Abstract
Purpose:
A relevant challenge for the improvement of clear cell renal cell carcinoma management could derive from the identification of novel molecular biomarkers that could greatly improve the diagnosis, prognosis, and treatment choice of these neoplasms. In this study, we investigate whether quantitative parameters obtained from computed tomography texture analysis may correlate with the expression of selected oncogenic microRNAs.
Methods:
In a retrospective single-center study, multiphasic computed tomography examination (with arterial, portal, and urographic phases) was performed on 20 patients with clear cell renal cell carcinoma and computed tomography texture analysis parameters such as entropy, kurtosis, skewness, mean, and standard deviation of pixel distribution were measured using multiple filter settings. These quantitative data were correlated with the expression of selected microRNAs (miR-21-5p, miR-210-3p, miR-185-5p, miR-221-3p, miR-145-5p). Both the evaluations (microRNAs and computed tomography texture analysis) were performed on matched tumor and normal corticomedullar tissues of the same patients cohort.
Results:
In this pilot study, we evidenced that computed tomography texture analysis has robust parameters (eg, entropy, mean, standard deviation) to distinguish normal from pathological tissues. Moreover, a higher coefficient of determination between entropy and miR-21-5p expression was evidenced in tumor versus normal tissue. Interestingly, entropy and miR-21-5p show promising correlation in clear cell renal cell carcinoma opening to a radiogenomic strategy to improve clear cell renal cell carcinoma management.
Conclusion:
In this pilot study, a promising correlation between microRNAs and computed tomography texture analysis has been found in clear cell renal cell carcinoma. A clear cell renal cell carcinoma can benefit from noninvasive evaluation of texture parameters in adjunction to biopsy results. In particular, a promising correlation between entropy and miR-21-5p was found.
Keywords: texture analysis, microRNA, renal cell carcinoma, radiogenomics
Introduction
Clear cell renal cell carcinoma (ccRCC) is a group of chemotherapy-resistant tumors representing 2% to 3% of all adult malignancies and is the third most common urological cancer after prostate and bladder cancer. There is a wide prevalence of clear cell histotype (ccRCC, 80%-90% of all RCCs), which has, also, the highest mortality rate.1,2 Deeper molecular characterization could improve ccRCC diagnosis and management, as well as prognosis and treatment choice. In this scenario, microRNAs (miRNAs) are emerging as interesting biomarkers for several tumors.3,4 They are small noncoding RNAs that have an important role in the regulation of carcinogenesis. The miRNAs expression is indeed deregulated in neoplastic tissue compared with corresponding normal tissue. MicroRNAs may regulate crucial break points during carcinogenesis.5,6 Recently, they have been addressed as part of ccRCC tumorigenesis and progression,7,8 so that a “miRNA signature” in ccRCC has been described and significantly correlated with patients’ outcome.9,10 miR-21 and miR-210, as well as miR-185 and miR-221, showed functional relevance for ccRCC tumorigenesis.11,12
The grade of expression of miRNAs is cancer- and tissue-specific; for this reason, the expression profile of miRNAs in ccRCC can be helpful to differentiate healthy from pathological tissue, to identify slightly differentiated cancers that would otherwise be undetermined with the use of conventional histology and immunohistochemistry, and, lastly, to recognize tumors with different histotypes within the same organ.9 Further, we tried to understand if ccRCC-associated miRNAs had a corresponding phenotype in radiological examinations (radiophenotype), looking for a connection between genotype and radiophenotype.
Radiogenomics refers to the correlation between cancer imaging features and gene expression: The most interesting results have been obtained in onco-imaging field. Recently, some authors have evaluated the correlation between the imaging characteristics and molecular features of malignancies.13-15
An emerging potentially useful imaging biomarker is computed tomography (CT) tumor texture analysis (CTTA), which has shown promising results in predicting patient outcome, overall survival, and response to therapy for multiple tumors, including RCC.13,16
Computed tomography texture analysis is a quantitative technique that allows to characterize the heterogeneity of a lesion inside a region of interest (ROI) by analyzing the distribution and relationship of pixel gray levels using both unfiltered and frequency-filtered images, deriving quantitative texture parameters based on attributes of the pixels and the image histogram.16
To the best of our knowledge, a possible correlation between CT texture parameters and miRNAs expression in ccRCC was not investigated yet. To this end in this study, we investigate whether quantitative parameters obtained from CTTA correlate with different grades of expression of miRNAs in patients affected by ccRCC.
Material and Methods
Study Population
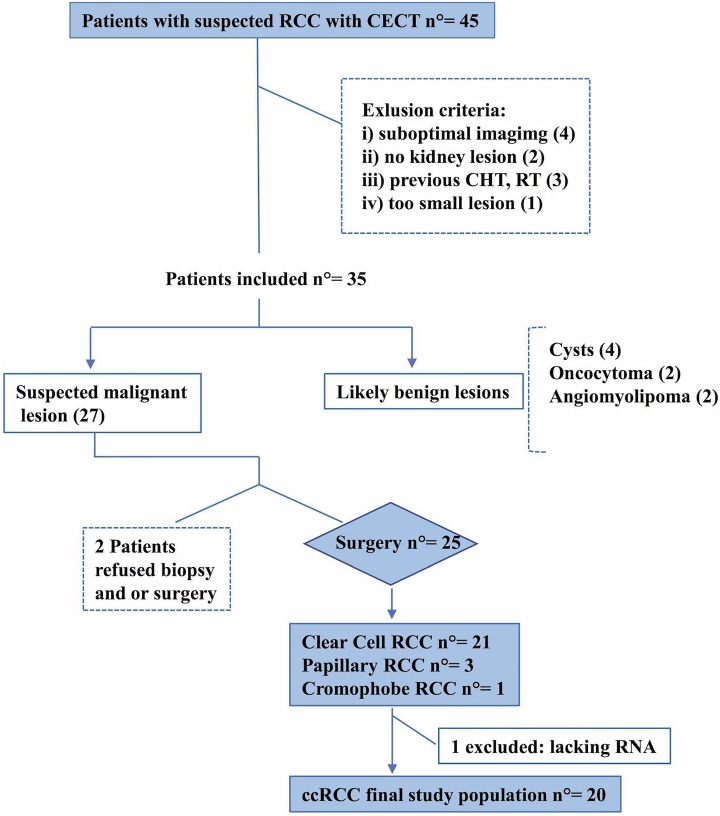
We designed a retrospective single-center study conducted on patients who had contrast-enhanced computed tomography (CECT) of the abdomen and pelvis for suspected ccRCC between April 1, 2014, and June 1, 2016. Patients’ recruitment was based on the Standards for Reporting of Diagnostic Accuracy initiative as reported in accrual flowchart (Figure 1). Institutional review board approved the study protocol and written consent was acquired from each patient. Clinical indications for CT included (1) clinical suspicion for ccRCC based on the patient’s clinical history and/or elevated tumor markers levels or (2) patients known to have a suspicious renal lesion on the basis of the results of prior imaging studies, such as ultrasonography, unenhanced CT, or magnetic resonance imaging (MRI). Patients characteristics are summarized in Table 1.
Figure 1.
Flowchart of patients’ selection.
Table 1.
Patient Clinical and Histological Characteristics.
| Patient Demographics and Characteristics at Diagnosis | |
|---|---|
| Number of patients | 20 |
| Age, years | 65 ± 3 |
| Gender | |
| Male | 14 |
| Female | 6 |
| Body mass index, kg/m2 | 27 ± 4.37 |
| Serum creatinine, mg/dL | 1.45 ± 0.72 |
| Smoking habit | |
| Yes | 13 |
| No | 7 |
| Pathological tumor stage (pT) | |
| pT1a | 11 |
| pT1b | 4 |
| pT2a | 1 |
| pT2b | 1 |
| pT3 | 3 |
| Surgical procedure | |
| Tumorectomy | 7 |
| Radical nephrectomy | 13 |
| Clear cell tumor grade | |
| 1 | 3 |
| 2 | 15 |
| 3 | 2 |
| Tumor maximum diameter, cm | 4.2 ± 2.4 |
No authors are employees of or consultants for industry or had control of inclusion of any data and information that could represent a conflict of interest. There was no industry support specifically for this study.
Forty-five consecutive patients who underwent CECT of the abdomen and pelvis for suspected ccRCC between April 1, 2014, and June 1, 2016, were primarily included.
Ten patients were considered not eligible for this study due to (1) the CT examination resulted in no kidneys lesions; (2) patients underwent previously chemotherapy, radiotherapy, or immunotherapy; (3) too small lesion for good CTTA (<2 cm), (4) inadequate image quality due to suboptimal injection technique, poor timing for the acquisition of the urographic phase, or deviations from the routine CT protocol (eg, inappropriate selection of kV or reconstruction kernel by the CT technologist). Based on CT and clinical presurgery evaluations, we could include 35 patients, 8 of which were proved to have benign lesions (basing on other imaging evaluations, follow-up, or percutaneous biopsy). Finally, 2 patients with suspicious RCC on the base of CT characteristics refused biopsy or surgery due to old age and suboptimal global clinical conditions. Among these 27 patients, 6 underwent biopsy and 26 direct surgeries (nephrectomy or partial nephrectomy based on tumor size). After histopathological examination, only 21 ccRCC were found; 1 of them was not eligible for the study because of no complete miRNAs extraction for tumor and matched normal tissue.
Our final study population included 20 patients (14 males, 6 females; mean age 65 ± 13 years, range 35-87 years; mean body mass index 27 kg/m2 ± 4.37; range 20.8-40.3 kg/m2) with 20 ccRCC suitable for CTTA examination and miRNAs extraction (Figure 1 and Table 1).
Histopathological Analysis, RNA Extraction, and MiRNA Expression Analysis
Eight samples of fresh-frozen (FF) tissues from 8 ccRCC lesions were analyzed and homogenized by gentle dissociator (Miltenyi Biotec, USA) in 700 μL of Qiazol (Qiagen, Chatsworth, California); RNA was extracted using the manufacturer’s instructions.
A cohort of 12 ccRCC formalin-fixed paraffin-embedded (FFPE) tissue samples from 12 patients was analyzed. RNA was extracted using the miRneasy FFPE kit (Qiagen) following the manufacturer’s instructions.
For each patient, a matched normal tissue sample was collected. For both cohorts, the concentration and purity of total RNA were assessed using a Nanodrop 1000 spectrophotometer (Nanodrop Technologies, Wilmington, Delaware). RNA from FF tissues and FFPE tissues showed comparable quality. A quantity of 150 ng of total RNA was reverse transcribed in 8 µL using miScript II real-time (RT) kit (Qiagen), and 1 µL of complementary DNA (cDNA) dilution (1:4) was used for quantitative RT polymerase chain reaction (PCR) experiments. A PCR quantification analysis of the SCARNA17 SNORD61, SNORD68, RNU6-2, and miRNAs such as miR-21-5p, miR-210-3p, miR-185-5p, miR-221-3p, and miR-145-5p was performed using the miScript SYBR Green PCR kit (Qiagen) with the miScript Primer assay Hs-SCARNA17 (#MS00014014), SNORD61 (#MS00033705), SNORD68 (#MS00033712), RNU6B-2 (#MS00033740), Hs-miR-21-5p (#MS00009079), Hs-miR-210-3p (#MS00003801), Hs-miR-185-5p (#MS00003647), Hs-miR-221-3p (#MS00003857), and Hs-miR-145-5p (#MS00003528; Qiagen).
The expression analyses of RNU19 and RNU66 were performed using TaqMan miRNA RT assay and TaqMan miRNA assays (RNU19 #001003 and RNU66 #001002; Applied Biosystems, Foster City, California) according to the manufacturer’s protocol. All reactions were performed in duplicate. Data were analyzed by quantification relatively to a standard curve. The standard curve was prepared with serial dilutions of a reference cDNA obtained from RNA extracted from a tumor sample. Z scores were calculated for all expression values to standardize the data. Subsequently, z score values of RNU66, RNU19, and SCARNA17 were averaged and used to normalize the expression values of each miRNA in FFPE samples, whereas z score values of SNORD61, SNORD68, and RNU6-2 were averaged and used to normalize the expression values of each miRNA in FF samples (Table 2).
Table 2.
Evaluation of MicroRNAs Levels in Patients With ccRCC.
| Mean Values MicroRNAs Levels (±SD) | |||||
|---|---|---|---|---|---|
| miR-21-5p | miR-210-3p | miR-221-3p | miR-185-5p | miR-145-5p | |
| Normal tissue | 0.60 (±0.51) | 1.18 (±0.94) | 0.71 (±0.32) | 1.08 (±0.96) | 1.04 (±0.33) |
| ccRCC | 0.94 (±0.61) | 1.57 (±1.31) | 1.13 (±0.74) | 1.03 (±0.47) | 1.14 (±0.65) |
| P value | <.05 | <.05 | <.05 | >.05 | >.05 |
Abbreviations: ccRCC, clear cell renal cell carcinoma; SD, standard deviation.
MDCT Acquisition Protocol
All scans were performed with a 64-row multiple-detector computed tomography (MDCT) scanner (Lightspeed VCT, GE Medical Systems, Waukesha, Wisconsin) using the following parameters: kV, 120; beam pitch, 1.375:1; detector configuration, 64 mm × 0.625 mm; and reconstructed section thickness, 1.25 mm.
A z-axis tube current modulation was used, with a noise index of 28 (min/max mA: 200/600), which was recommended by the manufacturer for standard abdominal CT in all cases. All examinations were performed using a multiphase MDCT protocol (Table 3).
Table 3.
CT Acquisition and Reconstruction Parameters.
| MDCT Parameters | |
|---|---|
| Detector configuration, mm | 64 × 0.625 |
| Tube voltage, kV | 120 |
| Automatic exposure control (AEC) | On |
| Current tube modulation, mAs | 200/600 |
| Noise index | 28 |
| Field of view, cm | 50 |
| Rotation time, seconds | 0.5 |
| Pitch | 1.375 |
| Reconstruction kernel | Medium-Smooth |
| Reconstruction algorithm | Q30 |
| Iterative reconstruction algorithm, % | 40 |
| Slice thickness, mm | 1.25 |
Abbreviations: CT, computed tomography; MDCT, multiple-detector computed tomography.
All patients received an average of 120 mL of an intravenous nonionic contrast medium (CM) with an iodine concentration of 350 mg iodine/mL (Iomeprol, Iomeron 350; Bracco, Milan, Italy). The bolus of CM was injected through an 18 or 20 gauge cannula inserted into an antecubital vein using a dual-chamber peristaltic injector (CT Exprès, Bracco, Milan, Italy) at a flow rate of 3.5 mL/s.
All patients were positioned supine with head first on the scanning table. The scanning protocol started with the acquisition of anteroposterior and lateral digital localizer radiographs. The acquisitions of the abdomen and pelvis were performed in the craniocaudal direction after iodine intravenous injection of CM in arterial, portal, and urographic phases.
A bolus tracking technique was used to minimize the influence of cardiac output; CM detection was monitored in an ROI placed in the aorta at the level of the diaphragm; the threshold for the start of the scan was set at 100 HU. A late arterial phase was acquired 18 seconds after reaching the threshold; a portal phase 35 seconds after the end of arterial phase; an urographic phase was acquired after 12 minutes.
Image Reconstruction
Imaging reconstructions were obtained using 40% of an iterative reconstruction algorithm (ASiR, GE Healthcare, Milwaukee, Wisconsin) as recommended by the manufactory. The use of 40% iterative reconstruction algorithm and 60% of filtered back projection algorithm (standard algorithm) allows to obtain high image quality, with low image noise, even when a low-radiation dose acquisition protocol is used, as in our study.
Images were reconstructed using a medium-smooth kernel (Q30) at 1.25 mm reconstructed section thickness (Table 3).
Imaging Data Analysis
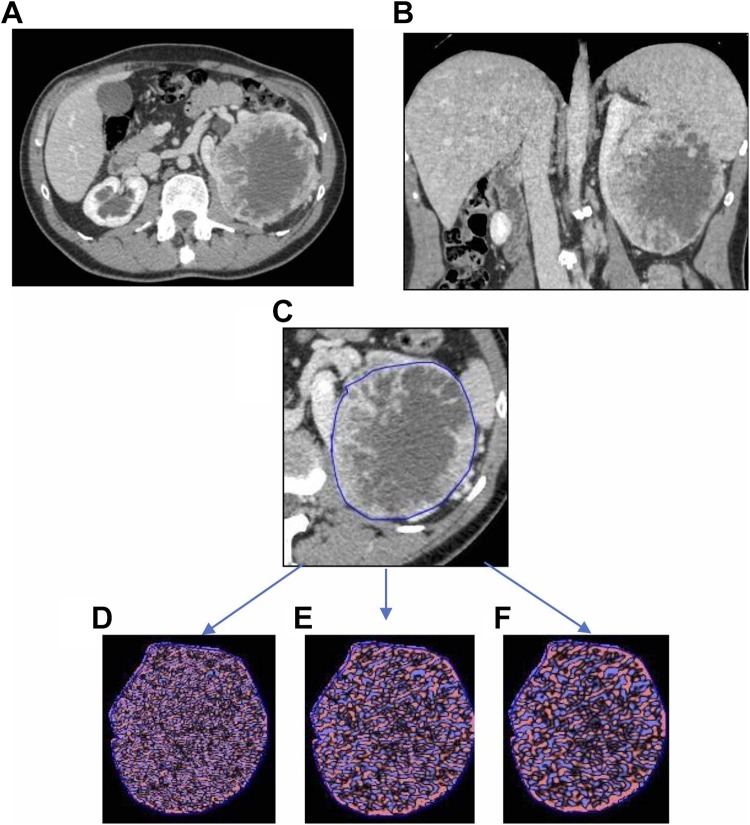
All analyses were performed using a commercially available CTTA research software platform (version 1.1; TexRAD Ltd, Somerset, United Kingdom) on a dedicated workstation. Computed tomography texture analysis was performed on 20 ccRCCs drawing manually 2 different polygonal ROIs at 3 different axial levels in arterial, portal, and urographic phases (Figure 2) (1) into the lesion (including the majority of the lesion in that plane and excluding margins) and (2) in the normal parenchyma of the kidney, adjacent to the tumor, including cortical and medullar layers.
Figure 2.
A 58-year-old man with a large ccRCC on the left side. A-B, Portal venous phase contrast-enhanced transverse (A) and coronal (B) CT images showing tumor (arrows). C, Texture analysis image showing ROI (blue line) outlining cancer. D-F, Color texture overlays of cancer outlined by an ROI (blue line) showing images with a fine filtering (SSF1) (D), medium filtering (SSF1.5) (E), and coarse filtering (SSF2) (F). ccRCC indicates clear cell renal cell carcinoma; ROI, region of interest; SSF, spatial scaling factor.
All images were reviewed, and slices were selected by 2 readers (radiologist with 10 years of experience and radiology resident with 3 years of experience on abdominal imaging).
The technique consisted of a preliminary filtration step, followed by quantification of the texture within the filtered images.
The filtration step comprised Laplacian of Gaussian spatial band-pass filter used to produce multiple series of derived images extracting and enhancing features at different anatomical spatial scales; this resulted in a series of derived images from fine to coarse texture within an ROI identified with “spatial scaling factor” (SSF). The scale was selected by tuning the filter parameter between SSF0 and SSF2.0, where SSF1.0 indicates fine texture (features of approximately 2 pixels in width), SSF1.5, SSF1.8, and SSF2.0 indicate degrees of medium textures (features of approximately 6, 8, and 10 pixels in width, respectively).
This is followed by quantification of statistical parameters in a histogram-based statistical approach (first-order, second-order, or higher order parameters). The software output includes mean pixel attenuation (M, mean), standard deviation of the pixel distribution histogram (standard deviation [SD], dispersion from the mean), entropy (E, irregularity in terms of randomness of distribution of pixels), mean of positive pixels (MPP), skewness of the pixel histogram (S, asymmetry), kurtosis (K, sharpness) of the pixel histogram, and the percentage of positive pixels; all these histogram-based parameters are provided for each SSF.
Statistical Analysis
All data were described as mean ± SD. Computed tomography texture analysis was performed for each of the 6 parameters at each SSF (in number of 5) and for all postcontrast CT phases acquired (ie, normal renal parenchyma, arterial phase, portal phase, and urographic phase), with a total of 120 variables.
A matrix of data on MatLab have been elaborated from 5 different miRNAs (miR-145-5p, miR-185-5p, miR-221-3p, miR-21-5p, and miR-210-3p), 6 texture parameters (mean, SD, entropy, MPP, skewness, and kurtosis) in 5 different SSFs (SSF0, SSF1, SSF1.5, SSF1.8, and SSF2.0). All data were elaborated for normal kidney parenchyma and for 3 different ROIs on tumor volume, finally producing a total of more than 500 graphics of correlation. Normality of each continuous variable was tested with Z test. Differences between normal tissue and tumor in all miRNAs expression and all CTTA parameters were assessed using paired Student t test. First step of statistical analysis was aimed to assess statistically significant differences between the 2 groups of lesions’ samples: FFPE and FF.
Differences in the evaluations of CTTA parameters between the 2 operators were calculated (interobserver agreement with Cohen κ, with κ ≤ .40 poor agreement, κ = .40-.75 good agreement, κ ≥ .76 excellent agreement).
Secondary, all lesions were evaluated all together, since no statistically significant difference in miRNAs expression was found between the 2 groups and between the 2 operators for CTTA. In addition, statistically significant difference was assessed for (1) miRNA normal tissue versus tumor samples and (2) CTTA parameters in normal tissue versus tumor samples.
For comparison of miRNA versus CTTA parameters was then used: (1) the Pearson correlation coefficient (r): r was interpreted as follows: A negative value or less than 0.20 indicated poor agreement; a value of 0.21 to 0.40, fair agreement; a value of 0.41 to 0.60, moderate agreement; a value of 0.61 to 0.80, substantial agreement; and a value of 0.81 to 1.00, almost perfect agreement; (2) the coefficient of determination (R2) in a polynomial interpolation graphic type of order 2. R2 quantifies the amount of variance of the variable that is explained by the selected polynomial with respect to the average of the data. R2 was interpreted as follows: A negative value or less than 0.20 indicated poor agreement; a value of 0.21 to 0.40, fair agreement; a value of 0.41 to 0.60, moderate agreement; a value of 0.61 to 0.80, substantial agreement; and a value of 0.81 to 1.00, almost perfect agreement.
For all comparisons, statistical significance was assumed to be P < .05. All statistical analyses were performed using a commercially available statistical software package SPSS version 21.0 (SPSSInc, Chicago, Missouri).
Results
A total of 20 matched ccRCC and adjacent normal tissue samples were collected and analyzed for the expression of miR-21-5p, miR-210-3p, miR-185-5p, miR-221-3p, and miR-145-5p included in our previous study on ccRCC.17 According to previous reports, miR-21-5p, miR-210-3p, and miR-221-3p resulted significantly upregulated in ccRCC (0.94 ± 0.61, 1.57 ± 1.32, and 1.13 ± 0.74, respectively) versus normal (0.60 ± 0.52, 1.18 ± 0.94, and 0.71 ± 0.33, respectively) tissues (P < .05; Table 2). miR-185-5p and miR-145-5p did not show any statistically significant difference between tumor and normal tissues (Table 2).
Regarding CTTA acquisition, all scans were performed with a 64-row MDCT scanner following acquisition and reconstruction parameters summarized in Table 3. No differences were found between the 2 operators in collected CTTA parameters (κ = .84). The analyses shown below were elaborated choosing data from the best operator results (best correlations found). When considering texture parameters alone, differences between healthy and pathological tissue within the same SSF and the same contrast phase were consistent and statistically significant for the majority of parameters (entropy, MPP, SD, and mean) when a medium texture filter setting (SSF1 or SSF1.5) was used (P < .05); for these parameters, we observed a statistically significant difference between normal tissue ROIs and pathological tissues, in all contrast phases (Table 4 and Figure 3).
Table 4.
Expression of CTTA Texture Parameters in Normal Tissue and Tumor in Arterial Phase.a
| Arterial Phase SSF1.5 | Normal Tissue | Tumor | P Value | P Value Venous Phase | P Value Urographic Phase |
|---|---|---|---|---|---|
| Mean | −1.23 (±14.7) | 23.31 (±17.38) | <.001 | <.05 | <.05 |
| SD | 101.2 (±37.8) | 120.9 (±32.7) | <.05 | .09 | .06 |
| Entropy | 4.71 (±0.20) | 5.76 (±0.40) | <.001 | <.05 | <.05 |
| MPP | 85.39 (±38.75) | 107.17 (±33.1) | <.05 | <.05 | <.05 |
| Skewness | 0.25 (±0.42) | 0.36 (±0.49) | .45 | .52 | .30 |
| Kurtosis | −0.04 (±0.63) | 1.67 (±5.50) | .17 | .23 | .10 |
Abbreviations: CTTA, computed tomography texture analysis; MPP, mean of positive pixels; SD, standard deviation; SSF, spatial scaling factor.
a P values are shown for arterial, venous, and urographic phase.
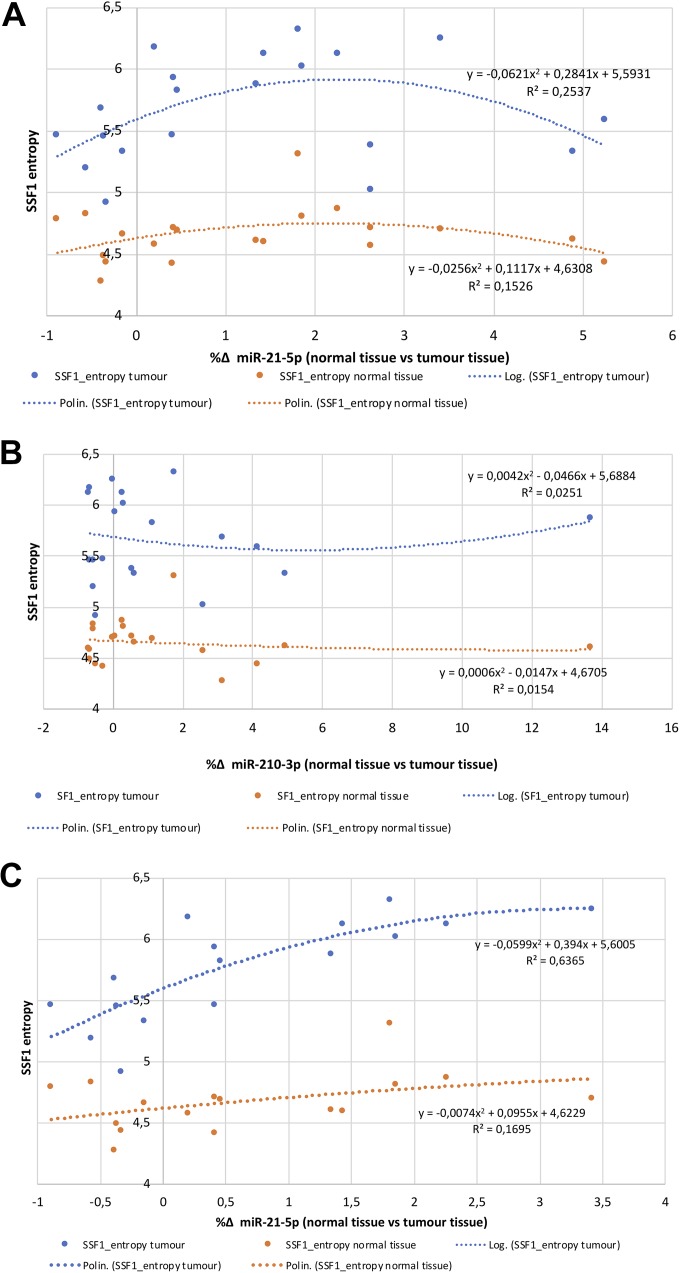
Figure 3.
Graphics of relation and R2 value between microRNAs and CTTA features in ccRCC. A, Comparison between miR-21-5p (expressed as %Δ normal vs tumor) and tissue entropy (SSF1). B, Comparison between miR-210-3p (expressed as %Δ normal vs tumor) and tissue entropy (SSF1). C, Comparison between miR-21-5p (expressed as %Δ normal vs tumor) and tissue entropy excluding 4 patients with extremely overexpressed miR-21-5p in tumor samples (too far from median values). ccRCC indicates clear cell renal cell carcinoma; CECT, contrast-enhanced computed tomography; CTTA, computed tomography texture analysis; SSF, spatial scaling factor.
Moreover, comparing different contrast phases within the same SSF or the same contrast phase among different SSF, no significant differences were found in particular at medium filters (SSF 1.5–1.8–2.0). In specific entropy, values in the arterial phase in normal tissue were 4.66 ± 0.21 (SF1), 4.71 ± 0.21 (SF1.5), 4.72 ± 0.19 (SF1.8), and 4.73 ± 0.19(SF2.0), while in tumor ROI entropy was 5.67 ± 0.41 (SF1), 5.76 ± 0.40 (SF1.5); 5.79 ± 0.39 (SF1.8), and 5.80 ± 0.40 (SF 2.0).
MicroRNAs expression in normal tissue didn’t correlate significantly with any CTTA parameter (P > .05). Analysis of tumor samples evidenced only a trend of positive correlation between miRNAs (miR-21-5p and miR-210-3p) and some CTTA parameters. The best trends were found when delta of percent (%Δ) of expression between healthy tissue and pathological tissue were used to express our data (Table 5).
Table 5.
CTTA Parameters and MiRNAs Pearson Correlation (Arterial Phase; SSF1.5).
| Mean | SD | Entropy | MPP | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|
| Normal tissue | ||||||
| Δ (%) miR-21 | 0.30 | −0.14 | 0.06 | −0.10 | −0.21 | 0.09 |
| Δ (%) miR-210 | −0.09 | −0.26 | 0.23 | −0.27 | −0.28 | −0.21 |
| Tumor | ||||||
| Δ (%) miR-21 | −0.24 | 0.07 | 0.16 | 0.007 | 0.04 | −0.09 |
| Δ (%) miR-210 | 0.11 | 0.15 | 0.09 | 0.17 | −0.01 | −0.13 |
Abbreviations: CTTA, computed tomography texture analysis; MPP, mean of positive pixels; SD, standard deviation; SSF, spatial scaling factor.
When comparing CT texture parameters and miRNAs expression in a polynomial interpolation graphic type of order 2, we found a dispersion of data in the graphics of comparison, showing poor agreement with miRNAs expressivity for most of the parameters (data shown in Supplementary material).
Interestingly, entropy showed the best agreement between miRNAs expression and CT texture parameters. For example, as shown by nonlinear graphic analysis between %Δ of miRNA expression (matched tumor vs normal tissues) and entropy values (Figure 3A), in tumor tissues a higher coefficient of determination is present between entropy and the %Δ of miR-21-5p expression (R2 = 0.25) compared to normal tissues (R2 = 0.15; Figure 3A). On the contrary, a similar effect was not observed for miR-210-3p (R2 = 0.025 in tumor vs R2 = 0.014 in normal; Figure 3B). These results were confirmed using all SSF and all postcontrast phases (data not shown). Moreover, a nonlinear graphic analysis between %Δ of miR-21-5p expression (matched tumor vs normal tissues) and skewness values didn’t show any correlation (R2 = 0.0020 in tumor vs R2 = 0.072 in normal; data not shown). Analyzing data, we recognized 4 patients with particularly increased miR-21-5p expression in tumor versus normal tissues but only slight increase in entropy values (as compared to normal tissue): Their texture parameters were not far from the median values of the texture parameters of the other patients. Interestingly, excluding them from the analysis, we found again that normal tissue entropy was not related to miR-21-5p (R2 = 0.17), while an excellent relation of entropy in tumor samples with miR-21-5p was found (R2 = 0.64; Figure 3C).
Discussion
Nowadays, the evaluation of a renal lesion using CT or MRI is based on the radiologist’s 2-dimensional examination of lesion morphology and enhancement, and this routinely determines subsequent patient management. Both CT and MRI are the most used imaging techniques in the staging of ccRCC before treatment; however, this type of tumors is characterized by genetic, epigenetic, and pathologic heterogeneity, which makes accurate diagnosis or prognosis prediction difficult. Many efforts have been done by radiologists and experts, looking for imaging biomarkers to characterize tumors and manage therapies.
Computed tomography texture analysis is a relatively new tool with a great potential; it can be of great help for the radiologist to better characterize lesions and to identify parameters of response to treatment. However, it is still soon to consider all done. Computed tomography texture analysis allows quantification of lesion heterogeneity based on the distribution of pixel intensities within an ROI.
In our study, most of CTTA parameters showed significant differences comparing normal corticomedullar tissue with ccRCC. Computed tomography texture analysis has robust parameters to distinguish normal tissue from ccRCC (eg, entropy, mean, SD). This could be of great future help for imaging characterization of renal lesion, even in terms of survival rates and clinical outcome. The surprising result of not significant increase in skewness and kurtosis in our cohort of ccRCC, in contrast with previous studies on other solid tumors (eg, rectal or lung adenocarcinoma),18-21 supports the fact that different tumors display different radiophenotypes and address the attention to more research in this field,22,23 with the aim of looking for a “texture signature” typical for every tumor type.
In the field of urological carcinoma, miRNAs are acquiring a role as biomarkers.24-26 In particular, miRNA-21-5p is the most overexpressed in renal cancer: It interacts with Ras phosphoinositide-3-kinase (PI-3K)/PTEN/AKT apoptosis pathways, so it may be considered a clinical biomarker in RCC. In a recent publication, Petrozza et al found interesting results on overexpression of miR-210-3p on urinary samples of patients with ccRCC and they also prove the significant reduction in concentration of this miRNA after surgery and during follow-up.27 This finding seems to be very interesting, opening new doors for genetic biomarkers in ready and “easy-to-collect” tissue samples.
In a radiogenomics point of view, we decided to compare CT texture parameters and miRNAs expression profile of ccRCC looking for correlation between them.
First, we confirmed deregulation of specific miRNAs in our group of 20 ccRCC (miR-21-5p, miR-210-3p, miR-185-5p, miR-145-5p, and miR-221-3p), according to what found previously.17
Regarding the comparison between expression of miRNA and modification of CTTA in tumor samples, a surprisingly poor positive association was found among the majority of them in ccRCC with poor statistical significance. Interesting results were found comparing entropy and miR-21-5p expression: No relation was found with normal tissue, while tumor entropy is slightly positively correlated with miR-21-5p. The results are almost stable moving into different contrast phases and different spatial filters, giving to the correlation more importance.
The correlation was a bit limited by data from 4 (20%) patients, who had a significant upregulation of miR-21-5p but a slight increase of entropy in lesions as compared to normal tissues. The reason why these patients showed such results in still not clear. We couldn’t find a reasonable factor to explain this behavior, since patients performed the same CT protocol and their tissue samples were either FF (for 2) or FFPE (for the others). Furthermore, they were not different neither in terms of tumor–node–metastasis or grading, without any apparent difference in comparison with the others. The only interesting common factor was that they were among the youngest in the cohort of patients (<60 years of age), and this could have a role in the significant hyperexpression of miRNAs in tumor samples. More data are needed to support this conclusion. Nevertheless, without those 4 patients, we found more significant results with very good correlation.
Texture analysis is efficient, reproducible, and can be considered complementary to 2-dimensional imaging evaluation of ccRCC on MDCT, because it maximizes the information obtained from the lesion and has the potential to become a tool for prediction of prognosis.
There are some limitations to the present study, for example, the small number of patients. Moreover, we didn’t perform analysis on prognostic value of the parameters because of lack of follow-up outcome data.
Conclusion
Both miRNAs and CTTA show promising correlations in ccRCC and no correlations with normal renal tissues. Texture parameters, in adjunction to biopsy results, can be considered useful tools for the noninvasive evaluation of ccRCC. In particular, our study demonstrated good correlation between entropy and miR-21-5p, one of the most important miRNA involved in tumorigenesis, and good correlation between texture parameters (mean and entropy) and ccRCC.
Supplemental Material
Appendix_1 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Supplemental Material
Appendix_2 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Supplemental Material
Appendix_3 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Supplemental Material
Appendix_4 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Supplemental Material
Appendix_5 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Supplemental Material
Appendix_6 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Acknowledgments
The authors thank greatly appreciated Prof Giorgio Bellotti (Department of Engineering, “Roma Tre” University, Rome, Italy) for his contribution toward data analysis and interpretation.
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- cDNA
complementary DNA
- CECT
contrast-enhanced computed tomography
- CM
contrast medium
- CT
computed tomography
- CTTA
computed tomography texture analysis
- FF
fresh-frozen
- FFPE
formalin-fixed paraffin-embedded
- MDCT
multiple-detector computed tomography
- miRNA
microRNA
- MPP
mean of positive pixels
- MRI
magnetic resonance imaging
- PCR
polymerase chain reaction
- ROI
region of interest
- RT
real-time
- SD
standard deviation
- SSF
spatial scaling factor
Authors’ Note: V.P. and A.L. made substantial contributions to conception and design; C.M. and F.F. made substantial contributions to acquisition of data analysis and interpretation of data; S.B., M.R., D.C., A.L.P., C.T., S.M., G.P., and A.C. made substantial contributions to acquisition of data; C.M., F.F., S.B., and D.B. have been involved in drafting the manuscript. All authors have been involved in revising the manuscript critically for important intellectual content, gave final approval of the version to be published, agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the IRB/Commissione Ricerca Sapienza Università di Roma (approval no. RM116154BAA09F2A). All patients provided written informed consent prior to enrollment in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Contribution of Sapienza University of Rome to V.P. and A.L. was greatly appreciated.
ORCID iD: Chiara Marigliano  https://orcid.org/0000-0002-3592-4073
https://orcid.org/0000-0002-3592-4073
Marco Rengo  https://orcid.org/0000-0001-9087-5692
https://orcid.org/0000-0001-9087-5692
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2. Jonasch E, Futreal PA, Davis IJ, et al. State of the science: an update on renal cell carcinoma. Mol Cancer Res. 2012;10(7):859–880. doi:10.1158/1541-7786.MCR-12-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schaefer A, Stephan C, Busch J, Yousef GM, Jung K. Diagnostic, prognostic and therapeutic implications of microRNAs in urologic tumors. Nat Rev Urol. 2010;7(5):286–297. doi:10.1038/nrurol.2010.45. [DOI] [PubMed] [Google Scholar]
- 4. Fridman E, Dotan Z, Barshack I, et al. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn. 2010;12(5):687–696. doi:10.2353/jmoldx.2010.090187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joosten SC, Smits KM, Aarts MJ, et al. Epigenetics in renal cell cancer: mechanisms and clinical applications. Nat Rev Urol. 2018;15(7):430–451. doi:10.1038/s41585-018-0023-z. [DOI] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi:10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Osanto S, Qin Y, Buermans HP, et al. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PLoS One. 2012;7(6):e38298 doi:10.1371/journal.pone.0038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gowrishankar B, Ibragimova I, Zhou Y, et al. MicroRNA expression signatures of stage, grade, and progression in clear cell RCC. Cancer Biol Ther. 2014;15(3):329–341. doi:10.4161/cbt.27314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ge YZ, Wu R, Xin H, et al. A tumor-specific microRNA signature predicts survival in clear cell renal cell carcinoma. J Cancer Res Clin Oncol. 2015;141(7):1291–1299. doi:10.1007/s00432-015-1927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang K, Xu H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: an integrated miRNA expression profiling analysis. Sci Rep, 2015;5:10272 doi:10.1038/srep10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu GJ, Dong YQ, Zhang QM, et al. MiRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8(5):5224–5229. [PMC free article] [PubMed] [Google Scholar]
- 13. Jain R, Poisson LM, Gutman D, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272(2):484–493. doi:10.1148/radiol.14131691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shinagare AB, Vikram R, Jaffe C, et al. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of the Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group. Abdom Imaging, 2015;40(6):1684–1692. doi:10.1007/s00261-015-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X, Zhou Z, Hannan R, et al. Reliable gene mutation prediction in clear cell renal cell carcinoma through multi-classifier multi-objective radiogenomics model. Phys Med Biol. 2018;63(21):215008 doi:10.1088/1361-6560/aae5cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng. 2008;55:1822–1830. doi:10.1109/TBME.2008.919735. [DOI] [PubMed] [Google Scholar]
- 17. Petrozza V, Carbone A, Bellissimo T, et al. Oncogenic microRNAs characterization in clear cell renal cell carcinoma. Int J Mol Sci. 2015;16(12):29219–29225. doi:10.3390/ijms161226160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rizzo S, Petrella F, Buscarino V, et al. CT Radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol. 2016;26(1):32–42. doi:10.1007/s00330-015-3814-0. [DOI] [PubMed] [Google Scholar]
- 19. De Cecco CN, Ganeshan B, Ciolina M, et al. Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Invest Radiol. 2015;50(4):239–245. doi:10.1097/RLI.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 20. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006 doi:10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sacconi B, Anzidei M, Leonardi A, et al. Analysis of CT features and quantitative texture analysis in patients with lung adenocarcinoma: a correlation with EGFR mutations and survival rates. Clin Radiol. 2017;72(6):443–450. doi:10.1016/j.crad.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 22. Lubner MG, Stabo N, Abel EJ, et al. CT textural analysis of large primary renal cell carcinomas: pretreatment tumor heterogeneity correlates with histologic findings and clinical outcomes. AJR Am J Roentgenol. 2016;207(1):96–105. doi:10.2214/AJR.15.15451. [DOI] [PubMed] [Google Scholar]
- 23. Karlo CA, Di Paolo PL, Chaim J, et al. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270(2):464–471. doi:10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol. 2016;13(12):734–752. doi:10.1038/nrurol.2016.193. [DOI] [PubMed] [Google Scholar]
- 25. Lou N, Ruan AM, Qiu B, et al. MiR-144-3p as a novel plasma diagnostic biomarker for clear cell renal cell carcinoma. Urol Oncol. 2017;35(1):36.e37–36.e44. doi:10.1016/j.urolonc.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 26. Mlcochova H, Hezova R, Stanik M, Slaby O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Oncol. 2014;32(1):41.e41–49. doi:10.1016/j.urolonc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 27. Petrozza V, Pastore AL, Palleschi G, et al. Secreted miR-210-3p as non-invasive biomarker in clear cell renal cell carcinoma. Oncotarget. 2017;8(41):69551–69558. doi:10.18632/oncotarget.18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix_1 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Appendix_2 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Appendix_3 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Appendix_4 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Appendix_5 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment
Appendix_6 for Radiogenomics in Clear Cell Renal Cell Carcinoma: Correlations Between Advanced CT Imaging (Texture Analysis) and MicroRNAs Expression by Chiara Marigliano, Stefano Badia, Davide Bellini, Marco Rengo, Damiano Caruso, Claudia Tito, Selenia Miglietta, Giovanni Palleschi, Antonio Luigi Pastore, Antonio Carbone, Francesco Fazi, Vincenzo Petrozza and Andrea Laghi in Technology in Cancer Research & Treatment