Abstract
Objective
Aphids harbor a nutritional obligate endosymbiont in specialized cells called bacteriocytes, which aggregate to form an organ known as the bacteriome. Aphid bacteriomes display distinct gene expression profiles that facilitate the symbiotic relationship. Currently, the mechanisms that regulate these patterns of gene expression are unknown. Recently using computational pipelines, we identified miRNAs that are conserved in expression in the bacteriomes of two aphid species and proposed that they function as important regulators of bacteriocyte gene expression. Here using a dual luciferase assay in mouse NIH/3T3 cell culture, we aimed to experimentally validate the computationally predicted interaction between Myzus persicae miR-92a and the predicted target region of M. persicae bacteriocyte-specific secreted protein 1 (SP1) mRNA.
Results
In the dual luciferase assay, miR-92a interacted with the SP1 target region resulting in a significant downregulation of the luciferase signal. Our results demonstrate that miR-92a interacts with SP1 to alter expression in a heterologous expression system, thereby supporting our earlier assertion that miRNAs are regulators of the aphid/Buchnera symbiotic interaction.
Keywords: Aphid, miRNA, Symbiosis, Bacteriocyte, SP1, Dual luciferase assay
Introduction
Aphids are obligately dependent on their ancient endosymbiotic relationship with the gamma-proteobacterium Buchnera aphidicola [1, 2]. The symbiont, Buchnera, is housed in a specialized organ called the bacteriome, inside specialized host cells called bacteriocytes [1–4]. Bacteriomes are enriched in expression of genes associated with functions that include amino acid biosynthesis and metabolism, and transporters that mediate metabolite exchange between aphid and Buchnera [5–8]. Bacteriome gene expression profiles also feature expression of two groups of aphid orphan genes: bacteriocyte-specific cysteine-rich proteins and aphid-specific putative secreted proteins [9]. One putative secreted protein is secreted protein 1 (SP1), a gene whose expression is restricted to bacteriocytes. The lineage specificity of SP1, coupled with its tissue-specific expression suggests that this orphan gene may have contributed to the evolution of aphid-specific traits, i.e. the symbiosis with Buchnera [9].
Recently using two aphid species, the pea aphid, Acyrthosiphon pisum, and the green peach aphid, Myzus persicae, we identified 14 evolutionary conserved microRNAs (miRNAs) that were bacteriome-specific and/or bacteriome-enriched and were predicted to regulate 103 aphid genes, many of which have known importance to the aphid/Buchnera symbiosis [10]. Among those predictions, miR-92a was significantly upregulated in bacteriocytes and predicted to target the bacteriocyte-specific SP1 (Fig. 1) [10]. Remarkably, miR-92a has been shown to be important in a great diversity of host/microbe interactions that include host/virus interactions in a mosquito [11] and a fall armyworm [12], and host/pathogen interactions in a mosquito [13], marine filter feeders [14, 15], a spider mite [16], and a fish [17]. Here, we experimentally interrogate our computationally predicted interaction of M. persicae mpe-miR-92a with SP1.
Fig. 1.
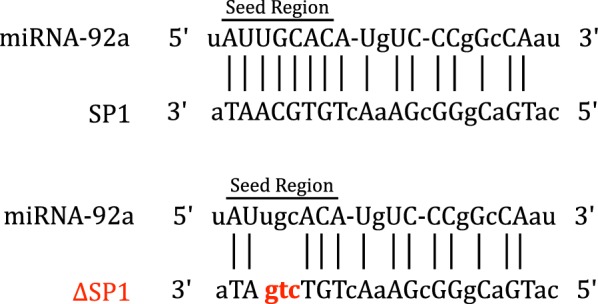
Mpe-miR-92a is predicted to regulate secreted protein 1 (SP1). The predicted base pairing between mpe-miR-92a and the target region in the 3′ UTR of the SP1 transcript. The seed regions of mpe-miR-92a are indicated; the mutated target regions are highlighted in red
Main text
Methods
Target prediction
In our previous study we used miRanda [18], PITA [19] and RNAhybrid [20] to predict potential miRNA::mRNA interactions [10]. All three algorithms predicted the same seed region. The target site predicted by miRanda was the largest and spanned the target sites predicted by PITA and RNAhybrid (Additional file 1: Figure S1), thus, we designed our dual luciferase assay using the miRanda target site prediction.
Mus musculus embryonic fibroblast NIH/3T3 cell culture
We maintained NIH/3T3 cells under sterile conditions at 37 °C with 5% CO2. Cells were cultured in the ATCC-formulated Dulbecco’s Modified Eagle’s Medium with bovine calf serum (Gibco, USA) at 10%, gentamycin at 0.5% (v/v) and penicillin–streptomycin at 1% (v/v).
Plasmid preparation and miRNA mimic synthesis
To validate the predicted miRNA::mRNA interaction, we utilized pmirGLO Dual-Luciferase miRNA target expression vector (pmirGLO) (Promega, USA) and miRNA mimics. The pmirGLO vector expresses two luciferases: the firefly luciferase (an experimental reporter that can be subject to the effect of miRNA regulation) and the Renilla luciferase (an internal control). Using pmirGLO, we prepared an experimental plasmid, a negative and a positive control plasmid. The experimental plasmid, pmirGLO-SP1, contained a synthesized miR-92a::SP1 target region corresponding to the miR-92a binding site on the SP1 3′ UTR of M. persicae (Additional file 2: Table S1) [10]. The negative control plasmid, pmirGLO-ΔSP1, contained a synthesized mutated SP1 (ΔSP1) that was designed based on the M. persicae miR-92a::SP1 target region using the Illegitimate microRNA predictor (Additional file 2: Table S1) [21]. We obtained the positive control plasmid, pmirGLO-miR21T, that includes the M. musculus miR-21 target site from Promega, USA. Our experiments used two miRNA mimics, a miR-92a mimic (Fig. 1) and a non-specific negative control siRNA i.e. AllStars Negative Control siRNA from QIAGEN, USA (Cat#: SI03650318). AllStars Negative Control siRNA has a proprietary sequence with no homology to any known mammalian gene.
Cell transfection
We performed the transient cell transfection experiment three times. For the first two experiments, we used the Effectene Transfection Reagent (Qiagen, USA). Briefly, 400 ng DNA plasmid and/or 300 nM miRNA mimics were used to transfect/co-transfect 4 × 105 cells/well in 6-well plates for 24 h. Then, we harvested cells at 48 h for the dual luciferase assay. In the third experiment, we used the Attractene Transfection Reagent (Qiagen, USA). Briefly, 400 ng DNA plasmids and/or 6 pmol miRNA mimics were used to transfect/co-transfect 1.6 × 105 cells/well in 24-well plates. Cells were transfected for 48 h and harvested for the dual luciferase assay.
Dual luciferase assay
Transfected cells were assayed using the Dual-GLO® Luciferase Assay System (Promega, USA). For each sample, the firefly and Renilla luciferase activities were measured sequentially by collecting emitted luminescence from the entire visible spectrum (300–700 nm) on a Synergy H1 Multi-Mode Reader (BioTek, USA). Briefly, the firefly luciferase activity was measured 10 min after induction of cell lysis and provision of the firefly luciferase substrate. Then, we quenched the firefly luciferase reaction and provided the Renilla luciferase substrate. Ten minutes later we captured the Renilla luciferase activity. The luminescence measurement for each well represents the average of 12 serial luminescence readings.
Each plate included four technical replicates of each treatment, plus four control technical replicates (cells were exposed only to the transfection reagents) to allow background luminescence subtraction. Following background subtraction, we calculated the ratios of firefly/Renilla luminescences. To compare data across the three experiments we normalized data within each experiment to the empty pmirGLO control treatment by dividing each firefly/Renilla ratio by the mean firefly/Renilla ratio of the empty pmirGLO control treatment.
Statistical analyses
We tested for differences in the normalized firefly/Renilla ratios among treatments using one-way ANOVA with a fixed factor of treatment and a block effect of experiment, followed by a Tukey HSD post hoc test for multiple comparisons in SPSS v.24.
Results
miR-92a interacts with the predicted target region of SP1 mRNA in NIH/3T3 cells
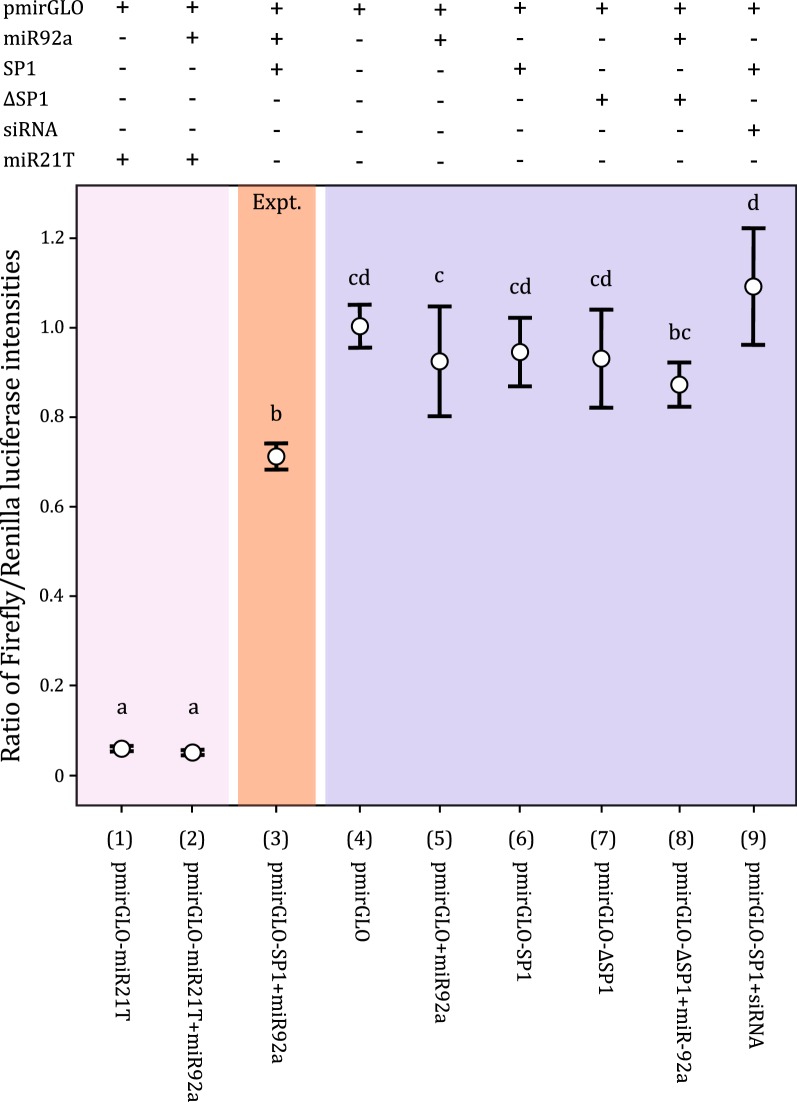
To test the predicted miR-92a::SP1 interaction, we performed a dual luciferase assay in NIH/3T3 cells that we transfected with a pmirGLO-SP1 construct together with mature miR-92a (pmirGLO-SP1 + miR-92a, treatment 3 in Fig. 2). In parallel we performed a series of controls that included (i) cells transfected with an NIH/3T3 endogenous miRNA construct: pmirGLO-miR21T (treatment 1, Fig. 2), (ii) a pmirGLO-miR21T construct + miR-92a (treatment 2, Fig. 2), (iii) a pmirGLO empty construct (treatment 4, Fig. 2), (iv) a pmirGLO empty construct + miR-92a (treatment 5, Fig. 2), (v) a pmirGLO-SP1 construct (treatment 6, Fig. 2), (vi) a pmirGLO-ΔSP1 construct (treatment 7, Fig. 2), (vii) a pmirGLO-ΔSP1 construct + miR-92a (treatment 8, Fig. 2), and (viii) a pmirGLO-SP1 construct + siRNA (treatment 9, Fig. 2).
Fig. 2.
Dual luciferase assay of mpe-miR-92a::SP1 interactions. The ratios of the firefly luciferase versus the Renilla luciferase activities were compared across different treatments. “+” means the presence of the element in the treatment and “−” means the absence of the element in the treatment. Expt.: Experimental treatment. The data was tested using 1-way ANOVA, controlling for random block effects, followed by Tukey HSD post hoc analysis. The lowercase letters above each whisker (a, b, bc, c, cd, d) denote statistically significant differences between treatments. Error bar = ± standard error (n = 12) from three experimental replicates
In the dual luciferase assay, we validated that mpe-miR-92a specifically interacts with the predicted SP1 target region. After we removed any random block effects (Table 1: experiment, F(2) = 0.948, p = 0.391), we observed significant differences in firefly/Renilla ratios between groups under different treatment conditions (Table 1: treatment, F(8, 11) = 114.567, p < 0.0001; Fig. 2). First, the significant difference between the pmirGLO-miR21T treatments and the empty pmirGLO (p < 0.0001) indicated that the dual luciferase assay was working properly (Fig. 2, treatments 1 vs 4, 2 vs 4). Second, the treatment of pmirGLO-SP1 + miR-92a was also significantly different from empty pmirGLO (p < 0.0001, Fig. 2, treatments 3 vs 4) and pmirGLO-SP1 only (p = 0.001, Fig. 2, treatments 3 vs 6), indicating that mpe-miR-92a specifically interacted with the predicted SP1 target region, resulting in significant downregulation of the luciferase signal. Third, we found no difference between treatments of pmirGLO-SP1 only and empty pmirGLO (p = 0.976, Fig. 2, treatments 4 vs 6) indicating an absence of endogenous NIH/3T3 miRNA interactions with the predicted SP1 target region. Fourth, we observed a significant difference between the pmirGLO-SP1 + miR-92a and the pmirGLO + miR-92a (p = 0.003, Fig. 2, treatments 3 vs 5) indicating that the miR-92a downregulation of pmirGLO-SP1 was not the result of interactions between miR-92a and the pmirGLO vector. Fifth, we found no significant difference between the pmirGLO-SP1 + miR-92a and pmirGLO-ΔSP1 + miR-92a treatments (p = 0.072, Fig. 2, treatments 3 vs 8), suggesting that miR-92a can interact with ΔSP1. However, we found no significant difference between the pmirGLO-ΔSP1 and pmirGLO-ΔSP1 + miR-92a (p = 0.973, Fig. 2, treatments 7 vs 8), suggesting that the interaction between miR-92a and ΔSP1 was not as strong as the interaction between miR-92a and the bona fide SP1 target region (Fig. 2). We suspect that the interactions between miR-92a and ΔSP1 may result from (i) possible G-U wobble base-pairings between the mutated nucleotides and miR-92a; and (ii) extensive base-pairing in the non-seed region of miR-92a (Fig. 1), because the non-seed region of miRNAs (nucleotides 12–17) have been shown in mammalian cells to be important for miRNA targeting [22, 23]. Lastly, we found a significant difference in signal between the pmirGLO-SP1 + miR-92a and pmirGLO-SP1 + siRNA (Fig. 2, Treatments 3 vs 9), and no difference in signal between the pmirGLO-SP1 + siRNA, empty pmirGLO, and pmirGLO-SP1 treatments (Fig. 2, Treatments 4 vs 6; 4 vs 9; 6 vs 9), suggesting that the interaction between miR-92a and SP1 is sequence specific.
Table 1.
ANOVA statistics of mpe-miR-92a and SP1 dual luciferase assays
| Source | Type III Sum of squares |
df | Mean square | F | Sig. |
|---|---|---|---|---|---|
| Corrected model | 15.156a | 10 | 1.516 | 91.843 | 0.000 |
| Intercept | 57.933 | 1 | 57.933 | 3510.743 | 0.000 |
| Treatment | 15.124 | 8 | 1.891 | 114.567 | 0.000 |
| Experiment | 0.031 | 2 | 0.016 | 0.948 | 0.391 |
| Error | 1.601 | 97 | 0.017 | ||
| Total | 74.690 | 108 | |||
| Corrected total | 16.756 | 107 |
aR Squared = 0.904 (Adjusted R Squared = 0.895)
Discussion
Aphid miR-92a interacts with SP1
In our dual luciferase assay in NIH/3T3 cells, mpe-miR-92a physically interacted with the predicted SP1 target region, resulting in significant downregulation of gene expression (Fig. 2). This assay validates our earlier computational prediction that SP1 is regulated by miR-92a [10] and further, highlights both the role of miRNAs as regulators of gene expression in aphid bacteriomes, and the potential of targeting the miR-92a::SP1 interaction for pest aphid control [24, 25].
miRNAs regulate gene expression in aphid bacteriocytes
Aphid gene expression in bacteriomes is crucial to the function of the aphid/Buchnera symbiosis. What remains elusive are the mechanisms by which the expression of these bacteriocyte-specific genes are regulated.
The abundance of proteins in a cell results from the dynamic interplay of transcriptional, post-transcriptional (e.g. miRNA regulation), translational, and post-translational regulation [26–28]. The identification of transcriptional regulation in aphids has been limited to studies of aphid development and regulation of Buchnera gene expression. Three transcription factors: Distal-less, Engrailed, and Ultrabithorax/Abdominal-A have been implicated in bacteriocyte specification and development in aphids [29]. While in Buchnera, studies have demonstrated limited transcriptional regulation of the expression of heat shock [30–33] and amino acid biosynthesis genes [34, 35]. More recently, a remarkable example of post-translational regulation of amino acid biosynthesis in bacteriocytes has been proposed in A. pisum by glutamine transporter, ApGLNT1 [36]. ApGLNT1 localizes to the bacteriocyte plasma membrane where it transports glutamine from aphid hemolymph into bacteriocyte cells. Importantly, glutamine transport is competitively inhibited by a Buchnera-synthesized essential amino acid end-product, arginine. Thus ApGLNT1 regulates the transport of glutamine, a host-supplied amino acid precursor, by an endosymbiont-synthesized end-product via substrate feedback inhibition at the post-translational level [36].
In other work, post-transcriptional regulation of gene expression has been suggested to be important for regulation of the aphid/Buchnera endosymbiosis. For example, comparison of Buchnera gene expression in embryonic and maternal bacteriocytes found no differences in mRNA abundance, but differences in protein abundance that have been attributed to a Buchnera encoded set of conserved small RNAs [37, 38]. In addition, we recently identified a set of evolutionarily conserved aphid miRNAs that are bacteriome-specific and/or bacteriome-enriched in M. persicae and A. pisum. Notably, many of the conserved miRNAs were predicted to target bacteriocyte-specific genes of known importance to aphid/Buchnera symbiosis [10]. Here using a heterologous expression system, we validated one of our predicted miRNA::mRNA interactions, the miR-92a::SP1 interaction. Our validation of the miR-92a::SP1 interaction, coupled with our earlier genome-wide analyses, highlight miRNAs as post-transcriptional regulators in the aphid/Buchnera symbiosis.
miR-92a and its targets are potential targets for aphid control
Recent attempts have been made to develop miRNAs as tools for pest control (reviewed in [25]), either by engineering miRNAs for insecticidal activities [39] or by silencing insect defensive miRNAs [40]. Here we have validated the role of miR-92a in regulation of the orphan, bacteriocyte-expressed gene SP1, a gene that encodes a secreted protein that has been argued to be important for symbiotic function. Since aphids lacking a stable Buchnera symbiosis fail to reproduce [41–43], it follows that miR-92a offers promise as a target for control of pest aphid populations.
Limitations
We validated the M. persicae miR-92a::SP1 interaction in a heterologous expression system, the interaction remains elusive in vivo.
Supplementary information
Additional file 1: Figure S1. Prediction of the miR-92a::SP1 interaction by miRanda, PITA, and RNAhybrid in M. persicae (10). The target region predicted by miRanda includes the regions predicted by PITA and RNAhybrid. Thus, the sequence of the miRanda prediction was used to construct the experimental plasmid, pmirGLO-SP1. Grey shaded area marks the seed region of the mature miRNA.
Additional file 2: Table S1. Oligonucleotides used for preparing pmirGLO-target plasmids.
Acknowledgements
We thank Nicolas Durand and Linda White for help establishing cell cultures, Leonidas Bachas, Megan Gillespie and Elsayed Zahran for plate reader access, Michelle Afkhami for advice on data analysis.
Abbreviations
- miRNA
microRNA
- SP1
secreted protein 1
- ΔSP1
mutated secreted protein 1
- ApGLNT1
Acyrthosiphon pisum glutamine transporter 1
- pmirGLO
pmirGLO Dual-Luciferase miRNA target expression vector
Authors’ contributions
HF and ACCW conceived of the study. HF, JSP, RGZ, and ACCW designed the experiments. HF performed the experiments. HF and ACCW contributed to data analysis. HF and ACCW wrote the manuscript. All authors contributed to preparation of the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Science Foundation Awards IOS-1121847 and IOS-354154, and a University of Miami Provost Award to ACCW, and by National Institutes of Health (NIH) R56NS095893 to RGZ. HF was supported by a University of Miami Maytag Fellowship, the William H. Evoy Graduate Research Support Fund, and a Molecular Biosciences Graduate Research Award from the Department of Biology at the University of Miami. Funding agencies had no role in the design of the study and clollection, analysis, and interpretation of the data, or writing of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Honglin Feng, Email: h.feng2@umiami.edu.
Joun S. Park, Email: jpark@med.miami.edu
R. Grace Zhai, Email: gzhai@med.miami.edu.
Alexandra C. C. Wilson, Email: acwilson@miami.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13104-019-4665-6.
References
- 1.Baumann L, Baumann P. Growth kinetics of the endosymbiont Buchnera aphidicola in the aphid Schizaphis graminum. Appl Environ Microbiol. 1994;60(9):3440–3443. doi: 10.1128/aem.60.9.3440-3443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchner P. Endosymbiosis of animals with plant microorganisms. Geneva: Interscience Publishers; 1965. p. xvii, 909.
- 3.Douglas AE, Dixon AFG. The mycetocyte symbiosis of aphids—variation with age and morph in virginoparae of Megoura viciae and Acyrthosiphon pisum. J Insect Physiol. 1987;33(2):109–113. doi: 10.1016/0022-1910(87)90082-5. [DOI] [Google Scholar]
- 4.Mclean DL, Houk EJ. Phase contrast and electron microscopy of the mycetocytes and symbiotes of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1973;19:625–633. doi: 10.1016/0022-1910(73)90071-1. [DOI] [Google Scholar]
- 5.Duncan RP, Husnik F, Van Leuven JT, Gilbert DG, Davalos LM, McCutcheon JP, et al. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol Ecol. 2014;23(6):1608–1623. doi: 10.1111/mec.12627. [DOI] [PubMed] [Google Scholar]
- 6.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108(7):2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102(15):5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, et al. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteom. 2011;10(6):M110.007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shigenobu S, Stern DL. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc R Soc B Biol Sci. 2013;280(1750):2012.1952. doi: 10.1098/rspb.2012.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng H, Wang L, Wuchty S, Wilson ACC. microRNA regulation in an ancient obligate endosymbiosis. Mol Ecol. 2018;27(8):1777–1793. doi: 10.1111/mec.14464. [DOI] [PubMed] [Google Scholar]
- 11.Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 2010;11(1):119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehrabadi M, Hussain M, Asgari S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J Gen Virol. 2013;94(Pt 6):1385–1397. doi: 10.1099/vir.0.051060-0. [DOI] [PubMed] [Google Scholar]
- 13.Mayoral JG, Etebari K, Hussain M, Khromykh AA, Asgari S. Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS ONE. 2014;9(4):e96107. doi: 10.1371/journal.pone.0096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Li C, Shao Y, Chen X, Li Y, Su X, et al. Identification and characterization of miR-92a and its targets modulating Vibrio splendidus challenged Apostichopus japonicus. Fish Shellfish Immunol. 2014;38(2):383–388. doi: 10.1016/j.fsi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Jin P, Li S, Sun L, Lv C, Ma F. Transcriptome-wide analysis of microRNAs in Branchiostoma belcheri upon Vibrio parahemolyticus infection. Dev Comp Immunol. 2017;74:243–252. doi: 10.1016/j.dci.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Rong X, Zhang YK, Zhang KJ, Hong XY. Identification of Wolbachia-responsive microRNAs in the two-spotted spider mite, Tetranychus urticae. BMC Genom. 2014;15:1122. doi: 10.1186/1471-2164-15-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiang J, Tao YF, He J, Li HX, Xu P, Bao JW, et al. Inhibition of miR-92d-3p enhances inflammation responses in genetically improved farmed tilapia (GIFT, Oreochromis niloticus) with Streptococcus iniae infection by modulating complement C3. Fish Shellfish Immunol. 2017;63:367–375. doi: 10.1016/j.fsi.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 18.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2004;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 20.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan BC, Werner TS, Howard PL, Chow RL. ImiRP: a computational approach to microRNA target site mutation. BMC Bioinform. 2016;17:190. doi: 10.1186/s12859-016-1057-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the seed supports microRNA targeting specificity. Mol Cell. 2016;64(2):320–333. doi: 10.1016/j.molcel.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu XD, Liu ZC, Huang SL, Chen ZQ, Sun YW, Duan PF, et al. RNAi-mediated plant protection against aphids. Pest Manag Sci. 2016;72(6):1090–1098. doi: 10.1002/ps.4258. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Wong AYP, Wang S, Jia Q, Chuang WP, Bendena WG, et al. miRNA-mediated interactions in and between plants and insects. Int J Mol Sci. 2018;19(10):3239. doi: 10.3390/ijms19103239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gokhale S, Nyayanit D, Gadgil C. A systems view of the protein expression process. Syst Synth Biol. 2011;5(3–4):139–150. doi: 10.1007/s11693-011-9088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165(3):535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1(1):70–76. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilcox JL, Dunbar HE, Wolfinger RD, Moran NA. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 2003;48(6):1491–1500. doi: 10.1046/j.1365-2958.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson ACC, Dunbar HE, Davis GK, Hunter WB, Stern DL, Moran NA. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomic. 2006;7:50. doi: 10.1186/1471-2164-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunbar HE, Wilson ACC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007;5(5):1006–1015. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke GR, McLaughlin HJ, Simon JC, Moran NA. Dynamics of a recurrent Buchnera mutation that affects thermal tolerance of pea aphid hosts. Genetics. 2010;186(1):367–U577. doi: 10.1534/genetics.110.117440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005;187(12):4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reymond N, Calevro F, Vinuelas J, Morin N, Rahbe Y, Febvay G, et al. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl Environ Microbiol. 2006;72(12):7760–7766. doi: 10.1128/AEM.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price DR, Feng H, Baker JD, Bavan S, Luetje CW, Wilson ACC. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111(1):320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen AK, Degnan PH. Widespread expression of conserved small RNAs in small symbiont genomes. ISME J. 2014;8:2490. doi: 10.1038/ismej.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thairu MW, Cheng S, Hansen AK. A sRNA in a reduced mutualistic symbiont genome regulates its own gene expression. Mol Ecol. 2017;27:1766–1776. doi: 10.1111/mec.14424. [DOI] [PubMed] [Google Scholar]
- 39.Agrawal A, Rajamani V, Reddy VS, Mukherjee SK, Bhatnagar RK. Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: an alternative to Bt-toxin technology. Transgenic Res. 2015;24(5):791–801. doi: 10.1007/s11248-015-9880-x. [DOI] [PubMed] [Google Scholar]
- 40.Ran ZH, Shi XJ, Han FT, Li JB, Zhang YY, Zhou YJ, et al. Expressing microRNA bantam sponge drastically improves the insecticidal activity of baculovirus via increasing the level of ecdysteroid hormone in Spodoptera exigua larvae. Front Microbiol. 2018;9:1824. doi: 10.3389/fmicb.2018.01824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki T, Hayashi H, Ishikawa H. Growth and reproduction of the symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum maintained on artificial diets. Journal of Insect Physiology. 1991;37(10):749–756. doi: 10.1016/0022-1910(91)90109-D. [DOI] [Google Scholar]
- 42.Douglas AE. Reproductive failure and the free amino acid pools in pea aphids (Acyrthosiphon pisum) lacking symbiotic bacteria. J Insect Physiol. 1996;42(3):247–255. doi: 10.1016/0022-1910(95)00105-0. [DOI] [Google Scholar]
- 43.Caillaud CM, Rahbe Y. Aposymbiosis in a cereal aphid: reproductive failure and influence on plant utilization. Ecol Entomol. 1999;24(1):111–114. doi: 10.1046/j.1365-2311.1999.00179.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Prediction of the miR-92a::SP1 interaction by miRanda, PITA, and RNAhybrid in M. persicae (10). The target region predicted by miRanda includes the regions predicted by PITA and RNAhybrid. Thus, the sequence of the miRanda prediction was used to construct the experimental plasmid, pmirGLO-SP1. Grey shaded area marks the seed region of the mature miRNA.
Additional file 2: Table S1. Oligonucleotides used for preparing pmirGLO-target plasmids.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.