Daunt, 2017). There was no significant change in the Isle of May puffin population between 2013 and 2017 (Newell et al., 2017), so the distribution of occupied burrows here is likely representative of our study period. Census counts were divided among 27 different areas of the island, and we standardized each count by dividing by the area of the relevant region in hectares.
2.3. Mouse life history and physiology
We recorded sex, total body mass and snout–vent length (SVL) of each individual immediately after death. The eyes were removed and fixed in 10% formalin. We later dissected the eyes to remove the lenses which we dried at 60°C for 48 hr or until they showed no further weight loss (Rowe, Bradfield, Quy, & Swinney, 1985). The dry mass of each pair of lenses was used to estimate mouse age in days using the method from Rowe et al. (1985).
As a measure of condition, we calculated the Scaled Mass Index (SMI) from the body mass and SVL for each individual using the method described by Peig and Green (2009). SMI represents the equivalent value for body mass after allometric scaling to a standard body length and therefore represents excess or shortfall of mass for a given size. SMI correlates positively with the size of nutritional reserves (Peig & Green, 2009).
Circulating leptin levels were used as a further biomarker of physiological status and body condition (Abolins et al., 2018; Abolins, Pocock, Hafalla, Riley, & Viney, 2011). Serum samples were processed in duplicate with a custom Bio‐Rad Bio‐Plex mouse cytokine reagent kit according to manufacturer's protocol (Bio‐Rad). Along with detection antibodies for leptin, we also included detection antibodies for a number of cytokines in the multiplex assay (see “Immune markers” below).
Following incubation, the reaction mixture was analysed using a Bio‐Plex 200 Luminex‐based multiplex analysis system (Bio‐Rad). Unknown cytokine concentrations were calculated by Bio‐Plex Manager Software using standard curves derived from recombinant cytokine standards. Data that were below the assay's range of detection were assigned values of 0.001 (Abolins et al., 2017).
2.4. Parasite counts
The digestive tract was removed from each culled animal and stored individually in 70% ethanol. It was later dissected, and both gut contents and mucosa were examined for gastrointestinal helminths under a dissecting microscope. Helminths were identified to species level based on morphology. Two species were found in our samples: the pinworm Syphacia obvelata and the whipworm Trichuris muris. Juvenile and adult life stages of both sexes were all recorded and pooled as a single count.
2.5. Immune markers
We carried out qPCR to measure normalized mRNA expression of a suite of genes reflecting different functional arms of the immune system in spleen (14 genes) and MLN tissue (11 genes; for details, see Table S1). The spleen and MLNs were removed immediately following death and placed in RNAlater solution (Life Technologies). Samples were kept at 4°C for 24 hr; then, the supernatant was removed, and samples were stored at −80°C until extraction.
RNA was extracted from up to 30 mg spleen and MLN tissue using the NucleoSpin RNA kit (Macherey‐Nagel) following the manufacturers' protocol. The purity, concentration and integrity of RNA were assessed following Robertson, Bradley, and MacColl (2016). Synthesis of cDNA was performed on up to 2 µg of total RNA using the nanoScript2 Reverse Transcription Kit (Primerdesign), using a combination of oligo‐dT and random nonamer primers, following the manufacturers' protocol. All cDNA samples were diluted with nuclease‐free water (1:10 for spleen samples and 1:5 for MLN) and stored at −20°C before further use.
qPCRs were performed as described in Robertson et al. (2016), with primers designed and validated by Primerdesign (Southampton, UK). All samples were run in duplicate, with each plate also containing negative controls and a pooled reference sample. Due to technical issues, expression levels of some genes are only available from one tissue type (Table S1).
Six candidate endogenous “housekeeping” genes (Actb, Gapdh, Rn18s, Rpl13a, Sdha and Ubc) were assessed for stability by a geNorm assay (SYBR Green Kit; Primerdesign) against 15 randomly selected spleen or MLN cDNA samples. Sdha and Ubc were selected as the most stably expressed of the reference genes. The expression of the immunological target genes was normalized against the reference cDNA using the 2−ΔΔ C T method (Pfaffl, 2001; Vandesompele et al., 2002).
In addition to the mRNA measurements, we measured circulating serum concentrations of nine cytokine molecules (IFN‐γ, IL‐1β, IL‐5, IL‐6, IL‐10, IL‐12β, IL‐13, IL‐17 and TNF‐α; see Table S1 for functions) using multiplex bead assay, following the method described for leptin under “Mouse life history and physiology” above.
2.6. Stable isotope analysis
Leg muscle tissue was taken from euthanized mice for use in SIA. Muscle tissue was used because it accurately reflects the isotopic composition of an animal's diet over several weeks or months, while tissues with a higher metabolic rate (e.g. liver) will reflect a shorter dietary period (Hobson & Clark, 1992; Kurle & Worthy, 2002; Tieszen et al., 1983). In the case of mice, the half‐life for carbon and nitrogen isotopes in muscle tissue is approximately 3–4 weeks (MacAvoy, Macko, & Arneson, 2005). This integration period ensured that our isotopic data were not too sensitive to noise caused by short‐term dietary variation, but reflected an “average” diet consumed over recent weeks.
All samples were kept frozen prior to drying and were then freeze‐dried at −50°C for approximately 12 hr before being ground to a fine powder with a mortar and pestle. Lipids were removed by soaking in a 2:1 chloroform:methanol solution (Cherry, Derocher, Hobson, Stirling, & Thiemann, 2011; Folch, Lees, & Sloane Stanley, 1957). Lipids typically have less 13C (DeNiro & Epstein, 1977), so lipid extraction reduces the risk of significant bias in δ13C values (Post et al., 2007; Tieszen et al., 1983).
Approximately 0.6 mg of prepared tissue from each sample was used in SIA. The isotope ratio mass spectrometry took place at the NERC Isotope Geosciences Facility (British Geological Survey, UK), measured on a continuous flow‐elemental analyser (Flash/EA) coupled to a Thermo Finnigan Delta Plus XL via a ConFlo III interface (all from Thermo Scientific). Isotope results were expressed as delta (δ) values, reported in per mil (‰) relative to international standards for δ13C (Vienna Pee Dee Belemnite (VPDB)) and δ15N (atmospheric nitrogen (AIR)), according to the following equation:
where X is either 13C or 15N, and R_sample and R_standard are the 13C:12C or 15N:14N ratios of the sample or standard, respectively. δ13C and δ15N ratios were calibrated using an in‐house reference material M1360p (powdered gelatine from British Drug Houses, Poole, UK) with expected delta values of –20.32‰ (calibrated against CH7, IAEA) and + 8.12‰ (calibrated against N‐1 and N‐2, IAEA) for C and N, respectively. δ13C and δ15N analyses were undertaken in duplicate, and the average standard deviation of these pairs was δ15N = ±0.11‰ and δ13C = ±0.22‰. The 1σ reproducibility for mass spectrometry controls for these analyses was better than ±0.2‰ for both isotopes.
In our analysis, we used δ13C and δ15N values as proxies for dietary variation, without inferring specific details about the identity of the dietary sources. With sufficient data on the isotope values of potential food sources, it is possible to estimate the proportions consumed by each consumer (Parnell, Inger, Bearhop, & Jackson, 2010; Phillips, 2001). However, we are aware that δ15N values in island vegetation, and consequently further up the food chain, can be strongly influenced by sampling location over a fine spatial scale, at least in part due to the input of nitrogen from seabird guano (Cocks, Balfour, & Stock, 1998; Drever, Blight, Hobson, & Bertram, 2000; Mizutani & Wada, 1988; Wainright, Haney, Kerr, Golovkin, & Flint, 1998). We were therefore unable to infer the composition of an individual mouse's diet with any degree of certainty without taking large numbers of samples of each potential source type from each trapping location around the island. While such detailed sampling would undoubtedly have been informative, it was unfortunately not within the scope of the present study.
Instead, we have accounted for geographical variation in our models where necessary, such that remaining variation in δ15N and δ13C values is not attributable to the trapping location. We therefore made the reasonable assumption that the majority of the remaining variation corresponded to mice that are using food sources in different proportions, without identifying specifically what those sources were.
2.7. Statistical analysis
We analysed the relationship between ecological variables, including diet, and immune markers using redundancy analyses (Legendre & Legendre, 1998). Immune variables were divided into three groups: expression of immune genes in the spleen, expression of immune genes in the MLNs and cytokine protein concentrations measured from the blood. Each group was used as the set of response variables for a separate redundancy analysis. The immune variables were (log + 1) transformed to bring them close to a normal distribution, which ensured that the construction of ordination axes was not excessively influenced by extreme data points (Legendre & Legendre, 1998). Each redundancy analysis used the same set of predictor variables, all loosely related to the individual's ecology: δ13C, δ15N, age, sex, leptin concentration and presence/absence of S. obvelata and T. muris. Tests of significance for these ecological predictor variables were carried out against 9999 random permutations of the data and therefore did not depend on parametric assumptions (Legendre & Legendre, 1998).
To assess the relationship between diet and body condition, we conducted two linear mixed models with leptin concentration and SMI as the respective response variables, and δ13C and δ15N as predictors in each. Age, sex and presence/absence of S. obvelata and T. muris were also included as fixed effects and location as a random effect. We confirmed normality of residuals by inspection of a quantile–quantile plot and confirmed homoscedasticity on a scale‐location plot. We found no evidence for nonlinear relationships between continuous predictors (δ13C, δ15N and age) and response variables when examining plots of model residuals against predictor values. We also found no evidence for spatial correlation at the scale of sampling sites after plotting variograms of the model residuals against latitude and longitude coordinates (Legendre & Legendre, 1998). The contribution of the random effect was assessed via a likelihood ratio test by comparison with a linear model omitting the random term, and the random term was only retained if significant at p < 0.05. After this, we assessed all possible combinations of fixed effects using the R package MuMIn (Barton, 2016) and took the average of any resulting models for which ΔAIC (Akaike information criterion) < 2 as compared to the model with the lowest AIC value.
To examine associations between dietary variation and other ecological factors, we conducted two linear mixed models with δ13C and δ15N as the respective response variables. Fixed effects were age, sex, dominant vegetation, puffin density and presence/absence of T. muris and S. obvelata, with a random effect of sample site. Model checking, simplification and averaging were carried out as above.
2.8. Software
Statistical analysis was carried out in r version 3.4.4 (R Core Team, 2018), using packages tidyverse for data manipulation and visualization (Wickham, 2016), vegan for redundancy analysis (Oksanen et al. 2016), lme4 for linear mixed modelling (Bates, Mächler, Bolker, & Walker, 2015) and nlme for construction of variograms (Pinheiro et al. 2018).
3. RESULTS
3.1. Diet and immune response: low carbon isotope values are associated with stronger immune response in the MLNs
We conducted stable isotope analysis on 74 individual mice (21 females and 53 males; 24 juvenile and 50 mature). We found that variation in immune markers from the blood and the spleen was not explained by variation in ecological predictors (δ13C, δ15N, age, sex, leptin, S. obvelata and T. muris) (redundancy analysis; blood: R 2 = 0.13, Adjusted R 2 = 0.035, p = 0.16, n = 74; spleen: R 2 = 0.092, Adjusted R 2 = −0.024, p = 0.68, n = 63). However, we did find a significant association between MLN immune markers and our ecological predictors (R 2 = 0.20, Adjusted R 2 = 0.082, p = 0.019, n = 55). This association is driven by the δ13C values (pseudo‐F = 5.14, degrees of freedom = 1, 47, p = 0.0007); none of the other predictors contributed significantly.
To examine how δ13C value relates to gene expression in the MLNs in more detail, we examined the loadings of RDA1, which correlated very strongly with δ13C value (Figure 2, Table 1). RDA1 represented 11% of the measured variation in gene expression and 53% of the explained (as opposed to residual) variation and correlated positively with almost all of the genes included. In particular, five of the six most strongly correlated genes (r > 0.3) were linked to pro‐inflammatory or Th1 pathways (Il1b, Il6, Tbx21, Irf5 and Ifng), and the three with the lowest correlation (Gata3, Il10 and Il13) were linked to Th2 or anti‐inflammatory pathways. RDA1 also correlated strongly, but in the opposite direction, with δ13C. Therefore, mice with lower δ13C values tended to show higher levels of immune signalling in the MLNs, especially expression of genes with a connection to anti‐microbial inflammatory response.
Figure 2.
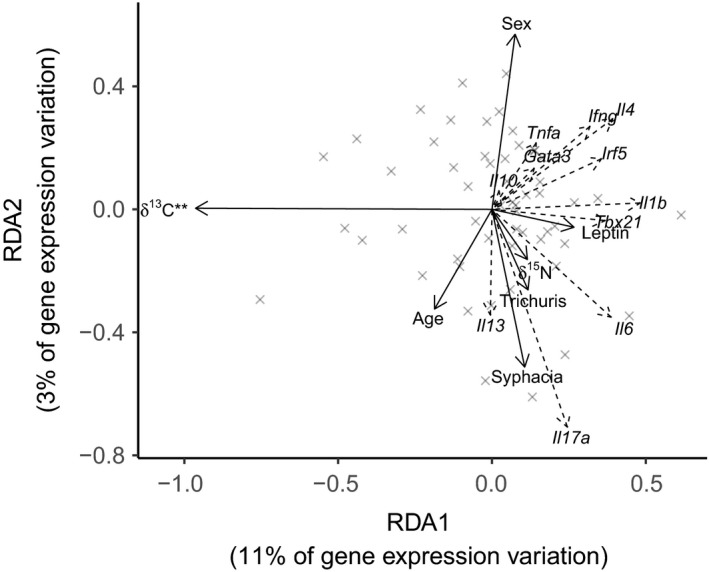
Triplot from a redundancy analysis of expression of immune‐related genes in the MLNs. Solid arrows indicate predictor variables, with **p < 0.001. Dashed arrows and italic labels indicate response variables. Grey crosses represent individual mice
Table 1.
Correlations of predictor (ecological) and response (MLN gene expression) variables with RDA1 from a redundancy analysis. Variables are ordered by the magnitude of their correlation
| Predictor | RDA1 | Gene | RDA1 |
|---|---|---|---|
| δ13C | −0.964 | Il1b | 0.482 |
| Leptin | 0.267 | Il4 | 0.393 |
| Age (days) | −0.187 | Il6 | 0.387 |
| Trichuris muris (present) | 0.117 | Tbx21 | 0.368 |
| δ15N | 0.115 | Irf5 | 0.356 |
| Syphacia obvelata (present) | 0.106 | Ifng | 0.318 |
| Sex (male) | 0.075 | Il17a | 0.245 |
| Tnfa | 0.144 | ||
| Gata3 | 0.137 | ||
| Il10 | 0.023 | ||
| Il13 | −0.005 |
3.2. Diet and condition: low carbon isotope values are associated with higher levels of circulating leptin
Although both leptin concentration in the blood and SMI can be considered indices of condition, they did not correlate with one another (Kendall's tau = 0.097, p = 0.22). We found a negative association between leptin concentration and δ13C value (coefficient (Coef) = −0.351, 95% confidence interval (CI) = −0.628 to −0.075; Figure 3 and Table 2), but no association with other predictors. We found no associations between SMI and isotope values, or other predictor variables (Table 2). The random effect of location was dropped from both the leptin and SMI models as it did not significantly improve fit (leptin ΔAIC = −0.82, p = 0.28; SMI: ΔAIC = 0.03, p = 0.15).
Figure 3.
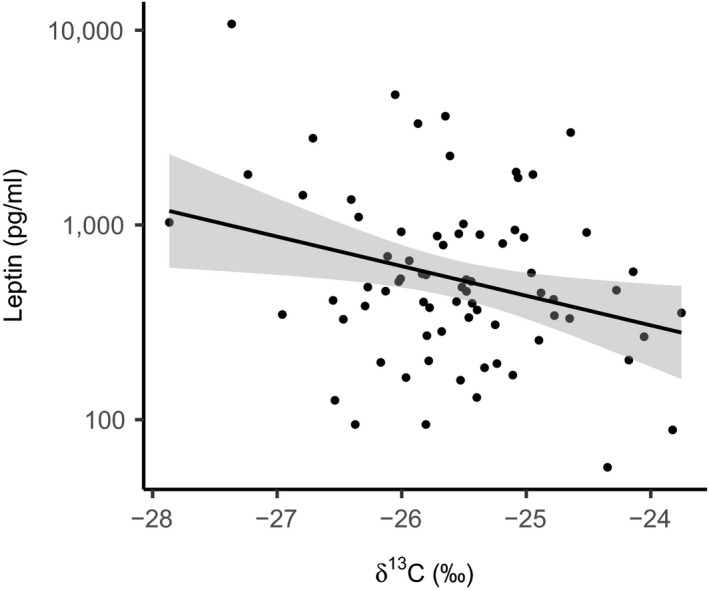
Variation in circulating leptin concentration with carbon isotope values. Points represent individual mice, and line shows model prediction with grey shading for ±SE
Table 2.
Final coefficients for linear models of two condition variables (leptin concentration and Scaled Mass Index (SMI)) after model selection and averaging. Coefficients highlighted in bold are those for which the 95% confidence interval does not include zero
| Term | Leptin | SMI | ||||||
|---|---|---|---|---|---|---|---|---|
| Coefficient | LCI | UCI | Weight | Coefficient | LCI | UCI | Weight | |
| Intercept | −2.69 | −9.78 | 4.40 | NA | 20.2 | 14.3 | 26.1 | NA |
| δ13C | −0.351 | −0.628 | −0.075 | 1 | −0.012 | −0.218 | 0.194 | 0.086 |
| δ15N | −0.034 | −0.197 | 0.129 | 0.252 | ||||
| Sex | −0.114 | −0.798 | 0.571 | 0.201 | ||||
| Age | 0.0005 | −0.0026 | 0.0037 | 0.185 | 0.0003 | −0.0035 | 0.0041 | 0.091 |
| Syphacia obvelata | 0.018 | −0.171 | 0.206 | 0.139 | 0.23 | −0.65 | 1.10 | 0.345 |
| Trichuris muris | −0.180 | −0.676 | 0.315 | 0.5 | ||||
LCI: lower 95% confidence interval; UCI: upper 95% confidence interval.
3.3. Nitrogen isotope values vary with location
The tested ecological variables did not explain any dietary variation in terms of carbon isotope values (Table 3). Trapping location was not retained in the δ13C model as it did not contribute to an improved fit (ΔAIC = −2, p = 1). In the case of δ15N, there was significant variation by trapping location (ΔAIC = 7.31, p = 0.0023; Figure 4, Table 3) but this was not associated with either puffin burrow density (Coef = 0.00001, CI = −0.00011 to 0.00014) or the dominant vegetation type (Coef for S. uniflora = −0.018, CI = −0.185 to 0.150). In addition, δ15N values were significantly higher in males (Coef = 1.14, CI = 0.19 to 2.09) and in individuals infected with S. obvelata (Coef = 1.03, CI = 0.04 to 2.01).
Table 3.
Final coefficients for linear models of δ13C and δ15N after model selection and averaging. Coefficients highlighted in bold are those for which the 95% confidence interval does not include zero
| Term | δ13C | δ15N | ||||||
|---|---|---|---|---|---|---|---|---|
| Coefficient | LCI | UCI | Weight | Coefficient | LCI | UCI | Weight | |
| Intercept | −25.6 | −25.8 | −25.3 | NA | 12.4 | 10.7 | 14.1 | NA |
| Sex | −0.014 | −0.184 | 0.156 | 0.139 | 1.140 | 0.190 | 2.090 | 1.00 |
| Age | 0.00037 | −0.00192 | 0.00265 | 0.194 | 0 | |||
| Puffins | 0.00001 | −0.00011 | 0.00014 | 0.151 | 0 | |||
| Vegetation (Silene uniflora) | −0.018 | −0.185 | 0.150 | 0.151 | 1.10 | −0.96 | 3.15 | 0.65 |
| Syphacia obvelata | 0 | 1.03 | 0.04 | 2.01 | 1.00 | |||
| Trichuris muris | 0 | 0 | ||||||
LCI: lower 95% confidence interval; UCI: upper 95% confidence interval.
Figure 4.
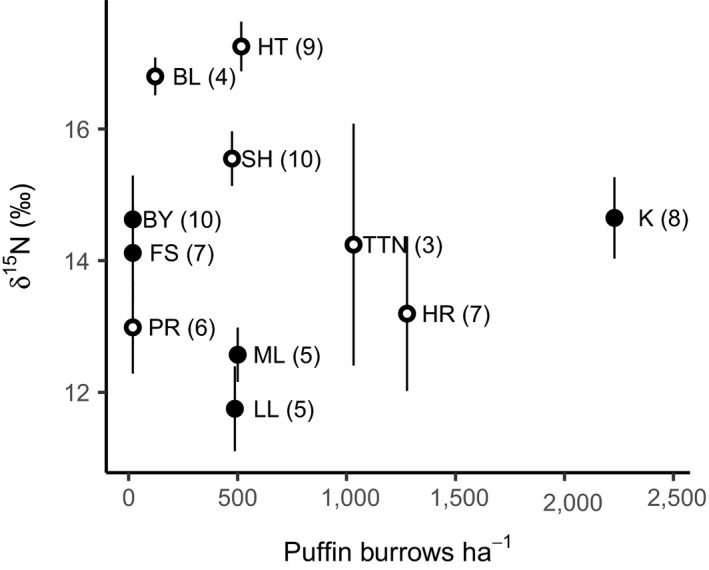
Variation in mouse nitrogen isotope values among trapping locations. Points show the mean value for each of the 11 locations, and vertical lines show ±SE. Letter codes refer to the sampling locations detailed in Figure 1, with sample size in brackets. Also shown are the density of occupied puffin burrows (x‐axis) and the dominant vegetation type (Silene uniflora, open circles; Holcus lanatus, filled circles). Of the variables shown here, only location explained a significant proportion of variation in δ15N values
4. DISCUSSION
Here, we have shown that stable isotope values, very likely linked to natural dietary variation, are associated with the levels of stimulation for a wild mouse's immune system. Specifically, individuals with low values for δ13C show increased expression of immune‐related genes, particularly those associated with inflammatory responses. The effects appear to be local to the gut, as we observed these changes in the MLNs, but failed to find evidence for similar effects in the spleen or circulating blood. We found that the individuals with low δ13C also tended to have higher concentrations of leptin in the blood, although leptin alone did not explain the change in immune state. We found microgeographical variation in nitrogen but not carbon isotope values.
Our data support the prediction that diet is an important determinant of immune state. This adds to a recent body of work seeking to establish the sources of variation in immune state in wild animals (Abolins et al., 2018; Arriero et al., 2017; Pedersen & Babayan, 2011; Turner, Begon, Jackson, Bradley, & Paterson, 2011). We know that genetic variation underpins some variation in cytokine concentrations (Turner et al., 2011), but that ecological variables such as season and body condition also play a major role in determining immune state (Abolins et al., 2018). Given the known association between condition and immune state (Abolins et al., 2018) and evidence from food supplementation experiments (e.g. Forbes et al., 2016; Strandin, Babayan, & Forbes, 2018), it is unsurprising that dietary variation among individuals should be associated with the state of the immune system; nonetheless, to our knowledge our study is the first to show this under natural conditions in a non‐human animal (for humans, see, e.g., Barbaresko et al., 2013).
We note that, beyond the significant association described above, a large proportion (approximately 80% for the MLN data) of the variation in immune parameters remains unexplained in our model. Inevitably, in a wild observational study such as this one, there are myriad possible variables that might influence immune state, only some of which we can account for. Furthermore, cytokine concentrations can change over the course of days or even hours (Scheiermann, Kunisaki, & Frenette, 2013), whereas our isotopic data are reflective of dietary variation over the course of several weeks (MacAvoy et al., 2005). Despite this difference in time‐scale, we still observe an association, which suggests that changes to the immune state in this context may be somewhat persistent rather than acute responses to isolated antigens. An interesting future extension would be to examine the acute response in these wild mice directly, by challenging mice with deliberate and controlled introduction of antigens. Further immunological measures, such as functional blood cell counts, might also be informative as to the detailed nature of the immune changes observed.
Similarly to stable isotope studies in other species (e.g. Graves et al., 2012; Robertson, McDonald, Delahay, Kelly, & Bearhop, 2015; Mangipane et al., 2018), we found an association between isotope values and nutritional status, measured in our case by concentration of circulating leptin. However, it is often observed that different biomarkers of nutritional status yield conflicting results (Graves et al., 2012; Mangipane et al., 2018). While we found circulating leptin concentration correlated negatively with δ13C, the same was not true for SMI. The potential for inconsistency among biomarkers of nutritional status is well recognized, as different indices can reflect subtly different aspects of an animal's condition (Labocha, Schutz, & Hayes, 2014). Of the two measures used in our study, SMI may primarily reflect variation in mass of protein and water (Schulte‐Hostedde, Millar, & Hickling, 2001) while leptin is expected to correlate more closely with fat content (Frederich et al., 1995).
A connection between diet and inflammation has been well studied in mice in the laboratory due to associations with important aspects of human health such as obesity and diabetes (Murphy et al., 2015). The gut microbiota plays a pivotal role in low‐level gut inflammation (Cani et al., 2008), with high‐fat diets causing an increase in the proportion of bacteria of the phylum Firmicutes and stimulation of Toll‐like receptor 4, triggering inflammatory pathways (Kim, Gu, Lee, Joh, & Kim, 2012). We know that dietary variation also influences the gut microbial community in wild mice (Wang et al., 2014), and therefore, it is possible that the microbiota plays a role in mediating our observed association between diet and inflammation.
In theory, it is also possible that other gut organisms could provide a link between diet and immune response. For example, helminths (including S. obvelata and T. muris present in this study) can be acquired through feeding (Baker, 2007) and elicit a characteristic immune response from the host (Pritchard, Hewitt, & Moqbel, 1997), but in the case of this study we did not find any evidence for a difference in immune state between infected and uninfected individuals.
From our observational data on diet, condition and immune state, it is not possible to draw firm conclusions regarding causal relationships, as there are several different possible scenarios that are compatible with our data. For example, diet could affect immune state via changes in the individual's nutritional status (Forbes et al., 2016), or it could be that diet affects both immune state and nutritional status, but via largely independent mechanisms. It is even possible that immune state might be the cause of changes in feeding behaviour (Kyriazakis, Tolkamp, & Hutchings, 1998). In our opinion, given that the observed association appears to be specific to the MLNs rather than system‐wide, the most likely explanation is that dietary intake has separate effects on both immune state in the gut and nutritional status. Experimental evidence will be required to separate these hypotheses.
Unfortunately, a limitation of the present study is that we were not able to collect sufficient isotope data from potential food sources to characterize dietary composition: although we can use isotope values as a proxy for dietary variation, we can only hypothesize as to the food items involved. Variation in δ13C values is typically associated with differences in the photosynthetic origin of carbon in the food chain, for example discriminating between C3 and C4 producers (Ben‐David & Flaherty, 2012; Peterson & Fry, 1987). While there are no C4‐using plant species in this study system (Holcus lanatus is a C3 grass), in coastal or island locations such as here, low values of δ13C may indicate terrestrial producers and high values marine producers (Hobson, 1987). Therefore, the mice with high δ13C values that show poorer condition and lower levels of immune activity may have an unusually high proportion of “marine” food sources in their diet. The most likely candidate in this case would be seabird material: mice on the Isle of May have been observed scavenging on seabird carcasses (D. Steel and M. Newell, pers. comm. 2015), and both direct predation and scavenging of seabirds by mice have been recorded on subantarctic islands (Angel et al., 2009; Cuthbert & Hilton, 2004). We speculate that these marine food sources may be of poorer quality than terrestrial sources such as plants or invertebrates, leading to the poorer condition of the individual mice that consume them. Alternatively, mice already in poor condition might be subject to intraspecific competition forcing them to switch to consuming the marine food sources, as observed in another island population of M. musculus (Cuthbert et al., 2016). Further study is required to determine the dietary sources in more detail.
It is worth emphasizing that our observations of immune state were not consistent among tissues, in that the effects observed in the MLNs were not reflected in the spleen or blood data. We must therefore consider that any outcomes for the individual in terms of altered disease susceptibility are likely also to be primarily local to the gut region.
AUTHORS' CONTRIBUTIONS
S.Y., J.F., A.D.C.M. and J.E.B. conceived the ideas and designed methodology; S.Y., J.F., A.E.L. and B.P. carried out fieldwork and immunological assays; A.L.L. carried out stable isotope analysis; C.H.T. and S.Y. conducted statistical analysis of the data; C.H.T. led the writing of the manuscript; and C.H.T. and S.Y. contributed equally to this publication and should be considered joint first authors. All authors contributed critically to the drafts and gave final approval for publication.
DATA ACCESSIBILITY
Data deposited in the Dryad Digital Repository: https://doi.org/10.5061/dryad.3ng4kr8 (Taylor et al. 2019).
Supporting information
ACKNOWLEDGEMENTS
The authors would like to thank Scottish National Heritage for permission to carry out work on the Isle of May; David Steel (Scottish National Heritage) and Mark Newell (Centre for Ecology and Hydrology) for support with fieldwork; René van der Wal for providing data on vegetation distributions on the island; and Kathryn Else, Andrew Wolfenden and three anonymous reviewers for very helpful comments on the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/J014508/1], a Doctoral Training Programme studentship awarded to S.Y. and J.F. This work was approved by the University of Nottingham Animal Welfare and Ethical Review Body and complies with the UK's Animals (Scientific Procedures) Act of 1986.
Taylor CH, Young S, Fenn J, et al. Immune state is associated with natural dietary variation in wild mice Mus musculus domesticus . Funct Ecol. 2019;33:1425–1435. 10.1111/1365-2435.13354
REFERENCES
- Abolins, S. , King, E. C. , Lazarou, L. , Weldon, L. , Hughes, L. , Drescher, P. , … Riley, E. M. (2017). The comparative immunology of wild and laboratory mice, Mus musculus domesticus . Nature Communications, 8, 14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abolins, S. , Lazarou, L. , Weldon, L. , Hughes, L. , King, E. C. , Drescher, P. , … Viney, M. (2018). The ecology of immune state in a wild mammal, Mus musculus domesticus . PLoS Biology, 16, e2003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abolins, S. R. , Pocock, M. J. O. , Hafalla, J. C. R. , Riley, E. M. , & Viney, M. E. (2011). Measures of immune function of wild mice, Mus musculus . Molecular Ecology, 20, 881–892. [DOI] [PubMed] [Google Scholar]
- Angel, A. , Wanless, R. M. , & Cooper, J. (2009). Review of impacts of the introduced house mouse on islands in the Southern Ocean: Are mice equivalent to rats? Biological Invasions, 11, 1743–1754. [Google Scholar]
- Arriero, E. , Wanelik, K. M. , Birtles, R. J. , Bradley, J. E. , Jackson, J. A. , Paterson, S. , & Begon, M. (2017). From the animal house to the field: Are there consistent individual differences in immunological profile in wild populations of field voles (Microtus agrestis)? PLoS One, 12, e0183450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. G. (2007). Flynn's parasites of laboratory animals, 2nd ed Ames, IA: Blackwell. [Google Scholar]
- Barbaresko, J. , Koch, M. , Schulze, M. B. , & Nöthlings, U. (2013). Dietary pattern analysis and biomarkers of low‐grade inflammation: A systematic literature review. Nutrition Reviews, 71, 511–527. [DOI] [PubMed] [Google Scholar]
- Barton, K. (2016). MuMIn: Multi‐model inference. R package version 1.15.6.
- Bates, D. , Mächler, M. , Bolker, B. , & Walker, S. (2015). Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Ben‐David, M. , & Flaherty, E. A. (2012). Stable isotopes in mammalian research: A beginner's guide. Journal of Mammalogy, 93, 312–328. [Google Scholar]
- Cani, P. D. , Bibiloni, R. , Knauf, C. , Waget, A. , Neyrinck, A. M. , Delzenne, N. M. , & Burcelin, R. (2008). Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes, 57, 1470–1481. [DOI] [PubMed] [Google Scholar]
- Cherry, S. G. , Derocher, A. E. , Hobson, K. A. , Stirling, I. , & Thiemann, G. W. (2011). Quantifying dietary pathways of proteins and lipids to tissues of a marine predator. Journal of Applied Ecology, 48, 373–381. [Google Scholar]
- Cocks, M. P. , Balfour, D. A. , & Stock, W. D. (1998). On the uptake of ornithogenic products by plants on the inland mountains of Dronning Maud Land, Antarctica, using stable isotopes. Polar Biology, 20, 107–111. [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Cuthbert, R. , & Hilton, G. (2004). Introduced house mice Mus musculus: A significant predator of threatened and endemic birds on Gough Island, South Atlantic Ocean? Biological Conservation, 117, 483–489. [Google Scholar]
- Cuthbert, R. J. , Wanless, R. M. , Angel, A. , Burle, M.‐H. , Hilton, G. M. , Louw, H. , … Ryan, P. G. (2016). Drivers of predatory behavior and extreme size in house mice Mus musculus on Gough Island. Journal of Mammalogy, 97, 533–544. [Google Scholar]
- DeNiro, M. J. , & Epstein, S. (1977). Mechanism of carbon isotope fractionation associated with lipid synthesis. Science, 197, 261–263. [DOI] [PubMed] [Google Scholar]
- DeNiro, M. J. , & Epstein, S. (1978). Influence of diet on the distribution of carbon isotopes in animals. Geochimica Et Cosmochimica Acta, 42, 495–506. [Google Scholar]
- DeNiro, M. J. , & Epstein, S. (1981). Influence of diet on the distribution of nitrogen isotopes in animals. Geochimica Et Cosmochimica Acta, 45, 341–351. [Google Scholar]
- Drever, M. C. , Blight, L. K. , Hobson, K. A. , & Bertram, D. F. (2000). Predation on seabird eggs by Keen's mice (Peromyscus keeni): Using stable isotopes to decipher the diet of a terrestrial omnivore on a remote offshore island. Canadian Journal of Zoology, 78, 2010–2018. [Google Scholar]
- Folch, J. , Lees, M. , & Sloane Stanley, G. (1957). A simple method for the isolation and purification of total lipids from animal tissues. Journal of Biological Chemistry, 226, 497–509. [PubMed] [Google Scholar]
- Forbes, K. M. , Mappes, T. , Sironen, T. , Strandin, T. , Stuart, P. , Meri, S. , … Huitu, O. (2016). Food limitation constrains host immune responses to nematode infections. Biology Letters, 12: 20160471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederich, R. C. , Hamann, A. , Anderson, S. , Löllmann, B. , Lowell, B. B. , & Flier, J. S. (1995). Leptin levels reflect body lipid content in mice: Evidence for diet‐induced resistance to leptin action. Nature Medicine, 1, 1311–1314. [DOI] [PubMed] [Google Scholar]
- Graves, G. R. , Newsome, S. D. , Willard, D. E. , Grosshuesch, D. A. , Wurzel, W. W. , & Fogel, M. L. (2012). Nutritional stress and body condition in the Great Gray Owl (Strix nebulosa) during winter irruptive migrations. Canadian Journal of Zoology, 90, 787–797. [Google Scholar]
- Hobson, K. A. (1987). Use of stable‐carbon isotope analysis to estimate marine and terrestrial protein content in gull diets. Canadian Journal of Zoology, 65, 1210–1213. [Google Scholar]
- Hobson, K. A. , & Clark, R. G. (1992). Assessing avian diets using stable isotopes I: Turnover of 13C in tissues. The Condor, 94, 181–188. [Google Scholar]
- Jackson, J. A. , Hall, A. J. , Friberg, I. M. , Ralli, C. , Lowe, A. , Zawadzka, M. , … Begon, M. (2014). An immunological marker of tolerance to infection in wild rodents. PLoS Biology, 12, e1001901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J. F. (2000). Stable isotopes of carbon and nitrogen in the study of avian and mammalian trophic ecology. Canadian Journal of Zoology, 78, 1–27. [Google Scholar]
- Kim, K. A. , Gu, W. , Lee, I. A. , Joh, E. H. , & Kim, D. H. (2012). High fat diet‐induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One, 7, e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurle, C. M. , & Worthy, G. A. J. (2002). Stable nitrogen and carbon isotope ratios in multiple tissues of the northern fur seal Callorhinus ursinus: Implications for dietary and migratory reconstructions. Marine Ecology Progress Series, 236, 289–300. [Google Scholar]
- Kyriazakis, I. , Tolkamp, B. J. , & Hutchings, M. R. (1998). Towards a functional explanation for the occurrence of anorexia during parasitic infections. Animal Behaviour, 56, 265–274. [DOI] [PubMed] [Google Scholar]
- Labocha, M. K. , Schutz, H. , & Hayes, J. P. (2014). Which body condition index is best? Oikos, 123, 111–119. [Google Scholar]
- Legendre, P. , & Legendre, L. (1998). Numerical ecology, 2nd English ed Amsterdam, the Netherlands: Elsevier. [Google Scholar]
- Lochmiller, R. L. , & Deerenberg, C. (2003). Trade‐offs in evolutionary immunology: Just what is the cost of immunity? Oikos, 88, 87–98. [Google Scholar]
- MacAvoy, S. E. , Macko, S. A. , & Arneson, L. S. (2005). Growth versus metabolic tissue replacement in mouse tissues determined by stable carbon and nitrogen isotope analysis. Canadian Journal of Zoology, 83, 631–641. [Google Scholar]
- Mangipane, L. S. , Belant, J. L. , Lafferty, D. J. R. , Gustine, D. D. , Hiller, T. L. , Colvin, M. E. , … Hilderbrand, G. V. (2018). Dietary plasticity in a nutrient‐rich system does not influence brown bear (Ursus arctos) body condition or denning. Polar Biology, 41, 763–772. [Google Scholar]
- Minagawa, M. , & Wada, E. (1984). Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochimica Et Cosmochimica Acta, 48, 1135–1140. [Google Scholar]
- Mizutani, H. , & Wada, E. (1988). Nitrogen and carbon isotope ratios in seabird rookeries and their ecological implications. Ecology, 69, 340–349. [Google Scholar]
- Murphy, E. A. , Velazquez, K. T. , & Herbert, K. M. (2015). Influence of high‐fat‐diet on gut microbiota: A driving force for chronic disease risk. Current Opinion in Clinical Nutrition and Metabolic Care, 18, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell, M. , Harris, M. P. , Burthe, S. J. , & Daunt, F. (2017). Status of the Atlantic Puffin Fratercula arctica on the Isle of May National Nature Reserve in 2017. Penicuik, UK: NERC Centre for Ecology and Hydrology. [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , … Wagner, H. (2016) vegan: Community Ecology Package. R package version 2.4‐1.
- Parnell, A. C. , Inger, R. , Bearhop, S. , & Jackson, A. L. (2010). Source partitioning using stable isotopes: Coping with too much variation. PLoS One, 5, e9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, A. B. , & Babayan, S. A. (2011). Wild immunology. Molecular Ecology, 20, 872–880. [DOI] [PubMed] [Google Scholar]
- Peig, J. , & Green, A. J. (2009). New perspectives for estimating body condition from mass/length data: The scaled mass index as an alternative method. Oikos, 118, 1883–1891. [Google Scholar]
- Perrow, M. , & Jowitt, A. (1995). What future for the harvest mouse? British Wildlife, 6, 356–356. [Google Scholar]
- Peterson, B. J. , & Fry, B. (1987). Stable isotopes in ecosystem studies. Annual Review of Ecology and Systematics, 18, 293–320. [Google Scholar]
- Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real‐time RT–PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, D. L. (2001). Mixing models in analyses of diet using multiple stable isotopes: A critique. Oecologia, 127, 166–170. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. , Bates, D. , DebRoy, S. , Sarkar, D. , & Core Team, R. (2018) nlme: linear and nonlinear mixed effects models. R package version 3.1‐137.
- Post, D. M. , Layman, C. A. , Arrington, D. A. , Takimoto, G. , Quattrochi, J. , & Montana, C. G. (2007). Getting to the fat of the matter: Models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia, 152, 179–189. [DOI] [PubMed] [Google Scholar]
- Pritchard, D. I. , Hewitt, C. , & Moqbel, R. (1997). The relationship between immunological responsiveness controlled by T‐helper 2 lymphocytes and infections with parasitic helminths. Parasitology, 115, 33–44. [DOI] [PubMed] [Google Scholar]
- Robertson, A. , McDonald, R. A. , Delahay, R. J. , Kelly, S. D. , & Bearhop, S. (2015). Resource availability affects individual niche variation and its consequences in group‐living European badgers Meles meles . Oecologia, 178, 31–43. [DOI] [PubMed] [Google Scholar]
- Robertson, S. , Bradley, J. E. , & MacColl, A. D. C. (2016). Measuring the immune system of the three‐spined stickleback – investigating natural variation by quantifying immune expression in the laboratory and the wild. Molecular Ecology Resources, 16, 701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart, S. P. , Vassallo, B. G. , Angeletti, D. , Hutchinson, D. S. , Morgan, A. P. , Takeda, K. , … Rehermann, B. (2017). Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell, 171, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, F. P. , Bradfield, A. , Quy, R. J. , & Swinney, T. (1985). Relationship between eye lens weight and age in the wild house mouse (Mus musculus). Journal of Applied Ecology, 22, 55–61. [Google Scholar]
- Sage, R. D. (1981). Wild mice In Foster H. (Ed.), The Mouse in biomedical research history, genetics and wild mice (pp. 40–90). San Diego, CA: Academic Press. [Google Scholar]
- Saino, N. , Ferrari, R. , Romano, M. , Martinelli, R. , & Møller, A. P. (2003). Experimental manipulation of egg carotenoids affects immunity of barn swallow nestlings. Proceedings of the Royal Society of London. Series B: Biological Sciences, 270, 2485–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann, C. , Kunisaki, Y. , & Frenette, P. S. (2013). Circadian control of the immune system. Nature Reviews Immunology, 13, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeninger, M. J. , DeNiro, M. J. , & Tauber, H. (1983). Stable nitrogen isotope ratios of bone collagen reflect marine and terrestrial components of prehistoric human diet. Science, 220, 1381–1383. [DOI] [PubMed] [Google Scholar]
- Schulte‐Hostedde, A. I. , Millar, J. S. , & Hickling, G. J. (2001). Evaluating body condition in small mammals. Canadian Journal of Zoology, 79, 1021–1029. [Google Scholar]
- Stapp, P. (2002). Stable isotopes reveal evidence of predation by ship rats on seabirds on the Shiant Islands, Scotland. Journal of Applied Ecology, 39, 831–840. [Google Scholar]
- Strandin, T. , Babayan, S. A. , & Forbes, K. M. (2018). Reviewing the effects of food provisioning on wildlife immunity. Philosophical Transactions of the Royal Society B: Biological Sciences, 373, 20170088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C. H. , Young, S. , Fenn, J. , Lamb, A. L. , Lowe, A. E. , Poulin, B. , … Bradley, J. E. (2019). Data from: Immune state is associated with natural dietary variation in wild mice Mus musculus domesticus . Dryad Digital Repository, 10.5061/dryad.3ng4kr8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieszen, L. L. , Boutton, T. W. , Tesdahl, K. G. , & Slade, N. A. (1983). Fractionation and turnover of stable carbon isotopes in animal tissues: Implications for δ13C analysis of diet. Oecologia, 57, 32–37. [DOI] [PubMed] [Google Scholar]
- Triggs, G. S. (1991). The population ecology of house mice (Mus domesticus) on the Isle of May, Scotland. Journal of Zoology, 225, 449–468. [Google Scholar]
- Turner, A. K. , Begon, M. , Jackson, J. A. , Bradley, J. E. , & Paterson, S. (2011). Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genetics, 7, e1002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. , & Speleman, F. (2002). Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright, S. C. , Haney, J. C. , Kerr, C. , Golovkin, A. N. , & Flint, M. V. (1998). Utilization of nitrogen derived from seabird guano by terrestrial and marine plants at St. Paul, Pribilof Islands, Bering Sea. Alaska. Marine Biology, 131, 63–71. [Google Scholar]
- Wang, J. , Linnenbrink, M. , Künzel, S. , Fernandes, R. , Nadeau, M.‐J. , Rosenstiel, P. , & Baines, J. F. (2014). Dietary history contributes to enterotype‐like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proceedings of the National Academy of Sciences, 111, E2703–E2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. L. , McNeilly, T. N. , Watt, K. A. , Pemberton, J. M. , Pilkington, J. G. , Waterfall, M. , … Nussey, D. H. (2016). Cellular and humoral immunity in a wild mammal: Variation with age & sex and association with overwinter survival. Ecology and Evolution, 6, 8695–8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, R. E. , Leslie, D. M. , Lochmiller, R. L. , & Masters, R. E. (2003). Immune function and hematology of male cotton rats (Sigmodon hispidus) in response to food supplementation and methionine. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 136, 577–589. [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2016). tidyverse: Easily Install and Load "Tidyverse" Packages. R package version 1.0.0.
- Wright, D. G. , van der Wal, R. , Wanless, S. , & Bardgett, R. D. (2010). The influence of seabird nutrient enrichment and grazing on the structure and function of island soil food webs. Soil Biology and Biochemistry, 42, 592–600. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited in the Dryad Digital Repository: https://doi.org/10.5061/dryad.3ng4kr8 (Taylor et al. 2019).


