Abstract
Enzymes with a protein kinase fold transfer phosphate from ATP to substrates in a process known as phosphorylation. Here we show that the Legionella meta-effector SidJ adopts a protein kinase fold, yet unexpectedly catalyzes protein polyglutamylation. SidJ is activated by host cell calmodulin to polyglutamylate the SidE-family of ubiquitin (Ub) ligases. Crystal structures of the SidJ-calmodulin complex reveal a protein kinase fold that catalyzes ATP-dependent isopeptide bond formation between the amino group of free glutamate and the γ-carboxyl group of an active site glutamate in SidE. We show that SidJ polyglutamylation of SidE and the consequent inactivation of Ub ligase activity is required for successful Legionella replication in a viable eukaryotic host cell.
One Sentence Summary:
Polyglutamylation by the protein kinase fold
In search of divergent members of the protein kinase superfamily, we analyzed the type 4 secretion system (T4SS) effector repertoire from the genus Legionella, a group of Gram-negative bacteria that includes Legionella pneumophila, the causative agent of Legionnaires disease. L. pneumophila secretes more than 300 proteins via the T4SS to target host cell signaling and establish a replicative niche known as the Legionella-containing vacuole (LCV) (1–3).
Using the sequence profile method Fold and Function Assignment System (FFAS) (4), we determined that the bacterial effector SidJ (lpg2155) bears borderline sequence similarity to protein kinases. SidJ acts as a “meta-effector” to antagonize four functionally-redundant effector paralogs, which share > 40% sequence identity (SdeA, SdeB, SdeC, and SidE; collectively known as the SidE family) (5, 6). The SidE effectors are required for Legionella replication in host cells and SdeA can functionally complement a deletion of all four SidE-family members (7).
The SidE effectors successively employ ADP-Ribosyltransferase (ART) and phosphodiesterase (PDE) activities to catalyze covalent ligation of ubiquitin (Ub) to Ser residues on substrate proteins independent of E1 or E2 Ub-activating enzymes (8–10). The ART domain of SidE ADP-ribosylates (ADPR) Ub on Arg42, which is subsequently converted by the PDE domain of SidE to phosphoribosyl (pR) Ub or linked to serine residues of target proteins through a Ser-pR-Ub linkage (Fig. S1). Ubiquitination of host proteins, including Rab33 (8) and Rtn4 (9), is required for proper formation of the LCV (11). However, unrestrained SidE activity is harmful to the host, poisoning the cellular Ub pool by phosphoribosylation and possibly blocking the action of other Legionella effectors that manipulate the host Ub machinery (5, 6, 10). As it would be detrimental to the bacterium to damage its host, Legionella uses SidJ to modulate the potent SidE family. While the SidE effectors are translocated into the host cell during the early stages of infection, SidJ levels rise slowly, permitting temporal control of SidE activity as the LCV matures (6, 12).
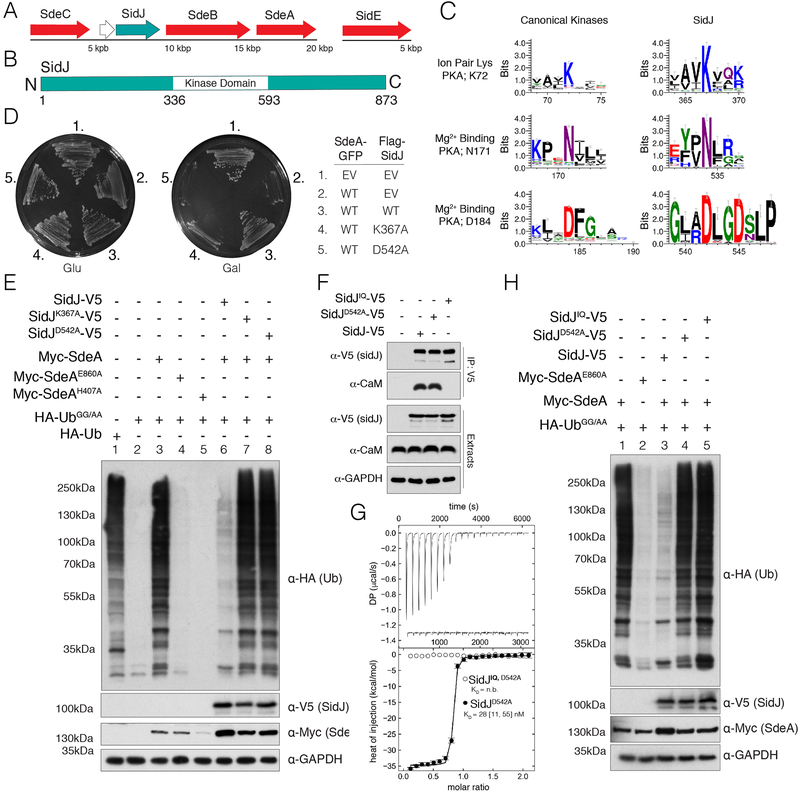
SidJ resides in a genomic locus with SdeA, SdeB, and SdeC and can suppress the toxicity of each of them, as well as SidE, which resides in a distinct genomic locus (Fig. 1A). We identified a putative protein kinase domain in SidJ spanning residues 336–593 (Fig. 1B). Although we could tentatively assign the active site ion pair residue K72 (PKA nomenclature, SidJ; K367) and the metal-binding residues N171 and D184 (SidJ; N534 and D542) (Fig. 1C), the locations of the Gly-rich loop and the catalytic loop were ambiguous, suggesting SidJ may be a pseudokinase. Notably, the putative metal-binding D542 lies within a 542DXXD motif, previously shown to be required for successful Legionella replication in amoeba (6) and mammalian cells (12).
Fig. 1. A putative kinase domain and CaM binding are required for SidJ suppression of SdeA toxicity and non-canonical ubiquitination.
(A) Organization of the SidE family (red) and SidJ (teal) effectors in the genome of L. pneumophila. (B) Domain architecture of L. pneumophila SidJ depicting the location of the predicted kinase domain. (C) Sequence logos (weblogos) highlighting conserved kinase active site residues in 106 SidJ homologs and 3,998 homologs of typical protein kinases (Pfam domain PF00069). The height of the amino acid stack is proportional to the sequence conservation at that position. (D) Growth inhibition assay depicting the growth of S. cerevisiae expressing SdeA-GFP and Flag-SidJ, or the predicted inactive K367A and D542A mutants. EV; empty vector. (E) Protein immunoblotting of total extracts from HEK293A cells expressing HA-Ub, HA-UbGG/AA, Myc-SdeA and SidJ-V5, or the indicated mutants. GAPDH is shown as a loading control. (F) Protein immunoblotting of V5-immunoprecipitates and cell extracts from HEK293A cells expressing V5 tagged SidJ, SidJD542A, or the SidJIQ mutant. Immunoprecipitates and cell extracts were analyzed for CaM and SidJ. GAPDH is shown as a loading control. (G) Isotherms depicting the binding of human CaM to SidJD542AΔNC. The top panel shows the SVD-reconstructed thermograms (DP=differential power), the lower panel shows the isotherms. Results are reported as best fit with boundaries of 68.3% confidence interval. (H) Protein immunoblotting of HEK293A total cell extracts expressing HA-tagged UbGG/AA, Myc-SdeA, V5 tagged SidJ, SidJD542A, or the SidJIQ mutant. GAPDH is shown as a loading control.
Expression of SdeA or SdeC caused a strong growth inhibition phenotype in yeast that was partially suppressed by co-expression of SidJ but not the K367A and D542A mutants (Fig. 1D & Fig. S2A). Host cell E1 enzymes activate Ub for transfer by adenylating its C-terminal glycine residue (13); however, this residue is not required for SidE catalyzed ubiquitination (8). We mutated the carboxy terminal GG motif of Ub to AA (UbGG/AA) for expression in mammalian cells to interrogate SidE-catalyzed ubiquitination. Expression of hemagglutinin (HA)-tagged WT Ub but not UbGG/AA resulted in the modification of several host proteins by the endogenous ubiquitination machinery (Fig. 1E, lanes 1 and 2). When UbGG/AA was co-expressed with Myc-tagged SdeA, but not the E860A (ART domain) or the H407A (PDE domain) mutants, we detected abundant ubiquitination of host proteins (Fig. 1E, lanes 3–5). Co-expression of SidJ, but not the K367A or D542A mutants markedly inhibited SdeA-catalyzed ubiquitination (Fig. 1E, lanes 6–8). Likewise, deletion of SidJ in Legionella led to an accumulation of pR-Ub in infected mammalian cells, which was ameliorated by complementation with WT SidJ but not the K367A mutant (Fig. S2B).
We expressed Flag-epitope tagged fusions of SidJ (Flag-SidJ) in yeast. Analysis of anti-Flag immunoprecipitates by LC-MS/MS identified calmodulin (CaM) in SidJ and SidJK367A, but not in control immunoprecipitates (Fig. S3A). A region within SidJ (841–851) roughly follows the consensus sequence of a CaM interacting “IQ” motif (Fig. S3B). We generated mutations that we predicted would disrupt CaM binding by substituting I841, Q842, R843 and R846 with acidic residues (Fig. S3B, SidJIQ). SidJ immunoprecipitated endogenous CaM when expressed in mammalian cells, but the SidJIQ mutant did not (Fig. 1F). A stable truncation of SidJ (59–851, SidJΔNC), which was suitable for purification in E. coli, suppressed SdeA-catalyzed ubiquitination (Fig. S3C), formed a stable complex with CaM during gel filtration chromatography (Fig. S3D), and bound CaM with a KD of ~30 nM (Fig. 1G). CaM binding was required for SidJ to inhibit SdeA-catalyzed ubiquitination in mammalian cells (Fig. 1H). Thus, SidJ requires kinase catalytic residues and CaM binding to counteract SidE.
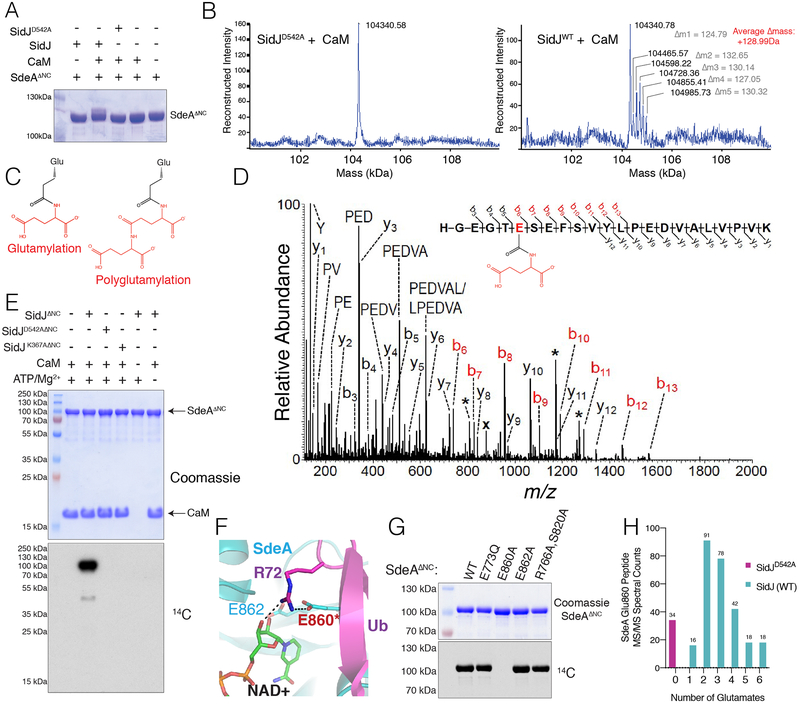
Although we were unable to observe phosphorylation of SdeA by SidJ in vitro, we observed an electrophoretic mobility shift of SdeA 178–1100 (SdeAΔNC, PDE and ART domains) that formed only when WT SidJ and CaM were co-expressed (Fig. 2A). Intact mass analysis revealed a series of mass increases in increments of ~128.99 Da (Fig. 2B). Searching a database of mass tags (14), we observed a match to glutamylation, a post-translation modification of the γ-carboxyl of a glutamate side chain modified with the amino group of free glutamate forming an isopeptide bond (Fig. 2C). LC-MS/MS analysis identified peptides of SdeA modified by one or two glutamates (Fig. 2D & Fig. S4A). Using [U-14C] Glu, we reconstituted glutamylation in vitro. The reaction required CaM, ATP/Mg2+ and the kinase residues in SidJ (Fig. 2E). Glutamylation of SdeAΔNC was time-dependent (Fig. S4B) and preferred Glu over Gln, Asp, Lys, or Gly (Fig. S4C). SidJ also glutamylated SdeB, SdeC and SidE (Fig. S4D). While both yeast and human CaM activated SidJ to glutamylate SdeA, a related protein, Troponin C, did not (Fig. S4E). Furthermore, SidJ purified from Legionella also glutamylated SdeAΔNC (Fig. S4F).
Fig. 2. SidJ polyglutamylates the SidE effectors.
(A) SDS-PAGE and Coomassie staining of SdeAΔNC isolated from E. coli following co-expression with CaM, SidJ, or the indicated mutants. (B) Intact mass LC/MS spectra of SdeAΔNC from (A). (C) Structures depicting glutamylation and polyglutamylation. (D) MS/MS spectrum of monoglutamylated (Glu) SdeAΔNC peptide ion HGEGTE(Glu)SEFSVYLPEDVALVPVK. The precursor ion, m/z 878.10 (3+) and labeled with (x), was subjected to HCD fragmentation to generate the MS/MS spectrum shown. Fragment b-ions containing the modified glutamate residue show a mass shift consistent with the addition of one Glu group (+129.043 Da) (red labels). Peaks labeled with a single asterisk (*) correspond to loss of water (−18 Da) from fragment ions. (E) Incorporation of 14C-Glu into SdeAΔNC by SidJΔNC. Reaction products were separated by SDS PAGE and visualized by Coomassie staining (upper) and autoradiography (lower). (F) Close-up of the NAD+ binding pocket of SdeA (PDB 5YIJ (16)). (G) Incorporation of 14C-Glu into WT SdeAΔNC, but not the E860A mutant. Reaction products analyzed as in (E). (H) Histogram depicting the MS/MS spectral matches to unmodified, glutamylated, and polyglutamylated E860-containing tryptic peptides of Myc-tagged SdeA immunopurified from HEK293 cells expressing SidJ-V5 or the D542A mutant.
LC-MS/MS and mutagenesis revealed the catalytic E860 of SdeA as the major site of glutamylation in vitro (Fig. 2F & G). Likewise, Myc-tagged SdeA isolated from mammalian cells co-expressing WT SidJ but not the D542A mutant was polyglutamylated on E860 (Fig. 2H). We co-expressed Myc-tagged SdeA with FcγRIIa and challenged HEK293 cells with antibody-opsonized Legionella (15). SdeA isolated from cells challenged with the WT strain (Lp02) but not the ΔsidJ (Fig. S5A–C) or T4SS mutants, was glutamylated on E860 as judged by LC-MS/MS (Fig. S5D). Thus, SidJ is a CaM-dependent protein polyglutamylase that modifies a catalytic glutamate residue in the ART active site of the SidE effectors in vitro and during infection.
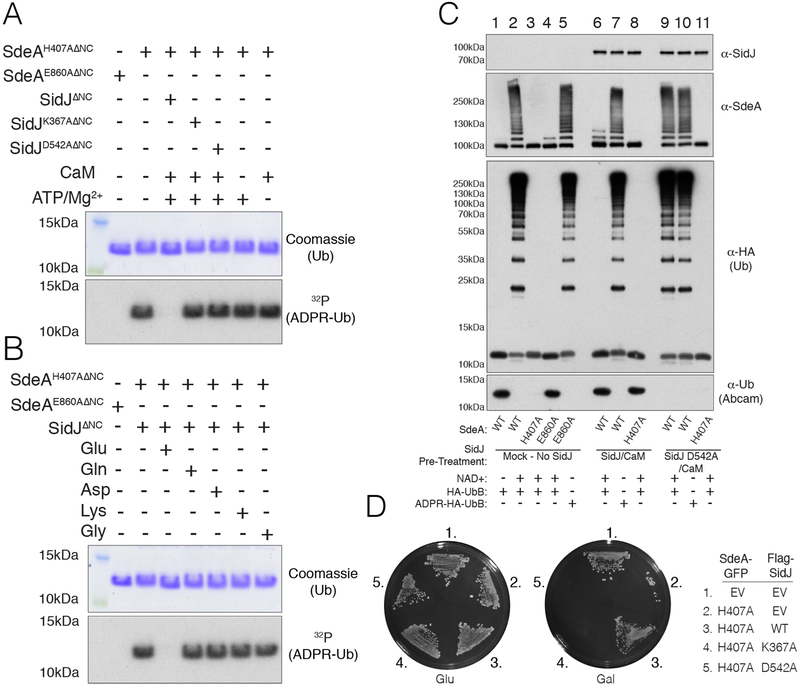
Glu860 is part of the conserved catalytic R-S-E860XE motif present in Arg-specific ARTs. Structures of SdeA and SidE with Ub revealed that E860 plays a key role in catalysis (Fig. 2F) (16–18). We used the SdeA H407A mutant, which inactivates the PDE domain and allows for specific analysis of ART activity (10). Glutamylation of SdeAH407A ΔNC inhibited its ability to ADP-ribosylate Ub (Fig. 3A). Inactivation of SdeAH407A ΔNC required CaM, ATP/Mg2+, Glu and K367 and D542 of SidJ. Substituting Glu with Gln, Asp, Lys, or Gly had no effect on SdeAH407A ΔNC ART activity (Fig. 3B).
Fig. 3. SidJ inactivates SdeA Ub ligase activity by polyglutamylating an active site residue in the ART domain.
(A) SDS PAGE and autoradiography depicting the incorporation of 32P-ADPR into HA-Ub from [α−32P]NAD+ by SdeAH407AΔNC. SdeAH407AΔNC or the E860A mutant were pretreated in glutamylation assays with SidJΔNC or the indicated mutants (−/+ ATP/Mg2+ or CaM) and SdeAH407AΔNC activity was subsequently analyzed. Reaction products were analyzed as in Fig. 2E. (B) SDS PAGE and autoradiography depicting the incorporation of 32P-ADPR into HA-Ub from [α−32P]NAD+ by SdeAH407AΔNC. SdeAH407AΔNC or the E860A mutant were pretreated with SidJΔNC, CaM, ATP/Mg2+ or Glu, Gln, Asp, Lys or Gly and SdeAH407AΔNC activity was analyzed as in A. (C) Protein immunoblotting following in vitro ubiquitination assays with the indicated mutants of SdeAΔNC. SdeAΔNC and mutants were pretreated in glutamylation reactions in the absence (lanes 1–5) or presence (lanes 6–8) of SidJΔNC/CaM, or SidJD542AΔNC/CaM (lanes 9–11). Ubiquitination reactions were started by the addition of NAD+ and HA-Ub or HA-ADPR Ub. The reaction components were resolved by SDS-PAGE and SidJΔNC, SdeAΔNC and Ub (using anti-HA or Abcam antibodies. which do not recognize Arg42 modified Ub) were detected by immunoblotting. (D) Growth inhibition assay depicting the growth of S. cerevisiae expressing SdeAH407A-GFP and Flag-SidJ, or the predicted catalytically inactive K367A and D542A mutants. EV; empty vector.
Glutamylated SdeAΔNC behaved identically to SdeAΔNCE860A in ubiquitination assays, losing both its ability to ladder and to pR-Ub, yet was unaffected when ADPR-Ub was used as a substrate to bypass the ART reaction (Fig. 3C). Moreover, SidJ suppressed the toxicity of SdeAH407A and SdeCH416A in yeast (Fig. 3D & Fig. S6), indicating that SidJ targets the SidE effectors at the point of ART activity. Thus, SidJ inhibits SidE by inactivating ART activity.
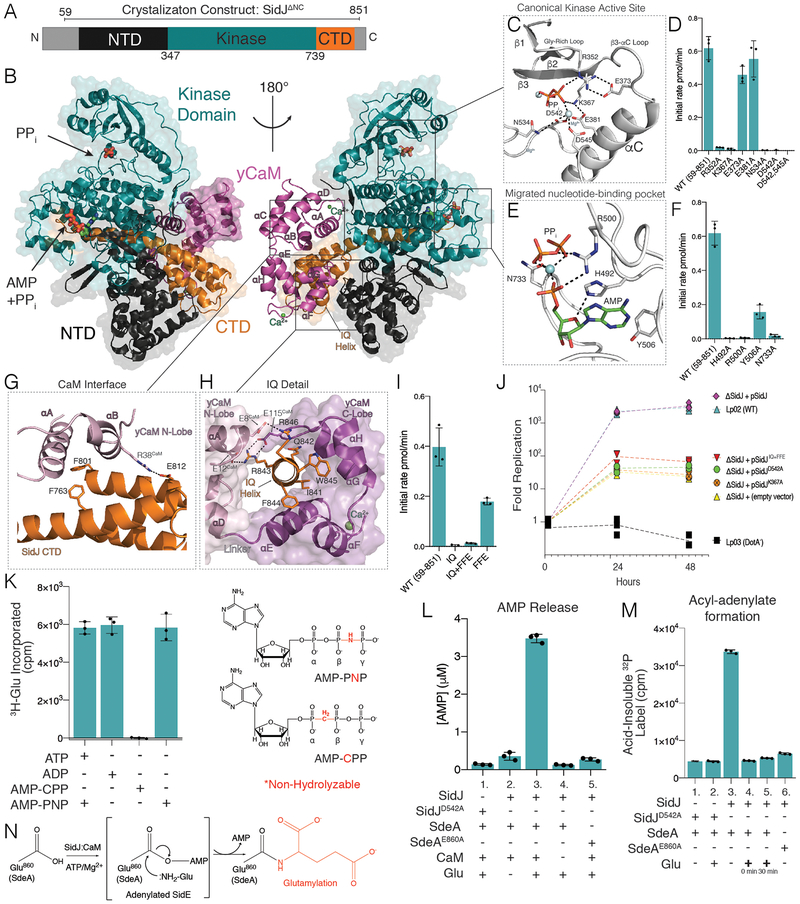
We solved the structure of SidJΔNC bound to yeast CaM (yCaM) to a resolution of 2.1Å (Fig. 4A, B, S7 &Table S1). The structure resolves an N-terminal domain (NTD, black), the kinase domain (KD, teal) and a C-terminal domain (CTD, orange) of SidJ. The α-helical NTD and CTD are nestled beneath the C-lobe of the KD, and yCaM is bound to the ‘back’ of SidJ (Fig. 4B). yCaM adopts a closed conformation with Ca2+ ions coordinated in EF1 and EF3 (Fig. S8A). The SidJ KD exhibits a β-strand-rich N-lobe containing the regulatory αC helix, and an α-helical-rich C-lobe. Structural homology searches (19) on the SidJ KD identify the Histone H3 kinase Haspin (20) and the AMPylating selenoprotein-O (SelO) (21) as the closest structural homologs (Fig. S8B). The SidJ KD is bound to AMP, Mg2+, and pyrophosphate (PPi). In typical kinases, the nucleotide is positioned in a pocket between the N- and C-lobes. Surprisingly, in the SidJ structure, AMP, PPi, and Mg2+ are bound in a migrated pocket formed by an insertion in the KD catalytic loop (Fig. S8C). This insertion contains highly-conserved residues which bury the adenosine ring in a tight cleft near the base of the SidJ C-lobe.
Fig. 4. Crystal structure of the SidJ-CaM complex uncovers insight into the mechanism of SidJ-catalyzed polyglutamylation.
(A) Domain architecture of SidJ depicting the N-terminal domain (NTD; black), the C-terminal domain (CTD; orange) and the kinase domain (KD; teal). Residues corresponding to SidJΔNC are indicated above the schematic. (B) Overall structure of the SidJΔNC-yCaM complex. yCaM is in magenta, and SidJ is colored as in A. (C) Close-up view of the canonical kinase active site of SidJ showing the interactions (dashed lines) involved in PPi binding. The PPi is shown in stick and the Mg2+ ions are in light blue spheres. (D) Glutamylation activity of SidJ and mutants using SdeAΔNC and [3H]Glu as substrates. Reaction products were resolved by SDS PAGE and radioactive gel bands were excised and 3H incorporation into SdeAΔNC was quantified by scintillation counting. (E) Close-up view of the migrated SidJ nucleotide-binding site depicting the interactions involved in AMP and PPi binding. The AMP and PPi are shown in stick and the Mg2+ ion as a light blue sphere. (F) Glutamylation activity of SidJ and kinase active site mutants using SdeAΔNC and [3H]Glu as substrates. Reaction products were analyzed as in D. (G) Close-up view of the interaction between the N-lobe of CaM and residues in the CTD of SidJ. SidJ is colored as in A and the yCaM N-lobe and C lobes are in pink and magenta, respectively. (H) Close-up view of the IQ helix of SidJ and its interactions with the N- and C-lobes of yCaM. Color coding as in G. (I) Glutamylation activity of SidJ and CaM binding mutants. Reaction products were analyzed as in D. (J) Replication of L. pneumophila strains in A. castellanii. Infected amoeba cells were lysed at the indicated timepoints and bacterial replication was quantified by plating serial dilutions of lysates. Results are representative of three independent experiments. (K) Glutamylation activity of SidJ using SdeAΔNC and [3H]Glu as substrates with different nucleotide analogs. Reactions went to completion and products were analyzed as in (D). The chemical structures of AMP-PNP and AMP-CPP are also shown with the non-hydrolysable bonds in red. (L) HPLC-MS/MS quantification of AMP released during SidJ-catalyzed glutamylation of SdeAΔNC. (M) Quantification of acyl-adenylate formation following reactions with SidJΔNC, SdeAΔNC and [α−32P]ATP. The reactions were terminated by the addition of TCA and the SdeA acyl-adenylate was detected by scintillation counting of the acid-insoluble material. In lanes 4 and 5, Glu was added at time 0 or after 30 minutes. (N) Schematic representation of the proposed SidJ-catalyzed glutamylation reaction with the acyl-AMP SidE intermediate in brackets.
The SidJ canonical kinase active site contains pyrophosphate (PPi) and 2 Mg2+ ions, which are coordinated by standard kinase catalytic residues (Fig. 4C). In addition to the K367A and D542A mutants, Ala substitutions of R352 and N534 inactivated SidJ (Fig. 4D). Within the migrated nucleotide-binding pocket, Ala substitutions of the invariant H492 and Y506 also inactivated SidJ (Fig. 4E & F). Arg500 and N733 also coordinate the phosphate group of AMP and face into solvent; their mutagenesis to Ala markedly inhibited glutamylation activity.
The interactions between SidJ and yCaM span more than 2000 Å2 of surface area and involve both lobes of yCaM (Fig. 4G & H). Within the IQ helix, I841 is buried in a hydrophobic cleft of the yCaM C-lobe. R843 and R846 contact Glu’s, pinching the yCaM molecule around the IQ helix (Fig. 4H). The αA and αB helices of the yCaM N-lobe are engaged in a groove formed by the CTD of SidJ (Fig. 4G). To disrupt the binding of the N-lobe of CaM to the CTD of SidJ, we mutated F763, F801 and E812 to Ala, Glu, Ala, respectively, to generate the mutant “SidJFFE”. SidJFFE bound to CaM with 100-fold lower affinity than WT SidJ and, SidJIQ+FFE failed to bind CaM entirely (Fig. S9A–D). The CaM-binding mutants were also significantly impaired in their ability to glutamylate SdeAΔNC in vitro (Fig. 4I). The SidJ mutants which lost glutamylation activity and CaM binding in vitro also lost the ability to suppress SdeA-catalyzed ubiquitination of host proteins in mammalian cells (Fig. S9E).
SidJ is one of only a handful of T4SS effectors which produce a growth inhibition phenotype when ablated from the L. pneumophila genome (22, 23). We complemented the ΔsidJ Legionella strain with SidJ mutants (Fig. S5A–C) and monitored growth within the environmental host Acanthamoeba castellanii. Deletion of SidJ resulted in a marked growth defect, consistent with previous reports (6) (Fig. 4J). Complementation of the ΔsidJ Legionella strain with WT SidJ, but not the K367A, D542A or the SidJIQ+FFE mutants, restored repliocation. Thus, SidJ requires kinase and CaM binding residues to facilitate Legionella infection by inactivating SidE.
Amidoligase reactions typically proceed through an acyl-phosphate or acyl-AMP intermediate. (24). Although SidJ prefers ATP (Fig. S10), it was able to use ADP and AMP-PNP, but not AMP-CPP, as co-substrates in glutamylation reactions (Fig. 4K). We detected AMP, but not ADP release, during the SidJ reaction but only when Glu was present, suggesting the formation of an acyl adenylate intermediate (Fig. 4L). Therefore, we monitored [32P]AMP incorporation from [α−32P]ATP during glutamylation reactions in the absence of Glu. Because the intermediate contains an alkali unstable acyl-adenylate, reactions were terminated with trichloroacetic acid (TCA) and the acyl-adenylate was detected as 32P in TCA-insoluble material. SidJ adenylated SdeA but not the SdeAE860A mutant (Fig. 4M). Addition of Glu decreased acid-insoluble label, suggesting that AMP is liberated by the formation of the Glu-Glu isopeptide bond (Fig. 4M).
We propose a catalytic mechanism for SidJ glutamylation of SdeA whereby CaM binds SidJ to stabilize an active conformation, which allows the canonical kinase-like active site of SidJ to bind ATP and transfer AMP to E860 on SdeA (Fig. 4N). Adenylated SdeA then binds the migrated nucleotide binding pocket in SidJ, which positions the acyl-adenylate and glutamate for glutamylation and inactivation of the SidE effector. Because CaM is a eukaryotic specific protein, its requirement for SidJ activity prevents premature inactivation of the SidE effectors in the bacterium. While our results are consistent with many previous conclusions regarding SidJ, others have shown that SidJ functions as a deubiquitinase (DUB) to deconjugate SidE-catalyzed Ub chains (12). However, our structural and biochemical analyses failed to detect any features that would suggest SidJ can function as a DUB (Fig. S11).
In summary, our results underscore the diversity and catalytic versatility of the protein kinase superfamily. We propose that ATP-dependent ligation reactions may be a common feature among the vast diversity of eukaryotic protein kinase-like enzymes found in nature (25). There are over 500 protein kinases in humans and our results suggest they should also be examined for alternative activities.
Supplementary Material
Acknowledgments:
We thank Drs. M. Phillips, J. Goldstein, K. Orth, N. Alto and members of the Tagliabracci laboratory for discussions. We thank Drs. R. Isberg for Legionella strains, C. Brautigan for help with ITC and A. Lemoff and N. Williams for help with mass spectrometry. Results shown in this report are derived from work performed at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source.
Funding: Work was funded by NIH Grants R00DK099254 (V.S.T.), F30HL143859–01(M.H.B.), Welch Foundation Grants I-1911, a CPRIT grant RP170674 (V.S.T) and the Polish National Science Centre grant 2014/15/B/NZ1/03559 (K.P.). V.S.T. is the Michael L. Rosenberg Scholar in Medical Research, CPRIT Scholar (RR150033) and Searle Scholar.
Footnotes
Competing Interests:N/A.
Data and Material Availability: All materials developed in this manuscript will be made available upon request. The structure will be deposited in the PDB ####.
References and Notes
- 1.Cornejo E, Schlaermann P, Mukherjee S, How to rewire the host cell: A home improvement guide for intracellular bacteria. J Cell Biol 216, 3931–3948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherwood RK, Roy CR, Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection. Annu Rev Microbiol 70, 413–433 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Isberg RR, O’Connor TJ, Heidtman M, The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7, 13–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu D, Jaroszewski L, Li Z, Godzik A, FFAS-3D: improving fold recognition by including optimized structural features and template re-ranking. Bioinformatics 30, 660–667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havey JC, Roy CR, Toxicity and SidJ-Mediated Suppression of Toxicity Require Distinct Regions in the SidE Family of Legionella pneumophila Effectors. Infect Immun 83, 3506–3514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong KC, Sexton JA, Vogel JP, Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11, e1004695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardill JP, Miller JL, Vogel JP, IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56, 90–103 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Qiu J et al. , Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotewicz KM et al. , A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 21, 169–181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhogaraju S et al. , Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 167, 1636–1649 e1613 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Kalayil S et al. , Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557, 734–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu J et al. , A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res 27, 865–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershko A, Ciechanover A, The ubiquitin system. Annu Rev Biochem 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 14.abrf.org/delta-mass.
- 15.Mukherjee S et al. , Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y et al. , Structural basis of ubiquitin modification by the Legionella effector SdeA. Nature 557, 674–678 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Wang Y et al. , Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 173, 1231–1243 e1216 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Akturk A et al. , Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector. Nature 557, 729–733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L, Rosenstrom P, Dali server: conservation mapping in 3D. Nucleic Acids Res 38, W545–549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eswaran J et al. , Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A 106, 20198–20203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sreelatha A et al. , Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell 175, 809–821 e819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR, Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108, 14733–14740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Luo ZQ, The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75, 592–603 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L, Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol Biosyst 5, 1636–1660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G, Structural and functional diversity of the microbial kinome. PLoS Biol 5, e17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomon D et al. , Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat Commun 4, 2973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger KH, Isberg RR, Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7, 7–19 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR, Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun 65, 2497–2501 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sexton JA et al. , The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol 186, 1658–1666 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffat JF, Tompkins LS, A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun 60, 296–301 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller S et al. , High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem 84, 5066–5073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent UM, Purification of antibodies using ammonium sulfate fractionation or gel filtration. Methods Mol Biol 115, 11–18 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Wollweber L, Harlow E and Lane D (Editors), Antibodies: A Laboratory Manual. XIII + 726 S., 50 Abb., 62 Tab. Cold Spring Harbor 1988. Cold Spring Harbor Laboratory. $50.00. ISBN: 0-87969-314-2. Journal of Basic Microbiology 30, 164–164 (1990). [Google Scholar]
- 34.McCarthy JS et al. , Safety, tolerability, pharmacokinetics, and activity of the novel long-acting antimalarial DSM265: a two-part first-in-human phase 1a/1b randomised study. Lancet Infect Dis 17, 626–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann L et al. , A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Drozdetskiy A, Cole C, Procter J, Barton GJ, JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43, W389–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn RD et al. , HMMER web server: 2015 update. Nucleic Acids Res 43, W30–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooks GE, Hon G, Chandonia JM, Brenner SE, WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dereeper A et al. , Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Borek D, Minor W, Otwinowski Z, Measurement errors and their consequences in protein crystallography. Acta Crystallogr D Biol Crystallogr 59, 2031–2038 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Borek D, Majewski W, Minor W, Multiparametric scaling of diffraction intensities. Acta Crystallogr A 59, 228–234 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Borek D, Cymborowski M, Machius M, Minor W, Otwinowski Z, Diffraction data analysis in the presence of radiation damage. Acta Crystallogr D Biol Crystallogr 66, 426–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borek D, Dauter Z, Otwinowski Z, Identification of patterns in diffraction intensities affected by radiation exposure. J Synchrotron Radiat 20, 37–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider TR, Sheldrick GM, Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, in CCP4 Study Weekend, Wolf M, Evans PR, Leslie AGW, Eds. (Daresbury Laboratory: Science & Engineering Research Council, 1991), pp. 80–86. [Google Scholar]
- 50.Cowtan K, Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cowtan K, The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.