Abstract
A chemoselective primary amine modification strategy which enables the three-component, one-pot bioconjugation is described. The specifically designed, mercaptobenzaldehyde-based bifunctional linker achieves highly selective and robust amine labeling under biocompatible conditions. This linker demonstrates wide functional group tolerance and is simple to prepare which allowed facile payload incorporation. Finally, our studies have shown that the introduction of linker does not impair the function of modified protein such as insulin.
Graphical Abstract
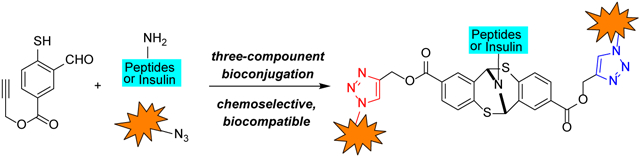
Efficient chemical modification of peptide or protein systems with multiple components has been shown to be essential towards achieving important biological and therapeutic functions.[1] A number of therapeutic applications have recently been reported where proteins have been functionalized with different bioactive components.[2] As the case in point, the usage of multiple antigen modified trivalent peptides to trigger a potent immune response,[3] linkage of polypeptide and biotin in a three-component fashion to facilitate elucidation of intracellular interactome with pull-down assays.[4,5] Despite the aforementioned examples, convergent synthetic strategies to achieve multi-component peptide assemblies in a one-pot reaction remain challenging.
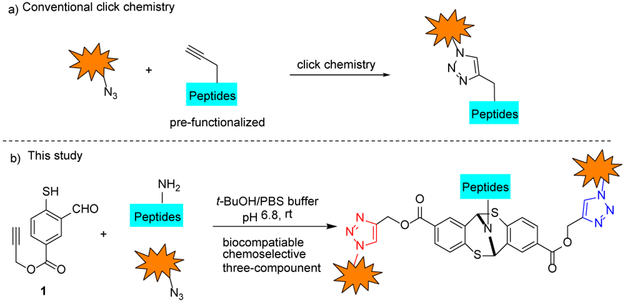
A number of approaches have emerged in recent years.[6] Most notably, Francis et al. reported a three-component Mannich-type transformation for selective tyrosine modification.[7] Meanwhile, Ball and co-workers recently described an organometallic approach for three-component tyrosine bioconjugation.[8] Herein, we report a one-pot three-component protocol that is specifically targeting primary amine position at native polypeptides and proteins under mild biocompatible conditions. The described approach builds upon the conventional ‘click’ modification[9] (Figure 1a) and recently developed bioorthogonal strategies that entail genetic incorporation of unnatural amino acid residues and typically result in mono-functionalized peptides.[10]
Figure 1.
Three-component one-pot bioconjugation.
Our approach is based on the our understanding of the unique reactivity of the thiol group towards imines.[11] We hypothesized that alkynyl mercaptobenzoate 1 will be a suitable heterobifunctional linker which can be concisely synthesized (Figure 1b). The mercaptobenzoate end of the molecule will selectively react with primary amines whereas alkynyl group could facilitate a robust copper-catalyzed azide alkyne cycloaddition (CuAAC)[12] within the protein framework. The proposed chemoselective amine conjugation requires no exogenous additives or reagents, concurrent conjugation of primary amines and CuAAC may be carried out at the same time in a one-pot manner. Lastly, due to the planar structure of aryl ring, the shown modification is unlikely to impair the native functions and activity of the targeted peptides or proteins.[13]
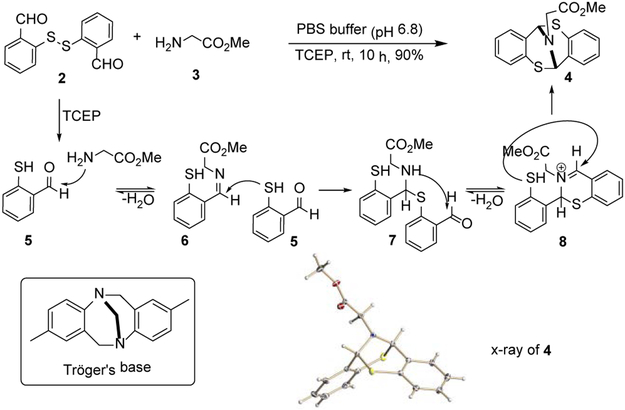
To test our hypothesis, we carried out a model study shown in Scheme 1. A known disulfide substrate 2 was obtained from the commercially available mercapto-benzyl alcohol in two steps. The reaction of 2 and glycine methyl ester under the conditions (TCEP, THF/PBS buffer (1:9)) at neutral pH (6.8) generated 4 containing a unique [3.3.1] bicyclo nonane structure in 90% yield. Previously, compound 4 has been reported as the analogue of Tröger's base.[14] The pioneering work by Toste and co-workers described synthesis of 4 in non-aqueous solution and at the elevated temperature (80 °C).[15] We found that the transformation could be carried out at room temperature and in a biocompatible PBS buffer (Scheme 1). We believe that the reaction began with reduction of the disulfide bond of 2 to liberate mercaptobenzaldehyde 5. Amination of 5 with the glycine methyl ester 3 generated imine 6 which was trapped by a second equivalent of 5. The adduct 7 subsequently underwent intramolecular amination and yielded imine 8. Another intramolecular thiol addition to the iminium intermediate afforded bicyclic product 4, which was confirmed by X-ray analysis.
Scheme 1. Synthesis of bicyclic [3.3.1] framework under mild conditions.a.
[a] Reaction conditions: compound 2 (0.364 mmol) was dissolved in THF (0.4 mL), THF/PBS buffer (1: 9, v/v), rt, 10 h.
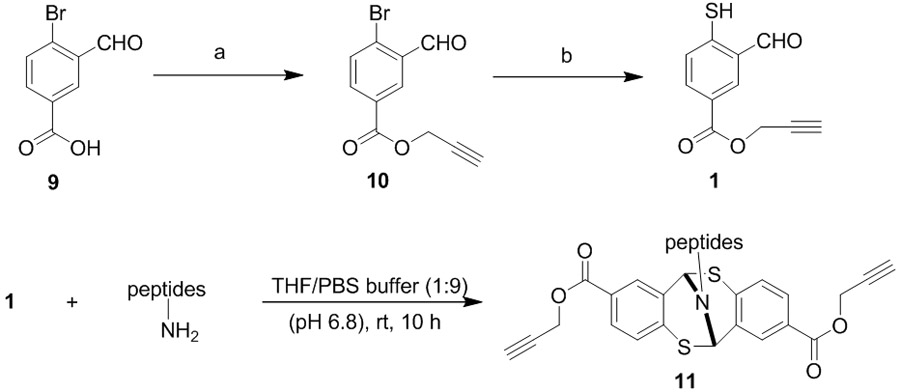
Intrigued by the benign reaction conditions and excellent yields, we hypothesized that the mercaptobenzaldehyde derivatives could be applied toward the modification of primary amines within the protein’s framework. However, utilization of the disulfide 2 would not be ideal. The substrate would require disulfide reduction using TCEP which could also disrupt the protein’s disulfide linkages, thereby denaturing the protein. On the other hand, a motif with a similar structure of the substrate 5 would be preferred. Addition of an alkynyl group facilitates a secondary site of conjugation using well-established CuAAC chemistry. Synthesis of the heterobifunctional linker was carried out in two steps using readily available 4-bromo-3-formylbenzoic acid 9, as shown in Table 1. O-propargylation furnished bromo-benzaldehyde 10 in 95% yield, which was treated with sodium sulfide in dimethylformamide afforded 1 in 90% yield.
Table 1.
Scope of peptides in conjugation with bifunctional linker 1.
 | |||||
|---|---|---|---|---|---|
| peptides | yieldsc | peptides | yields | ||
| 11a | Fmoc-Glu-Gly-Lys-Asn-Ala-Glu-Gly | 83% | 11b | Pro-Ala-Lys-Met-Gln-His-Gly | 88% |
| 11c | Pro-Glu-Asn-Leu-Lys-Tyr | 90% | 11d | Pro-Tyr-Gly-Lys-Gln-Leu-Glu-Gly | 84% |
| 11e | Pro-Gly-Lys-Glu-Asn-Ala-Glu-Gly | 90% | 11f | Pro-Ala-Glu-Lys-Gly-Glu-Asn-Tyr-Gly | 86% |
| 11g | Pro-Glu-Phe-Glu-Ala-Lys-Asn-Leu-Gly-Tyr | 88% | 11h | Val-Glu-Lys-Gln-Ile-Asn-Tyrd | 82% |
| 11i | Val-Glu-Ala-Lys-Asn-Leu-Glu-Glyd | 78% | 11j | Val-Leu-Glu-Lys-Asn-Gly-Glu-Gly-Gln-Tyrd | 71% |
| 11k | Ala-Gln-Ser-Gly-Cys-Glu-Gly | 85% | 11l | Ala-Gln-Leu-Glu-Tyr | 81% |
| 11m | Gly-Tyr-Glu-Ala-Gln-Gly | 75% | 11n | Gly-Glu-Ala-Trp-Leu-Arg-Gly | 89% |
Reaction conditions: propargyl bromide, K2CO3, DMF, rt, 2 h, 95%
Reaction conditions: Na2S, DMF, rt, 5 h, 90%. Compound 1 was dissolved in in THF (80 μL), THF/PBS buffer (1: 9, v/v), final concentration of peptides and compound 1 is 0.01 M and 0.022 M.
Isolated yields.
Structure was confirmed by tandem MS spectrum.
With the desired compound in hand, we next investigated the reactivity of 1 in the bioconjugation settings. Under the standard reaction conditions, the coupling of linker 1 and peptide proceeded smoothly and the bicyclic adduct 11 was produced in high yield in THF/PBS buffer. The small amount of THF was necessary to achieve complete solubility of 1. A small library of peptides was prepared and subjected to the standard bioconjugation conditions (Table 1). It is notable that the described chemistry is compatable with functional groups that are relevant to proteins and polypeptide. Fmoc protected peptides 11a furnished lysine modification products in 83% yield. Hydrophobic and hydrophilic sidechains are well tolerated; lysine could be modified in the presence of adjacent carboxylic acid or amide groups. Secondary amine of proline N-terminus will not engage bioconjugation and lysine was modified with excellent yields (11b-11g). We next examined the chemoselectivity between peptidyl N-terminus and the lysine ε-amino group (entries 11h-11j). Under neutral reaction medium, lysine was selectively modified when the polypeptides constitute sterically demanding amino acids such as valine residues at the N-terminus. The polypeptides bearing either alanine or glycine residues at the N-terminus can also be efficiently functionalized with compound 1 (entries 11k-11n). Overall, the linker 1 demonstrated an exclusive preference for primary amines in the polypeptides. The reaction conditions are compatible with all type of amino acid residues. Commonly known nucleophilic peptidyl sidechains such as His, Cys, Trp, and Ser are compatible with our bioconjugation approach. Other amino acids with polar sidechains (Asn, Glu, Tyr, Met and Arg) would not interfere with the described conjugations.
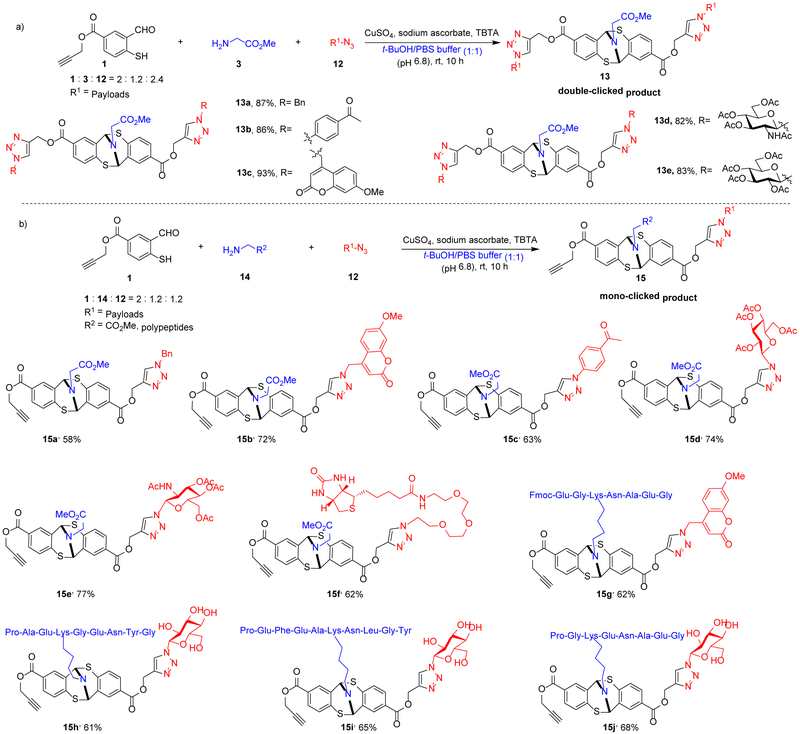
We subsequently investigated one-pot three-component bioconjugation using compound 1 under the CuAAC conditions (CuSO4, sodium ascorbate, TBTA, t-BuOH/H2O (1:1)). The reaction involving linker 1, glycine methylester 3, and the azido cargos 12 generated the double-clicked adducts 13. In one step, through linker 1 and produced 13a and 13b in 87% and 86% yields respectively. Fluorophore azido-methoxycoumarin and benzyl azide and acetophenone azide were connected to glycine monosaccharide azides were evaluated, and 13c-13e were obtained in good to excellent yield.
We have discovered that the level of cargo installation can be controlled using azido compounds as limiting reagents. The one-pot reaction exclusively yielded double-clicked products when the 2.4 equivalents of azido compounds were used (13a-13e) (Scheme 2a). On the other hand, when 1.2 equivalents of azido compounds were used, the mono-clicked adducts were produced (Scheme 2b). Under the mono-clicked conjugation conditions, fluorophore azido-methoxycoumarin was furnished in 72% yield (15b). In addition, we evaluated a variety of azides, such as monosaccharide azides (15d-15e), and a biotinylation reagent azide-PEG3-biotin (15f). these examples successfully modified target amino acids or peptides in good yields in a one-pot fashion. Moreover, one-pot labeling experiments were extended to polypeptides without the erosion of the yields. Methoxycoumarin and glucose was connected to the peptides in good yields under bioconjugation conditions (15h-15j).
Scheme 2. One-pot bioconjugation with different azido compounds.a, b.
[a] Reaction conditions: 0.1 equivalent of CuSO4, 0.2 equivalent of sodium ascorbate, 0.1 equivalent of TBTA and 2 equivalents of 1, 1.2 equivalent of glycine methyl ester hydrochloride and 1.2 or 2.4 equivalent of azide were used. [b] Isolated yields.
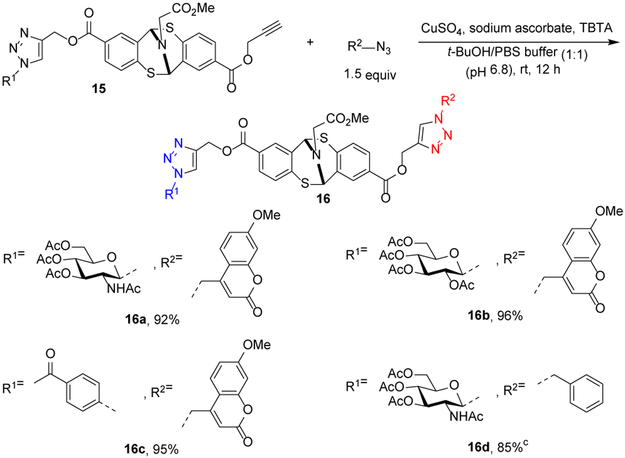
Furthermore, mono-clicked constructs generated in Scheme 2b retained an unreacted alkynyl group which potentially could be used to intergrade a different labeling group. We continued with our investigation with these considerations (Scheme 3). A consequent conjugation of mono-functionalized compounds 15c-15e was achieved under standard reaction conditions. Formation of the heterobifunctional compounds 16 was realized. The conjugate successfully placed both N-acetylglucosamine and methoxycoumarin on the molecule in excellent yield (16a). Glucose, benzylphenone and methoxycoumarin were combined to furnish 16b and 16c with almost quantitative yields. The generation of 16d required longer reaction time and provided an 85% yield. In principle, the highly diversified functional groups could be introduced to the amino sites of the protein.
Scheme 3. Second cargo incorporation of mono-clicked products.a, b.
[a] Reaction conditions: 0.1 equivalent of CuSO4, 0.2 equivalent of sodium ascorbate, 0.1 equivalent of TBTA and 1.5 equivalents of azido compound were used. [b] Isolated yields. [c] Reaction was stirred for 30 h.
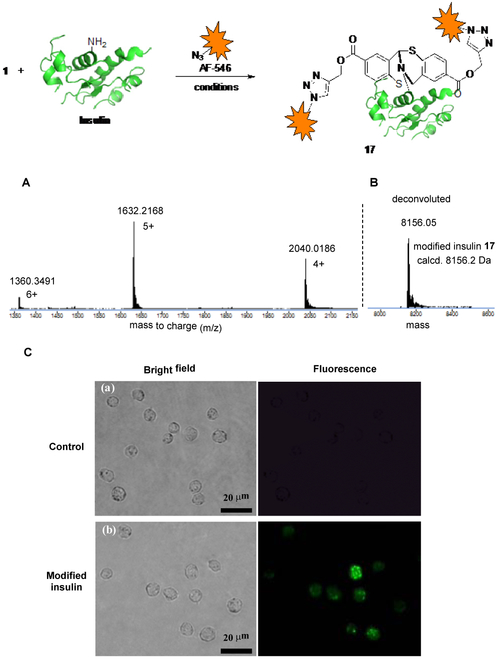
To demonstrate the practicality of our three-component bioconjugation strategy, we performed the modification of a more sophisticated protein. The recombinant human insulin was selected due to its well-established biological activities. Structurally, insulin consists of two peptide chains, the 21 amino acid A chain and the 30 amino acid residue B chain which are connected by three pairs of disulfide bonds. Lys residue is located at B53. The N-terminus of insulin was chosen for modification, as it would cause the least perturbation to its activity.[13] Considering the insulin could form insoluble fibrils upon stirring,[16] the optimized conditions were used to chemoselectively modify insulin at the N-terminus (Figure 2a). The linkage between insulin and AF546 azide was accomplished in one-pot setting. The modification of the insulin site specifically occurred at the both N-termini of A chain and B chain, which was validated by the extensive LC-MS/MS analysis.[17] The N-termini selectivity might be attributed to either the lower pKa of N-terminus compared to lysine amino group or the steric of insulin shielding the lysine side chain.[16,18] More interestingly, the disulfide bonds remained intact during the modification process which suggested that our linker is compatible with commonly observed protein disulfide linkages.
Figure 2.
Three-component conjugation of human insulin. Reagents and conditions: 100 μL THF/PBS (1:4, v/v), pH 5.5, 0.15 equivalent of CuSO4, 0.3 equivalent of sodium ascorbate, 0.3 equivalent of TBTA and 9 equivalents of AF546 azide were used, final concentration of insulin is 1.72 mM, incubated for 4 h. Examination of modified insulin 17 function on THP1 cells. (a) Bright field and fluorescence images of THP1 cells incubated with 0.06 mM AF546 azide for 4 h. (b) Bright field and fluorescence images of THP1 cells incubated with 0.06 mM modified insulin 17 for 4 h. Scale bar: 20 μm.
As illustrated in Figure 2c, the modified insulin is well-suited for cell biology studies. Labeling of insulin with the AF-546 fluorophore showed no obvious perturbation to the insulin’s functions. THP1 cells incubated with the modified insulin show the expected strong fluorescence, whereas the control cells incubated with the fluorophore and native insulin demonstrated negative fluorescence. Further examination of the fluorescent cells found that fluorescence is not uniform across the cells but rather forms clusters near cellular nuclei. That might be due to the rapid internalization of insulin-receptor being transported to endosomal apparatus of the cell.[19] Such a phenomenon implies that the insulin activity was not impaired by the conjugation.
In summary, we have established a general protocol for chemo-selective conjugation of primary amine groups within the protein framework using alkynyl mercaptobenzaldhyde 1. This bioorthogonal method allows native peptide and protein conjugation with two different functional groups, thus facilitating rapid modification of biomolecules with great flexibility. We have shown that the method is compatible with a variety of amino acid side-chains and disulfide linkages, which allows facile attachment of functional payloads, such as glycans, fluorescent groups and affinity handles in a one-pot fashion. Moreover, the modification of the mercaptobenzaldhyde aryl ring could introduce a variety of moieties other than alkynyl ester, and it is reasonable to speculate that diversified functional groups[20] could be selected as the suitable linkers. The practicality of our approach was illustrated by modifying insulin with fluorescent groups and carried out cell imaging experiments which suggested that the protein’s functions were not impaired. Further investigations of the bifunctional linker towards other protein modification will be reported in due course.
Supplementary Material
ACKNOWLEDGMENT
Support for this work was provided by the National Science Foundation (CHE 1710174), and the University at Albany-SUNY to Q.Z. This work was partially supported by National Institute of Health (R01GM12898401) and NYSTEM (C32574GG) to J.W. The early work performed by Dr. Gu Zhan (University at Albany–SUNY) is acknowledged. Dr. Zheng Wei (University at Albany–SUNY) is acknowledged for assistance with X-ray analysis.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Complete experimental procedures and characterization data for all new compounds (PDF)
Accession Codes
CCDC 1825270 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
The authors declare no competing financial interest.
REFERENCES
- 1.For selected reviews: (a) deGruyter JN; Malins LR; Baran PS Residue-Specific Peptide Modification: A Chemist's Guide. Biochemistry, 2017, 56, 3863–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tamura T; Hamachi I Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems. J. Am. Chem. Soc 2019, 141, 2782–2799 [DOI] [PubMed] [Google Scholar]; (c) Spicer CD; Davis BG Selective chemical protein modification. Nat. Commun 2014, 5, 4740–4754 [DOI] [PubMed] [Google Scholar]; (d) Lang K; Chin JW Bioorthogonal Reactions for Labeling Proteins. ACS Chem. Biol 2014, 9,16–20 [DOI] [PubMed] [Google Scholar]; (e) Sletten EM; Bertozzi CR Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed 2009, 48, 6974–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Krall N; Da Cruz FP; Boutureira O; Bernardes GJ Site-selective protein-modification chemistry for basic biology and drug development. L. Nat. Chem 2016, 8, 103–113 [DOI] [PubMed] [Google Scholar]; (g) Omar Boutureira O; Bernardes GJ Advances in Chemical Protein Modification. Chem. Rev 2015, 115, 2174–2195 [DOI] [PubMed] [Google Scholar]; (h) Silvestri AP; Cistrone PA; Dawson PE Adapting the Glaser Reaction for Bioconjugation: Robust Access to Structurally Simple, Rigid Linkers. Angew. Chem. Int. Ed 2017, 56, 10438–10442 [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Devaraj NK The Future of Bioorthogonal Chemistry. ACS Cent. Sci, 2018, 4, 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Reguera L; Yanira Méndez Y; Humpierre AR; Valdés O; Rivera DG Multicomponent Reactions in Ligation and Bioconjugation Chemistry. Acc. Chem. Res, 2018, 51, 1475–1486 [DOI] [PubMed] [Google Scholar]; (k) Li J; Chen PR Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol 2016, 12, 129–137 [DOI] [PubMed] [Google Scholar]; (l) Konievab O; Wagner A Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev, 2015, 44, 5495–5551 [DOI] [PubMed] [Google Scholar]; (m) Blagg J; Workman P Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell 2017, 32, 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher SA; Baker AEG; Shoichet MS Designing Peptide and Protein Modified Hydrogels: Selecting the Optimal Conjugation Strategy. J. Am. Chem. Soc 2017, 139, 7416–7427. [DOI] [PubMed] [Google Scholar]
- 3.Cai H; Zhang R; Orwenyo J; Giddens J; Yang Q; LaBranche CC; Montefiori DC; Wang L-X Multivalent Antigen Presentation Enhances the Immunogenicity of a Synthetic Three-Component HIV-1 V3 Glycopeptide Vaccine. ACS Cent. Sci 2018, 4, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. (a).Wright MH; Sieber SA Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep, 2016, 33, 681–708 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Leitner A Cross-linking and other structural proteomics techniques: how chemistry is enabling mass spectrometry applications in structural biology. Chem. Sci, 2016, 7, 4792–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) MacKinnon AL; Taunton J Target Identification by Diazirine Photo-Cross-linking and Click Chemistry. Curr. Protoc. Chem. Biol 2009, 1, 55–73 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Carrico IS Chemoselective modification of proteins: hitting the target. Chem. Soc. Rev, 2008, 37, 1423–1431. [DOI] [PubMed] [Google Scholar]
- 5. (a).Mu J; Liu F; Shafiq Rajab M; Shi M; Li S; Goh C; Lu L; Xu Q; Liu B; Ng L; Xing B-G A Small-Molecule FRET Reporter for the Real-Time Visualization of Cell-Surface Proteolytic Enzyme Functions. Angew. Chem. Int. Ed 2014, 53, 14357–14362 [DOI] [PubMed] [Google Scholar]; (b) Weinstain R; Savariar EN; Felsen CN; Tsien RY In Vivo Targeting of Hydrogen Peroxide by Activatable Cell-Penetrating Peptides. J. Am. Chem. Soc 2014, 136, 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.For selected examples: (a) Tung CL; Wong CTT; Fung EYM; Li XC Traceless and Chemoselective Amine Bioconjugation via Phthalimidine Formation in Native Protein Modification. Org. Lett, 2016, 18, 2600–2603 [DOI] [PubMed] [Google Scholar]; (b) Raj M; Wu H; Blosser S; Vittoria M; Arora P Aldehyde capture ligation for synthesis of native peptide bonds. J. Am. Chem. Soc 2015, 137, 6932–6940 [DOI] [PubMed] [Google Scholar]; (c) Zhang C; Dai P; Vinogradov AA; Gates ZP; Pentelute BL Site-Selective Cysteine-Cyclooctyne Conjugation. Angew. Chem. Int. Ed 2018, 57, 6459–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bloom S; Liu C; Kölmel DK; Qiao JX; Zhang Y; Poss MA; Ewing WR; MacMillan DWC Decarboxylative alkylation for site-selective bioconjugation of native proteins via oxidation potentials. Nat. Chem, 2018, 10, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lin S; Yang X; Jia S; Weeks AM; Hornsby M; Lee PS; Nichiporuk RV; Iavarone AT; Wells JA; Toste FD; Chang CJ Redox-based reagents for chemoselective methionine bioconjugation. Science, 2017, 355, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Taylor MT; Nelson JE; Suero MG; Gaunt MJ A protein functionalization platform based on selective reactions at methionine residues. Nature, 2018, 562, 563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Bertran-Vicente J; Penkert M; Nieto-Garcia O; Jeckelmann J-M; Schmieder P; Krause E; Hackenberger CPR Chemoselective synthesis and analysis of naturally occurring phosphorylated cysteine peptides. Nat. Comm, 2016, 7, 12703–12712 [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) McCarthy KA; Kelly MA; Li K; Cambray S; Hosseini AS; van Opijnen T; Gao J-M Phage Display of Dynamic Covalent Binding Motifs Enables Facile Development of Targeted Antibiotics. J. Am. Chem. Soc 2018, 140, 6137–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Bandyopadhyay A; Cambray S; Gao J-M Fast and selective labeling of N-terminal cysteines at neutral pH via thiazolidino boronate formation. Chem. Sci, 2016, 7, 4589–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Diehl KL; Kolesnichenko IV; Robotham SA; Bachman JL; Zhong Y; Brodbelt JS; Anslyn EV Click and chemically triggered declick reactions through reversible amine and thiol coupling via a conjugate acceptor. Nat. Chem, 2016, 8, 968–973 [Google Scholar]; (k) Méndez Y; Chang J; Humpierre AR; Zanuy A; Garrido R; Vasco AV; Pedroso J; Santana D; Rodŕıguez LM; Garćıa-Rivera D; Valdés Y; Vérez-Bencomo V; Rivera DG Multicomponent polysaccharide-protein bioconjugation in the development of antibacterial glycoconjugate vaccine candidates. Chem. Sci, 2018, 9, 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Matos MJ; Oliveira BL; Martínez-Sáez N; Guerreiro A; Cal PM; Bertoldo J; Maneiro M; Perkins E; Howard J; Deery MJ; Chalker JM; Corzana F; Jiménez-Osés G; Bernardes GJL Chemo- and Regioselective Lysine Modification on Native Proteins. J. Am. Chem. Soc 2018, 140, 4004–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Kubota K; Dai P; Pentelute BL; Buchwald SL Palladium Oxidative Addition Complexes for Peptide and Protein Cross-linking. J. Am. Chem. Soc 2018, 140, 3128–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Boutureira O; Brindle KM; Neves AA; Corzana F; Bernardes GJL Site‐Selective Modification of Proteins with Oxetanes Chem. Eur. J 2017, 23, 6483–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Row RD; Shih H-W; Alexander AT; Mehl RA; Prescher JA Cyclopropenones for Metabolic Targeting and Sequential Bioorthogonal Labeling. J. Am. Chem. Soc 2017, 139, 7370–7375 [DOI] [PubMed] [Google Scholar]; (p) Zhang Y; Zhou X; Xie Y; Greenberg MM; Xi Z; Zhou C Thiol Specific and Tracelessly Removable Bioconjugation via Michael Addition to 5-Methylene Pyrrolones. J. Am. Chem. Soc 2017, 139, 6146–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]; (q) Chan AO-Y Ho C-M; Chong H-C; Leung Y-C; Huang J-S; Wong M-K; Che C-M Modification of N-terminal α-amino groups of peptides and proteins using ketenes. J. Am. Chem. Soc 2012, 134, 2589–2598 [DOI] [PubMed] [Google Scholar]; (r) Ariyasu S; Hayashi H; Xing B; Chiba S Site-Specific Dual Functionalization of Cysteine Residue in Peptides and Proteins with 2-Azidoacrylates. Bioconjugate Chem 2017, 28, 897–902 [DOI] [PubMed] [Google Scholar]; (s) Hacker Stephan M., S. M; Backus KM; Lazear MR; Forli S; Correia BE; Cravatt BF Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem, 2017, 9, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]; (t) Adusumalli SR; Rawale DG; Singh U; Tripathi P; Paul R; Kalra N; Mishra RK; Shukla S; Rai V Single-site labeling of native proteins enabled by a chemoselective and site-selective chemical technology. J. Am. Chem. Soc 2018, 140, 15114–15123. [DOI] [PubMed] [Google Scholar]
- 7.Joshi NS; Whitaker LR; Francis MB A three-component Mannich-type reaction for selective tyrosine bioconjugation. J. Am. Chem. Soc 2004, 126, 15942–15943. [DOI] [PubMed] [Google Scholar]
- 8.Ohata J; Miller MK; Mountain CM; Vohidov F; Ball ZT A Three-Component Organometallic Tyrosine Bioconjugation. Angew. Chem., Int. Ed 2018, 57, 2827–2830. [DOI] [PubMed] [Google Scholar]
- 9.Moses JE; Moorhouse AD The growing applications of click chemistry. Chem. Soc. Rev, 2007, 36, 1249–1262. [DOI] [PubMed] [Google Scholar]
- 10. (a).Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J. Am. Chem. Soc 2012, 134, 10317–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu DS; Tangpeerachaikul A; Selvaraj R; Taylor MT; Fox JM; Ting AY Diels-Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J. Am. Chem. Soc 2012, 134, 792–795 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tang W; Becker ML “Click” reactions: a versatile toolbox for the synthesis of peptide-conjugates. Chem. Soc. Rev, 2014, 43, 7013–7039. [DOI] [PubMed] [Google Scholar]
- 11.Wu X; Li X; Danishefsky SJ Thio-mediated two-component coupling reaction of carboxylic acids and isonitriles under mild conditions Tetrahedron Lett 2009, 50,1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. (a).Kolb HC; Finn MG; Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed 2001, 40, 2004–2021 [DOI] [PubMed] [Google Scholar]; (b) Presolski SI; Hong VP; Finn MG Copper-Catalyzed Azide–Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol 2011, 3, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kappe CO; Van der Eycken E Click chemistry under non-classical reaction conditions. Chem. Soc. Rev, 2010, 39, 1280–1290 [DOI] [PubMed] [Google Scholar]; (d) Singh MS; Chowdhury S; Koley S Advances of azide-alkyne cycloaddition-click chemistry over the recent decade Tetrahedron 2016, 72, 5257–5283. [Google Scholar]
- 13.Chen D; Disotuar MM; Xiong X; Wang Y; Chou D H-C. Selective N-terminal functionalization of native peptides and proteins. Chem. Sci, 2017, 8, 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spielman MA The Structure of Troeger’s Base J. Am. Chem. Soc,1935, 57, 583–585. [Google Scholar]
- 15.Still I; Natividad-Preyra R; Toste FD A versatile synthetic route to 1,5-dithiocins from o-mercapto aromatic aldehydes. Can. J. Chem 1999, 77, 113–121. [Google Scholar]
- 16.Ichiishi N; Caldwell JP; Lin M; Zhong W; Zhu X; Streckfuss E; Kim H-Y; Parish CA; Krska SW Protecting group free radical C–H trifluoromethylation of peptides. Chem. Sci, 2018, 9, 4168–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See Supporting Information for details of the structure elucidation.
- 18.Ivanova MI; Sievers SA; Sawaya MR; Wall JS; Eisenberg D Molecular basis for insulin fibril assembly. Proc. Natl. Acad. Sci. USA, 2009, 106, 18990–18995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Guglielmo GM; Drake PG; Baass PC; Authier F; Posner BI; Bergeron JJ Insulin receptor internalization and signalling. Mol Cell Biochem 1998, 182, 59–64. [PubMed] [Google Scholar]
- 20. (a).Anderson GW; Zimmerman JF; Callahan FM N-Hydroxysuccinimide Esters in Peptide Synthesis. J. Am. Chem. Soc 1963, 85, 3039–3039 [Google Scholar]; (b) Oliveira BL; Guoa Z; Bernardes GJL Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev, 2017, 46, 4895–4950 [DOI] [PubMed] [Google Scholar]; (c) Darko A; Wallace S; Dmitrenko O; Machovina MM; Mehl RA; Chin JW; Foxa JM Conformationally strained trans-cyclooctene with improved stability and excellent reactivity in tetrazine ligation. Chem Sci, 2014, 5, 3770–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Jewett JC; Bertozzi CR Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev, 2010, 39, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.