Abstract
Background
Bio-based production of organic acids promises to be an attractive alternative for the chemicals industry to substitute petrochemicals as building-block chemicals. In recent years, itaconic acid (IA, methylenesuccinic acid) has been established as a sustainable building-block chemical for the manufacture of various products such as synthetic resins, coatings, and biofuels. The natural IA producer Aspergillus terreus is currently used for industrial IA production; however, the filamentous fungus Aspergillus niger has been suggested to be a more suitable host for this purpose. In our previous report, we communicated the overexpression of a putative cytosolic citrate synthase citB in an A. niger strain carrying the full IA biosynthesis gene cluster from A. terreus, which resulted in the highest final titer reported for A. niger (26.2 g/L IA). In this research, we have attempted to improve this pathway by increasing the cytosolic acetyl-CoA pool. Additionally, we have also performed fermentation optimization by varying the nitrogen source and concentration.
Results
To increase the cytosolic acetyl-CoA pool, we have overexpressed genes acl1 and acl2 that together encode for ATP-citrate lyase (ACL). Metabolic engineering of ACL resulted in improved IA production through an apparent increase in glycolytic flux. Strains that overexpress acl12 show an increased yield, titer and productivity in comparison with parental strain CitB#99. Furthermore, IA fermentation conditions were improved by nitrogen supplementation, which resulted in alkalization of the medium and thereby reducing IA-induced weak-acid stress. In turn, the alkalizing effect of nitrogen supplementation enabled an elongated idiophase and allowed final titers up to 42.7 g/L to be reached at a productivity of 0.18 g/L/h and yield of 0.26 g/g in 10-L bioreactors.
Conclusion
Ultimately, this study shows that metabolic engineering of ACL in our rewired IA biosynthesis pathway leads to improved IA production in A. niger due to an increase in glycolytic flux. Furthermore, IA fermentation conditions were improved by nitrogen supplementation that alleviates IA induced weak-acid stress and extends the idiophase.
Keywords: Itaconic acid, Aspergillus niger, Transcriptome analysis, Metabolic engineering, ATP-citrate lyase, Fermentation optimization
Introduction
A shortage of fossil fuels in the near future as well as their contribution to global carbon emission increases demand for renewable energy sources. Bio-based production promises to be an attractive alternative for the chemicals industry to substitute petrochemicals as building-block chemicals. In a landmark study in 2004, the US Department of Energy evaluated 300 molecules that could be made from biomass [1]. Twelve of these molecules were recognized as promising new building-block chemicals with high potential to be produced by biotechnological means, 8 of which belong to the organic acids. This list has been revisited by Bozell et al. [2] more recently. These biotechnologically produced organic acids are used, among others, in the food industry as food additive (e.g., citric acid) or in the chemicals industry as promising substitutes of petrochemicals (e.g., itaconic acid). In recent years, itaconic acid (IA, methylenesuccinic acid) has been established as a sustainable building-block chemical for the manufacture of various synthetic resins, coatings, and polymers and for application in thickeners, binders and adhesives [3–6]. In addition, IA has great potential to replace petroleum-based acrylic or methacrylic acid, used for synthesis of methyl methacrylate and its polymer polymethyl methacrylate (i.e., acrylic glass), and is suitable for synthesis of 3-methyltetrahydrofuran, a potential biofuel [4]. Its flexibility as a building-block chemical is expected to provide an increasing market for IA production.
The natural IA producer Aspergillus terreus is currently used for industrial IA production. Up to now, under laboratory conditions, this fungus has been shown to achieve a maximum productivity of 1.9 g/L/h with a final titer of 160 g/L IA in 1.5-L scale fermentations [7]. However, the conditions used in this study are not industrially scalable due to the use of specialized columns to remove trace amounts of Mn2+ from the medium, for which A. terreus is highly sensitive [8]. Industrial levels reported are in the range of 80–100 g/L IA [9]. IA can also be synthesized chemically, but this requires a multi-step modification process along with high substrate cost and relatively low yields and is, therefore, not preferred over bio-based production [3]. The world market of IA is around 40 kT per annum, with market prices between $1.5 and $2.0 per kg [10, 11].
Previously, the filamentous fungus A. niger has been suggested to be a suitable host for the industrial production of IA because of its optimized pathways towards organic acids, highlighted by its current status as key production host in industrial citric acid production with an estimated annual production exceeding 2 million tons (data from 2015) [12, 13]. Unlike A. terreus, A. niger is not able to produce IA and requires genetic modification to enable IA production. However, for several products, fermentation with A. niger has received the GRAS status (generally recognized as safe) from the US Food and Drug Administration, in contrast to A. terreus [14]. Another significant advantage of A. niger over A. terreus is its superior tolerance towards impurities that are abundantly present in industrial cultivation media [15, 16].
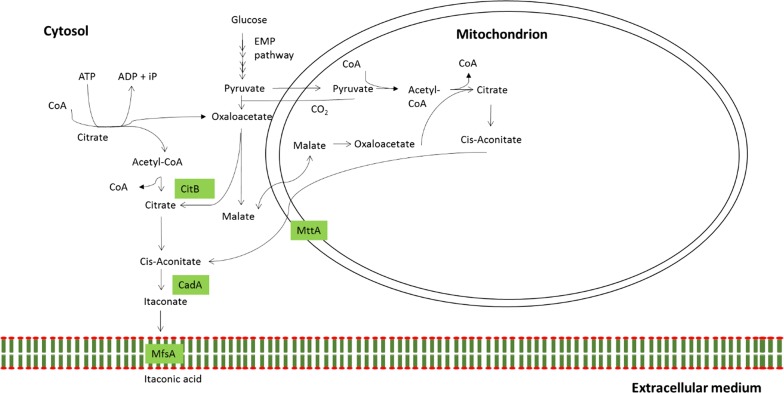
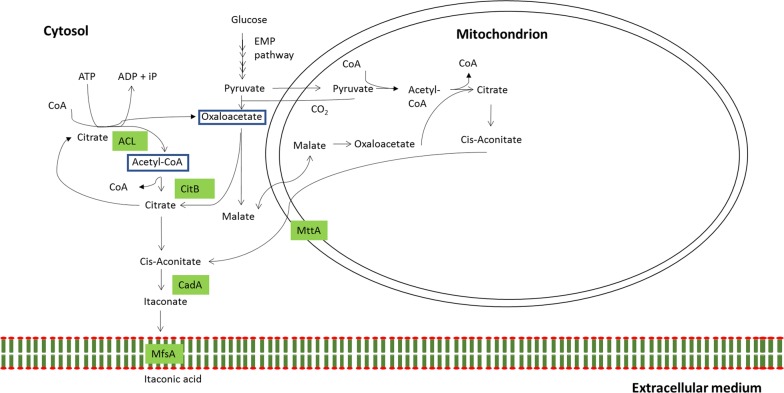
In our previous report, we communicated the overexpression of a putative cytosolic citrate synthase citB in an A. niger strain carrying the full IA biosynthesis gene cluster from A. terreus, where we suggest that rewiring of CitB to heterologous IA production leads to the formation of a cytosolic IA biosynthetic pathway in which cytosolic acetyl-CoA and oxaloacetate are converted into citrate and ultimately converted into IA by CadA (Fig. 1) [17]. Remarkably, this resulted in the highest final titer reported for A. niger (26.2 g/L IA with a maximum productivity of 0.35 g/L/h in 5-L scale fermentations) [17]. We also hypothesized that cytosolic precursors for citrate synthesis (i.e., oxaloacetate and acetyl-CoA) might be a limiting factor in our rewired pathway, resulting in limited IA production. Whereas cytosolic oxaloacetate is mainly synthesized by the activity of pyruvate carboxylase, the synthesis and supply of cytosolic acetyl-CoA is more of an unknown factor [18, 19]. In the research presented here we have designed an approach to increase heterologous IA production rates by increasing cytosolic acetyl-CoA levels.
Fig. 1.
Proposed rewired IA biosynthesis pathway in A. niger. MttA (mitochondrial tricarboxylate transporter) transports the TCA-cycle intermediate cis-aconitate from the mitochondrion into the cytosol where CadA (cis-aconitate decarboxylase) decarboxylates cis-aconitate and thereby forms itaconate. Itaconate is transporter over the cellular membrane by action of MfsA (major facilitator superfamily protein). CitB (cytosolic citrate synthase) catalyzes the synthesis of citrate in the cytosol
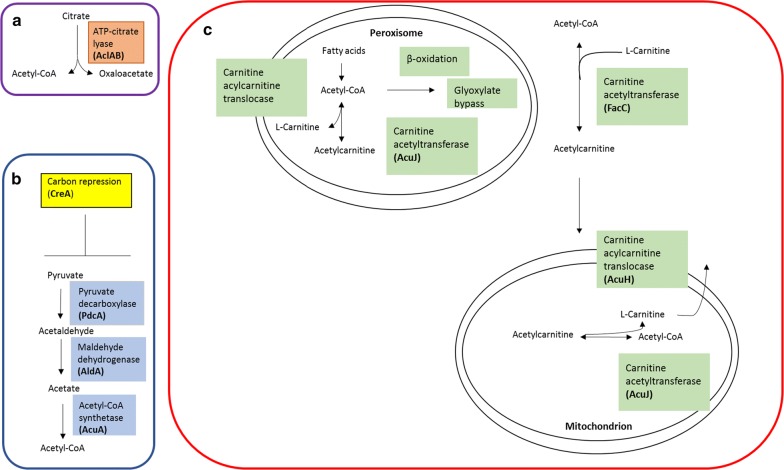
In literature, three pathways are described that can supply or synthesize cytosolic acetyl-CoA in filamentous fungi: through ATP-citrate lyase (ACL), the pyruvate–acetaldehyde–acetate pathway (PAA) and the carnitine acetyltransferase (CAT) system [18, 19] (Fig. 2). ACL has been described and characterized as an essential enzyme for the production of cytosolic acetyl-CoA in filamentous fungi and in particular Aspergillus [19, 20]. ACL (EC 2.3.3.8) consists of two different subunits which cleave citrate into its constituent components oxaloacetate and acetyl-CoA at the cost of one ATP (Fig. 2a). Increased activity of ACL was shown to increase the cytosolic pool of acetyl-CoA by Weyda et al. [21] in Aspergillus carbonarius, a species closely related to A. niger. In the yeast Saccharomyces cerevisiae, only the PAA pathway has been reported to be a significant source of cytoplasmic acetyl-CoA synthesis, while the ATP-citrate lyase route is completely absent [22]. In the PAA pathway, pyruvate is decarboxylated to acetaldehyde by action of pyruvate decarboxylase (PdcA) (EC 4.1.1.1). Acetaldehyde is acted upon by aldehyde dehydrogenase (ALD) (EC 1.2.1.3) to synthesize acetate which in turn is converted into acetyl-CoA by acetyl-CoA synthetase(ACS) (EC 6.2.1.1) (Fig. 2b). The PAA pathway may be an important pathway for cytosolic acetyl-CoA synthesis in S. cerevisiae; however, in A. niger, this pathway is an unlikely source of acetyl-CoA due to the low-level intracellular concentrations of the precursor acetate [23, 24]. Moreover, under glucose-grown conditions, the PAA pathway is repressed by the global carbon catabolite repressor CreA at the transcriptional level in the closely related fungal model organism Aspergillus nidulans [25]. The third possible pathway for cytosolic acetyl-CoA generation, the CAT system, consists of the mitochondrial and peroxisomal carnitine acetyltransferase AcuJ (EC 2.3.1.7) that catalyzes the reversible reaction of acetyl-CoA into acetylcarnitine. The cytosolic carnitine acetyltransferase FacC (EC 2.3.1.7) can catalyze the same reversible reaction in the cytosol. Together with the mitochondrial carrier protein AcuH, the CAT system provides a means to shuttle acetyl-CoA between the peroxisome, mitochondria and cytosol [18, 26, 27] (Fig. 2c). However, like the PAA pathway, the CAT system is not active under glucose grown conditions but only during growth on acetate or fatty acids [27].
Fig. 2.
Cytosolic acetyl-CoA synthesis pathways described in literature. a ATP-citrate lyase (ACL) cleaves citrate into its constituent components oxaloacetate and acetyl-CoA at the expense of one ATP. b Pyruvate-Acetaldehyde-Acetate (PAA) pathway that converts pyruvate into acetyl-CoA by action of PdcA, AldA and AcuA. This pathway is directly repressed at the transcriptional level by CreA. c Carnitine acetyltransferase (CAT) system that consists of mitochondrial and peroxisomal carnitine acetyltransferase AcuJ or cytosolic FacC that catalyzes the reversible reaction of acetyl-CoA into acetylcarnitine. Acetylcarnitine in turn can be transported to and from the mitochondrion by carnitine acylcarnitine translocase (AcuH) or by a yet unknown translocase from the peroxisome
Next to genetic engineering to improve the biosynthetic pathway, we have also focused our attention on optimizing the culture medium used for IA production. Current IA producing conditions were tailored to the first-generation low IA producing A. niger strains that were generated when cadA was first expressed in A. niger by Li et al. [28]. For this purpose, we have looked into literature to identify the IA producing conditions and medium components reported for A. terreus [8, 29]. Based on these reported conditions, we have designed a new IA production medium and fermentation protocol.
Materials and methods
Strains and culture conditions
Strains of Aspergillus niger used in this study have an AB1.13 background and are listed in Table 1 [30]. All strains were stored in 20% glycerol at − 80 °C and maintained on minimal medium (MM) agar [31]. Growth medium was supplemented with 10-mM uridine and 10-mM uracil when required. Spore suspensions were prepared by harvesting conidia from MM agar after 3–6 days of growth at 33 °C using 0.9% (w/v) NaCl and a sterile cotton stick. Spore suspensions were stored at 4 °C for up to 1 month without loss of viability.
Table 1.
Strains of Aspergillus niger used in this study
| Strain of A. niger | Abbreviation | Description |
|---|---|---|
| AB1.13 pyrG+ | AB1.13 | Uridine prototroph of AB1.13 pyrG- [32] |
| AB1.13 CAD 4.1 | AB1.13 CAD | Selected pyrG + transformant of cadA expressing transformant (CAD10.1) of AB1.13 [28] |
| AB1.13 CAD + MFS + MTT + CitB #99 | CitB#99 | citB overexpressing strain of AB1.13 CAD + MFS + MTT #49B [17] |
|
AB1.13 CAD + MFS + MTT + CitB #99-acl A4 R AB1.13 CAD + MFS + MTT + CitB #99-acl C3 R AB1.13 CAD + MFS + MTT + CitB #99-acl D9 R AB1.13 CAD + MFS + MTT + CitB #99-acl E11 Z AB1.13 CAD + MFS + MTT + CitB #99-acl F11 Z AB1.13 CAD + MFS + MTT + CitB #99-acl G1 Z AB1.13 CAD + MFS + MTT + CitB #99-acl G8 Z AB1.13 CAD + MFS + MTT + CitB #99-acl G11 Z AB1.13 CAD + MFS + MTT + CitB #99-acl H4 Z AB1.13 CAD + MFS + MTT + CitB #99-acl A9 G AB1.13 CAD + MFS + MTT + CitB #99-acl A12 G AB1.13 CAD + MFS + MTT + CitB #99-acl B11 G AB1.13 CAD + MFS + MTT + CitB #99-acl D10 G AB1.13 CAD + MFS + MTT + CitB #99-acl D12 G |
ACL A4 R ACL C3 R ACL D9 R ACL E11 Z ACL F11 Z ACL G1 Z ACL G8 Z ACL G11 Z ACL H4 Z ACL A9 Z ACL A12 G ACL B11 G ACL D10 G ACL D12 G |
acl1 and acl2 overexpressing strain of AB1.13 CAD + MFS + MTT + CitB #99 (this study) |
Molecular biological techniques
General cloning procedures in E. coli were according to Sambrook and Russell [33]. Restriction enzymes and buffers were obtained from Thermo Scientific or New England Biolabs and used according to the manufacturer’s instructions. DNA fragments were extracted from agarose gel using the QIAquick Gel Extraction Kit (QIAGEN) according to the manufacturer’s instructions. For amplification of plasmid DNA, chemically competent cells of E. coli strain DH5α (i.e., Thermo Scientific Subcloning Efficiency DH5α Competent Cells or Thermo Scientific Library Efficiency DH5α Competent Cells when required) were transformed by heat shock according to the manufacturer’s instructions. E. coli was grown in lysogeny broth (LB) at 37 °C. Plasmid DNA was isolated by miniprep using the GeneJET Plasmid Miniprep Kit (Thermo Scientific) or by maxiprep using the Plasmid Plus Maxi Kit (QIAGEN) according to the manufacturer’s instructions.
Fungal genomic DNA was isolated from mycelium cultivated in 1-mL liquid complete medium (CM) (MM + 2.5 g/L yeast extract) in 96-well microtiter plates, using the mag mini DNA extraction from µL blood kit (LGC) according to the manufacturer’s instructions. When high yield of genomic DNA was required, mycelium from 250-mL CM shake flask cultures was frozen in liquid nitrogen and ground to powder using a sterile mortar and pestle before extraction with the mag mini DNA extraction from µL blood kit. DNA concentration and purity were determined using the NanoDrop Onec UV–Vis Spectrophotometer (Thermo Scientific). PCR was performed on an Alpha Cycler 4 (PCRmax) using DreamTaq DNA Polymerase (Thermo Scientific) or Phusion Hot Start II DNA Polymerase (Thermo Scientific) when required. Amplified DNA from PCR was purified using the QIAquick PCR Purification Kit (QIAGEN) according to manufacturer’s instructions. Primers were obtained from Eurogentec and are listed in Table 2. DNA sequencing was carried out by BaseClear. Isolation of RNA, library preparation, sequencing and transcriptome data analysis are described in Hossain et al. [17].
Table 2.
Primers used in this study
| Primer name | F/R | Sequence 5′–3′ | Template |
|---|---|---|---|
| 332 AclA-3,600 F | F | TCAGCCCAGGTCT-TGTTCAG | A. carbonarius acl2 ORF |
| 333 AclA-4,237 R | R | CCCTCCATGTCCA-ATGATAAGGA | A. carbonarius tef1 promoter |
| 334 AclB-3,910 F | F | TCCCTTCCCTCGCT-TCTCTC | A. nidulans tef1 promoter |
| 335 AclB-4,497 R | R | ATGGCCTTGCTGA-CATCCTG | A. carbonarius acl1 ORF |
Cloning, transformation and cultivation of A. niger
The acl overexpression vectors pSBi415aACLoe and pSBi415bACLoe, containing the acl1 and acl2 expression cassettes as designed by Weyda et al. [21], were introduced into E. coli for amplification of plasmid DNA (Additional file 1: Figure S1).
Auxotrophic pyrE-mutants of CitB#99 were generated by cultivation on selective MM plates containing 5-fluoroorotic acid for 3–5 days at 33 °C until colonies appeared [34]. Organic acid production of CitB#99 pyrE-mutants was examined by cultivation in a 1-mL liquid culture of IA production medium M12++ (1.43 g/L NH4NO3, 0.11 g/L KH2PO4, 0.5 g/L MgSO4 × 7H2O, 0.005 g/L CuSO4 × 5H2O, 0.0006 g/L FeCl3 × 6H2O, 0.0006 g/L ZnSO4 × 7H2O, 0.074 g/L NaCl, 0.13 g/L CaCl2 × 2H2O, 100 g/L glucose, set at pH 2.3 with 10 M H2SO4), adapted from Li et al. [28]) in a 96-well deep-well plate that was incubated for 5 days at 33 °C, 850 rpm. To create an acl12 overexpression strain of CitB#99, one of the generated CitB#99 pyrE-mutants was transformed with both pSBi415aACLoe and pSBi415bACLoe simultaneously, by co-transformation with pJET1.2-pyrE, harbouring the pyrE auxotrophic marker (i.e., gene encoding orotate P-ribosyl transferase) from Aspergillus oryzae, in a ratio of 1:10 (5 µg of each construct:0.5 µg marker). Fungal transformation procedures were according to Arentshorst et al. [35]. Transformants were selected for their ability to grow on MM agar containing 115 g/L sucrose or 109.3 g/L sorbitol without uridine/uracil (i.e., uracil prototrophy).
Transformant screening
Individual colonies from transformation plates were transferred to a 48-well plate containing selective MM agar using a sterile toothpick that in turn was used to inoculate a 1-mL liquid culture of complete medium (CM), consisting of MM with the addition of 0.5% (w/v) yeast extract and 0.1% (w/v) casamino acids, or M12++ medium in a 96-well deep-well plate that was incubated for 3–5 days at 33 °C, 850 rpm. Supernatant was removed when mycelia were grown sufficiently, followed by isolation of genomic DNA as described in “Molecular biological techniques”. Isolated genomic DNA was used as template in diagnostic PCR to screen for transformants containing the constructs of interest, using primers 332 + 333 for pSBi415aACLoe and primers 334 + 335 for pSBi415bACLoe. Positive transformants were streaked from the 48-well plate on selective MM agar using 0.9% (w/v) NaCl and a sterile cotton stick to obtain pure single colony transformants, which were used to prepare spore suspensions as described in ‘Strains and culture conditions’.
Shake flask cultivation and related analyses
For analysis of organic acid production and glucose consumption of acl12 positive transformants, shake flasks (500 mL) containing 100 mL M12++ medium or MRA medium (3 g/L NH4NO3, 0.11 g/L KH2PO4, 1 g/L MgSO4 × 7H2O, 0.005 g/L CuSO4 × 5H2O, 0.0016 g/L FeCl3 × 6H2O, 0.0006 g/L ZnSO4 × 7H2O, 0.074 g/L NaCl, 0.13 g/L CaCl2 × 2H2O, 100 g/L glucose, set at pH 2.3 with 10 M H2SO4) were inoculated with 1.0 × 106 spores/mL and cultivated for up to 2 weeks at 33 °C, 250 rpm. A 200-µL sample was filtered over a 0.22-µm filter daily to determine organic acid and glucose concentrations in the extracellular medium using High-Performance Liquid Chromatography (HPLC). Cultures were weighed before sampling to account for evaporation of the culture medium. Shake flask cultures were supplemented with 17.87 mM NH4NO3, 35.74 mM NH4Cl or 35.74 mM NaNO3 before titers reached 20 g/L IA during experiments on nitrogen supplementation. Supplementation continued beyond this titer at time intervals of 2–3 days when nitrogen was estimated to be depleted. Equimolar amounts of nitrogen were supplemented at once for all nitrogen sources.
Analysis of metabolite production was performed by HPLC using a WATERS e2695 separations module equipped with an Aminex HPX-87H column (Bio-Rad) and 5-mM H2SO4 as eluent. Column temperature was set at 60 °C and eluent flow was 600 µL/min. Peak detection occurred simultaneously by a refractive index detector (WATERS 2414) and a dual-wavelength detector (WATERS UV/Vis 2489). Empower Pro software (Empower 2 Software, copyright 2005–2008, Waters Corporation) was used for data processing.
Southern blotting
For Southern blotting of CitB#99-acl transformants, shake flask cultures containing 100-mL CM (pH 4.5) were inoculated with 1.0 × 105 spores/mL and grown for 2 days at 33 °C, 200 rpm. Mycelia were harvested using a sterile Miracloth filter, dried using paper towels and frozen in liquid nitrogen. Genomic DNA was isolated from 300-µg ground mycelium per strain as described in “Molecular biologic techniques”. Quality of isolated DNA was tested by running on 0.8% (w/v) agarose gel. 1-µg genomic DNA was digested per strain using PvuII and run on agarose gel O/N. Blotting occurred on nitrocellulose Hybond-N + blotting paper (Amersham Biosciences) using SSC solution (175.5-g NaCl and 88.2-g Na-Citrate in 1 L; 20 × stock). After blotting, the membranes were treated in a UV-chamber to cross-link DNA. Digoxigenin-labeled (DIG) probes for acl1 and acl2 were created by PCR from pSBi415bACLoe using primers 334 + 335 and from pSBi415aACLoe using primers 332 + 333, respectively, using the PCR DIG Probe Synthesis Kit (Roche Life Sciences) according to the manufacturer’s instructions. Labelling and detection were performed using the DIG Easy Hyb labelling and detection kit for Southern blotting purposes (Roche Life Sciences) according to the manufacturer’s instructions. The ChemiDoc XRS + Imaging System (Bio-Rad) was used for imaging of Southern blots.
Controlled fed-batch cultivations and related analyses
Controlled fed-batch cultivations in 16-L stainless steel stirred tank bioreactors (NLF22, Bioengineering, Switzerland) were conducted at 33 °C with a working volume of 10 L. Cultivation occurred in M2 medium that consisted of 2.29 g/L NH4NO3, 0.18 g/L KH2PO4, 0.80 g/L MgSO4 × 7H2O, 0.008 g/L CuSO4 × 5H2O, 0.001 g/L FeCl3 × 6H2O, 0.001 g/L ZnSO4 × 7H2O, 0.118 g/L NaCl, 0.21 g/L CaCl2 × 2H2O and 160 g/L glucose. Dissolved oxygen levels were maintained above 30% by controlling the stirrer speed and aeration rate between 450 and 800 rpm and 3–8 L/min, respectively. The pH was maintained at 3.0 after 48-h EFT by continuous addition of 3-M NH4OH. Additionally, the pH was controlled offline twice per day to exclude errors of the online sensor. Bioreactor inoculum was prepared by filling a 1-L non-baffled shake flask with 200-mL M12++ medium and inoculating with 1.0*106 spores/mL and cultivated for 96 h at 33 °C and 250 RPM. A sterile 700 g/L glucose solution was used for addition of sugar during runs. For determination of the cell dry weight, biomass was separated from fermentation broth using vacuum filtration and washed with deionized water, then dried at 105 °C for 48 h. The exhaust gas of the fermentation was analyzed with a BioPAT Xgas analyzer (Sartorius Stedim Biotech GmbH, powered by BlueSens) on %CO2 and %O2.
For quantification of sugars, organic acids and nitrogen in the media, samples were filtered through a 0.45-µm filter (Minisart RC, Sartorius, Germany) and analyzed by HPLC (1100 Series, Agilent Technologies, USA). Total nitrogen and ammonia concentrations were determined by Kjeldahl and distillation–titration methods, respectively [36]. Phosphate content was analyzed by a colorimetric method using LCK 349 Phosphate Kit (HACH).
Results
Analysis of cytosolic acetyl-CoA generating pathways
Rewiring CitB to heterologous IA production resulted in increased production titers (26.2 g/L IA). From our previous research, it was hypothesized that acetyl-CoA might be a limiting factor for cytosolic IA production in the rewired pathway [17]. Therefore, transcriptome analysis of possible acetyl-CoA pathways was carried out in the AB1.13 WT strain and AB1.13 CAD strain to understand which pathways predominantly generate cytosolic acetyl-CoA in A. niger. As described above from literature, it was discerned that three pathways are identified as cytosolic acetyl-CoA generating pathways in fungi: through ACL that cleaves cytosolic citrate into oxaloacetate and acetyl-CoA, the PAA pathway that converts pyruvate into acetyl-CoA and the CAT system that shuttles acetyl-CoA in the form of acetylcarnitine from and between the peroxisome and mitochondrion into the cytosol (Fig. 2) [18, 19, 37]. Interestingly, our transcriptome analysis shows that the expression of PAA pathway gene pdcA is strongly downregulated in the IA producing AB1.13 CAD strain compared to its parental strain AB1.13 (Table 3). It has been shown in a study conducted by Meijer et al. [38] that the PdcA enzyme was inactive in oxygen-limiting conditions. The reason for the low expression of pdcA in AB1.13 CAD and its link with heterologous IA production is, however, at the moment, not clear. Furthermore, from our transcriptome data (Table 3), we also see that the ACS coding gene (acuA) is expressed at low levels, in accordance with findings that this gene is under control of carbon catabolite repression [39, 40].
Table 3.
Transcriptome data of known cytosolic acetyl-CoA generating pathways and the genes related to heterologous IA production in A. niger
| Locus tag | Enzyme | Old locus tag | Localization | AB1.13 WT | AB1.13 CAD | ||
|---|---|---|---|---|---|---|---|
| RPKM | RPKM | ||||||
| ATP-citrate lyase | |||||||
| ANI_1_76094 | ATP-citrate lyase subunit 1 (aclA) | An11g00510 | Cyto | 325.61 | 325.45 | 506.32 | 505.54 |
| ANI_1_78094 | ATP-citrate lyase subunit 2 (aclB) | An11g00530 | Cyto | 331.2 | 332.23 | 587.4 | 587.02 |
| CAT system | |||||||
| ANI_1_724074 | Carnitine acetyl transferase (facC) | An08g04990 | Cyto/Nucleus | 28.78 | 29.17 | 24.89 | 24.77 |
| ANI-1-192164 | Carnitine acetyl transferase (acuJ) | An18g01590 | Per/Mito | 45.97 | 47.7 | 38.2 | 38.08 |
| ANI_1_388034 | Carnitine/acyl carnitine carrier (acuH) | An03g03360 | Cyto/Mito | 31.08 | 30.93 | 29.01 | 28.95 |
| PAA pathway | |||||||
| ANI_1_936024 | Pyruvate decarboxylase (pdcA) | An02g06820 | Cyto | 5910.75 | 5915.54 | 304.97 | 306.49 |
| ANI_1_1024084 | Pyruvate decarboxylase (pdcB) | An09g01030 | Cyto | 38.82 | 38.64 | 47.95 | 48.26 |
| ANI_1_2276014 | Pyruvate decarboxylase | An01g01590 | Nucleus | 0.35 | 0.45 | 0.17 | 0.18 |
| ANI_1_796114 | Pyruvate decarboxylase | An13g03320 | Mito/Cyto | 0 | 0 | 0.03 | 0.03 |
| ANI_1_1024074 | Aldehyde dehydrogenase (aldA) | An08g07290 | Cyto | 143.48 | 142.19 | 178.07 | 177.72 |
| ANI_1_226174 | Aldehyde dehydrogenase | An10g00850 | Per | 1.59 | 1.35 | 1.69 | 2.21 |
| ANI_1_1748184 | Aldehyde dehydrogenase | An04g03400 | Cyto | 54.25 | 54.85 | 53.22 | 54.15 |
| ANI_1_924184 | Acetyl-CoA synthetase (acuA) | An04g05620 | Cyto | 139.46 | 141.21 | 96.66 | 96.09 |
| ANI_1_938144 | Acetyl-CoA hydrolase (ach1) | An16g07110 | Mito | 98.53 | 99.38 | 22.36 | 22.93 |
| ANI_1_878024 | Acetate kinase | An02g06420 | Cyto/Mito | 42.53 | 43.41 | 13.72 | 13.75 |
| Mitochondrial acetyl-CoA synthesis | |||||||
| ANI_1_1206064 | Pyruvate dehydrogenase E1 component subunit alpha (pda1) | An07g09530 | Mito | 263.81 | 264.72 | 205.40 | 207.09 |
| ANI_1_622094 | Pyruvate dehydrogenase E1 component subunit alpha | An11g04550 | Mito | 10.81 | 9.47 | 10.54 | 9.75 |
| ANI_1_12014 | Pyruvate dehydrogenase E1 component subunit beta | An01g00100 | Mito | 215.18 | 213.19 | 177.40 | 176.43 |
| ANI_1_274064 | Pyruvate dehydrogenase E2 component | An07g02180 | Mito | 299.81 | 301.04 | 286.65 | 284.54 |
| Citrate synthases | |||||||
| ANI_1_876084 | Citrate synthase (citA) | An09g06680 | Mito | 478.40 | 482.85 | 428.47 | 426.67 |
| ANI_1_1226134 | Methylcitrate synthase (mcsA) | An15g01920 | Mito | 51.49 | 51.55 | 25.69 | 24.11 |
| ANI_1_1474074 | Citrate synthase (citB) | An08g10920 | Cyto | 58.09 | 57.90 | 522.44 | 521.45 |
| ANI_1_2950014 | Citrate synthase (citC) | An01g09940 | Cyto | 3.86 | 3.52 | 0.91 | 0.96 |
| Aconitases | |||||||
| ANI_1_1410074 | Aconitate hydratase (aco1) | An08g10530 | Mito | 234.09 | 234.83 | 397.94 | 397.68 |
| ANI_1_470084 | Aconitate hydratase | An09g03870 | Mito | 64.88 | 64.41 | 26.01 | 26.07 |
| ANI_1_3018024 | Aconitate hydratase | An02g11040 | Cyto | 0.04 | 0.04 | 0.00 | 0.00 |
| ANI_1_1808144 | Aconitate hydratase | An16g05760 | Cyto | 0.85 | 0.85 | 0.98 | 0.99 |
| ANI_1_578044 | Aconitase | An05g02230 | Cyto | 6.29 | 6.38 | 14.13 | 14.28 |
| ANI_1_1802134 | Aconitase | An15g07730 | Cyto | 26.63 | 26.21 | 30.62 | 30.72 |
| Itaconate biosynthesis | |||||||
| cis-aconitate decarboxylase (cadA) | Cyto | 1.27 | 1.20 | 4287.14 | 4300.13 | ||
The picture that derives from transcriptome analysis indicates that the ACL pathway may actually be the major cytosolic acetyl-CoA generating pathway in A. niger (Table 3). This observation is also in accordance with the observations from Pfitzner et al. [24]. They have previously shown that ACL is ubiquitously present in A. niger cell lysates independent of the C-sources tested. Moreover, acetate, an intermediate in the PAA pathway, was below detection limit in cellular extracts; whereas, citrate was always present in detectable amounts [24]. These results are in accordance with our observation of significant expression of acl1 and acl2. Therefore, and together with the results presented by Weyda et al. [21], we decided to overexpress acl1 and acl2 for increased cytosolic acetyl-CoA generation.
Overexpression of acl12
To enhance IA production beyond titers reached with CitB#99, overexpression of acl12 (i.e., overexpression of the A. carbonarius orthologues of the A. niger ACL subunit encoding genes aclA and aclB) was established in this strain. Presence of both acl1 and acl2 overexpression vectors in the genomes of 14 out of 144 transformants was confirmed by diagnostic PCR (data not shown). HPLC analysis of shake flask cultivations with acl12 overexpressing CitB#99 transformants and the parental strain CitB#99 in M12++ medium with 125 g/L glucose showed that 9 CitB#99-acl transformants had an increase in max. IA titer compared to CitB#99, but most apparent was the increase in max. productivity of all transformants, excluding two strains with impaired IA production (i.e., ACL F11 Z and A12 G) (Table 4). In none of the strains, significant levels of other organic acids were detected. These results, particularly the increase in max. IA productivity, are in accordance with the expected increase in glycolytic flux upon overexpression of the ACL encoding genes. ACL G1 Z showed the highest increase (32%) in max. titer (20.6 g/L IA) of all CitB#99-acl transformants compared to the parental strain (15.6 g/L IA) (Table 4). This transformant outperformed all other transformants in overall IA production, also given its max. productivity of 0.20 g/L/h and yield of 0.30 g/g compared to the max. productivity and yield of the parental strain CitB#99 (0.13 g/L/h and 0.23 g/g) (Table 4).
Table 4.
Itaconic acid production of acl12 overexpressing CitB#99 transformants
| Strain | Max. titer (g/L) | Max. productivity (g/L/h) | Yield (g/g glucose) |
|---|---|---|---|
| CitB#99 | 15.6 | 0.13 | 0.23 |
| CitB #99 ACL A12 G | 2.2 | 0.02 | 0.04 |
| CitB #99 ACL F11 Z | 10.5 | 0.11 | 0.15 |
| CitB #99 ACL D12 G | 14.2 | 0.14 | 0.22 |
| CitB #99 ACL A4 R | 15.2 | 0.14 | 0.23 |
| CitB #99 ACL D10 G | 15.3 | 0.14 | 0.23 |
| CitB #99 ACL H4 Z | 15.7 | 0.15 | 0.24 |
| CitB #99 ACL G11 Z | 16.2 | 0.15 | 0.24 |
| CitB #99 ACL G8 Z | 16.30 | 0.16 | 0.25 |
| CitB #99 ACL C3 R | 16.6 | 0.16 | 0.22 |
| CitB #99 ACL A9 G | 17.9 | 0.18 | 0.25 |
| CitB #99 ACL B11 G | 18.0 | 0.17 | 0.26 |
| CitB #99 ACL E11 Z | 18.4 | 0.17 | 0.26 |
| CitB #99 ACL D9 R | 18.4 | 0.17 | 0.28 |
| CitB #99 ACL G1 Z | 20.6 | 0.20 | 0.30 |
Results are from a single representative shake flask experiment for each transformant
Notably, ACL G1 Z reached a max. titer of 24.8 g/L IA in a similar shake flask cultivation, along with a max. productivity of 0.28 g/L/h and a yield of 0.33 g/g (Additional file 1: Figure S2). Southern blot analysis of the five best IA producing CitB#99-acl transformants and parental strain CitB#99 showed that ACL G1 Z did not possess a 1:1 ratio of acl1 and acl2, harboring one copy of acl1 and three copies of acl2 (Additional file 1: Table S1). Overall, no correlation was found between IA production and number and/or ratio of gene copies of acl1 and acl2 present in the genomes of the CitB#99-acl transformants that were analyzed by Southern blotting (Table 4; Additional file 1: Table S1). Results in Additional file 1: Table S1 are derived from Southern blots shown in Additional file 1: Figure S3.
Culture optimization and alkalizing nitrogen source supplementation
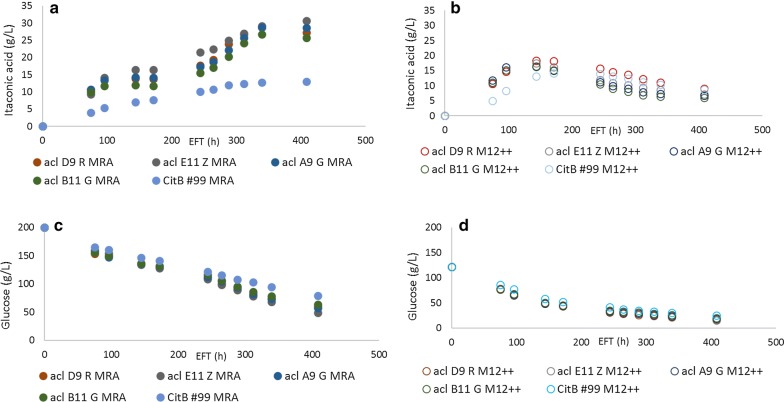
Having established a new generation of improved IA producing A. niger strains, we have subsequently focused our attention on optimizing the culture medium used for IA production. Adjustments to the M12++ medium previously developed included an increase in glucose, nitrogen, magnesium and iron (III) concentration, based on a published IA production medium used for A. terreus [8, 29]. The resulting medium was termed MRA and its exact composition is described in the Materials and Methods. HPLC analysis of shake flask cultivations with acl transformants ACL D9 R, ACL E11 Z, ACL A9 G and ACL B11 G showed that these transformants reached titers ranging from 26.7 to 30.6 g/L IA when cultivated in MRA medium with 200 g/L glucose (Fig. 3). The average increase in max. titer was 58% compared to cultivation in M12++ medium with 125 g/L glucose. Although max. titers improved significantly upon cultivation in MRA medium compared to M12++ medium, both overall yield and max. productivity decreased under these culture conditions with average decreases of 16% and 13%, respectively. Note that although less glucose was provided here during cultivations in M12++ medium, this does not explain the lower titers reached given that glucose was not yet depleted when IA production stalled. Interestingly, it appears that in MRA medium, IA production shows two stages of production with an intermediary lag-phase in between the two production phases. From inoculation up to 96-h EFT, IA was being produced at a constant rate of 0.14 ± 0.05 g/L/h in MRA medium and 0.15 ± 0.04 g/L/h in M12++ medium. Between 96-h and 168-h EFT, IA production appears to have reached a plateau and no production appears to occur in both media. However, after 244-h EFT, IA production commenced further in MRA medium but not in M12++ medium, where a clear decline in IA titer can be observed. We have observed in the past that IA titers beyond 20 g/L can have a toxic effect on mycelia and thereby severely limit growth and biomass formation [17]. The observation that additional NH4NO3, among others, was able to induce IA production beyond titers that are normally toxic and inhibitory for A. niger is interesting and was further investigated by feeding different nitrogen sources during shake flask cultivation.
Fig. 3.
IA production of CitB#99 and CitB#99-acl overexpressing strains. a IA production in MRA medium. b IA production in M12++ medium. c Glucose consumption in MRA medium. d Glucose consumption in M12++ medium. Results depicted are from a single representative shake flask experiment for each transformant
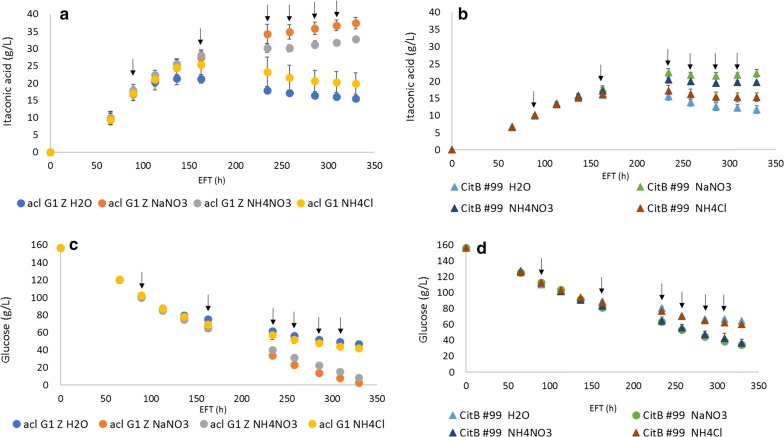
To determine whether a specific nitrogen source was responsible for this effect, shake flask cultures with M12++ medium were supplemented with either NH4NO3, NaNO3 or NH4Cl upon reaching inhibitory conditions at titers of 15–20 g/L IA. Pulse feed was chosen over an increased initial nitrogen concentration in the culture medium to prevent any inhibition of IA production by excess nitrogen availability [4, 42].
HPLC analysis results of nitrogen fed shake flask cultivations of ACL G1 Z and its parental strain CitB#99 are shown in Fig. 4. Interestingly, it was found that strains supplemented with NH4NO3 or NaNO3 extended their production phase till glucose was fully consumed, while strains not supplemented (dH2O) or supplemented with NH4Cl stopped producing at titers around 20 g/L IA (Fig. 4), although pulse feed with NH4Cl caused a slight increase in IA production compared to pulse feed with dH2O. This effect was observed for both strains ACL G1 Z and CitB#99. ACL G1 Z reached an average max. titer of 25.3 g/L IA when the culture medium was repeatedly supplemented with NH4Cl, after which IA titers went into decline, while IA production still continued to titers of 32.8 and 37.5 g/L IA when the culture medium was repeatedly supplemented with NH4NO3 or NaNO3, respectively (Fig. 4). These results show that availability of NO3− and to a lesser degree NH4+ is important for A. niger to cope with toxic titers of IA, being able to extend the production phase beyond inhibitory conditions. Note that even though nitrogen sources were pulse fed, productivity decreased upon increasing IA titers suggesting depletion of other limiting compounds. These findings were taken into account during further research of ACL G1 Z under controlled fermentation conditions.
Fig. 4.
Nitrogen supplementation of CitB#99 and ACL G1 Z during cultivation in M12++ medium. Cultivation flasks inoculated with strains CitB#99 and ACL G1 Z were pulse fed with dH2O, NaNO3, NH4NO3 or NH4Cl (pulse feed indicated by arrow). a IA production of ACL G1 Z. b IA production of CitB#99. c Glucose consumption of ACL G1 Z. d Glucose consumption of CitB#99. Experiment was performed in duplicate. Error bars represent SEM (Standard Error of the Mean). Note that in those cases where the error bar is not visible it falls within the size of the datapoints
Controlled fed-batch cultivations
In nitrogen fed shake flask experiments we have observed higher IA production when compared to non-fed flasks, as shown above. In particular, shake flasks fed with a N-source that has an alkalizing effect on the medium (i.e., NaNO3 and NH4NO3), had a positive effect on IA titer (Fig. 4). To further test IA production of ACL G1 Z, this strain was cultivated in controlled 10-L fed-batch fermentations where the pH was maintained at 3.0 using 3-M NH4OH. IA production, biomass formation and sugar consumption are shown in Fig. 5. Glucose concentration rapidly dropped from 160 to 100 g/L after which glucose was pulse fed. IA concentration at moment of feed was 18 g/L and biomass dry weight was 10 g/L. IA production further continued to a max. titer of 42.7 g/L after 240-h EFT, after which IA titer did not increase further. The overall productivity and yield at which IA was produced in this experiment was 0.18 g/L/h and 0.26 g IA/g glucose, respectively. During the course of fermentation, some citric acid formation was observed (max. titer 9.7 g/L; Fig. 5). The carbon balance accounted for 89.9% of total carbon that was added to the fermentor (Additional file 1: Table S2). The pH was steadily maintained at 3.0 after 48-h EFT by addition of 3-M NH4OH (Additional file 1: Figure S4). Although we attempted to keep the DO at minimal 30% by manually adjusting the airflow, at some points during the cultivation, the DO dropped below 30% (Additional file 1: Figure S4).
Fig. 5.
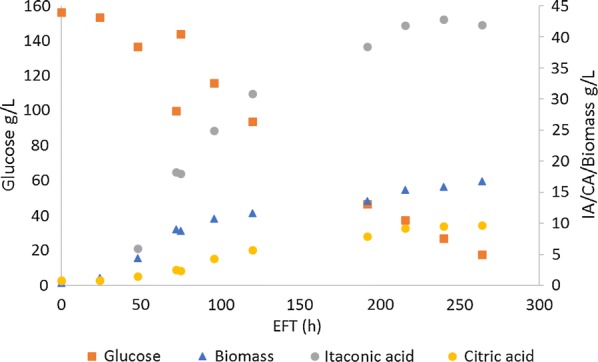
Controlled fed-batch cultivation of ACL G1 Z. IA production, CA production, biomass formation and glucose consumption is depicted. Performed as single experiment
Biomass increased from 10 g/L after 60 h EFT to 16 g/L at the end of fermentation (264-h EFT). Nitrogen concentration in the media decreased in the first 48 h in accordance with the cell growth phase; however, after 48-h EFT nitrogen accumulation occurred, due to pH control with NH4OH addition (Additional file 1: Figure S5).
Discussion
This research aimed to improve the biological production of IA by targeting its heterologous biosynthesis in A. niger. IA biosynthesis could be improved by overexpression of the ATP-citrate lyase encoding genes acl1 and acl2. Furthermore, alkalizing nitrogen source availability turned out to be crucial for achieving high IA titers, which were greatly improved by the use of an IA production medium with increased nitrogen concentration or by supplementation of the culture medium with alkalizing nitrogen sources, i.e., nitrate and ammonia, extending the IA production phase.
The products of ACL are direct precursors for the generation of citrate by CitB and would, therefore, seemingly benefit the flux towards IA biosynthesis by facilitating the CitB precursor pool. However, it should be noted that the combined reactions of ACL and CitB constitute a futile cycle in which citrate is both substrate and product (Fig. 6). We interpret the increase in IA max. titer and max. productivity as a function of an increase in glycolytic flux that results from ATP consumption during the catabolic activity of ACL, which is supported by Wellen et al. [43] who showed that silencing the ACL encoding genes in mammalian cells leads to a significant decrease in cellular glycolytic activity. Additionally, Yin et al. [44] have shown that an ACL-mediated futile cycle of citrate cleavage and synthesis during idiophase is of importance to achieve high-yield citric acid production in A. niger. The increase in glycolytic flux ultimately benefits the flux towards IA biosynthesis by increasing precursor formation via the conversion of glucose to pyruvate and finally to oxaloacetate by pyruvate carboxylase in the cytosol and/or synthesis of cis-aconitate in the TCA cycle. This could arguably prevent the flux of potential IA precursors towards metabolic pathways other than for IA biosynthesis, explaining the observed increase in yield of the better IA producing CitB#99-acl transformants (Table 4).
Fig. 6.
Proposed rewired IA biosynthesis pathway in A. niger that contains a futile cycle of citrate synthesis and cleavage through action of CitB and ACL, respectively. See also Fig. 1
A correlation between IA production and gene copy number of acl1 and acl2 in the genomes of the CitB#99-acl transformants was not found (Table 4, Additional file 1: Table S1). Because both genes encode one subunit of the ACL enzyme, the optimal ratio was expected to be 1:1 for acl1 and acl2. The best IA producing CitB#99-acl transformant (i.e., ACL G1 Z) was not necessarily the closest one to this ratio compared to the other transformants analyzed by Southern blotting. However, this does not exclude the possibility that, of all acl12 overexpression transformants, this strain is the closest to a 1:1 ratio in terms of expression of both genes, since besides number of gene copies, gene expression is also highly dependent on their site of integration in the genome.
Besides linking carbon metabolism to fatty acid synthesis, it was shown by Wellen et al. [43] that ACL also plays a central role in protein acetylation, including histone acetylation in the nucleus. In detail, these researchers found that ACL-dependent acetylation contributes to the selective regulation of genes involved in glucose metabolism of mammalian cells. It would be interesting to speculate on the regulatory effects of acl12 overexpression on glycolysis and ultimately IA production.
Furthermore, this research showed the importance of aligning fermentation optimization with genetic engineering. While overexpression of acl12 improved overall IA production in M12++ medium, the use of MRA medium showed that elevated levels of NH4NO3, magnesium and iron (III) already had significant impact on IA titers that could be reached by this new strain lineage. Subsequent efforts on fermentation optimization revealed the importance of NO3− and NH4+ availability during IA production. However, extensive research on this topic suggests that IA production should remain at nitrogen limiting conditions, which would direct global gene expression towards overflow metabolism of nitrogen-free compounds such as IA [4, 42]. Therefore, nitrogen concentration has to be tightly regulated during the course of cultivation in order to ensure optimal IA production.
In our previous experiments, we have observed that CitB#99 and acl12 overexpressing transformants start to degrade IA at titers between 15 and 20 g/L (Fig. 3) [45]. Interestingly, by maintaining NO3− and NH4+ availability, both cessation of IA degradation and elevated titers could be established as shown by the cultivations in MRA medium as well as pulse-feeding experiments (Figs. 3, 4), suggesting a vital role of these compounds in IA detoxification. Literature describes the central role of these nitrogen sources in a specific pathway on which acid-tolerant microorganisms rely heavily to reduce the cytotoxic effects of weak acids in acidic environments [46, 47]. This pathway is referred to as the glutamate-dependent acid resistance mechanism (GDARM), in which NH4+ and/or NO3− are essential precursors for generation of glutamate allowing a proton-consuming decarboxylation reaction into γ-aminobutyric acid (GABA) to ultimately increase the alkalinity of the cytoplasm [46, 48–52]. Kubicek et al. [53] found that A. niger accumulates GABA during acidogenesis, suggesting that A. niger also relies on this system to cope with acidic environments. Supported by our results, maintaining NH4+ or NO3− availability, therefore, seems essential for A. niger to cope with high IA titers, continue IA production and prevent its degradation in response to weak-acid stress. Note that in this pathway, NO3− first has to be converted into NH4+ to function as precursor for glutamate. This energy-draining process in the form of NADPH consumption causes an increase in glycolytic flux, in addition to the fact that ammonium salts further decrease the extracellular pH as they are consumed while nitrate does not, together might explain the bigger impact of NO3− over NH4+ observed during the pulse-feed experiments. Note that in the shake flask pulse-feed experiments acidifying NH4+ sources have been used in the form of NH4Cl and NH4NO3; whereas in the controlled fed-batch cultivations NH4OH, which is not an acidifying ammonium source, has been used to maintain the extracellular pH at 3.0 and feed NH4+. The results presented here show that the fermentation protocol with the ACL G1 Z strain that was designed is robust, in the sense that applying these conditions in different fermentation labs and different equipment (see Additional file 1: Figure S6) resulted in similarly high IA titer, yield and productivity (56.6 g/L IA; 0.28 g IA/g glucose; 0.18 g IA/L/h, respectively). Moreover in this specific case, no CA was detected (Additional file 1: Figure S6) and the carbon balance accounted for 96.6% of total carbon that was added to the fermenter (Additional file 1: Table S3).
Simultaneously with the findings on NO3−/NH4+ availability in relation to IA degradation, we have observed upregulation of the IA bioconversion pathway in A. niger responsible for detoxifying the extracellular medium of IA during high producing conditions, resembling the IA degrading pathway of A. terreus described by Chen et al. [45, 54]. Knock-out of the key pathway-associated genes in CitB#99 resulted in cessation of IA bioconversion and elevated titers [45]. However, as these high IA titers have shown to result in stronger growth effects possibly also due to weak-acid stress, combining this knock-out of the IA degradation pathway in combination with NH4+/NO3− balancing conditions is a clear target for ongoing research to improve IA production in A. niger.
Conclusions
Ultimately, this study showed that the targeting of genes that improve glycolytic flux, more specifically the genes encoding ATP-citrate lyase acl12, constitutes a successful strategy for the improvement of IA production. This improvement manifests in increased IA titer, yield and productivity. Furthermore, this study also highlights the importance of medium composition and culture conditions for IA production. In shake flask cultivation, optimization of the cultivation conditions by supplementation of an alkalizing nitrogen source showed great potential for improvement of IA production in A. niger and these results were also shown in controlled fed-batch cultivations at 10-L scale. These strategies all yield great promise for future improvement of biological IA production to ensure its viability in the rising bio-based economy.
Supplementary information
Additional file 1: Figure S1. Design of acl1 and acl2 expression cassette. Figure S2. Shake flask cultivation of CitB#99 ACL G1 Z. Table S1. Relative copy numbers of introduced acl12 and IA titers achieved in shake flask cultivations. Figure S3. Southern-Blot results of selected CitB#99-acl transformants. Table S2. Carbon balance of 10L fed-batch bioreactor cultivation of ACL G1 Z. Figure S4. Dissolved oxygen profile and pH profile during 10 L controlled fed-batch cultivation of strain CitB#99-ACL G1 Z. Figure S5. Total nitrogen concentration during 10 L controlled fed-batch cultivation of ACL G1 Z. Figure S6. Repeat fed-batch cultivation of ACL G1 Z performed in 5 L BioFlo 320 (Eppendorf) controlled bioreactors. Table S3. Carbon balance of repeat fed-batch 5 L bioreactor cultivation of ACL G1 Z.
Acknowledgements
Not applicable.
Authors’ contributions
AH and PJP designed the experiments and analyzed the results; AH and RVG performed the experiments; AH, RVG and PJP wrote the manuscript. PSL designed and created the overexpression cassettes. KMO, HT and MT designed and performed the experiments and analyzed the results of the controlled fed-batch cultivations. KMO performed the C-balance analysis. All authors read and approved the final manuscript.
Funding
This research was fully funded by Dutch DNA Biotech BV. Pakmaya’s contribution has been partially funded by TUBITAK (The Scientific and Technological Research Council of Turkey), within International Industrial R&D Projects Grant Programme (Teydeb 1509 - Project number: 9150147) under ERA-IB-2 6th Call (Project number: ERA-IB-15-011).
Availability of data and materials
Transcriptome data will be uploaded on GEO.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abeer H. Hossain and Roy van Gerven contributed equally to this work
Contributor Information
Abeer H. Hossain, Email: abeer.hossain@ddna-biotech.com
Roy van Gerven, Email: roy.vangerven@ddna-biotech.com.
Karin M. Overkamp, Email: karin.overkamp@ddna-biotech.com
Peter S. Lübeck, Email: psl@bio.aau.dk
Hatice Taşpınar, Email: hatice.demir@pakmaya.com.
Mustafa Türker, Email: mustafa.turker@pakmaya.com.
Peter J. Punt, Email: peter.punt@ddna-biotech.com
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-019-1577-6.
References
- 1.Werpy T, Petersen G. Top value added chemicals from biomass volume I—results of screening for potential candidates from sugars and synthesis gas. Other Inf. PBD. 2004; Medium: ED; Size.
- 2.Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “Top 10” revisited. Green Chem. 2010;12:539. doi: 10.1039/b922014c. [DOI] [Google Scholar]
- 3.Willke T, Vorlop KD. Biotechnological production of itaconic acid. Appl Microbiol Biotechnol. 2001;56:289–295. doi: 10.1007/s002530100685. [DOI] [PubMed] [Google Scholar]
- 4.Voll A, Klement T, Gerhards G, Büchs J, Marquardt W. Metabolic modelling of itaconic acid fermentation with Ustilago maydis. Chem Eng Trans. 2012;27:367–372. [Google Scholar]
- 5.Robert T, Friebel S. Itaconic acid—a versatile building block for renewable polyesters with enhanced functionality. Green Chem. 2016;18:2922–2934. doi: 10.1039/C6GC00605A. [DOI] [Google Scholar]
- 6.Kumar S, Krishnan S, Samal SK, Mohanty S, Nayak SK. Itaconic acid used as a versatile building block for the synthesis of renewable resource-based resins and polyesters for future prospective: a review. Polym Int. 2017;66:1349–1363. doi: 10.1002/pi.5399. [DOI] [Google Scholar]
- 7.Krull S, Hevekerl A, Kuenz A, Prüße U. Process development of itaconic acid production by a natural wild type strain of Aspergillus terreus to reach industrially relevant final titers. Appl Microb Biotechnol. 2017;101:4063–4072. doi: 10.1007/s00253-017-8192-x. [DOI] [PubMed] [Google Scholar]
- 8.Karaffa L, Díaz R, Papp B, Fekete E, Sándor E, Kubicek C. A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl Microbiol Biotechnol. 2015;99:7937–7944. doi: 10.1007/s00253-015-6735-6. [DOI] [PubMed] [Google Scholar]
- 9.Okabe M, Lies D, Kanamasa S, Park EY. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol. 2009;84:597–606. doi: 10.1007/s00253-009-2132-3. [DOI] [PubMed] [Google Scholar]
- 10.De Carvalho JC, Magalhães AI, Soccol CR. Biobased itaconic acid market and research trends—is it really a promising chemical? Chim Oggi/Chem Today. 2018;36:56–58. [Google Scholar]
- 11.Weastra SRO. Determination of market potential for selected platform chemicals: itaconic acid, succinic adis, 2,5-furandicarboxylic acid. 2011:1–173.
- 12.Markit IHS. Citric acid-chemical economics handbook. 2015.
- 13.Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M. A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet Biol. 2011;48:602–611. doi: 10.1016/j.fgb.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 14.FDA/CFSAN U. Inventory of GRAS notices: summary of all GRAS notices. 2008.
- 15.Kuenz A, Krull S. Biotechnological production of itaconic acid—things you have to know. Appl Microbiol Biotechnol. 2018;102:3901–3914. doi: 10.1007/s00253-018-8895-7. [DOI] [PubMed] [Google Scholar]
- 16.Soccol CR, Vandenberghe LPS, Rodrigues C. New perspectives for citric acid production and application. Food Technol Biotechnol. 2006;44:141–149. [Google Scholar]
- 17.Hossain AH, Li A, Brickwedde A, Wilms L, Caspers M, Overkamp K, Punt PJ. Rewiring a secondary metabolite pathway towards itaconic acid production in Aspergillus niger. Microb Cell Fact. 2016;15:130. doi: 10.1186/s12934-016-0527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wynn JP, Hamid AA, Midgley M, Ratledge C. Short communication: widespread occurrence of ATP:citrate lyase and carnitine acetyltransferase in filamentous fungi. J Microbiol. 1998;14:145–147. [Google Scholar]
- 19.Hynes MJ, Murray SL. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot Cell. 2010;9:1039–1048. doi: 10.1128/EC.00080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulton C, Ratledge C. Correlation of lipid accumulation in yeasts with possession of ATP: citrate lyase. J Gen Microbiol. 1981;127:169–176. [Google Scholar]
- 21.Weyda I, Sinha M, Sørensen A, Peter S, Ahring BK. Increased production of free fatty acids and triglycerides in Aspergillus carbonarius by metabolic engineering of fatty acid biosynthesis and degradation pathways. JSM Microbiol. 2018;6:1–10. doi: 10.1186/s40168-017-0383-2. [DOI] [Google Scholar]
- 22.Fliksweert MT, van der Zanden L, Janssen WMTM, Yde Steensma H, van Dijken JP, Pronk JT. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3<247::AID-YEA911>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 23.Osmani SA, Scrutton MC. The sub-cellular localisation of pyruvate carboxylase and of some other enzymes in Aspergillus nidulans. Eur J Biochem. 1983;133:551–560. doi: 10.1111/j.1432-1033.1983.tb07499.x. [DOI] [PubMed] [Google Scholar]
- 24.Pfitzner A, Kubicek C, Rohr M. Presence and regulation of ATP:citrate lyase from the citric acid producing fungus Aspergillus niger. Arch Microbiol. 1987;147:88–91. doi: 10.1007/BF00492910. [DOI] [PubMed] [Google Scholar]
- 25.Katz ME, Hynes MJ. Isolation and analysis of the acetate regulatory gene, facB, from Aspergillus nidulans. Mol Cell Biol. 1989;9:5696–5701. doi: 10.1128/MCB.9.12.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Son H, Min K, Lee J, Choi GJ, Kim JC, Lee YW. Mitochondrial carnitine-dependent acetyl coenzyme a transport is required for normal sexual and asexual development of the ascomycete Gibberella zeae. Eukaryot Cell. 2012;11:1143–1153. doi: 10.1128/EC.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes MJ, Murray SL, Andrianopoulos A, Davis MA. Role of carnitine acetyltransferases in acetyl coenzyme A metabolism in Aspergillus nidulans. Eukaryot Cell. 2011;10:547–555. doi: 10.1128/EC.00295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li A, Pfelzer N, Zuijderwijk R, Punt P. Enhanced itaconic acid production in Aspergillus niger using genetic modification and medium optimization. BMC Biotechnol. 2012;12:57. doi: 10.1186/1472-6750-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hevekerl A, Kuenz A, Vorlop KD. Influence of the pH on the itaconic acid production with Aspergillus terreus. Appl Microbiol Biotechnol. 2014;98:6983–6989. doi: 10.1007/s00253-014-5743-2. [DOI] [PubMed] [Google Scholar]
- 30.Mattern IE, van Noort JM, van den Berg P, Archer DB, Roberts IN, van den Hondel CA. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol Gen Genet. 1992;234:332–336. doi: 10.1007/BF00283855. [DOI] [PubMed] [Google Scholar]
- 31.Bond J. More gene manipulations in fungi. Bennett WJ, Lasure LL, editors. London: Academic Press; 1991.
- 32.Li A, Pfelzer N, Zuijderwijk R, Brickwedde A, van Zeijl C, Punt P. Reduced by-product formation and modified oxygen availability improve itaconic acid production in Aspergillus niger. Appl Microbiol Biotechnol. 2013;97:3901–3911. doi: 10.1007/s00253-012-4684-x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW, David W. Molecular cloning : a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 34.d’Enfert C. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr Genet. 1996;30:76–82. doi: 10.1007/s002940050103. [DOI] [PubMed] [Google Scholar]
- 35.Arentshorst M, Ram AFJ, Meyer V. Plant fungal pathogens. In: Bolton MD, Thomma BPH, editors. 2012.
- 36.Bradstreet RB. The Kjeldahl method for organic nitrogen. London: Academic Press; 1965. [Google Scholar]
- 37.Rodriguez S, Denby CM, Van Vu T, Baidoo EEK, Wang G, Keasling JD. ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microb Cell Fact. 2016;15:48. doi: 10.1186/s12934-016-0447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meijer S, Panagiotou G, Olsson L, Nielsen J. Physiological characterization of xylose metabolism in aspergillus niger under oxygen-limited conditions. Biotechnol Bioeng. 2007;98:462–475. doi: 10.1002/bit.21397. [DOI] [PubMed] [Google Scholar]
- 39.Meijer S, Adriaan De Jongh W, Olsson L, Nielsen J. Physiological characterisation of acuB deletion in Aspergillus niger. Appl Microbiol Biotechnol. 2009;84:157–167. doi: 10.1007/s00253-009-2027-3. [DOI] [PubMed] [Google Scholar]
- 40.Ruijter GJG, van de Vondervoort PJI, Visser J. Oxalic acid production by Aspergillus niger: an oxalate-non-producing mutant produces citric acid at ph 5 and in the presence of manganese. Microbiology. 1999;145:2569–2576. doi: 10.1099/00221287-145-9-2569. [DOI] [PubMed] [Google Scholar]
- 41.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 42.Levinson WE, Kurtzman CP, Kuo TM. Production of itaconic acid by Pseudozyma antarctica NRRL Y-7808 under nitrogen-limited growth conditions. Enzym Microb Technol. 2006;39:824–827. doi: 10.1016/j.enzmictec.2006.01.005. [DOI] [Google Scholar]
- 43.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links metabolism to histone acetylation. Science. 2009;324:0–4. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin X, Shin H, Li J, Du G, Liu L, Chen J. Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci Rep. 2017;7:41040. doi: 10.1038/srep41040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hossain AH, Ter Beek A, Punt PJ. Itaconic acid degradation in Aspergillus niger: the role of unexpected bioconversion pathways. Fungal Biol Biotechnol. 2019;6:1. doi: 10.1186/s40694-018-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feehily C, Karatzas KAG. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x. [DOI] [PubMed] [Google Scholar]
- 47.Cotter PD, Ryan S, Gahan CGM, Hill C. Presence of GadD1 glutamate decarboxylase in selected listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Biotechnol. 2005;71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin J, Smith MP, Chapin KC, Suk Baik H, Bennett GN, Foster JW. Mechanisms of acid resistance in Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterman SR, Small P. Identification of dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 53.Kubicek CP, Hampel W. Manganese deficiency leads to elevated amino acid pools in citric acid accumulating Aspergillus niger. Arch Microbiol. 1979;79:73–79. doi: 10.1007/BF00403504. [DOI] [PubMed] [Google Scholar]
- 54.Chen M, Huang X, Zhong C, Li J, Lu X. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus. Appl Microbiol Biotechnol. 2016;100:7541–7548. doi: 10.1007/s00253-016-7554-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Design of acl1 and acl2 expression cassette. Figure S2. Shake flask cultivation of CitB#99 ACL G1 Z. Table S1. Relative copy numbers of introduced acl12 and IA titers achieved in shake flask cultivations. Figure S3. Southern-Blot results of selected CitB#99-acl transformants. Table S2. Carbon balance of 10L fed-batch bioreactor cultivation of ACL G1 Z. Figure S4. Dissolved oxygen profile and pH profile during 10 L controlled fed-batch cultivation of strain CitB#99-ACL G1 Z. Figure S5. Total nitrogen concentration during 10 L controlled fed-batch cultivation of ACL G1 Z. Figure S6. Repeat fed-batch cultivation of ACL G1 Z performed in 5 L BioFlo 320 (Eppendorf) controlled bioreactors. Table S3. Carbon balance of repeat fed-batch 5 L bioreactor cultivation of ACL G1 Z.
Data Availability Statement
Transcriptome data will be uploaded on GEO.