Abstract
Chronic infection with the hepatitis C virus (HCV) is a major cause of cirrhosis and hepatocellular carcinoma. In 2009, genome-wide association studies (GWAS) strongly linked genetic variants in the interferon lambda (IFN-λ) chromosomal region to HCV clearance. In 2013, discovery of the IFNL4 gene provided a functional explanation for those GWAS findings. The IFNL4-ΔG/TT (rs368234815) variant controls generation of the IFN-λ4 protein. Paradoxically, the IFNL4-TT allele, which abrogates IFN-λ4, associates with higher rates of spontaneous HCV clearance and better response to treatments for HCV infection. The finding that a “knock-out” allele for IFN-λ4 enhances HCV clearance challenges the paradigm of IFNs as antiviral cytokines. Genetic variants in the IFN-λ region have also been associated with hepatic inflammation and fibrosis from various etiologies, however, alleles that are linked with improved HCV clearance associates with worse inflammation and fibrosis. These studies demonstrate that GWAS of infectious diseases may yield important and unexpected biological insights.
Keywords: epidemiology, fibrosis, genetics, hepatocellular carcinoma, IFNL4, innate immunity, treatment response, viral clearance
Hepatitis C Virus Infection
Chronic infection with hepatitis C virus (HCV) affects ∼71 million people worldwide, or 1% of the global population (Polaris Observatory HCV Collaborators 2017). In the United States, at least 3.5 million people have chronic hepatitis C (CHC) (Edlin and others 2015). HCV is primarily transmitted through exposure to infected blood or blood products via injection drug use, unscreened blood transfusions or iatrogenic transmission (World Health Organization 2017). Approximately 70%–80% of individuals with an acute HCV infection are unable to clear the virus spontaneously and develop CHC. Among individuals with CHC, 10%–20% will develop advanced liver disease of whom, 1%–5% will go on to develop hepatocellular carcinoma (HCC) (Lavanchy 2011). Globally, CHC is responsible for 25% of cirrhosis and HCC cases (Perz and others 2006); in developed countries CHC is a leading indication for liver transplantation (Brown 2005).
CHC leads to liver injury through hepatocyte apoptosis and necrosis, driven by sustained inflammation and activation of immune mechanisms. Liver damage seen in HCV infection is likely caused by the host's immune response rather than the virus itself (Liang and others 2000). Prolonged inflammation of the liver can lead to the accumulation of excess connective tissue, resulting in fibrosis (Guidotti and Chisari 2006; Arzumanyan, Reis, and Feitelson 2013). Cirrhosis, the most advanced stage of fibrosis, is characterized by scarring of the liver and alteration of the normal liver structure into abnormal nodules (Anthony and others 1978). Inflammation, accompanied by oxidative stress and cellular proliferation, provokes somatic mutations and carcinogenesis. In patients with CHC, the risk of HCC is highly correlated with the severity of liver fibrosis (Forner and others 2012; Arzumanyan, Reis, and Feitelson 2013).
Previously, treatment with pegylated interferon alpha combined with the nucleoside inhibitor ribavirin (peg-IFNα/ribavirin) for 48 weeks was the standard of care for HCV treatment (Scheel and others 2013). Successful treatment cures HCV infection by producing a sustained viral response (SVR), defined as an undetectable level of HCV RNA in serum 12–24 weeks after the end of treatment. Treatment efficacy with the peg-IFNα/ribavirin regimen is modest and varies with the viral genotype (VGT) (McHutchison and others 1998). HCV strains are classified into 7 genotypes; VGT-1 and VGT-3 are the predominant genotypes globally and VGT-1 is the most prevalent strain in the United States (Messina and others 2015). Only 40%–50% of patients infected with VGT-1 and ∼70% infected with VGT-3 achieve SVR after peg-IFNα/ribavirin therapy (Fried and others 2002; Shiffman and others 2007).
In addition to viral factors, certain demographic and clinical factors are associated with better response to peg-IFNα/ribavirin (Reddy and others 1999). Race associates strongly with achieving SVR after peg-IFNα/ribavirin therapy and African American patients are less likely than white patients to have an SVR even after other factors are considered (Reddy and others 1999; Kinzie and others 2001; Layden-Almer and others 2003; Muir and others 2004; Conjeevaram and others 2006; Wilder and others 2016). Those findings supported a hypothesis that genetic factors play a role in response to treatment for CHC. Research efforts to address that question resulted in important discoveries with relevance to HCV infection and beyond.
IFN-λ Region Genetic Variants Strongly Associate with HCV Treatment Response and Spontaneous Clearance
Between 2009 and 2010, 4 groups of investigators independently reported results of genome-wide association studies (GWAS) that linked genetic variants in the IFN-λ chromosomal region with response to peg-IFNα/ribavirin therapy for CHC (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009; Rauch and others 2010). In a GWAS, individuals are tested for hundreds of thousands of single nucleotide polymorphisms (SNPs) that have been selected to cover all regions of the human genome with the goal of identifying regions that harbor functional genetic variants (Feero and others 2010). Ge and others (2009) found rs12979860 to be the SNP most significantly associated with SVR. Whereas most GWAS of noninfectious outcomes have yielded associations of modest effect size (Feero and others 2010), individuals with the rs12979860-CC genotype had a 2-fold greater rate of SVR than those with the rs12979860-CT or -TT genotypes. In addition, Ge and others (2009) reported that African American participants were less likely to have the favorable rs12979860-CC genotype, which offered an explanation for previous observations of poorer treatment response in that group. Genotype for rs12979860 also associated with the level of HCV RNA before treatment, but in a surprising fashion. Higher HCV RNA is generally associated with poorer treatment response (Ge and others 2009), however, the rs12979860-CC genotype associated with both higher pretreatment viral levels and a higher rate of SVR (Ge and others 2009).
In reports published almost simultaneously with that of Ge and others, 2 other groups reported consistent findings. The rs12979860 was not included on the array used by these investigators and their findings focused on a nearby SNP (rs8099917). Among individuals of European ancestry who were treated with peg-IFNα/ribavirin, Suppiah and others (2009) observed that the rs8099917 SNP was strongly related with a 2-fold association with SVR. Those with at least 1 copy of the rs8099917-G allele had higher SVR rates compared with noncarriers. The strongest associations were seen in homozygotes for the unfavorable genotype (rs8099917-TT) compared to individuals who carry the rs8099917-G allele (Suppiah and others 2009). In a Japanese population, Tanaka and others (2009) also found that individuals who were homozygous for the rs8099917-T allele had poorer response to IFNα-based therapy and were less likely to clear the virus than those who carry at least one copy of the rs8099917-G allele. The strong associations reported in these landmark studies suggested that the identified SNPs might have clinical utility for predicting response to treatment of CHC and established the IFN-λ region as playing a major role in HCV clearance.
Soon thereafter, studies linked these SNPs with spontaneous clearance of HCV. Thomas and others (2009) observed that rs12979860-CC genotype associated with spontaneous clearance in a manner similar to the findings for treatment response; homozygotes for the rs12979860-C allele were 3 times more likely to have cleared HCV as those who carried the rs12979860-T allele. Those investigators also were the first to provide a global map of the distribution of the rs12979860 variant, demonstrating dramatic differences in allele frequencies among African, European, and Asian populations. The favorable rs12979860-C is the major allele in Asians and Europeans, but the minor allele in Africans (Thomas and others 2009). In a Swiss cohort, Rauch and others (2010) showed that genotype for rs8099917 associated with both spontaneous HCV clearance and response to treatment with peg-IFNα/ribavirin.
At the time of those groundbreaking findings regarding HCV infection, the discovery of the IFN-λ region itself was still relatively recent. In 2003, 2 independent groups of investigators reported the discovery of 3 novel genes with similarity to type I IFNs, although with signaling via a receptor with expression that was much more limited than those for type I IFNs (Kotenko and others 2003; Sheppard and others 2003). Because IFN-λ was thought to be largely redundant to type I IFNs, the connection between genetic variation in the IFN-λ region and clearance of HCV infection was not obvious.
Discovery of IFNL4 Explains Association of IFN-λ Variants with HCV Clearance
Once a relevant chromosomal region is identified based on associations with SNPs in a GWAS, additional studies are needed to attempt to identify the functional genetic variant underlying that signal (Collins and others 1997; Feero and others 2010). Knowledge of linkage disequilibrium (LD), the nonrandom correlation between alleles at different loci (Slatkin 2008), is used to identify possible functional variants. It is noteworthy that although GWAS have implicated specific genetic regions for a wide range of disease phenotypes, in most cases a functional variant is yet to be identified (Feero and others 2010). The discovery of IFNL4 provides an exception to the common situation.
IFNL4 and the IFN-λ4 protein were discovered by our collaborative group during a search for that functional genetic variant (Prokunina-Olsson and others 2013). In discovering IFNL4, we showed that the rs12979860 SNP lies within intron 1 of IFNL4 and that rs8099917 lies nearer IFNL4 than IFNL3 (Fig. 1) (Prokunina-Olsson and others 2013). IL28B was the official gene symbol for IFNL3 at the time of the 2009 HCV GWAS articles (the current nomenclature was adapted upon the discovery of IFNL4 in 2013) and confusion on the location of these GWAS markers continues with these SNPs still frequently referred to as IFNL3 or “IL28B” variants. Unfortunately, this problem extends beyond semantics. There is a common misunderstanding that variation in IFNL3 underlies genetic associations with HCV clearance.
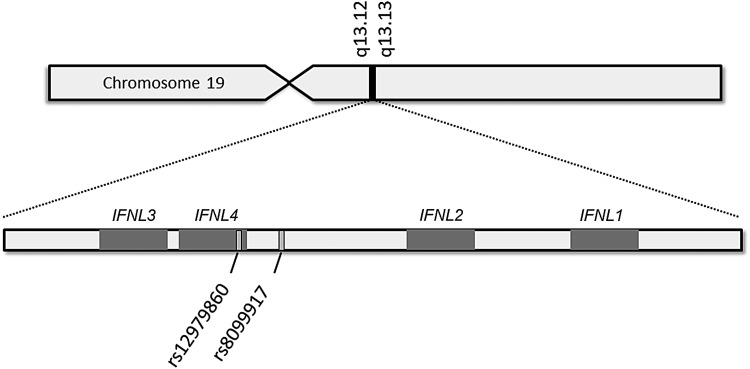
FIG. 1.
SNPs associated with response to peg-IFNα/ribavirin therapy and spontaneous clearance of HCV in GWAS (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009). rs12979860 lies within IFNL4 and rs8099917 lies nearer IFNL4 than IFNL3, therefore, these SNPs are properly termed variants of IFNL4, which was discovered subsequent to the GWAS reports (Prokunina-Olsson and others 2013). GWAS, genome-wide association studies; HCV, hepatitis C virus; IFN, interferon; SNP, single nucleotide polymorphism.
The rs12979860 and rs8099917 SNPs are in LD with a common dinucleotide frameshift variant that we call IFNL4-ΔG/TT (rs368234815; Fig. 2). This variant controls IFNL4, with the IFNL4-ΔG allele creating an open reading frame for full-length IFN-λ4 protein and the alternative allele (IFNL4-TT) abrogating the protein (Prokunina-Olsson and others 2013). Thus, IFNL4-ΔG is a functional variant that controls the generation of IFN-λ4 and IFN-λ4 is produced only by those with at least one copy of the ancestral IFNL4-ΔG allele.
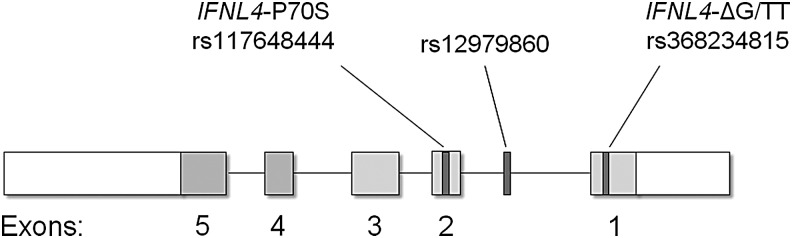
FIG. 2.
IFN-λ4 structure and genetic variants. The IFNL4-ΔG/TT variant (rs368234815), which controls generation of IFN-λ4, lies in exon 1; GWAS marker rs12979860 lies in intron 1; the IFNL4-P70S variant (rs117648444), which modifies IFN-λ4, lies in exon 2 (Prokunina-Olsson and others 2013).
Genotype for the IFNL4-ΔG/TT variant explains associations between IFN-λ variants and HCV clearance that were observed in GWAS. The IFNL4-ΔG allele is in high LD with the rs12979860-T allele in Asians (r2 = 1.00), correlates to a lesser extent in Europeans (r2 = 0.91), and has the weakest LD in Africans (r2 = 0.71) (Prokunina-Olsson and others 2013). To compare the relative strength of associations for IFNL4-ΔG/TT variant and the rs12979860 GWAS marker, we conducted analyses among African Americans, in whom LD between these variants is weaker than those of European ancestry. Examining decline in HCV RNA levels through the first 28 days of treatment with peg-IFNα/ribavirin among African American participants in the Virahep-C Trial, genotype for the IFNL4-ΔG/TT variant was a stronger predictor than IFNL4-rs12979860 genotype (P = 0.015; Fig. 3) and the IFNL4-ΔG allele, which generates the IFN-λ4 protein, was linked to worse response (Prokunina-Olsson and others 2013). We found consistent results in analyses for spontaneous HCV clearance among African American individuals (Prokunina-Olsson and others 2013; Aka and others 2014). Other investigators demonstrated that treatment outcomes were more strongly associated with IFNL4-ΔG/TT genotype than rs12979860 genotype in Europeans (Bibert and others 2013; Franco and others 2014). It seems paradoxical that expression of an additional IFN-λ interferon interferes with viral clearance (O'Brien and others 2014). Other articles in this special issue address this issue (Prokunina-Olsson and others, JICR submitted?).
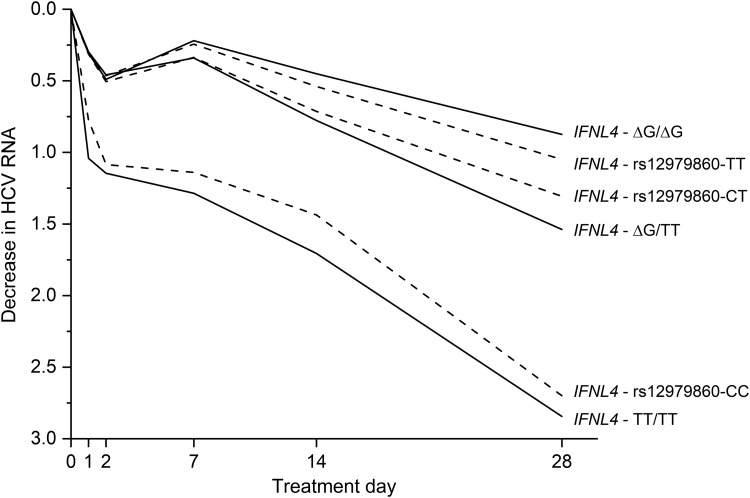
FIG. 3.
Decline in HCV RNA levels in response to treatment with peg-IFNα/ribavirin among African American participants in the Virahep-C Trial. Genotype for IFNL4-ΔG/TT was a stronger predictor than that for GWAS marker IFNL4-rs12979860 (P = 0.015) (Prokunina-Olsson and others 2013).
Nonsynonymous variant rs117648444 within exon 2 of IFNL4 is a second functional polymorphism that affects both the IFN-λ4 protein and HCV clearance (Fig. 3). This mutation alters amino acid position 70 of IFN-λ4, resulting in the substitution of serine (S70) for proline (P70). Compared to IFN-λ4 P70, the derived IFN-λ4 S70 form produces lower expression of intrahepatic IFN stimulating genes and diminished antiviral activity in vitro, yet individuals with a genotype that yields this weakened form of IFN-λ4 have higher rates of spontaneous HCV clearance and response to peg-IFN-α/ribavirin treatment than those with the P70 variant (Galmozzi and Aghemo 2014; Terczyńska-Dyla and others 2014), which is consistent with the overall finding that generation of IFN-λ4 impairs HCV clearance.
Studies of the protein altering IFNL4-ΔG/TT and IFNL4 P70S polymorphisms firmly establish IFNL4 as the primary gene involved in HCV clearance, however, a functional polymorphism found in the 3′ untranslated region of IFNL3 is also of interest. The rs4803217 SNP affects the degradation of IFNL3 mRNA and binding of HCV-induced microRNA (McFarland and others 2013). Similar to the relationships between the IFNL4-ΔG/TT and IFNL4-rs12979860 variants, LD between the IFNL3 rs4803217-T allele, which enhances IFNL3 mRNA degradation, and the IFNL4-ΔG allele is complete in Asians (r2 = 1.00) and strong in Europeans (r2 = 0.88), but weaker in Africans (r2 = 0.63) (Prokunina-Olsson and others 2013). Again, strong LD makes it difficult to differentiate a possible effect of IFNL3 rs4803217 from that of IFNL4-ΔG/TT in Asians or Europeans. To overcome that issue, we compared associations for closely linked genotypes in African Americans. We found that IFNL4-ΔG/TT was more strongly associated with both response to peg-IFNα/ribavirin therapy and spontaneous HCV clearance than rs4803217 (O'Brien and others 2015). Further, those with the IFNL4-ΔG:rs4803217-G haplotype, a combination that combines the unfavorable IFNL4 allele with the putatively favorable IFNL3 allele, was associated with the lowest SVR rates (O'Brien and others 2015). These results indicate that expression of the IFN-λ4 protein is the primary driver of HCV clearance. The possible contribution of IFNL3 rs4803217 to HCV clearance deserves further examination.
IFNL4 Genotypes Associate with Response to New Therapies for Hepatitis C
Direct-acting antiviral agents (DAAs) that attack viral replication and assembly were introduced in 2011(Scheel and others 2013) and, by 2014, highly effective regimens based on 2 or more second generation DAAs became available. These therapies, which target the HCV NS3/4A protease, NS5B RNA polymerase and NS5A viral proteins, produce cure rates >90% (Messina and others 2015) with fewer adverse effects than IFN-based therapies (Scheel and others 2013; Baumert and others 2019). DAAs have revolutionized the treatment of HCV and led to ambitious goals for reducing the prevalence of CHC and its associated morbidity and mortality worldwide.
Sofosbuvir, which inhibits the HCV NS5B protein, forms the backbone of several DAA regimens. Sofosbuvir can be combined with ribavirin for HCV treatment and Meissner and others (2014) showed that IFNL4-ΔG associated with slower viral decay and decreased drug efficacy in patients with HCV genotype 1 who were treated with this regimen. Sofosbuvir is more effective when combined with other DAAs. Both rs12979860-CC and rs8099917-TT were associated with higher SVR after 12 weeks treatment with a regimen that combined sofosbuvir with daclatasvir, which inhibits the HCV NS5A protein (Khan and others 2019). An assessment of treatment response to a variety of DAA regimens among HCV-1 infected patients treated in the Veterans Health Administration found that those with the IFNL4-ΔG/ΔG genotype had a much lower cure rate than either IFNL4-ΔG/TT or IFNL4-TT/TT genotypes (Backus and others 2018).
Standard treatment duration for sofosbuvir-based regimens is 12 weeks, however, a shorter course may be highly effective in patients with a favorable IFNL4 genotype. The ION-3 trial evaluated the combination of the NS5A inhibitor ledipasvir with sofosbuvir (Kowdley and others 2014). In reanalyzing data from that study, we found that only 2% of patients with the IFNL4-rs12979860-CC genotype failed to respond to treatment at 8 weeks compared to higher rates of failure among carriers of the unfavorable IFNL4-rs12979860-T (O'Brien and others 2017a). Similarly, we observed a 1% relapse rate for patients with the IFNL4-rs12979860-CC genotype who received 8 weeks of a regimen that combined sofosbuvir with the NS5A inhibitor velpatasvir and voxilaprevir, which inhibits HCV NS3/4A protease (O'Brien and others 2017b). These results demonstrate the potential utility of IFNL4 genotyping to inform decisions concerning the appropriate duration of therapy with sofosbuvir regimens. Shorter duration treatments could address the high cost of DAA regimens that is a barrier to fuller implementation of these curative treatments. Decisions regarding the appropriate duration of DAA treatment might be informed through an algorithm based on IFNL4 genotype and other relevant factors (O'Brien and others 2017a). Given patterns observed for spontaneous clearance and response to peg-IFN-α/ribavirin, an analysis of response to DAA treatment that considered both IFNL4-ΔG/TT and IFNL4 P70S might provide the best clinical prediction. Future studies should address that question. Alternatively, in regions such as East Asia where the frequency of the IFNL4-TT/TT genotype is very high, it might be cost saving to treat all patients for a shorter duration. Thus, knowledge regarding genotype for the IFNL4-ΔG/TT variant at either the individual or population level, might improve cost-effectiveness of treating the 71 million who are infected with HCV.
IFN-λ Region Variants Associate with Hepatic Inflammation and Fibrosis
Evidence that variants in the IFN-λ region associated with HCV clearance led to interest in whether these polymorphisms might also play a role in the progression of CHC. Necroinflammatory grade is thought to be the best predictor of fibrosis progression, which can lead to cirrhosis (Goodman 2007). In 2012, Bochud and others reported results from a study of ∼2,000 European patients with CHC. They found that the rs8099917-G allele, which is linked with impaired HCV clearance and was subsequently shown to be in LD with the IFNL4-ΔG allele, associated with decreased necroinflammatory activity, fibrosis, and fibrosis progression (Bochud and others 2012). Also among patients with CHC, Noureddin and others found rs12979860-CC genotype was associated with significantly higher portal inflammation and alanine aminotransferase levels. However, in a longitudinal analysis of paired biopsy results, genotype for rs12979860 was not associated with the frequency of fibrosis progression in that cohort (Noureddin and others 2013). In a subsequent study, Eslam and others examined the relationship between the IFNL4 rs12979860 marker and hepatic inflammation and fibrosis progression among 4,000 European-ancestry patients. Consistent with Bochud and others, they demonstrated that the rs12979860-CC genotype, which is associated with increased HCV clearance, associated with increased inflammation and fibrosis in CHC patients. Furthermore, they showed that this relationship extended to individuals with chronic hepatitis B and nonalcoholic fatty liver disease (NAFLD) (Eslam and others 2015). In 2017, Petta and others (2017) reported that the functional IFNL4 rs368234815-TT allele, which abrogates IFN-λ4, is associated with more severe fibrosis in NAFLD patients.
Taken together, those studies suggested that IFN-λ4 protects against hepatic inflammation and fibrosis of both infectious and noninfectious etiologies. However, results from a second article by Eslam and others challenged that hypothesis. Among ∼2000 patients of European ancestry with CHC, they examined associations of genotype for the IFNL4-ΔG/TT, IFNL4 rs12979860, and IFNL3 rs4803217 variants with hepatic inflammation, hepatic fibrosis, and inflammatory cell counts in liver biopsy specimens (Eslam and others 2017). Given the high LD between these genetic variants in that study population, they were unable to determine which polymorphisms had the strongest associations with these outcomes, however, they found no difference in inflammation or fibrosis between the variants that create the IFN-λ4 P70 and IFN-λ4 S70 proteins (Eslam and others 2017), a finding that was inconsistent with previous observations for HCV clearance (Galmozzi and Aghemo 2014; Terczyńska-Dyla and others 2014). Eslam and others also found that quantitative IFNL3 mRNA from liver biopsies of patients with CHC associated with IFNL4 rs12979860 genotype, hepatic inflammation, and hepatic fibrosis. On that basis, they concluded that IFN-λ3, rather than IFN-λ4, was the likely mediator of genetic associations with hepatic inflammation and fibrosis.
Because of the strong LD between IFNL3 and IFNL4 genetic variants in European populations, it remains an open question as to which genetic variant most strongly associates with hepatic inflammation/fibrosis (Park and others 2018). It is possible that the presence or absence of IFN-λ4 plays a role in hepatic inflammation, but the IFN-λ4 P70S does not affect that outcome. Future studies that include populations of African ancestry, in which LD between INF-λ region variants is less than that in Europeans, might help resolve this question. Better understanding of the mechanisms underlying the relationship between INF-λs and fibrosis progression could lead to interventions to prevent cirrhosis. Further, genetic factors could be used to improve risk stratification models for fibrosis progression (Tamaki and others 2015; Eslam and others 2016).
Future Directions—IFN-λ Variants in Infections and Other Conditions
Several lines of evidence suggest genetic variants in the IFN-λ region could play a role in a wider range of infections and other conditions. There was strong selection for the IFNL4-TT allele (which abrogates IFN-λ4), such that the IFNL4-ΔG/TT polymorphism is among the top 0.5% for differences between African and Asian populations differences genome-wide (Key and others 2014). The mutation that inactivated IFN-λ4 expression developed before the out-of-Africa migration and the ancestral IFNL4-ΔG allele is the most common form among Africans, however, subsequent selection resulted in IFNL4-TT becoming the major allele among Europeans and Asians (Prokunina-Olsson and others 2013), with allele frequencies for IFNL4-TT exceeding 90% in East Asian populations. HCV infection is a bloodborne infection transmitted primarily through use or misuse of medical devices, therefore, it was probably not a common infection before the modern era. Given the long period between initial infection and development of medical conditions that would impair reproduction, it is unlikely that HCV infection is the infectious driver behind the selective pressure of IFNL4 mutation. Therefore, loss of IFN-λ4 production may be advantageous in the context some of other infectious or inflammatory conditions (O'Brien and others 2014).
Recent studies in mice demonstrate that IFN-λs protect tissue barriers against a wide range of viral infections. Lazear and others (2015) demonstrated that exogenous IFN-λ3 tightens endothelial junctions of the blood brain barrier in response to West Nile Virus infection, thereby, reducing viral neuroinvasion. Mice lacking the IFN-λ receptor exhibit increased susceptibility to viral pathogens including norovirus (Nice and others 2015), rotavirus (Hernández and others 2015), influenza (Crotta and others 2013; Klinkhammer and others 2018), severe acute respiratory syndrome (Mahlakõiv and others 2012), and herpes simplex virus-2 (Ank and others 2006). Treatment of influenza-infected mice with IFN- λ therapy reduces viral titers and shedding (Crotta and others 2013; Galani and others 2017). Among human volunteers challenged with influenza (Egli and others 2014) or rhinovirus (Contoli and others 2006), reduced viral replication correlated with increased IFN-λ production. In contrast to these apparent benefits, IFN-λ increased bacterial levels of Staphylococcus and Pseudomonas in mice (Cohen and Prince 2013). Understanding the role of IFN-λ in these various conditions has important implications for potential treatment modalities.
In humans, genotype for the IFNL4-ΔG/TT variant has been associated with opportunistic infections in immunocompromised patients and certain cancers. Cytomegalovirus (CMV) retinitis causes visual impairment and blindness. Bibert and others (2014) found a 2-fold increase in the risk of developing CMV-related retinitis among HIV-positive patients with the IFNL4-ΔG/ΔG genotype compared to those with IFNL4-ΔG/TT and IFNL4-TT/TT genotypes. Similar results have been observed for the risk of CMV replication after solid-organ transplantation, whereby carrying the IFNL4-ΔG allele increases an individual's risk of infection (Manuel and others 2015). Human herpes-virus 8 is the causal agent for Kaposi's sarcoma. The haplotype that produces IFN-λ4 P70 was associated with an increased risk of Kaposi's sarcoma, while that which generates the less active IFN-λ4 S70 form was not (Bibert and others 2018). In a GWAS of mucinous ovarian carcinoma, a rare cancer sometimes confused with a metastasis to the ovary, the IFNL4-ΔG allele had a protective association (Kelemen and others 2015). Genotype for the IFNL4-ΔG/TT variant has also been associated with prostate cancer (Minas and others 2018; Tang and others 2018). Taken together, these findings support the hypothesis that genotype for IFNL4-ΔG/TT variant could affect a wide range of conditions. Further research in this area is clearly needed.
Acknowledgments
The authors would like to thank David Check for his assistance with the figures in the article. This research was supported by the Intramural Research Program of the National Institutes of Health (National Cancer Institute, Division of Cancer Epidemiology and Genetics).
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author Disclosure Statement
No competing financial interests exist.
References
- Aka PV, Kuniholm MH, Pfeiffer RM, Wang AS, Tang W, Chen S, Astemborski J, Plankey M, Villacres MC, Peters MG, Desai S, Seaberg EC, Edlin BR, Strickler HD, Thomas DL, Prokunina-Olsson L, Sharp GB, O'Brien TR. 2014. Association of the IFNL4-ΔG allele with impaired spontaneous clearance of hepatitis C virus. J Infect Dis 209(3):350–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. 2006. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol 80(9):4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony PP, Ishak KG, Nayak NC, Poulsen HE, Scheuer PJ, Sobin LH. 1978. The morphology of cirrhosis. Recommendations on definition, nomenclature, and classification by a working group sponsored by the World Health Organization. J Clin Pathol 31(5):395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzumanyan A, Reis HMGPV, Feitelson MA. 2013. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 13(2):123–135 [DOI] [PubMed] [Google Scholar]
- Backus LI, Shahoumian TA, Belperio PS, Winters M, Prokunina-Olsson L, O'Brien TR, Holodniy M. 2018. Impact of IFNL4-ΔG genotype on sustained virologic response in hepatitis C genotype 1 patients treated with direct-acting antivirals. Diagn Microbiol Infect Dis 92(1):34–36 [DOI] [PubMed] [Google Scholar]
- Baumert TF, Berg T, Lim JK, Nelson DR. 2019. Status of direct-acting antiviral therapy for hepatitis C virus infection and remaining challenges. Gastroenterology 156(2):431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FHT, Gerlach T, Malinverni R, Moradpour D, Negro F, Müllhaupt B, Bochud PY, Swiss Hepatitis C Cohort Study. 2013. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 210(6):1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibert S, Wojtowicz A, Taffé P, Manuel O, Bernasconi E, Furrer H, Günthard HF, Hoffmann M, Kaiser L, Osthoff M, Cavassini M, Bochud PY, Swiss HIV Cohort Study. 2014. The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS 28(13):1885–1889 [DOI] [PubMed] [Google Scholar]
- Bibert S, Wójtowicz A, Taffé P, Tarr PE, Bernasconi E, Furrer H, Günthard HF, Hoffmann M, Kaiser L, Osthoff M, Fellay J, Cavassini M, Bochud PY, Swiss HIV Cohort Study. 2018. Interferon lambda 3/4 polymorphisms are associated with AIDS-related Kaposi's sarcoma. AIDS 32(18):2759–2765 [DOI] [PubMed] [Google Scholar]
- Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Müllhaupt B, Gerlach T, Heim MH, Moradpour D, Cerny A, Malinverni R, Regenass S, Dollenmaier G, Hirsch H, Martinetti G, Gorgiewski M, Bourlière M, Poynard T, Theodorou I, Abel L, Pol S, Dufour JF, Negro F, Swiss Hepatitis C Cohort Study Group AN RS HC EP 26 Genoscan Study Group. 2012. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 55(2):384–394 [DOI] [PubMed] [Google Scholar]
- Brown RS. 2005. Hepatitis C and liver transplantation. Nature 436(7053):973–978 [DOI] [PubMed] [Google Scholar]
- Cohen TS, Prince AS. 2013. Bacterial pathogens activate a common inflammatory pathway through IFNλ regulation of PDCD4. PLoS Pathog 9(10):e1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A. 1997. Variations on a theme: cataloging human DNA sequence variation. Science 278(5343):1580–1581 [DOI] [PubMed] [Google Scholar]
- Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, Brown RS, Belle SH, Hoofnagle JH, Kleiner DE, Howell CD, Virahep-C Study Group. 2006. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology 131(2):470–477 [DOI] [PubMed] [Google Scholar]
- Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PAB, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, Slater L, Lewis-Antes A, Kon OM, Holgate ST, Davies DE, Kotenko SV, Papi A, Johnston SL. 2006. Role of deficient type III interferon-λ production in asthma exacerbations. Nat Med 12(9):1023–1026 [DOI] [PubMed] [Google Scholar]
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. 2013. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9(11):e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlin BR, Eckhardt BJ, Shu MA, Holmberg SD, Swan T. 2015. Toward a more accurate estimate of the prevalence of hepatitis C in the United States. Hepatology 62(5):1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli A, Santer DM, O'Shea D, Barakat K, Syedbasha M, Vollmer M, Baluch A, Bhat R, Groenendyk J, Joyce MA, Lisboa LF, Thomas BS, Battegay M, Khanna N, Mueller T, Tyrrell DL, Houghton M, Humar A, Kumar D. 2014. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog 10(12):e1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M, Hashem AM, Leung R, Romero-Gomez M, Berg T, Dore GJ, Chan HLK, Irving WL, Sheridan D, Abate ML, Adams LA, Mangia A, Weltman M, Bugianesi E, Spengler U, Shaker O, Fischer J, Mollison L, Cheng W, Powell E, Nattermann J, Riordan S, McLeod D, Armstrong NJ, Douglas MW, Liddle C, Booth DR, George J, Ahlenstiel G, International Hepatitis C Genetics Consortium (IHCGC). 2015. Interferon-λ rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun 6:6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M, Hashem AM, Romero-Gomez M, Berg T, Dore GJ, Mangia A, Chan HLY, Irving WL, Sheridan D, Abate ML, Adams LA, Weltman M, Bugianesi E, Spengler U, Shaker O, Fischer J, Mollison L, Cheng W, Nattermann J, Riordan S, Miele L, Kelaeng KS, Ampuero J, Ahlenstiel G, McLeod D, Powell E, Liddle C, Douglas MW, Booth DR, George J, Consortium International Liver Disease Genetics. 2016. FibroGENE: a gene-based model for staging liver fibrosis. J Hepatol 64(2):390–398 [DOI] [PubMed] [Google Scholar]
- Eslam M, McLeod D, Kelaeng KS, Mangia A, Berg T, Thabet K, Irving WL, Dore GJ, Sheridan D, Grønbæk H, Abate ML, Hartmann R, Bugianesi E, Spengler U, Rojas A, Booth DR, Weltman M, Mollison L, Cheng W, Riordan S, Mahajan H, Fischer J, Nattermann J, Douglas MW, Liddle C, Powell E, Romero-Gomez M, George J, Consortium the International Liver Disease Genetics. 2017. IFN-λ3, not IFN-λ4, likely mediates IFNL3–IFNL4 haplotype–dependent hepatic inflammation and fibrosis. Nat Genet 49(5):795–800 [DOI] [PubMed] [Google Scholar]
- Feero WG, Guttmacher AE, Collins FS. 2010. Genomic medicine—an updated primer. N Engl J Med 362(21):2001–2011 [DOI] [PubMed] [Google Scholar]
- Forner A, Llovet JM, Bruix J. 2012. Hepatocellular carcinoma. Lancet 379(9822):1245–1255 [DOI] [PubMed] [Google Scholar]
- Franco S, Aparicio E, Parera M, Clotet B, Tural C, Martinez MA. 2014. IFNL4 ss469415590 variant is a better predictor than rs12979860 of pegylated interferon-alpha/ribavirin therapy failure in hepatitis C virus/HIV-1 coinfected patients. AIDS 28(1):133–136 [DOI] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. 2002. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med 347(13):975–982 [DOI] [PubMed] [Google Scholar]
- Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, Thanos D, Doyle SE, Kotenko SV, Thanopoulou K, Andreakos E. 2017. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 46(5):875.e6–890.e6 [DOI] [PubMed] [Google Scholar]
- Galmozzi E, Aghemo A. 2014. Nonsynonymous variant Pro70Ser (rs117648444) in IFNL4 gene identifies carriers of the rs368234815 ΔG allele with higher HCV RNA decline during the first 4 weeks of pegylated interferon and ribavirin therapy in HCV-1 patients. J Clin Virol 59(4):274–275 [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461(7262):399–401 [DOI] [PubMed] [Google Scholar]
- Goodman ZD. 2007. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol 47(4):598–607 [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Chisari FV. 2006. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol 1:23–61 [DOI] [PubMed] [Google Scholar]
- Hernández PP, Mahlakõiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hoelscher C, Dumoutier L, Renauld J-C, Suerbaum S, Staeheli P, Diefenbach A. 2015. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 16(7):698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen LE, Lawrenson K, Tyrer J, Li Q, Lee JM, Seo JH, Phelan CM, Beesley J, Chen X, Spindler TJ, Aben KK, Anton-Culver H, Antonenkova N, Australian Cancer Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium. 2015. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat Genet 47(8):888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key FM, Peter B, Dennis MY, Huerta-Sánchez E, Tang W, Prokunina-Olsson L, Nielsen R, Andrés AM. 2014. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genet 10(10):e1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AJ, Aanand Saraswat V, Ranjan P, Parmar D, Singh Negi T, Mohindra S. 2019. Polymorphism in interferon λ3/interleukin-28B gene and risk to noncirrhotic chronic hepatitis C genotype 3 virus infection and its effect on the response to combined daclatasvir and sofosbuvir therapy. J Med Virol 91(4):659–667 [DOI] [PubMed] [Google Scholar]
- Kinzie JL, Naylor PH, Nathani MG, Peleman RR, Ehrinpreis MN, Lybik M, Turner JR, Janisse JJ, Massanari M, Mutchnick MG. 2001. African Americans with genotype 1 treated with interferon for chronic hepatitis C have a lower end of treatment response than Caucasians. J Viral Hepat 8(4):264–269 [DOI] [PubMed] [Google Scholar]
- Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakõiv T, Staeheli P. 2018. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 7:e33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4(1):69–77 [DOI] [PubMed] [Google Scholar]
- Kowdley KV, Gordon SC, Rajender Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M, Rustgi V, Chojkier M, Herring R, Di Bisceglie AM, Pockros PJ, Subramanian GM, Di An E, Svarovskaia E, Hyland RH, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Pound D, Fried MW ION-3 Investigators. 2014. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 370(20):1879–1888 [DOI] [PubMed] [Google Scholar]
- Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17(2):107–115 [DOI] [PubMed] [Google Scholar]
- Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. 2003. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology 37(6):1343–1350 [DOI] [PubMed] [Google Scholar]
- Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, Gale M, Klein RS, Diamond MS. 2015. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood–brain barrier. Sci Transl Med 7(284):284ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med 132(4):296–305 [DOI] [PubMed] [Google Scholar]
- Mahlakõiv T, Ritz D, Mordstein M, DeDiego ML, Enjuanes L, Müller MA, Drosten C, Staeheli P. 2012. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J Gen Virol 93(12):2601–2605 [DOI] [PubMed] [Google Scholar]
- Manuel O, Wójtowicz A, Bibert S, Mueller NJ, van Delden C, Hirsch HH, Steiger J, Stern M, Egli A, Garzoni C, Binet I, Weisser M, Berger C, Cusini A, Meylan P, Pascual M, Bochud PY, Swiss Transplant Cohort Study. 2015. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis 211(6):906–914 [DOI] [PubMed] [Google Scholar]
- McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M, Jr., Savan R. 2014. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus–induced microRNAs. Nat Immunol 15(1):72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling M-H, Cort S, Albrecht JK. 1998. Interferon Alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med 339(21):1485–1492 [DOI] [PubMed] [Google Scholar]
- Meissner EG, Bon D, Prokunina-Olsson L, Tang W, Masur H, O'Brien TR, Herrmann E, Kottilil S, Osinusi A. 2014. IFNL4-ΔG genotype is associated with slower viral clearance in hepatitis C, genotype-1 patients treated with sofosbuvir and ribavirin. J Infect Dis 209(11):1700–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61(1):77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas TZ, Tang W, Smith CJ, Onabajo OO, Obajemu A, Dorsey TH, Jordan SV, Obadi OM, Ryan BM, Prokunina-Olsson L, Loffredo CA, Ambs S. 2018. IFNL4-ΔG is associated with prostate cancer among men at increased risk of sexually transmitted infections. Commun Biol 1:191–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir AJ, Bornstein JD, Killenberg PG, Atlantic Coast Hepatitis Treatment Group. 2004. Peginterferon Alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med 350(22):2265–2271 [DOI] [PubMed] [Google Scholar]
- Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347(6219):269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, Zhao X, Conry-Cantilena C, Kleiner DE, Liang TJ, Ghany MG. 2013. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology 58(5):1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TR, Kottilil S, Feld JJ, Morgan TR, Pfeiffer RM. 2017a. Race or genetic makeup for hepatitis C virus treatment decisions? Hepatology 65(6):2124–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TR, Kottilil S, Pfeiffer RM. 2017b. IFNL4 genotype is associated with virologic relapse after 8-week treatment with sofosbuvir, velpatasvir, and voxilaprevir. Gastroenterology 153(6):1694–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TR, Pfeiffer RM, Paquin A, Lang Kuhs KAL, Chen S, Bonkovsky HL, Edlin BR, Howell CD, Kirk GD, Kuniholm MH, Morgan TR, Strickler HD, Thomas DL, Prokunina-Olsson L. 2015. Comparison of functional variants in IFNL4 and IFNL3 for association with HCV clearance. J Hepatol 63(5):1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TR, Prokunina-Olsson L, Donnelly RP. 2014. IFN-λ4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res 34(11):829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, O'Brien TR, Rehermann B. 2018. The role of genetics in hepatic fibrosis among hepatitis C virus patients. Hepatology 67(5):2043–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 45(4):529–538 [DOI] [PubMed] [Google Scholar]
- Petta S, Valenti L, Tuttolomondo A, Dongiovanni P, Pipitone RM, Cammà C, Cabibi D, Di Marco V, Fracanzani AL, Badiali S, Nobili V, Fargion S, Grimaudo S, Craxì A. 2017. Interferon lambda 4 rs368234815 TT>δG variant is associated with liver damage in patients with nonalcoholic fatty liver disease. Hepatology 66(6):1885–1893 [DOI] [PubMed] [Google Scholar]
- Polaris Observatory HCV Collaborators. 2017. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2(3):161–176 [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45(2):164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Günthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Müllhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY, Swiss Hepatitis C Cohort Study, Swiss HIV Cohort Study. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138(4):1338–1345, 1345.e1 [DOI] [PubMed] [Google Scholar]
- Reddy KR, Hoofnagle JH, Tong MJ, Lee WM, Pockros P, Heathcote EJ, Albert D, Joh T. 1999. Racial differences in responses to therapy with interferon in chronic hepatitis C. Consensus Interferon Study Group. Hepatology 30(3):787–793 [DOI] [PubMed] [Google Scholar]
- Scheel TKH, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19(7):837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4(1):63–68 [DOI] [PubMed] [Google Scholar]
- Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A, Zeuzem S, Accelerate Investigators. 2007. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3′. N Engl J Med 357(2):124–134 [DOI] [PubMed] [Google Scholar]
- Slatkin M. 2008. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat rev Genet 9(6):477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Müller T, Bahlo M, Stewart GJ, Booth DR, George J. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41(10):1100–1104 [DOI] [PubMed] [Google Scholar]
- Tamaki N, Kurosaki M, Higuchi M, Takada H, Nakakuki N, Yasui Y, Suzuki S, Tsuchiya K, Nakanishi H, Itakura J, Takahashi Y, Ogawa S, Tanaka Y, Asahina Y, Izumi N. 2015. Genetic polymorphisms of IL28B and PNPLA3 are predictive for HCV related rapid fibrosis progression and identify patients who require urgent antiviral treatment with new regimens. PLoS One 10(9):e0137351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. 2009. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet 41(10):1105–1109 [DOI] [PubMed] [Google Scholar]
- Tang W, Wallace TA, Yi M, Magi-Galluzzi C, Dorsey TH, Onabajo OO, Obajemu A, Jordan SV, Loffredo CA, Stephens RM, Silverman RH, Stark GR, Klein EA, Prokunina-Olsson L, Ambs S. 2018. IFNL4-ΔG allele is associated with an interferon signature in tumors and survival of African-American men with prostate cancer. Clin Cancer Res 24(21):5471–5481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terczyńska-Dyla E, Bibert S, Duong FHT, Krol I, Jørgensen S, Collinet E, Kutalik Z, Aubert V, Cerny A, Kaiser L, Malinverni R, Mangia A, Moradpour D, Müllhaupt B, Negro F, Santoro R, Semela D, Semmo N, Cohort Study Group Swiss Hepatitis, Heim MH, Bochud PY, Hartmann R. 2014. Reduced IFNλ4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nature Commun 5:5699. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461(7265):798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder J, Saraswathula A, Hasselblad V, Muir A. 2016. A systematic review of race and ethnicity in hepatitis c clinical trial enrollment. J Natl Med Assoc 108(1):24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2017. Global hepatitis report, 2017. Geneva: World Health Organization [Google Scholar]