Abstract
The purpose of our study was to investigate the underlying mechanism and functional role of microRNA-145 (miR-145) in cervical cancer. In this study, quantitative real-time PCR (qRT-PCR) was used to detect miR-145 and FSCN1 expression levels in tissues and HeLa cells. Western blotting was performed to determine the protein level of FSCN1. The luciferase assay was used to verify the direct target of miR-145. The CCK-8 assay and 2D colony formation assays were performed to determine the effects of miR-145 mimics or FSCN1 silencing on cell proliferation. miR-145 expression levels were significantly down-regulated, while FSCN1 expression levels were significantly up-regulated in the cervical carcinoma tissues compared with their matched non-cancerous tissues. In addition, FSCN1 expression levels were negatively correlated to miR-145 in tissues. Next, FSCN1 was verified as the direct target of miR-145 in HeLa cells. Moreover, overexpression of miR-145 dramatically inhibited the proliferation of HeLa cells. The silencing of FSCN1 exhibited the similar patterns on cell proliferation as miR-145 overexpression. The miR-145/ FSCN1 axis contributes to the progression of cervical cancer by inhibition of cervical cancer cell proliferation.
Keywords: microRNA (miRNA), cervical cancer, FSCN1
Introduction
Cervical cancer is one of the leading causes of cancer-related deaths worldwide, responsible for 9% and 8% of the total new cancer cases and cancer deaths, respectively1. Cervical cancer develops either from cervical intraepithelial neoplasia or squamous intraepithelial lesions2. Recently, the incidence of cervical cancer has been greatly reduced by introduction of new screening tests, diagnosis, and treatment. Despite these advances, thousands of women still suffer and die from the disease each year. Therefore, molecular biomarkers are urgently needed for cervical cancer, to open up the potential for molecular therapies.
Among molecular biomarkers, microRNAs (miRNAs) play a critical role in research. In recent years, miRNAs have become the center of interest in oncology. miRNAs, a class of short non-coding RNAs (ncRNAs) of ∼22 nucleotides in length, repress the expression of target mRNA by catalyzing mRNA cleavage or by inhibiting mRNA translation3. Numerous studies have demonstrated that the loss or gain of function of specific miRNAs may be key events in the cervical cancer process4. miR-145 was first found to be reduced in colorectal neoplasia. Then, miR-145 dysfunction was also showed in breast, ovarian, lung, nasopharyngeal, bladder, gastric, and prostate cancers2,4–10. However, the biological function and molecular mechanism of miR-145 in cervical cancer still remains unclear.
In this study, we demonstrated that miR-145 was significantly down-regulated, while FSCN1 was significantly up-regulated in the cervical carcinoma tissues. In addition, the expression of miR-145 is negatively correlated with FSCN1 levels in cervical cancer tissues. Next, we verified that FSCN1 is directly repressed by miR-145. Subsequently, we observed that overexpression of miR-145 leads to decreased cell proliferation of HeLa cells. Moreover, FSCN1 silencing showed a similar effect on cell proliferation as miR-145 overexpression. Our results imply that miR-145 may regulate the process of cervical cancer via targeting FSCN1.
Although miR-145/FSCN1 has been reported to be involved in the progression of cancer in different cancer types, this study provides the first evidence of the regulatory mechanisms of the miR 145/FSCN1 pathway in cervical cancer. Thus, our data indicate that the miR-145/FSCN1 axis may serve as a potential therapeutic candidate target for cervical cancer.
Materials and Methods
Cell Culture
HeLa cells were purchased from ATCC (Rockville, MD, USA) and cultured in RPMI 1640 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, penicillin and streptomycin (100 U/mL, and 100 μg/mL). The cells were incubated at 37°C in a humidified 5% CO2 and 95% air atmosphere.
qRT-PCR
qRT-PCR was used to determine the expression levels of miR-145 and FSCN1 in cells and tissues. Total RNA was extracted from HeLa cells and tissues with TRI Reagent (Molecular Research Center, Cincinnati, OH, USA). Residual DNA was removed using DNA-free DNase (Ambion, Austin, TX, USA). Reverse transcription was performed using the TaqManTM advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). The analysis of miR-145 levels was performed using the TaqMan Reverse Transcription Kit and TaqMan MicroRNA Assays Kit with U6 as an internal control (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions.
FSCN1 quantitation was determined by SYBR Green I detection with gene-specific primers (Forward primer: 5’-GGAGACCGACCAGGAGAC-3’, reverse: 5’-GATTGGACGCCCTCAGTG-3’) on an ABI 7500 system (Applied Biosystems). GAPDH (Forward Primer, 5′-CCTGCACCACCAACTGCTTA-3′. Reverse Primer, 5′-GGCCATCCACAGTCTTCTGAG-3′) was used as an internal control.
All independent PCR-based reactions were performed in triplicate. The selected internal controls for miRNA and gene were based on previous studies11,12. The amount of each gene and miRNA relative to their internal control was calculated using the comparative Ct method13.
3’ UTR Luciferase Assay
The 3’ UTR of FSCN1 identified by Targetscan was cloned into the pmirGLO vector (Promega, Madison, WI, USA). HeLa cells were seeded in a 96-well plate at the density of 2×104 per well. The cells were contransfected with miR-145 mimic or negative control (miR-Ctrl) and 3’ UTR vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with Opti-MEM (Thermo Fisher) as previously described14. Then a mutant 3′UTR plasmid was created by site-directed mutagenesis at the four miR-145 binding sites (Promega). After 48 h incubation, the cells were lysed for measurement of luciferase activities using the Dual Luciferase Reporter Assay System (Promega). The firefly luciferase signal was normalized to the Renilla luciferase activity.
miR-145 Overexpression
HeLa cells were seeded in the 6-well plates (1×105 per well) the day before transfection, and then transfected with 100 nM of miR-145 mimic or miR-Ctrl by the RNAiMAX Reagent according to manufacturer protocols (Invitrogen).
FSCN1 Silencing
Small interfering RNA for FSCN1 (si-FSCN1) and the respective control (si-Ctrl) were purchased from GenePharma (San Diego, CA, USA). The cells were transfected with100nM Si-FSCN1 and si-Ctrl by the Lipofectamine 2000 Reagent were according to manufacturer protocols (Invitrogen).
Western Blotting
The whole cells were lysed in lysis buffer (Thermo Fisher), and protein concentrations were quantified with Pierce BCA Protein Assay Kit (Thermo Fisher). Protein samples were separated with 4–15% polyacrylamide SDS gels and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The primary antibodies were used: anti-FSCN1 at 1:5000 dilution (ab49815, Abcam, Cambridge, UK) and anti-GAPDH at 1:500 (G9545, Sigma, St. Louis, MO, USA) followed by the HRP-conjugated secondary antibodies (Sigma). The signals were determined using an enhanced chemiluminescence (Thermo Fisher). All experiments were performed in triplicate. Separate blots were used for each independent experiment to avoid problems related to incomplete membrane stripping.
Cell Proliferation
Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Gaithersburg, MD, USA) allows sensitive colorimetric assays for the determination of the number of viable cells in cell proliferation. It is based on WST-8 reduced by dehydrogenases in cells to give an orange-colored product (formazan). The amount of the formazan dye produced in cells is directly proportional to the number of living cells. HeLa cells were seeded at the density of 5×103 cells per well in the 96-well plates. At the indicated time-point, CCK8 solution was added to each well and incubated for 1 h following the manufacturer’s protocol.
2D Colony Formation Assay
HeLa cells were released from clusters by trypsin and cell counts were made. The suspended cells were seeded in the 6-well plate at the density of 1,000 cells per well. After 14 days of culture, the cells were fixed in 80% ethanol and dyed with crystal violet solution (Millipore).
Statistical Analysis
SPSS17.0 for Windows (SPSS, Inc., Chicago, IL) was used to execute the statistical analysis. Data were presented as the mean ± standard errors from at least three independent experiments, and the difference between two conditions was analyzed by the two-tailed, unpaired Student’s t-test. The statistical significance was defined as p < 0.05. The Spearman’s rank correlation analysis was performed to analyze the correlation. p (probability) <0.05 was considered to indicate a statistically significant difference.
Results
miR-145 is Down-regulated in Cervical Cancer Tissues
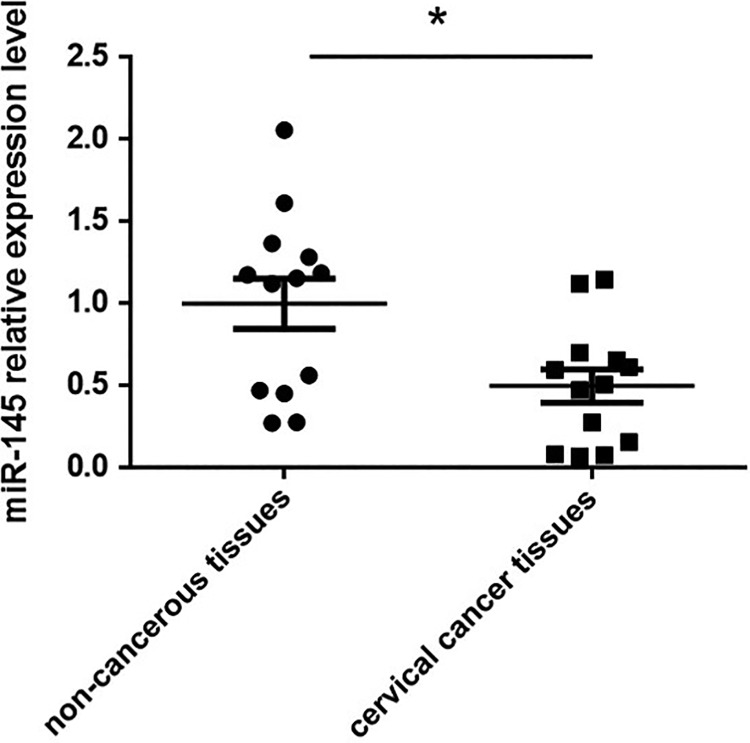
MiR-145 has been found to be tumor-suppressive in various cancer types15; however, data are still limited regarding miR145 and cervical cancer. In this study, we first assessed whether or not miR-145 was dysregulated in cervical cancer tissues. miR-145 expression levels from 13 pairs of cervical cancer tissues and their adjacent non-cancerous tissues were analyzed by qRT-PCR. We found significantly lower levels of miR-145 in cervical cancer tissues compared with their matched non-cancerous tissues (Fig. 1). These results suggested that miR-145 down-regulation could contribute to the tumor progression.
Fig. 1.
The expression levels of miR-145 were analyzed in cervical cancer tissues and paired non-cancerous tissues by qRT-PCR. *p < 0.05 vs non-cancerous tissues.
FSCN1 is Up-regulated and Negatively Correlated with miR-145 in Cervical Cancer Tissues
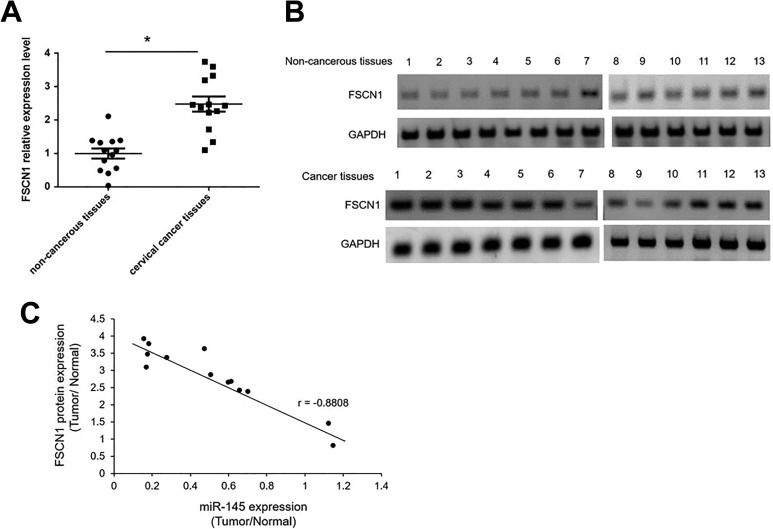
FSCN1 overexpression has been indicated in patients with breast cancer16. FSCN1 has also been verified as a direct target of miR-145 in other cancer types17,18. We determined FSCN1 expression levels in the 13 pairs of cervical cancer tissues and their adjacent non-cancerous tissues using qRT-PCR. As shown in Fig. 2A, FSCN1 mRNA expression levels are significantly up-regulated 2.5 fold in tumor tissues compared with non-cancerous tissues. The protein levels of FSCN1 were also higher in tumor tissues than in non-cancerous tissues (Fig. 2B). In cervical cancer tissues, miR-145 was down-regulated while FSCN1 was up-regulated. To elucidate whether FCSN1interacts with miR-145 in cervical cancer, we executed a correlation analysis between the expression of FSCN1 and miR-145. The protein expression of FSCN1 levels correlated inversely with miR-145 (Fig. 2C).
Fig. 2.
FSCN1 expression levels were negatively correlated with miR-145 levels in cervical cancer tissues (A) The expression levels of FSCN1 were analyzed in cervical cancer tissues and paired non-cancerous tissues by qRT-PCR. *p < 0.05 vs non-cancerous tissues. (B) The FSCN1 protein levels were analyzed by Western blotting in in cervical cancer tissues and paired non-cancerous tissues. (C) the correlation between the levels of miR-145 and FSCN1 protein from 13 pairs of cervical cancer and their matched non-cancerous tissues was determined by non-parametric correlation Spearman’s correlation test.
miR-145 Directly Suppresses FSCN1
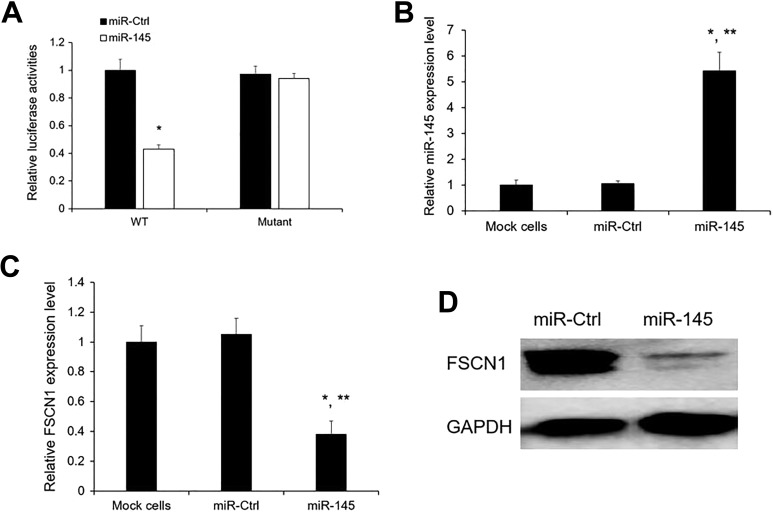
To elucidate whether FSCN1 is directly repressed by miR-145, HeLa cells were co-transfected with the FSCN1 3’-UTR and miR-145 mimic or miR-Ctrl. Luciferase reporter assay results showed that co-transfection with miR-145 obviously reduced the luciferase activity of the FSCN1 3’-UTR wild-type (WT) reporter gene, but not the mutant reporter gene (Fig. 3A), but miR-Ctrl had no effect on the luciferase activity of both WT and mutant (Fig. 3A). Next, we detected the expression level of FSCN1 after miR-145 overexpression (Fig. 3B) by qRT-PCR and Western blotting in HeLa cells. We noticed that the FSCN1 mRNA and protein levels were significantly decreased by miR-145 mimic compared with the miR-Ctl (Fig. 3C, D).
Fig. 3.
FSCN1 is the direct target of miR-145. (A) Relative luciferase activity of HeLa cells after co-transfection with wild-type (WT) or mutant FSCN1 3’ UTR reporter genes and miR-145 or miR-Ctrl. The relative luciferase activity was normalized to Renilla activities. *p < 0.05 vs miR-Ctrl. (B, C) miR-145 and FSCN1 expression levels after miR-145 overexpression were detected by qRT-PCR. (D) The FSCN1 protein levels were analyzed by Western blotting in cells overexpressing miR-Ctrl and miR-145 mimics. The results are presented as mean ± SE (n = 3). *p < 0.05 vs mock cells; **p < 0.05 vs miR-Ctrl.
miR-145 Inhibits Cell Proliferation Through Regulating FSCN1
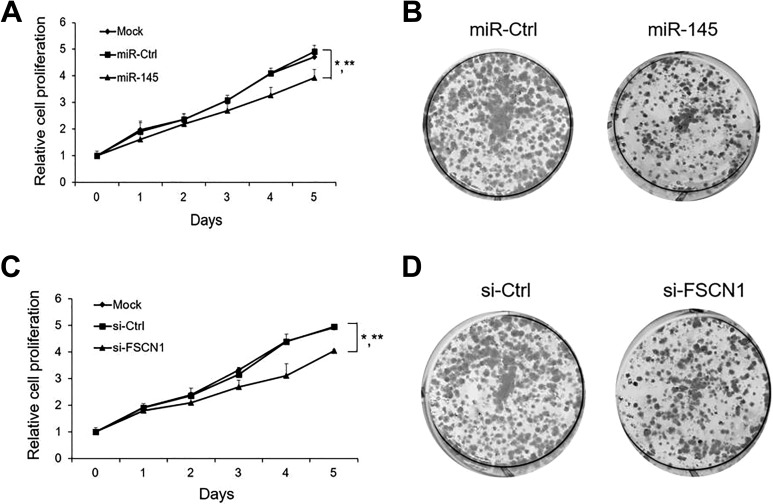
To decipher the biological role of miR-145/FSCN1 in the progression of cervical cancer, we monitored the cell proliferation rate after miR-145 overexpression. As shown in Fig. 4A, CCK-8 cell proliferation assays indicated that proliferation was inhibited when HeLa cells were transfected with miR-145 mimics compared with miR-Ctrl and mock cells. Furthermore, the 2D colony formation assay also revealed that miR-145 overexpression dramatically inhibited HeLa cell growth (Fig. 4B). Next, we silenced FSCN1 to assess whether FSCN1 also involved in the regulation network. We observed similar patterns of decreased cell proliferation after FSCN1 silencing in HeLa cells (Fig. 4C, D). Our data showed that miR-145/ FSCN1 signaling could significantly inhibit the proliferation of cervical cancer cells.
Fig. 4.
miR-145/ FSCN1 axis effect on the proliferation of HeLa cells (A) Relative cell proliferation rate of mock cells, miR-Ctrl or miR-145 transfected cells in vitro was monitored by CCK-8 assay.(B) representative images of the colony formation potential of HeLa cells overexpressing miR-Ctrl or miR-145. (C) Relative cell proliferation rate of mock cells, si-Ctrl or si-FSCN1 transfected cells in vitro was monitored by CCK-8 assay. (D) representative images of the colony formation potential of HeLa cells overexpressing si-Ctrl or si-FSCN1. The results are presented as mean ± SE (n = 3). *p < 0.05 vs mock cells; **p < 0.05 vs miR-Ctrl.
Discussion
A growing body of studies have suggested that miRNAs are important regulators in various cellular processes, such as cell cycle control, growth, apoptosis, and migration. Aberrant expression of miRNAs has been reported to be associated with a broad spectrum of human diseases, including cancer19–21. miR-145 has been frequently reported as a tumor suppressor in various kinds of cancers22,23. miR-145 up-regulation has been found to inhibit SW620 human colon cancer cells24. miR-145 has also been observed to inhibit invasion of bladder cancer cells by targeting PAK125. In addition, miR-145 has been shown to suppress human retinoblastoma cell proliferation and invasion by targeting ADAM1926. However, elucidating the features of expression and roles of miR-145 in cervical cancer remains an ongoing process. In this study, miR-145 was found to be down-regulated in cervical carcinoma tissues compared with the matched non-cancerous tissues. Moreover, the overexpression of miR-145 can inhibit HeLa cell proliferation.
In order to understand the effects of miR-145 on the progression of cervical cancer, we studied the target gene regulated by miR-145. Fascin actin-bundling protein 1 (FSCN1) was verified as the direct target of miR-145 in esophageal cancer14. FSCN1 has been reported to play significant roles in biological processes in cancers. Recently, FSCN1 has been found to promote epithelial–mesenchymal transition through increasing snail1 in ovarian cancer cells27. FSCN1 has also been shown to contribute to tumor suppression in gastric cancer cells28. Moreover, FSCN1 has been found as a biomarker for the development and progression of breast cancer16. In this study, FSCN1 was found to be up-regulated in cervical carcinoma tissues. In addition, FSCN1 was also identified as a direct target of miR-145 by 3’ UTR luciferase assay and Western blotting. Furthermore, FSCN1 silencing exhibited similar patterns of decreased cell proliferation as miR-145 overexpression. Future study will need to verify the in vitro results in an in vivo animal model.
In summary, this study suggests that miR-145 can inhibit cell growth in HeLa cells in vitro through directly regulating FSCN1. This newly identified miR 145/FSCN1 pathway implicates that tight regulation of the miR-145/FSCN1 pathway may be a basis for therapeutic applications in cervical cancer in the future.
Footnotes
Ethical Approval: The study was approved by the Institutional Ethics Committee of Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang, Henan, China.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the 2018 Henan Province medical science and technology research plan joint construction project (No.B2018108), Luoyang Central Hospital Incubation project(No.2018—FY17).
ORCID iD: Ling-Ling Li  https://orcid.org/0000-0001-9194-2527
https://orcid.org/0000-0001-9194-2527
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Park TW, Fujiwara H, Wright TC. Molecular biology of cervical cancer and its precursors. Cancer. 1995;76(10 Suppl):1902–1913. [DOI] [PubMed] [Google Scholar]
- 3. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774(2):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pardini B, De Maria D, Francavilla A, Di Gaetano C, Ronco G, Naccarati A. MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer. 2018;18(1):696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. [DOI] [PubMed] [Google Scholar]
- 6. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. [DOI] [PubMed] [Google Scholar]
- 7. Michael MZ, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 8. Takagi T, Iio AY. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77(1):12–21. [DOI] [PubMed] [Google Scholar]
- 9. Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–352. [DOI] [PubMed] [Google Scholar]
- 10. Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li H, DiRenzo J, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15(4):1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo M, Gao Z, Li H, Li Q, Zhang C, Xu W, Song S, Ma C, Wang S. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci Rep. 2018;8(1):4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su J, Zhang R, Dong J, Yang C. Evaluation of internal control genes for qRT-PCR normalization in tissues and cell culture for antiviral studies of grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2011;30(3):830–835. [DOI] [PubMed] [Google Scholar]
- 13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 14. Liu R, Liao J, Yang M, Sheng J, Yang H, Wang Y, Pan E, Guo W, Pu Y, Kim SJ, Yin L. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One. 2012;7(3):e33987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skjefstad K, Johannessen C, Grindstad T, Kilvaer T, Paulsen EE, Pedersen M, Donnem T, Andersen S, Bremnes R, Richardsen E, Al-Saad S, et al. A gender specific improved survival related to stromal miR-143 and miR-145 expression in non-small cell lung cancer. Sci Rep. 2018;8(1):8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang CQ, Tang CH, Wang Y, Jin L, Wang Q, Li X, Hu GN, Huang BF, Zhao YM, Su CM. FSCN1 gene polymorphisms: biomarkers for the development and progression of breast cancer. Sci Rep. 2017;7(1):15887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y-Q, He Y-M, Ren X-Y, Tang X-R, Xu Y-F, Wen X, Yang X-J, Ma J, Liu N. MiR-145 inhibits metastasis by targeting fascin actin-bundling protein 1 in nasopharyngeal carcinoma. Plos One. 2015;10(3):e0122228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Lin Q. MicroRNA-145 inhibits migration and invasion by down-regulating FSCN1 in lung cancer. Int J Clin Exp Med. 2015;8(6):8794–8802. [PMC free article] [PubMed] [Google Scholar]
- 19. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–R776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. James CD. Aberrant miRNA expression in brain tumors: a subject attracting an increasing amount of attention. Neuro Oncol. 2013;15(4):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montagnana M, Benati M, Danese E, Giudici S, Perfranceschi M, Ruzzenenete O, Salvagno GL, Bassi A, Gelati M, Paviati E, Guidi GC, et al. Aberrant microRNA expression in patients with endometrial cancer. Int J Gynecol Cancer. 2017;27(3);459–466. [DOI] [PubMed] [Google Scholar]
- 22. Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13(2):330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X, Yu J, Guan X, Jiang BH, Liu LZ. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim Biophys Acta. 2013;1829(2):239–247. [DOI] [PubMed] [Google Scholar]
- 24. Li C, Xu N, Li YQ, Wang Y, Zhu ZT. Inhibition of SW620 human colon cancer cells by upregulating mi RNA-145. World J Gastroenterol. 2016;22(9):2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ, Wang X, He D, Guo P. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol. 2014;32(6):846–854. [DOI] [PubMed] [Google Scholar]
- 26. Sun Z, Zhang A, Jiang T, Du Z, Che C, Wang F. miR-145 suppressed human retinoblastoma cell proliferation and invasion by targeting ADAM19. Int J Clin Exp Pathol. 2015;8(11):14521–14527. [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Lu J, Ye Z, Han X, Zheng X, Hou H, Chen W, Li X, Zhao L. 20(S)-Rg3 blocked epithelial-mesenchymal transition through DNMT3A/miR-145/FSCN1 in ovarian cancer. Oncotarget. 2017;8(32):53375–53386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang M, Dong BB, Lu M, Zheng MJ, Chen H, Ding JZ, Xu AM, Xu YH. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. Onco Targets Ther. 2016;9(Issue 1):1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]