Abstract
Background:
To identify the hub genes related to urothelial carcinoma of the bladder prognosis and to understand their underlying mechanism.
Methods:
The expression profiles of 18 pairs of urothelial carcinoma of the bladder patient tissue and paired adjacent tissue obtained from the Cancer Genome Atlas were performed. Weighted gene coexpression network analysis was employed to screen gene modules and hub genes with significant differential expressions in urothelial carcinoma of the bladder. The hub genes expression in urothelial carcinoma of the bladder tissues was validated by reverse transcription-quantitative polymerase chain reaction. The overall survival curve and disease-free survival curve of prognostic factor (LGALS4) were plotted using the Kaplan-Meier method. Furthermore, LGALS4 messenger RNA and protein expression were also assessed in 2 urothelial carcinoma of the bladder cell lines (T24 and 5637) by quantitative reverse transcription–polymerase chain reaction and Western blot. The functions of urothelial carcinoma of the bladder cells with transfected pcDNA3.1-LGALS4 were identified through MTT assay, plate clone formation assay, flow cytometry, and cell migration experiments.
Results:
LGALS4 was the hub gene of pink module and it was related to prognosis. Higher LGALS4 expression predicted higher probabilities of overall survival and disease-free survival. Overexpression of LGALS4 in urothelial carcinoma of the bladder cells suppressed cell viability and migration but induced apoptosis.
Conclusion:
LGALS4 played a critical role in the progression of urothelial carcinoma of the bladder and held a promise to be the biomarker for diagnosis and treatment of urothelial carcinoma of the bladder. It predicted good prognosis of urothelial carcinoma of the bladder and restrained the growth and migration of urothelial carcinoma of the bladder cells.
Keywords: urothelial carcinoma of bladder, WGCNA, prognosis, hub gene, LGALS4
Introduction
Bladder cancer is any of the several types of cancer arising from the tissues of the urinary bladder. The morbidity of bladder cancer is relatively high among common malignancies.1 By the end of 2015, bladder cancer had affected 3.4 million people and increases by 430 000 new cases each year.2 In addition, men are more prone to suffer from bladder cancer than women. It is reported that, in the United States, the number of new confirmed cases in 2017 is 79 030 with male accounting for 76.5% and female 23.5%; the death toll is about 16 870 including 12 240 men and 4630 women.3 Urothelial carcinoma of the bladder (UCB) accounts for more than 90% of bladder tumor.1 Therefore, as the main type of bladder cancer, UCB has become a huge threat to public health. Though UCB mortality rate (15%-21%) is not much higher than that of the other cancers, the recurrence rate is fairly high. Patients suffered from bladder cancer stand over 50% chances to relapse in 2 years following the procedure.4 In summary, finding effective therapeutic targets for UCB treatment is highly needed.
LGALS4, also known as galectin-4, belongs to galectins family of beta-galactoside-binding proteins which implicated in modulating cell–cell matrix interactions.5 Galectins family consists of 3 different subfamilies that are divided by the composition and recognition of the conserved carbohydrate-recognition domain (CRD). Galectin-4, 6, 8, 9, and 12 are similar galectins genes as they contain 2 CRDs. Those galectins widely present in various types of human cells and are involved in several cellular functions such as cell proliferation, apoptosis, signaling transduction, adhesion, immune response, and so on.6 For example, galectins could regulate cell death extracellularly7,8 and exert both suppressive and activation effect on integrin-mediated adhesion.9,10
When it comes to the influence of LGALS4 on human cancers, it was considered as a prognostic biomarker in hepatocellular carcinoma as low LGALS4 expression predicted more aggressive characteristics of hepatocellular cancer.11 It was also shown to exhibit a tumor-suppressive effect in colorectal carcinoma12 and pancreatic adenocarcinoma.13 In urothelial carcinoma, LGALS4 could inhibit cancer cell growth and invasiveness, and the promoter hypermethylation of LGALS4 was positively related to the inferior survival of patients.14 Previous studies indicated that LGALS4 might be an effective suppressor in various cancers. However, its function in UCB remains unclear.
Gene expression profile is an effective technique to analyze thousands of gene variations in different samples at the same time. Through bioinformatics analysis, Lu et al found that LGALS4 exhibited significant expression change in bladder cancer.15 A weighted gene coexpression network analysis (WGCNA) is a comparatively new method for gene expression analysis in bioinformatics, with the advantage of more convincing and meaningful outcomes. Zhang et al identified 5 hub genes through coexpression network analysis in bladder cancer, including LGALS4.16 The hub genes are the nodes with relatively high relationship with others in a network, thus their alterations are of top importance as a slight change in them may affect the situation as a whole. In addition to the bioinformatics analysis, survival analysis also helps researchers understand correlation between genes and diseases better. It integrates patients’ survival time, final state, gene expression, and so on to calculate the survival rate under different conditions.
Therefore, in this study, we performed WGCNA to discover the hub genes in UCB and investigated the association between the hub gene expression and the survival of patients. LGALS4 was identified as a hub gene in UCB, which not only predicted good prognosis but also restrained in vitro cell viability and mobility of UCB. To sum up, we considered LGALS4 could act as an effective therapeutic target and benefit patients suffered from UCB.
Methods and Materials
Differential Gene Expression Analysis
A total of 412 samples (412 UCB tissues and 18 adjacent normal tissues) from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/) were obtained for data mining. Among them, 18 pairs of UCB and matched adjacent normal samples were filtered for further differential analysis. The information of patient characteristics from TCGA database has been attached as supplemental material. In detail, crucial clinicopathological characteristics of 412 patients with UCB are gathered as shown in Table S1. The R software package DESeq2 was used to acquire the messenger RNA (mRNA) expression matrix and converted data into a log2 scale. Afterward, “baseMean,” “log2FoldChange,” “lfcSE,” “stat,” “P value,” and “adjusted P value” of the normal and tumor group were computed. Among them, P value was calculated by “Wald test” and adjusted using “BH” method. Genes with |log2FoldChange| >1 and adjusted P value <.05 were considered as differential expressed genes (DEGs).
WGCNA of UCB Tissues
The WGCNA was performed by package of R software.17 A weighted adjacency matrix in tumor group that provided continuous connection strength between 0 and 1 was established based on tumor group’s soft threshold β parameter. The parameter β was set 14 to satisfy network scale-free topology feature and ensure the high mean of genes adjacency functions simultaneously. Meanwhile, we built the coexpression matrix as well as topological overlap matrix.18,19 Gene module represents a collection of genes with high topological similarity.20 The connectivity between module and inside genes could be assessed using R software. The importance of the gene in a module could be reflected by module membership (MM), which refers to the correlation between the individual gene expression and the module Eigen gene (the expression pattern for a module).21 To identify unique module in tumor group, a Z score calculated by (Zdensity + Zconnectivity)/2 was used to calculate the preservation of modules between 2 groups. Modules with Z < 10 were regarded to be altered in tumor groups.22
Hub Genes and Prognostic Factors Identification
Hub genes are highly connected to their modules and play an important role in regulating the whole module. Empirically, hub genes are generally located in the middle of the network and usually of high correlation with other genes in the network. As all genes input were DEGs, the gene significance (correlation of individual gene expression with UCB) was high; we only took MM into consideration. By plotting the gene module network in Cytoscape software, we could easily locate the hub gene. Subsequently, hub genes were analyzed through Kaplan–Meier method based on gene expression data and clinical information from 412 samples (TCGA database, https://tcga-data.nci.nih.gov/tcga/), and of them prognostic factors were selected to generate overall survival (OS) and disease-free survival (DFS) curves. Kaplan-Meier plotter was implemented by R package “survival” with Log-rank (Mantel–Haenszel) tests, and the median value of LGALS4 expression was used as the cutoff.
Cell Lines and Cell Culture
Human bladder cancer cell lines (5637 and T24) and normal human ureteral epithelial cell line (SV-HUC-1) were purchased from BeNa Culture Collection (Beijing, China). Cell lines (5637 and T24) were maintained in ATCC-formulated RPMI-1640 Medium (Manassas, Virginia) with 10% of fetal bovine serum (FBS, Invitrogen, California), while SV-HUC-1 cells were maintained in 10% FBS and 90% high glucose Dulbecco’s Modified Eagle Medium (Invitrogen).
Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted by TRIzol® reagent (Invitrogen, Carlsbad, California, USA) and PureLink RNA Mini Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and reversely transcripted into complementary DNA by TIANScript II RT Kit (Tiangen, Beijing, China). Quantitative reverse transcription-polymerase chain reaction was performed by RealMasterMix SYBR Green Kit (Tiangen) and ABI7500 Applied Biosystems (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The relative mRNA expression was calculated by 2−△△CT method, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control. The primers were obtained from Sangon Biotech (Shanghai, China).
Cell Transfection
Urothelial carcinoma of the bladder cells were transfected with pcDNA3.1-LGALS4 constructed by LGALS4 and pcDNA3.1 (Thermo Fisher Scientific) by using Lipofectamine 3000 (Invitrogen) under instructions. Cells were cultured in 6-well plates for 24 hours before transfection. After being transfected with 0.5 μg pcDNA3.1-LGALS4 in each well, cells were further incubated for 48 hours before collection.
Western Blot
After being extracted radio immunoprecipitation assay (RIPA) lysis buffer (Solarbio, Shanghai, China) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins of UCB cells were transferred to 0.22 μM polyvinylidene fluoride membrane (Millipore, Billerica, Massachusetts, USA). The bovine serum albumin was used to block the membrane for 1 hour. The membrane was then incubated with rabbit anti-GAL4 (1:1000, ab229347) at 4°C overnight, washed by Tris-buffered saline with Tween 20 (TBS, 1 mL/L Tween-20), then incubated with horseradish peroxidase-labeled anti-rabbit IgG (1:2500, ab205718) at room temperature for 2 hours. The protein bands were visualized by ECL Western Blotting Detection Kit (GE Healthcare, Amersham, United Kingdom) and quantified by Image Lab (Bio-Rad, Hercules, California, USA). Rabbit anti-GAPDH (1:2500, ab9485) was the internal control. The antibodies were all obtained from Abcam (Cambridge, Massachusetts, USA).
MTT Assay
Cells were cultured in 96-well plates (3000 cells/100 μL/well) for 2 days before MTT assay. After incubation for 24, 48, and 72 hours, 10 μL MTT solution (5 mg/mL, pH = 7.4) was injected into each well to incubate the cells for 4 hours. Then the solution was replaced with 100 μL dimethyl sulfoxide to dissolve fonnazan crystals. A microplate reader was applied to measure the absorbance value at 490 nm.
Colony Formation Assay
The transfected UCB cells were incubated in 6-well plates (1000 cells/100 μL/well) for 2 to 3 weeks. Then the original culture medium was substituted for a fresh one. The cell clones were stained with 0.4% of crystal violet after being fixed with 10% methanol and 10% acetic acid. The ColCounte colony counter (Oxford Optronix Ltd, Abingdon, United Kingdom) was applied to count the stained clones.
Cell Apoptosis Detection
After being digested and resuspended by trypsin and binding buffer, respectively, the cells were collected at the density of 2 × 106 cells/mL. The cell suspension (100 μL) was stained with 5 μL Annexin V-APC and 5 μL propidium iodide (MULTI SCIENCES, Hangzhou, China) for 15 minutes. The flow cytometer FACS Calibur and FACS Diva from Becton Dickinson (Franklin lake, New Jersey, USA) were applied to detect the apoptotic cells and analyze the data, respectively.
Transwell Assay
The migration of UCB cells was investigated by the transwell (Millipore, Billerica, Massachusetts, USA) with 8-µm pore diameter. The upper chamber was placed with the transfected cells, while the lower one was filled with culture medium and 10% of FBS. After 24 hours, the remaining cells in the upper chamber were removed and the migrated cells in the lower chamber were fixed by methanol and stained by 0.1% crystal violet. The stained cells were observed by an inverted microscope.
Statistical Analysis
Data in this study were analyzed by GraphPad Prism 6.0 and expressed as mean ± standard deviation. The differences in data between 2 or multiple groups were analyzed by Student t test or one-way analysis of variance. The rank test was applied to analyze the heterogeneity of variance. All experiments were repeated for 3 times. Value of P < .05 was regarded as statistically significant.
Results
Weighted Gene Coexpression Networks of DEGs
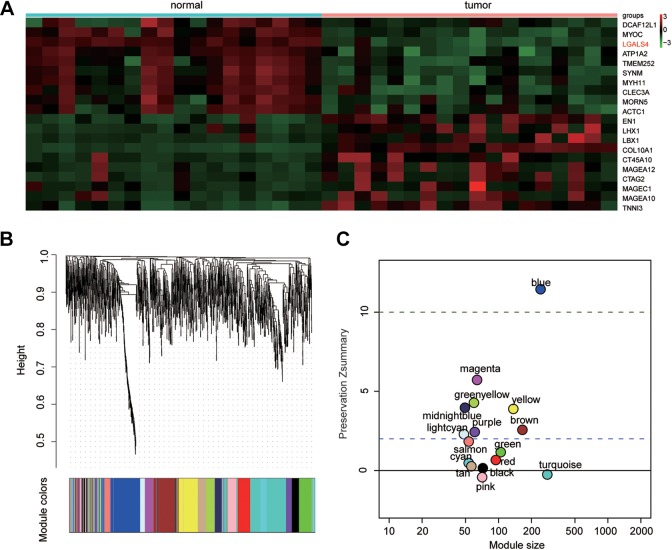
Totally, 1591 DEGs were detected dysregulated in UCB, including 564 overexpressed and 1027 lowexpressed genes. The heat map in Figure 1A delineated top 10 up- and downregulated genes in UCB. Those DEGs were then processed by WGCNA package to divide coexpression modules. A total of 17 modules were constructed in Figure 1B. Genes outside any other modules were finally collected in gray module, which would be discarded. Blue module was ruled out since its Z score exceeded 10, meaning there was little difference between UCB and normal group (Figure 1C).
Figure 1.
The differentially expressed gene expression analysis and the weighted gene coexpression network analysis modules of urothelial carcinoma of the bladder (UCB) tissues. A, Twenty most up- and downregulated messenger RNAs in UCB tissues were screened out by hierarchical clustering analysis. Totally, 1591 genes were identified dysregulated in tumor group compared with normal one. B, The gene coexpression networks of the tumor group showed that 17 modules were identified. The horizontal color bar represents different modules. C, Composite preservation statistics of the normal group and tumor group indicated that 15 out of the 17 modules were significantly nonconservative between the tumor and normal groups (gray module was excluded as it was a collection of genes not included in any other module). Colored points indicated different modules. Horizontal lines showed the thresholds of Zsummary (y-axis) = 2 and Zsummary (y-axis) = 10. Zsummary ≤ 10 represented poor preservation of the modules between the 2 groups.
Hub Gene LGALS4 Overexpression Predicted Better Prognosis of UCB
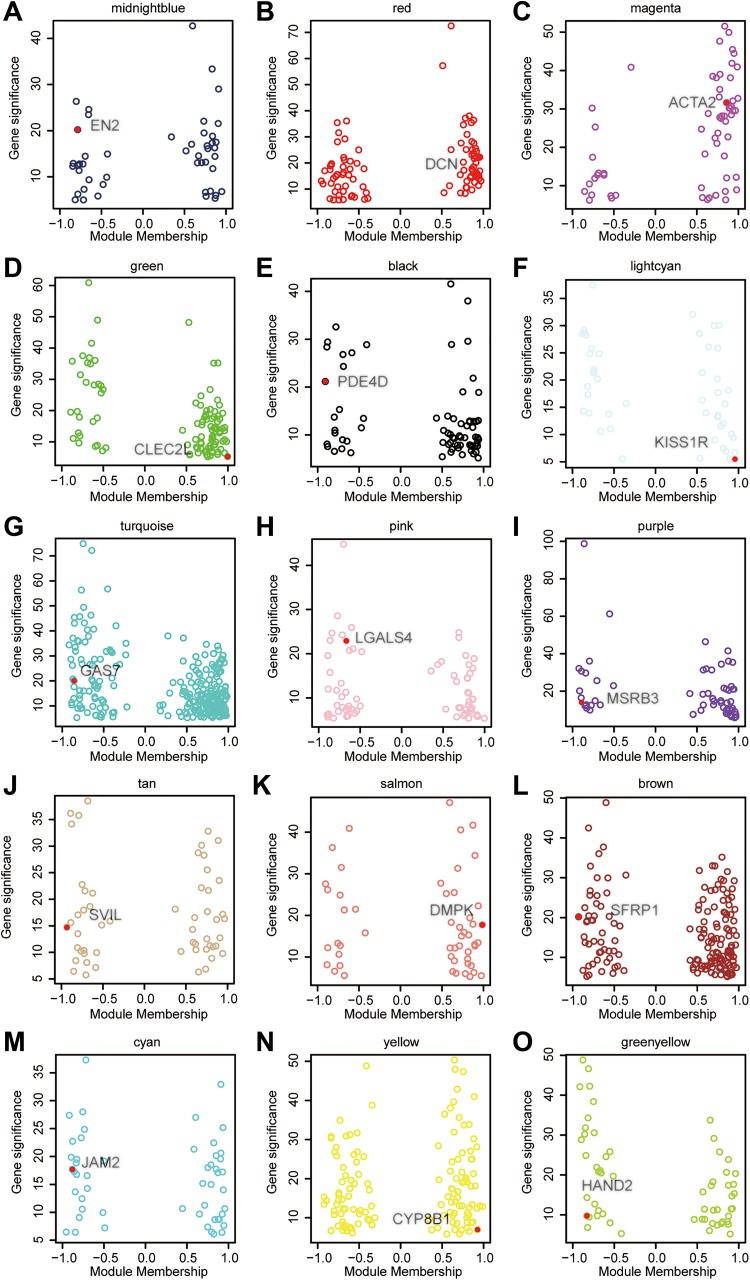
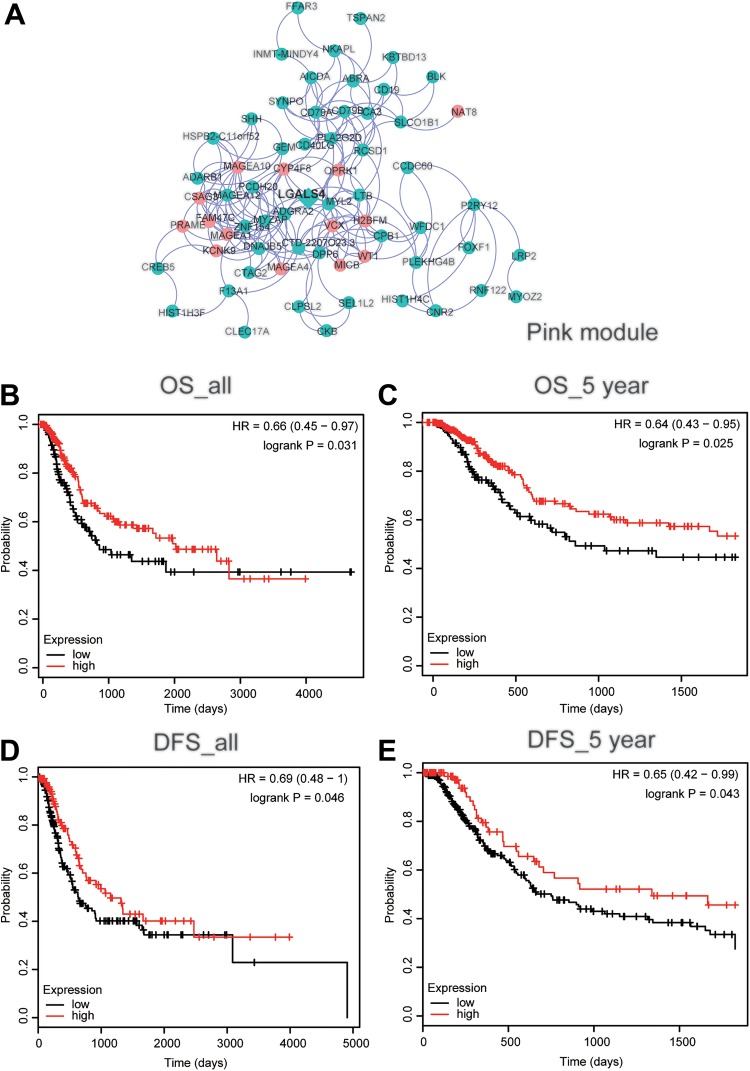
Using Cytoscape software, we drew the 15 left modules’ networks and marked their hub genes in Figure 2: EN2 (midnight blue), DCN (red), ACTA2 (magenta), CLEC2L (green), PDE4D (black), KISS1R (lightcyan), GAS7 (turquoise), LGALS4 (pink, Figure 3A), MSRB3 (purple), SVIL (tan), DMPK (salmon), SFRP1 (brown), JAM2 (cyan), CYP8B1 (yellow), and HAND2 (green-yellow). In the prognostic analysis, LGALS4 expression showed a huge correlation with patients’ survival. As shown in Figure 3B-C, the expression level of LGALS4 was positively related to OS and 5-year OS (P < .05). Moreover, higher LGALS4 expression also significantly improved the overall DFS and 5-year DFS of UCB patients (Figure 3D-E, P < .05).
Figure 2.
Correlation of the module membership (x-axis) and the gene significance (y-axis). The colored circles denoted genes in a module. The red dot indicated the hub gene of each module.
Figure 3.
LGALS4 overexpression predicted better prognosis of urothelial carcinoma of the bladder (UCB). (A) Interaction of gene weighted coexpression patterns in modules. For clarity, we set restricted threshold on the topological overlap matrix parameter in large modules to cut down the number of nodes and edges. The network was visualized using Cytoscape 3.5.1 software. The green and red nodes indicated downregulated and upregulated genes, respectively. The diamond denoted the hub genes in the pink modules. B-E, Kaplan–Meier’s survival curves depicted the prognostic significance of LGALS4 expression for UCB patients. (B) Overall survival (OS), (C) 5-year OS, (D) disease-free survival (DFS), and (E) 5-year DFS.
LGALS4 Overexpression Inhibited the Growth and Migration of UCB Cells
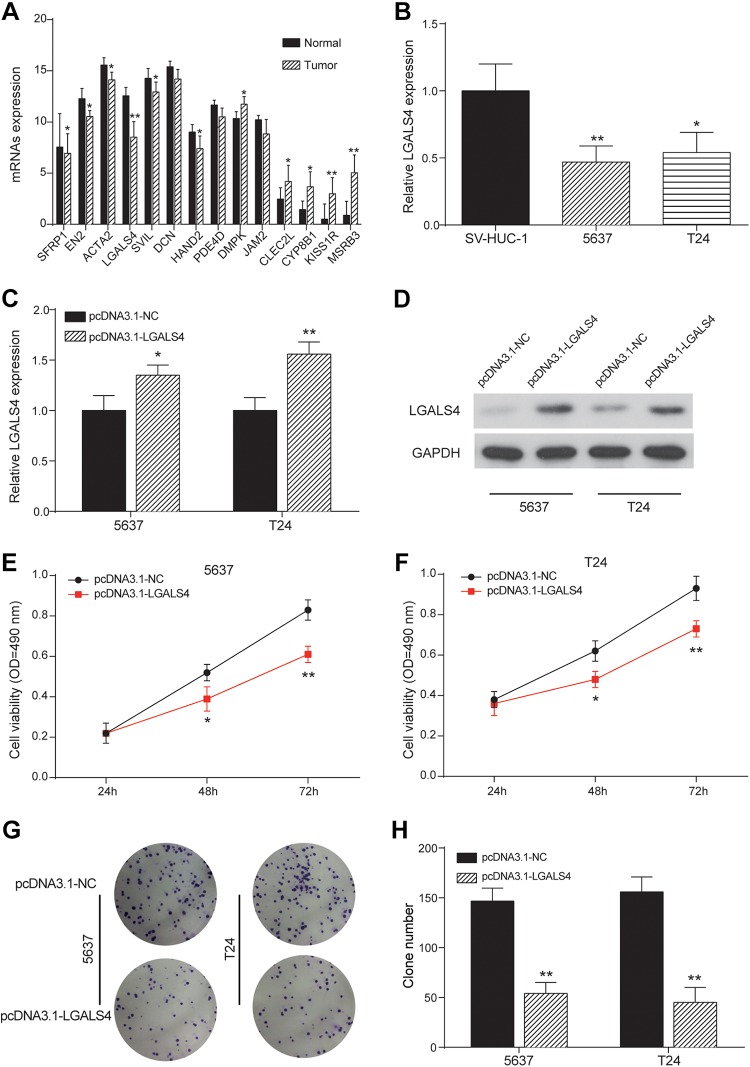
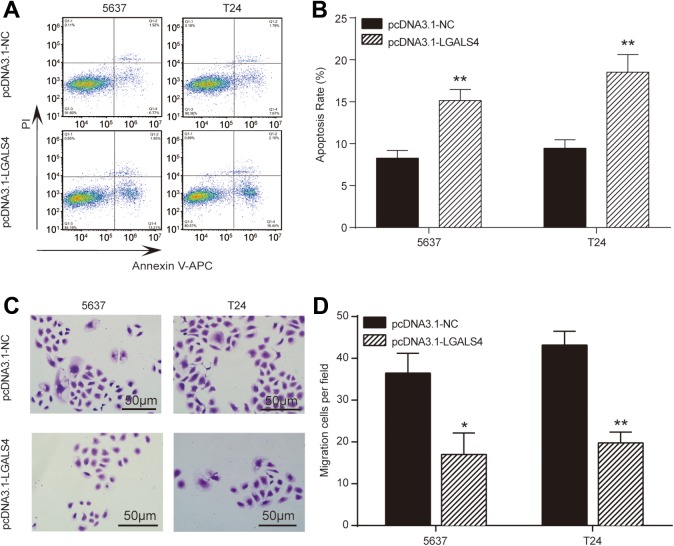
The significant downregulation of LGALS4 was found in UCB tissues (Figure 4A), which was also validated in cells experiment (Figure 4B). Moreover, we constructed 5637 and T24 cancer cells with overexpressed LGALS4 by introducing a vector containing LGALS4 cDNA (5637-LGALS4, T24-LGALS4). Then we assessed the effects of LGALS4 overexpression on proliferation, apoptosis, and migration to detect whether LGALS4 affects cells behavior. Vectors without the insert were transfected into cancer cells as controls (5637-NC and T24-NC). The results demonstrated that LGALS4 mRNA and protein was ectopically overexpressed in 5637-LGALS4 and T24-LGALS4 cell lines (Figure 4C-D). Cell viability analysis of these UCB cell lines indicated that 5637-LGALS4 and T24-LGALS4 cells exhibited a significant decline in cell proliferation compared to NC groups (Figure 4E-F). Meanwhile, colony formation of UCB cells with LGALS4 overexpression was also distinctively restrained (Figure 4G-H). Afterward, the UCB cells of each group were taken for detecting apoptosis by the flow cytometry. Figure 5A-B showed that the apoptosis of 5637 and T24 cells was remarkably enhanced after overexpression of LGALS4 as apoptogenic factor in pcDNA3.1-LGALS4 groups doubled compared to NC groups. Finally, using the transwell chamber assay, we examined whether expressed LGALS4 influenced the migration properties of UCB cells. Figure 5C-D manifested that 5637-LGALS4 and T24-LGALS4 displayed a significant reduction in their migration ability compared to NC groups. These results indicated that LGALS4 restricted the proliferation and migration abilities and activated apoptosis capability of UCB cells.
Figure 4.
LGALS4 overexpression suppressed proliferation of urothelial carcinoma of the bladder (UCB) cells. A, Expression pattern of hub genes in normal and tumor cells detected by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). LGALS4 was significantly downregulated in tumor group. *P < .05, **P < .01 compared with normal group. B, The relative expressions of LGALS4 detected by qRT-PCR in 2 bladder cancer cell lines 5637 and T24 were lower than that in normal cell line SV-HUC-1. *P < .05, **P < .01 compared with SV-HUC-1. C and D, The relative LGALS4 expression detected by qRT-PCR and protein expression detected by Western blot in 5637 and T24 cells were increased in pcDNA3.1-LGALS4 group. *P < .05, **P < .01 compared with pcDNA3.1-NC group. E and F, The cell viability of 5637 and T24 cells detected by MTT assay were decreased in pcDNA3.1-LGALS4 group. *P < .05, **P < .01 compared with pcDNA3.1-NC group. OD: optical density. G and H, The clone numbers of 5637 and T24 cells detected by colony formation assay were decreased in pcDNA3.1-LGALS4 group. **P < .01 compared with pcDNA3.1-NC group.
Figure 5.
LGALS4 overexpression induced apoptosis and suppressed migration of urothelial carcinoma of the bladder cells. A and B, The apoptosis rate of 5637 and T24 cells detected by flow cytometry were significantly increased in pcDNA3.1-LGALS4 group. C and D, The number of migrated 5637 and T24 cells detected by transwell assay was decreased in pcDNA3.1-LGALS4 group. *P < .05, **P < .01 compared with pcCDNA3.1-NC group.
Discussion
Based on the differentially expressed genes in UCB, we performed WGCNA and identified 15 modules with significant differences between UCB and normal tissues. Hub gene LGALS4 showed a strong correlation with the OS and DFS of UCB patients. High expression of LGALS4 predicted higher survival probability and better prognosis. In vitro experiments confirmed that LGALS4 could restrain the proliferation and migration of UCB cells and induce apoptosis.
Here, we totally screened out 1591 DEGs and constructed 15 modules which were significantly nonconservative and exhibited distinctive differences between UCB and normal samples. LGALS4 was the hub gene of the pink module. WGCNA is a powerful means to identify the hub genes that play vital roles in human cancers. To investigate the etiological agent of bladder cancer, Deng et al analyzed the differential coexpression networks and found that the DEGs in bladder cancer mainly involved in cellular physiological process and cellular metabolism.23 A complex expression pattern of the galectin network was revealed in urothelial carcinomas, and galectin-1, -2, -3 and -8 were considered as potential disease markers and possible targets.24 The expression change of LGALS4 was uncovered in the rodent model of bladder cancer and it was identified as upregulated.15 Meanwhile, through differential coexpression networks, LGALS4 was also suggested as one of the top hub genes with upregulation in bladder cancer.16 However, these results were not consistent with the network analysis performed in our study, in which LGALS4 was also the hub gene but downregulated in UCB. The probable reason for this might be the variations of samples as those used in this study were human UCB tissues, while those in previous studies were bladder tumor tissues of rats15 and human bladder cancer tissues.16 The results of bioinformatics analysis indicated that LGALS4 was of crucial importance in the progression of UCB, whereas its specific functions need to be verified by clinical analysis and laboratory experiments.
Therefore, we analyzed the survival situations of UCB patients and found that those with high expression of LGALS4 exhibited higher probabilities of OS and DFS in both 5-year and overall situations. The survival probability of patients is a meaning indicator which helps to analyze the outcomes of malignancies. Li et al applied WGCNA to DEGs of bladder cancer and identified 17 hub genes as candidate biomarkers related to the OS of bladder cancer patients.25 Yan et al pointed out that the overexpression of bromodomain 4 protein was a predictor of worse survival for UCB.26 Argonaute 2 protein was also positively correlated with the poorer OS of UCB.1 Other biomarkers that could predict poor prognosis of UCB included high expression of Fascin27 and Cdc25B28 and downregulation of miR-133b.29 However, the association between LGALS4 expression and patient survival of UCB has not been investigated before. We revealed the positive relationship between LGALS4 downregulation and poor OS and DFS of UCB for the first time. Based on this result, LGALS4 could be considered as a potential biomarker whose high expression predicts better prognosis of UCB.
Besides the clinical analysis, we also performed in vitro experiments to verify LGALS4’s function in UCB. Overexpression of LGALS4 successfully suppressed proliferation and migration but promoted apoptosis of UCB cells. Most studies suggested that LGALS4 could slow down the deterioration of cancers, especially for retarding metastasis, which was in line with our study. In colorectal cancer, LGALS4 was significantly downregulated and the abrogation of LGALS4 expression could promote the tumorigenesis of colorectal cancer.30 Forced expression of LGALS4 induced cell cycle arrest and retarded cell motility through the control of the Wnt signaling pathway in colorectal cancer.12 The modulation of Wnt/β-catenin signaling by LGALS4 through reducing the activation of Wnt target genes was also found in pancreatic adenocarcinoma.31 High LGALS4 expression could reduce the migration and metastasis of pancreatic cancer.13,31 The mobility of hepatocellular carcinoma cells were also significantly reduced through overexpression of LGALS4.11 Although the in vitro studies on LGALS4’s effect in UCB are limited, the low expression of LGALS4 and its suppressive effect in UCB was consistent with the result of WGCNA so that we could conclude that LGALS4 was an anti-oncogene of UCB.
Through WGCNA and in vitro experiments, we identified the suppressive effect of LGALS4 in UCB. However, the limitations in this study need to be thought over. For instance, in vivo experiments were not conducted here, and only in vitro experiments were not sufficient to verify the functions of LGALS4 in UCB. In addition, some studies showed that LGALS4 could regulate the Wnt signaling pathway in other types of cancers. Therefore, related signaling pathways in UCB could be studied in future to reveal the underlying regulatory mechanisms of LGALS4.
Conclusions
LGALS4 was the hub gene identified by WGCNA in UCB. High expression of LGALS4 predicted high probabilities of OS and DFS of UCB patients. Overexpression of LGALS4 suppressed viability and mobility of UCB cells and induced apoptosis. LGALS4 was a tumor-suppressive gene in UCB and might be a potential biomarker and target in UCB diagnosis and treatment.
Supplemental Material
Supplemental Material, Table_S1 for LGALS4 as a Prognostic Factor in Urothelial Carcinoma of Bladder Affects Cell Functions by Yu Ding, Qifeng Cao, Chen Wang, Huangqi Duan and Haibo Shen in Technology in Cancer Research & Treatment
Abbreviations
- CRD
carbohydrate-recognition domain
- DEG
differential expressed gene
- DFS
disease-free survival
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- MM
module membership
- mRNA
messenger RNA
- OS
overall survival
- TCGA
The Cancer Genome Atlas
- UCB
urothelial carcinoma of the bladder
- WGCNA
weighted gene coexpression network analysis
Footnotes
Authors’ Note: Yu Ding and Qifeng Cao contributed equally to this work. The study did not involve any ethical issues. The crucial clinicopathological characteristics of 412 patients shown in table S1 comes from TCGA (https://portal.gdc.cancer.gov/).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Haibo Shen  https://orcid.org/0000-0002-5302-6349
https://orcid.org/0000-0002-5302-6349
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Yang FQ, Huang JH, Liu M, et al. Argonaute 2 is up-regulated in tissues of urothelial carcinoma of bladder. Int J Clin Exp Pathol. 2014;7(1):340–347. [PMC free article] [PubMed] [Google Scholar]
- 2. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 5. Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572(2-3):209–231. [DOI] [PubMed] [Google Scholar]
- 6. Barrow H, Rhodes JM, Yu LG. Simultaneous determination of serum galectin-3 and -4 levels detects metastases in colorectal cancer patients. Cell Oncol (Dordr). 2013;36(1):9–13. [DOI] [PubMed] [Google Scholar]
- 7. Yang RY, Rabinovich GA, Liu FT. Galectins: structure, function and therapeutic potential. Expert Rev Mol Med. 2008;10:e17. [DOI] [PubMed] [Google Scholar]
- 8. Hernandez JD, Baum LG. Ah, sweet mystery of death! Galectins and control of cell fate. Glycobiology. 2002;12(10):127R–136R. [DOI] [PubMed] [Google Scholar]
- 9. Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482(7385):414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zick Y, Eisenstein M, Goren RA, Hadari YR, Levy Y, Ronen D. Role of galectin-8 as a modulator of cell adhesion and cell growth. Glycoconj J. 2002;19(7-9):517–526. [DOI] [PubMed] [Google Scholar]
- 11. Cai Z, Zeng Y, Xu B, et al. Galectin-4 serves as a prognostic biomarker for the early recurrence/metastasis of hepatocellular carcinoma. Cancer Sci. 2014;105(11):1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satelli A, Rao PS, Thirumala S, Rao US. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int J Cancer. 2011;129(4):799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belo AI, van der Sar AM, Tefsen B, van Die I. Galectin-4 reduces migration and metastasis formation of pancreatic cancer cells. PLoS One. 2013;8(6):e65957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu MM, Li CF, Lin LF, et al. Promoter hypermethylation of LGALS4 correlates with poor prognosis in patients with urothelial carcinoma. Oncotarget. 2017;8(14):23787–23802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu Y, Liu P, Wen W, et al. Cross-species comparison of orthologous gene expression in human bladder cancer and carcinogen-induced rodent models. Am J Transl Res. 2010;3(1):8–27. [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang DQ, Zhou CK, Chen SZ, Yang Y, Shi BK. Identification of hub genes and pathways associated with bladder cancer based on co-expression network analysis. Oncol Lett. 2017;14(1):1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yip AM, Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics. 2007;8(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao H, Cai W, Su S, Zhi D, Lu J, Liu S. Screening genes crucial for pediatric pilocytic astrocytoma using weighted gene coexpression network analysis combined with methylation data analysis. Cancer Gene Ther. 2014;21(10):448–455. [DOI] [PubMed] [Google Scholar]
- 19. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):1–43. [DOI] [PubMed] [Google Scholar]
- 20. Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24(5):719–720. [DOI] [PubMed] [Google Scholar]
- 21. Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1(1): 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng SP, Zhu L, Huang DS. Mining the bladder cancer-associated genes by an integrated strategy for the construction and analysis of differential co-expression networks. BMC Genomics. 2015;16(suppl 3):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langbein S, Brade J, Badawi JK, et al. Gene-expression signature of adhesion/growth-regulatory tissue lectins (galectins) in transitional cell cancer and its prognostic relevance. Histopathology. 2007;51(5):681–690. [DOI] [PubMed] [Google Scholar]
- 25. Li S, Liu X, Liu T, et al. Identification of biomarkers correlated with the TNM staging and overall survival of patients with bladder cancer. Front Physiol. 2017;8:947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan Y, Yang FQ, Zhang H, et al. Bromodomain 4 protein is a predictor of survival for urothelial carcinoma of bladder. Int J Clin Exp Pathol. 2014;7(7):4231–4238. [PMC free article] [PubMed] [Google Scholar]
- 27. Bi J, Chen X, Zhang Y, et al. Fascin is a predictor for invasiveness and recurrence of urothelial carcinoma of bladder. Urol Oncol. 2012;30(5):688–694. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z, Zhang G, Kong C. High expression of Cdc25B and low expression of 14-3-3sigma is associated with the development and poor prognosis in urothelial carcinoma of bladder. Tumour Biol. 2014;35(3):2503–2512. [DOI] [PubMed] [Google Scholar]
- 29. Chen X, Wu B, Xu Z, et al. Downregulation of miR-133b predict progression and poor prognosis in patients with urothelial carcinoma of bladder. Cancer Med. 2016;5(8):1856–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim SW, Park KC, Jeon SM, et al. Abrogation of galectin-4 expression promotes tumorigenesis in colorectal cancer. Cell Oncol (Dordr). 2013;36(2):169–178. [DOI] [PubMed] [Google Scholar]
- 31. Maftouh M, Belo AI, Avan A, et al. Galectin-4 expression is associated with reduced lymph node metastasis and modulation of Wnt/beta-catenin signalling in pancreatic adenocarcinoma. Oncotarget. 2014;5(14):5335–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Table_S1 for LGALS4 as a Prognostic Factor in Urothelial Carcinoma of Bladder Affects Cell Functions by Yu Ding, Qifeng Cao, Chen Wang, Huangqi Duan and Haibo Shen in Technology in Cancer Research & Treatment