Abstract
Cerebral organoids are an emerging cutting-edge technology to model human brain development and neurodevelopmental disorders, for which mouse models exhibit significant limitations. In the human brain, synaptic connections define neural circuits, and synaptic deficits account for various neurodevelopmental disorders. Thus, harnessing the full power of cerebral organoids for human brain modeling requires the ability to visualize and analyze synapses in cerebral organoids. Previously, we devised an optimized method to generate human cerebral organoids, and showed that optimal organoids express mature-neuron markers, including synaptic proteins and neurotransmitter receptors and transporters. Here, we give evidence for synaptogenesis in cerebral organoids, via microscopical visualization of synapses. We also describe multiple approaches to quantitatively analyze synapses in cerebral organoids. Collectively, our work provides sufficient evidence for the possibility of modeling synaptogenesis and synaptic disorders in cerebral organoids, and may help advance the use of cerebral organoids in molecular neuroscience and studies of neurodevelopmental disorders such as autism.
Keywords: cerebral organoids, human brain, stem cells, synapses, neurodevelopmental disorders, autism, synaptic deficits
Introduction
Cerebral organoids are stem-cell derived, three-dimensional (3D) neural-tissue-like structures, that recapitulate early (ca. first trimester) in vivo developing brain tissues. While an in vitro system, cerebral organoids perhaps provide the only portal to studies of human-specific brain development and disease in 3D. Cerebral organoids recapitulate lamination and organization of the human cortex1, diversity of the cell types observed in vivo (such as neuronal subtypes, glial cells, and neuroepithelial and neuroprogenitor cells2,3), and even the gene expression profiles observed during human fetal neocortical development as revealed by single-cell RNA-sequencing analyses4. The organoid technology is still in its rather early stages, but the promise it poses to the study of the human-specific developmental and disease neuroscience is enormous, which has encouraged ongoing efforts to further improve the technology and reduce its limitations, including attempts for circuitry reconstruction5–8, in order to enhance the organoids’ utility to study some aspects of human brain development or developmental disorders.
Human cerebral organoids enabled detection of disease-relevant phenotypes and characterization of disease mechanisms that could not be detected in traditional models1,9, including in vivo mouse models or the human two-dimensional model (stem-cell-derived neurons). Some disease-associated gene mutations that are responsible for severe phenotypes and devastating neurodevelopmental conditions in humans were found to exhibit no phenotype, or a marginal, barely significant, phenotype in the mouse brain. For example, mouse models for microcephaly-associated mutations (as in CDK5RAP2 (CDK5 (cyclin-dependent kinase 5) related activator protein 2 or CDK5 regulatory subunit-associated protein 2)) failed to show the severe microcephalic phenotype associated with these mutations in human patients1,9,10. Organoids were also used to investigate the pathological mechanisms of Zika virus infection and its relationship to dysregulation of neurogenesis and neuroinflammatory pathways and to microcephaly in humans11–14. Moreover, human forebrain organoids derived from induced pluripotent stem cells (iPSCs) of patients carrying a mutation in Disrupted-in-schizophrenia 1 (DISC1), a gene associated with multiple neuropsychiatric disorders such as schizophrenia, affective disorders and autism spectrum disorders (ASDs)15,16, revealed a role of this gene in cell cycle progression in radial glial cells and neurogenesis17. Arguably, cerebral organoids were also reported to recapitulate a neurodegenerative disease model. For example, cerebral organoids derived from a familial Alzheimer’s disease (FAD) patient’s iPSCs showed aggregation of amyloid-β and hyperphosphorylated tau18. Moreover, cerebral organoids harboring a frontotemporal dementia (FTD)-associated tau mutation showed evidence of a CDK5-dependent tau phosphorylation and a decrease in synaptophysin levels19.
Thus far, cerebral organoids have mostly been used for studies that are focused on functions of the radial glial and neural progenitor cells (including the aspects of proliferation and neurogenesis) 13,20–22,17 or on non-synaptic aspects of neuronal biology such as neuronal differentiation, neuronal migration, or gene expression and transcriptional pathways analyses7,8,23. However, synaptic function is at the very center of neuronal functions and is probably the most important neuronal function24–26. Moreover, analyzing synapses is essential to understanding the disease mechanisms of neurodevelopmental disorders that are caused by deficits in synapses27–30. Despite its immense importance, thorough synapse analyses have not yet been performed in cerebral organoids. Scarcely, some Ca2+ imaging experiments and electrophysiological analyses were performed on organoids1,5. While electrophysiological approaches, despite being highly tedious, provide important insights into synaptic connectivity, synaptic microscopical analyses (such as described here) complement the electrophysiological approaches by directly visualizing synapses and determining synapse counts31,32.
Therefore, in this study, we attempted to visualize synaptogenesis in cerebral organoids, via confocal microscopy. We have tested numerous approaches for analyzing synapses, and concisely report here multiple assays that were shown to be compatible with cerebral organoids. We provide ample procedural details and technical tips to allow researchers to successfully reproduce these assays. Thus, we expect this study to help advance the field of study of neurodevelopmental disorders and synaptogenesis in the human cerebral organoid model.
Materials and Methods
Chemicals and Reagents
Materials used in generation of organoids were previously described in details33. The following reagents were used for preparing the organoids for synapse analysis: Tissue-Tek® O.C.T. Compound (Cat # 25608-930; Sakura, Alphen aan den Rijn, The Netherlands), Tissue Path™ Disposable Base Molds (Cat # 22-363-553; Fisher Scientific, Hampton, NH, USA), Goat serum (Cat # 16210064; Invitrogen, Carlsbad, CA, USA), Vectashield 4′,6-diamidino-2-phenylindole (DAPI) mounting medium (Cat # H-1200; Vector Labs, Burlingame, CA, USA), Adhesive glass slides (Cat # I6172PLUS, Thermo Fisher, Waltham, MA, USA), Coverslips (Cat # 102222; Thermo Fisher), Kimwipes (Cat # 06-666; Fisher Scientific), and paraformaldehyde stock (32%) solution (Cat # 15714; Electron Microscopy Sciences (EMS), Hatfield, PA, USA).
Paraformaldehyde solution for tissue fixation (4% paraformaldehyde solution, pH 7.4) can be prepared by 1:8 dilution of the 32% stock solution in PBS pH 7.4; 4% paraformaldehyde solution is to be stored in amber-colored or light-proof containers at 4°C for 1–2 weeks.
The following solutions were used: Blocking Solution (5–10% goat serum, 0.1% BSA, 0.3% Triton X-100 in PBS), and Antibody Vehicle Solution (AVS) (1–2% goat serum, 0.1% Triton X-100 in PBS).
Primary Antibodies
The following primary antibodies were used: Synapsin 1 (SYN1) antibody, rabbit polyclonal antiserum (Cat # 106002; Synaptic Systems, Göttingen, Germany) was used at the dilution of 1:1000. Microtubule-Associated Protein 2 (MAP2) antibody, chicken polyclonal IgY (Cat # 188006; Synaptic Systems) was used at the dilution of 1:1000. Glutamate NMDA Receptor Subunit 1 (GluNR1) antibody, mouse monoclonal IgG (Cat # 114011; Synaptic Systems) was used at the dilution of 1:500. Vesicular Glutamate Transporter 1 (VGLUT1) antibody, Guinea pig polyclonal antiserum (Cat # AB5905; Millipore, Burlington, MA, USA) was used at the dilution of 1:1000.
Secondary Antibodies
Secondary antibodies conjugated with Alexa Fluor-488, -546 or -633 (Molecular Probes, Invitrogen) were used for confocal microscopy, as follows: Goat anti-rabbit IgG secondary antibody—Alexa Fluor-488 (Cat # A-11008; Invitrogen); Goat anti-mouse IgG secondary antibody—Alexa Fluor-546 (Cat # A-11030; Invitrogen); Goat anti-chicken IgY secondary antibody—Alexa Fluor-546 (Cat # A-11040; Invitrogen); Goat anti-Guinea pig IgG secondary antibody—Alexa Fluor-633 (Cat # A-21105; Invitrogen). Secondary antibodies were generally used at the dilution of 1:1000, except the anti-Guinea pig—Alexa Fluor-633 secondary antibodies, which were used at the dilution of 1:500.
Equipment
The following equipment was used: Liquid nitrogen tank and liquid nitrogen bucket, ImmEdge™ Hydrophobic Barrier Pen (Cat # 101098-065; VWR, Radnor, PA, USA), temperature-controlled microtome cryostat (Leica Research Cryostat, Cat # CM3050 S; Leica Biosystems, Buffalo Grove, IL, USA), Confocal microscope (Nikon, or Zeiss), NIS-ElementsTM software (Nikon, Tokyo, Japan) and sterile forceps.
Generation and Microscopy of Cerebral Organoids
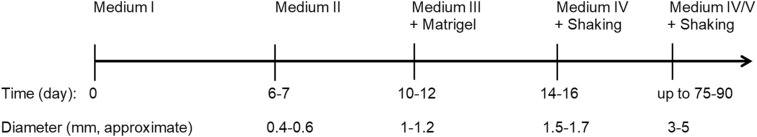
Generation of the human cerebral organoids was performed as summarized in Fig 1. Briefly, human cerebral organoids were generated from human embryonic stem cells (ESCs), (H1 or WA-01 cell line) in a stepwise procedure that encompasses five stages: (1) induction of embryoid bodies by seeding the ESCs dispersed in Medium I (contains Knockout serum replacement and basic fibroblast growth factor (bFGF)) on a low-attachment surface for 6–7 days; (2) induction of neuroectoderm for 4–5 days in the presence of Medium II (contains N2-supplement and heparin); (3) embedding the developing organoid in a drop of matrigel and growing it in a stationary culture containing Medium III (contains Neurobasal Medium and B27-supplement) for 4–5 days; (4) transferring the developing organoids to shaking cultures containing Medium IV (contains Neurobasal Medium, B27-supplement and retinoic acid); and (5) if desired, additional “maturating factors” (e.g. ascorbic acid, cyclic adenosine monophosphate (cAMP), brain-derived neurotropic factor (BDNF), glial cell line-derived neurotropic factor (GDNF), and transforming growth factor-β (TGFβ)) may be added to the organoids from day 60 post-induction of the embryoid bodies. Neuron-rich organoids can be observed within 40–50 days from the initial induction of embryoid bodies. Optimal organoids at the age of ∼75 days reach a diameter of ∼3–5 mm. Mature neurons could be reached by day 75–90 from the initial induction. Detailed composition of the various reagents was described previously33. Organoids analyzed in this study were used on day 80 post-induction. Three independent batches of organoids were analyzed; in each batch, 10–20 organoids were analyzed. From each organoid ∼10 sections were arbitrarily selected for staining and confocal microscopy.
Fig 1.
Overview of the cerebral organoid development procedure. Human ESCs were induced into cerebral organoids using the indicated multi-step procedure, where different media and treatments were introduced at each stage of the protocol. Timeline and approximate size measurements at each stage are also indicated.
To prepare the organoids for microscopy and synapse imaging, place the organoid in a sterile Petri dish. With the help of a surgical microscope, use a blunt tool such as blunt-end forceps or a 1000 μl pipette tip to carefully unwrap the organoid from the matrigel coat. Discard the matrigel wrap. Work carefully and quickly to avoid damaging the organoid structure. Immerse the matrigel-freed organoid in OCT CompoundTM in Tissue Path Disposable Base Molds. Make sure that there is a layer of OCT CompoundTM below and above the organoid (i.e. the organoid should be completely surrounded by OCT CompoundTM from all directions). If the organoid sinks down to the bottom of the mold, center it to the middle of the OCT CompoundTM using a blunt pipette tip gently. Snap-freeze the OCT CompoundTM-immersed organoid using liquid nitrogen (preferably) or ethanol-dry ice mix. Liquid nitrogen snap-frozen organoids can be stored for long periods of time (6–12 months, or longer) at –80°C.
Before sectioning the organoids, transfer the frozen organoids from –80°C to –20°C for at least 2 h or preferably overnight. We observed that gradually cooling and stabilizing the organoid at –20°C (which is the temperature for cryosectioning) gives better-quality sections that adhere to the slide better. Using a suitable cryostat at approximately –20°C (sometimes we found that –16°C to –17°C temperature may produce better quality sections than slightly lower temperatures), cut organoid sections at the desirable thickness. We found that a section thickness between 8 μm and 15 μm is ideal for on-slide staining and confocal microscopy. Collect the cryostat-cut sections on SuperfrostPlusTM glass slides. The tissue should adhere to the slide; if not, finely adjust the cryostat temperature as noted above, which we found helped reduce this problem. Air-dry the sections at room temperature for ∼15–20 min. Using a barrier pen, draw a line (hydrophobic barrier) around each section, to help save expensive reagents (such as antibodies) and reduce expenses.
Fix the sections on the slide using a suitable fixative. Testing various fixatives and fixation durations, we found that cold 4% PFA for 8–12 min is optimal. Remove the fixative solution off the section using a pipette, and add ∼100 μl PBS on the tissue. Perform this wash process twice, for 5 min each time, at room temperature. Block non-specific binding for 1 h at room temperature, by adding ∼100 μl (enough volume to cover the tissue) of Blocking Solution to the section. Incubate the sections with primary antibody (diluted in AVS) overnight at 4°C on a rotating shaker. Optimizing antibody dilution and exposure duration may be performed for the different antibodies. Wash the primary antibody off the sections, with PBS, two to three times, each for 5–20 min (the number of washes and the wash duration may be optimized for each antibody). Incubate the sections with fluorophore-conjugated secondary antibody diluted in AVS for 45–60 min at room temperature in the dark. Remove the secondary antibody off the sections and wash with PBS for three to four times, each for 15 min (numbers and duration of washes may be optimized depending on the secondary antibody to be used). Carefully remove any remaining moisture on the slide using Kimwipes without touching the sections. Mount the sections in DAPI-containing mounting medium, and cover the slide with the coverslip being cautious to not introduce air bubbles under the coverslip. Proceed to imaging using a suitable confocal microscope with a 100x (or at least 63x) magnification lens for best visualization of synapses, as performed here. Slides can be stored at room temperature in light-proof containers.
Results
Synapse Analysis Using Single Staining for SYN1 or GluNR1
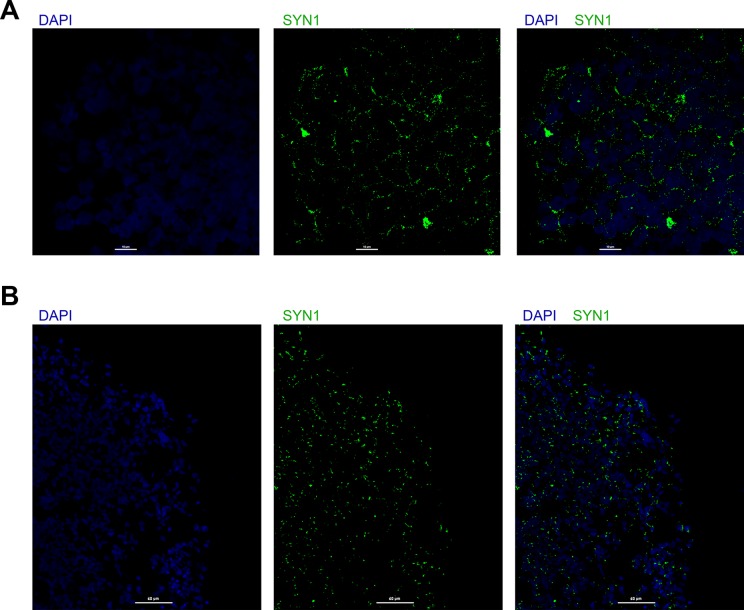
Organoid sections can be stained for SYN1 or GluNR1, via the procedure described above and using primary antibodies against SYN1 or GluNR1, and secondary antibodies conjugated with a suitable fluorophore, such as Alexa Fluor-488. Punctate SYN1 or GluNR1 staining denotes synapses, as shown in Fig 2.
Fig 2.
Synapse analysis using single staining for SYN1 or GluNR1. A. Organoid sections were stained for SYN1 (green) and nuclei (DAPI, blue), and imaged using confocal microscopy; scale bar 10 μm. B. Organoid sections were stained for GluNR1 (green) and nuclei (DAPI, blue) and imaged using confocal microscopy; scale bar 60 μm.
Synapse Analysis Using SYN1 and MAP2 Double Staining
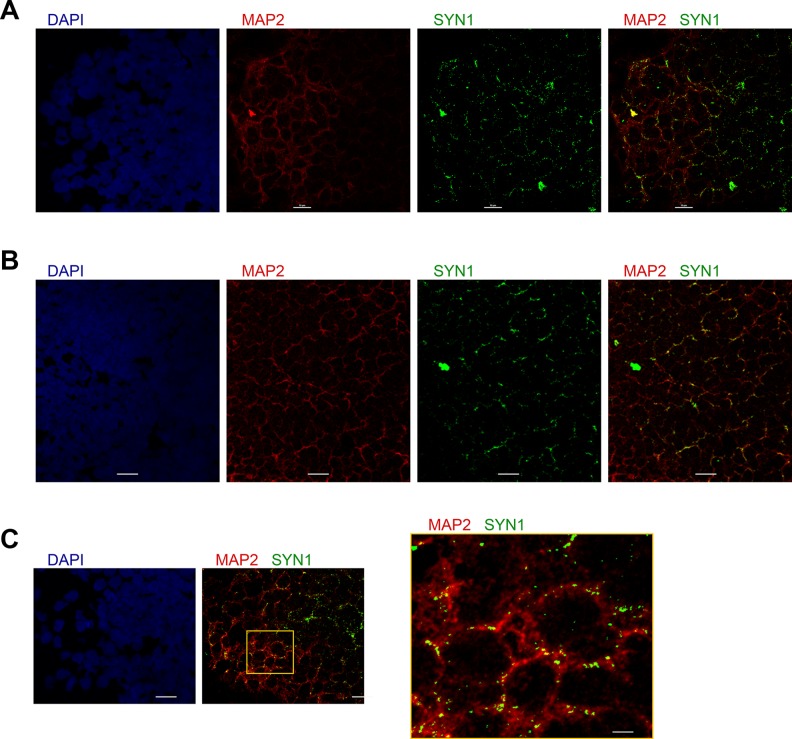
Organoid sections can be stained for MAP2 (stains neuronal dendrites) and SYN1 (stains synapses) via the procedure above, using primary antibodies against SYN1 and MAP2, and secondary antibodies conjugated with Alexa Fluor-488 (SYN1, green) and -546 (MAP2, red). Synapses can be observed as SYN1+ punctae that colocalize with MAP2 (Fig 3).
Fig 3.
Synapse analysis using double staining for SYN1 and MAP2. A, B. Various organoid sections stained for SYN1 (green), MAP2 (red), and nuclei (DAPI, blue); scale bar 10 μm. C. An organoid section stained for SYN1 and MAP2 (scale bar 10 μm); right panel shows a higher magnification of the box-enclosed area in the middle panel (scale bar 2 μm).
Synapse Analysis Using SYN1 and PSD95 Double Staining
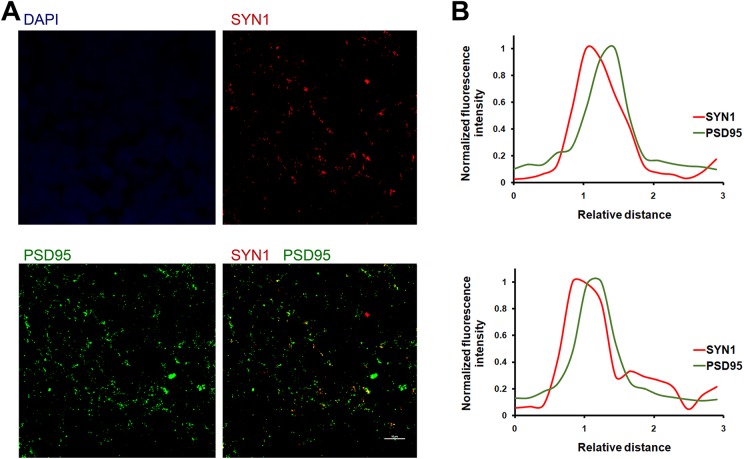
SYN1 is a presynaptic adaptor protein, while PSD95 is a postsynaptic adaptor present on the excitatory postsynapses. Thus, synapses appear as punctae that are double-positive for both SYN1 and PSD95. The organoid sections can be stained for SYN1 and PSD95 via the procedure above, using primary antibodies against SYN1 and PSD95, and Alexa Fluor-488- or -546-conjugated secondary antibodies. Fig 4A shows synapses in organoid sections that stain for both SYN1 and PSD95. Analysis of synaptic punctae should yield a Fluorescence-Relative distance curve, similar to that shown in Fig 4B, where the puncta shows that the presynapse and the postsynapse are spatially proximal yet separated, with SYN1 occupying the presynaptic localization and PSD95 occupying the postsynaptic localization. We analyzed a large number of synapses (∼100 synapses) throughout the section, and show a representative example in Fig 4B. This analysis can be performed using automated microscopy software (NIS-Elements; Nikon), which measures the distance of each fluorophore in a selected puncta relative to a reference point. The analysis confirms the spatial separation of the pre- and post-synaptic markers in the puncta; a bona-fide synapse should show that the presynaptic and the postsynaptic markers are spatially separated (relative distance > 0) and not superimposable (relative distance = 0).
Fig 4.
Synapse analysis using double staining for SYN1 and PSD95. A. An organoid section was stained for SYN1 (red), PSD95 (green), and nuclei (DAPI, blue); scale bar 10 μm. B. Fluorescence–Relative distance analysis for two random Syn1+/PSD95+ punctae in (A), showing the spatial separation of SYN1 and PSD95 peaks, denoting their presynaptic and postsynaptic localizations, respectively. Puncta analysis was performed using Nikon’s NIS-ElementsTM software.
Synapse Analysis Using SYN1 and GluNR1 Double Staining
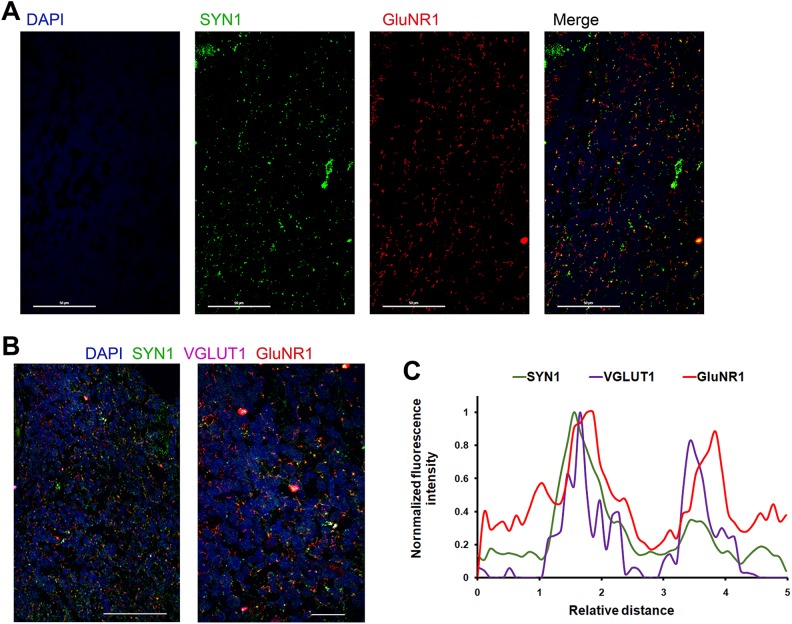
While SYN1 is a presynaptic marker, GluNR1 is a postsynaptic glutamate (NMDA) receptor on excitatory synapses. Thus, synapses appear as punctae double positive for both SYN1 and GluNR1. Organoid sections can be stained as usual, using primary antibodies against SYN1 and GluNR1, and Alexa Fluor-488 or -546-conjugated secondary antibodies. Synapses can be observed as SYN1+/GluNR1+ punctae as shown in Fig 5A.
Fig 5.
Synapse analysis using double and triple staining for SYN1, GluNR1, and VGLUT1. A. An organoid section stained for SYN1 (green), GluNR1 (red), and nuclei (DAPI, blue); scale bar 50 μm. B. An organoid section stained for SYN1 (green), GluNR1 (red), VGLUT1 (purple), and nuclei (DAPI, blue); scale bar 50 μm (left panel) or 10 μm (right panel). C. Fluorescence–Relative distance curves for two arbitrarily chosen SYN1+/VGLUT1+/GluNR1+ punctae in (B), showing spatial separation of SYN1, or VGLUT1, and GluNR1, and indicating the presynaptic localization of SYN1 or VGLUT1 and the postsynaptic localization of GluNR1. Analysis of the punctae was performed using Nikon’s NIS-ElementsTM software.
Synapse Analysis Using SYN1, VGLUT1 and GluNR1 Triple Staining
Synapses could be visualized using triple staining for SYN1, GluNR1 along with VGLUT1, a glutamate transporter present on the presynapse. Follow the staining Protocol above, using primary antibodies against SYN1, VGLUT1 and GluNR1, and Alexa Fluor-488, -546, and -633-conjugated secondary antibodies. Thus, synapses appear as punctae that are double and triple positive for SYN1, VGLUT1, and GluNR1, as shown in Fig 5B. Analysis of any synaptic puncta should yield a Fluorescence-Relative distance curve, similar to that shown in Fig 5C, where the punctae show that the presynapse and the postsynapse are spatially proximal but separated, with SYN1 and VGLUT1 localized presynaptically and GluNR1 localized postsynaptically.
Discussion
Synaptic function mediates circuitry development in the brain and regulates animal behavior. Deficits in circuitry development underlie devastating neurodevelopmental conditions, such as ASD, intellectual, communication and learning disorders, attention-deficit/hyperactivity disorder (ADHD), and other conditions. Harnessing the power of the organoid model to understand the mechanisms of these conditions requires the ability to analyze synapses in such a promising model. Here, we have described simple, immunofluorescence-based methods that we have tested and optimized to visualize and analyze synapses in sections of human cerebral organoids.
Synapses’ proteomic composition is very complex, comprising hundreds of proteins and protein isoforms34,35, and varies significantly between synapse subtypes35–37, and thus synapse analysis is not a simple task. Previous studies on the mouse cortex have attempted to label synapses using various presynaptic proteins that are common to all synapse subtypes as synaptic markers, such as synaptophysin, bassoon, and SYN1, but only SYN1, a phosphoprotein that is largely concentrated at the presynaptic boutons38,39, showed the most robust labeling, characterized by clearly visible punctae with very little nonspecific (non-synaptic) labeling40. Quite intriguingly, comparing the results of electron microscopy (EM) and immunofluorescence analyses in parallel has shown that the vast majority of synapses confirmed by EM stained positively for SYN1, further stressing the reliability of SYN1 immunofluorescence labeling for assaying synapses40. Learning from all those studies, we have devised here the confocal microscopical synapse analyses described, using SYN1 staining. SYN1 staining yields a non-cell-body punctate staining pattern characteristic of synapses24,40.
In addition to SYN1 on the presynapse, glutamate receptors decorate the excitatory postsyanpses34–36. Glutamate NMDA receptors are of cardinal importance for mediating Ca2+-dependent postsynaptic processes associated with long-term potentiation and depression and learning and memory41,42. Although presynaptic or extrasynaptic presence of NMDA receptors was suggested by some reports43, presynaptic or extrasynaptic expression of NMDA receptors was shown to be confined to a small subset of neurons in the mouse brain and mostly for the 2B subunit of the receptor (GluN2B)44–46. The overwhelming majority, however, of NMDA receptors are expressed at the excitatory postsynapses and have been used in the field as a reliable postsynaptic marker in mouse brain sections or neuron culture47–59. While multiple NMDA receptor subtypes exist, GluNR1 is the most important subunit as it is required for activity and is expressed mostly postsynaptically60, which subunit we have used in our assays here. GluNR1 staining in organoid sections yielded a punctate staining pattern as expected for synapses. Moreover, the spatial analysis of the SYN1/VGLUT1/GluNR1 synaptic punctae in Fig 5C clearly shows GluNR1’s postsynaptic localization and distance from the presynaptic proteins (SYN1 or VGLUT1); notice that SYN1 and VGLUT1 peaks are spatially superimposable or very close to one another, whereas the GluNR1 peak is significantly farther (denoting postsynaptic localization).
Furthermore, to add more confidence to SYN1 or GluNR1 single-staining-based immunofluorescence visualization of synapses, we have devised double and triple labeling involving SYN1 along with another presynaptic marker such as VGLUT1, or postsynaptic markers such as PSD95 and GluNR1. Colocalization of the presynaptic marker with the postsynaptic marker and the Fluorescence-Relative distance curves of those synapses collectively confirm the synaptic nature of the immunopositive punctae visualized microscopically.
Traditionally, besides immunofluorescence, synapses have been analyzed using EM. EM is probably the most accurate assay of visualizing synapses; however, EM approaches may have some limitations, such as being a low-throughput method, limiting the possibility of using it to compare a large number of experimental conditions61,62. We believe that the immunofluorescence- and confocal microscopy-based approaches described here will complement other approaches to visualize synapses in organoids representing multiple genetic or pharmacological conditions, and the conditions that seem promising via this immunofluorescence microscopical approach could be further validated and analyzed for synaptic connectivity and neurotransmitter release via electrophysiological approaches31,32. Overall, the work described here will enhance the utility of the human cerebral organoid model in studies of circuitry and molecular neuroscience and to investigate synaptic deficits that account for certain neurodevelopmental disorders.
Footnotes
Ethical Approval: Ethical approval is not applicable to this article.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: This article involves no human subjects; and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545(7652):48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jo J, Xiao Y, Sun AX, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19(2):248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112(51):15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian X, Nguyen HN, Song MM, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35(7):659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14(7):743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell. 2010;18:913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hattori N. Cerebral organoids model human brain development and microcephaly. Mov Disord. 2014;29(2):185. [DOI] [PubMed] [Google Scholar]
- 11. Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. [DOI] [PubMed] [Google Scholar]
- 12. Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19(2):258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, et al. Recent zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 2017;20(3):397–406.e5. [DOI] [PubMed] [Google Scholar]
- 14. Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, Kane LT, et al. Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from zika virus infection. Cell Stem Cell. 2016;19(6):703–708. [DOI] [PubMed] [Google Scholar]
- 15. Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders–cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69(2):428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomson PA, Malavasi EL, Grunewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol. 2013;8(1):1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RYC, Kim J, Song H, Ming GL, et al. DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis. Neuron. 2017;96(5):1041–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. Plos One. 2016;11(9):e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seo J, Kritskiy O, Watson LA, Barker SJ, Dey D, Raja WK, Lin YT, Ko T, Cho S, Penney J, Silva MC, et al. Inhibition of p25/Cdk5 Attenuates Tauopathy in Mouse and iPSC Models of Frontotemporal Dementia. J Neurosci. 2017;37(41):9917–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20(4):435–449.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mora-Bermúdez F, Badsha F, Kanton S, Camp JG, Vernot B, Köhler K, Voigt B, Okita K, Maricic T, He Z, Lachmann R, et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife. 2016;5:e18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171(4):877–889.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, Lachman HM. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism. 2017;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Missler M, Südhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol. 2012;4(4):a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pickel V, Segal M, (eds.). The Synapse- Structure and Function. New York: Academic Press; 2014. [Google Scholar]
- 26. Hell JW, Ehlers MD, (eds.). Structural and Functional Organization of the Synapse. New York: Springer US; 2008. [Google Scholar]
- 27. Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302(5646):826–830. [DOI] [PubMed] [Google Scholar]
- 28. Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4(3):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Etherton M, Földy C, Sharma M, Tabuchi K, Liu X, Shamloo M, Malenka RC, Südhof TC. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci U S A. 2011;108(33):13764–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aminoff MJ. Electrophysiology In: Textbook of Clinical Neurology (3rd). Philadelphia: Saunders; 2007. [Google Scholar]
- 32. Hofer SB, Ko H, Pichler B, Vogelstein J, Ros H, Zeng H, Lein E, Lesica NA, Mrsic-Flogel TD. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat Neurosci. 2011;14(8):1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yakoub AM, Sadek M. Development and characterization of human cerebral organoids: an optimized approach. Cell Transplant. 2018;27(3):393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grant S. Toward a molecular catalogue of synapses. Brain Res Rev. 2007;55(2):445–449. [DOI] [PubMed] [Google Scholar]
- 35. O’Rourke N, Weiler N, Micheva K, Smith SJ. Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat Rev Neurosci. 2012;13(6):365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheng M, Hoogenraad C. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. [DOI] [PubMed] [Google Scholar]
- 37. Sassoè-Pognetto M, Frola E, Pregno G, Briatore F, Patrizi A. Understanding the molecular diversity of gabaergic synapses. Front Cell Neurosci. 2011;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Camilli P, Cameron R, Greengard P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J Cell Biol. 1983;96(5):1337–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hilfiker S, Pieribone VA, Czernik AJ, Kao HT, Augustine GJ, Greengard P. Synapsins as regulators of neurotransmitter release. Philos Trans R Soc Lond B Biol Sci. 1999;354(1381):269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Micheva KD, Busse B, Weiler NC, O’Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68(4):639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lüscher C, Malenka RC. NMDA Receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb Perspect Biol. 2012;4(6):a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li F, Tsien JZ. Memory and the NMDA receptors. N Engl J Med. 2009;361(3):302–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park H, Popescu A, Poo MM. Essential role of presynaptic NMDA receptors in activity-dependent BDNF secretion and corticostriatal LTP. Neuron. 2014;84(5):1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Charton JP, Herkert M, Becker CM, Schröder H. Cellular and subcellular localization of the 2B-subunit of the NMDA receptor in the adult rat telencephalon. Brain Res. 1999;816(2):609–617. [DOI] [PubMed] [Google Scholar]
- 45. Brasier DJ, Feldman DE. Synapse-specific expression of functional presynaptic NMDA receptors in rat somatosensory cortex. J Neurosci. 2008;28(9):2199–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aoki C, Venkatesan C, Go CG, Mong JA, Dawson TM. Cellular and subcellular localization of NMDA-R1 subunit immunoreactivity in the visual cortex of adult and neonatal rats. J Neurosci. 1994;14(9):5202–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51(1):7–61. [PubMed] [Google Scholar]
- 48. Bear MF, Malenka RC. Synaptic plasticity: LTP and LTD. Curr Opin Neurobiol. 1994;4(3):389–399. [DOI] [PubMed] [Google Scholar]
- 49. Green MV, Thayer SA. NMDARs adapt to neurotoxic HIV protein tat downstream of a GluN2A-ubiquitin ligase signaling pathway. J Neurosci. 2016;36(50):12640–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mi R, Sia GM, Rosen K, Tang X, Moghekar A, Black JL, McEnery M, Huganir RL, O’Brien RJ. AMPA receptor-dependent clustering of synaptic NMDA receptors is mediated by Stargazin and NR2A/B in spinal neurons and hippocampal interneurons. Neuron. 2004;44(2):335–349. [DOI] [PubMed] [Google Scholar]
- 51. Hu JL, Liu G, Li YC, Gao WJ, Huang YQ. Dopamine D1 receptor-mediated NMDA receptor insertion depends on Fyn but not Src kinase pathway in prefrontal cortical neurons. Mol Brain. 2010;3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hughes EG, Peng X, Gleichman AJ, Lai M, Zhou L, Tsou R, Parsons TD, Lynch DR, Dalmau J, Balice-Gordon RJ. Cellular and synaptic mechanisms of anti-NMDA receptor encephalitis. J Neurosci. 2010;30(17):5866–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ferreira JS, Rooyakkers A, She K, Ribeiro L, Carvalho AL, Craig AM. Activity and protein kinase C regulate synaptic accumulation of N-methyl-D-aspartate (NMDA) receptors independently of GluN1 splice variant. J Biol Chem. 2011;286(32):28331–28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vastagh C, Gardoni F, Bagetta V, Stanic J, Zianni E, Giampà C, Picconi B, Calabresi P, Di Luca M. N-methyl-D-aspartate (NMDA) receptor composition modulates dendritic spine morphology in striatal medium spiny neurons. J Biol Chem. 2012;287(22):18103–18114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pickard L, Noël J, Henley JM, Collingridge GL, Molnar E. Developmental changes in synaptic AMPA and NMDA receptor distribution and AMPA receptor subunit composition in living hippocampal neurons. J Neurosci. 2000;20(21):7922–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin H, Hsu FC, Baumann BH, Coulter DA, Lynch DR. Cortical synaptic NMDA receptor deficits in α7 nicotinic acetylcholine receptor gene deletion models: implications for neuropsychiatric diseases. Neurobiol Dis. 2014;63:129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoey SE, Williams RJ, Perkinton MS. Synaptic NMDA receptor activation stimulates alpha-secretase amyloid precursor protein processing and inhibits amyloid-beta production. J Neurosci. 2009;29(14):4442–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161(4):805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pérez-Otaño I, Ehlers MD. Homeostatic plasticity and NMDA receptor trafficking. Trends Neurosci. 2005;28(5):229–238. [DOI] [PubMed] [Google Scholar]
- 60. Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14(6):383–400. [DOI] [PubMed] [Google Scholar]
- 61. Gray EG. Electron microscopy of excitatory and inhibitory synapses: a brief review. Prog Brain Res. 1969;31:141–155. [DOI] [PubMed] [Google Scholar]
- 62. Harris KM, Weinberg RJ. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb Perspect Biol. 2012;4(5):a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]