Abstract
The conserved structural motif D is an important determinant of the speed and fidelity of viral RNA-dependent RNA polymerases (RdRps). Structural and computational studies have suggested that conformational changes in the motif-D loop that help to reposition the catalytic lysine represent critical steps in nucleotide selection and incorporation. Conformations of the motif-D loop in the poliovirus RdRp are likely controlled in part by noncovalent interactions involving the motif-D residue Glu364. This residue swivels between making interactions with Lys228 and Asn370 to stabilize the open and closed loop conformations, respectively. We show here that we can rationally control the motif-D loop conformation by breaking these interactions. The K228A variant favors a more active closed conformation, leading to increased nucleotide incorporation rates and decreased nucleotide selectivity, and the N370A variant favors a less active open conformation, leading to decreased nucleotide incorporation rates and increased nucleotide selectivity. Similar competing interactions likely control nucleotide incorporation rates and fidelity in other viral RdRps. Rational engineering of these interactions may be important in the generation of live, attenuated vaccine strains, considering the established relationships between RdRp function and viral pathogenesis.
Graphical Abstract
INTRODUCTION
Global health systems continue to be challenged by new and reemerging viruses, including prominent RNA viruses such as Zika virus1, chikungunya2, West Nile virus3, and related viruses4–6. It has been suggested that a key determinant of RNA virus virulence is the accuracy with which they replicate their RNA genomes7–21. Genetic diversity within a population of viruses may be one way to surmount environmental obstacles and escape immune system challenges22–24. As such, viruses must balance genetic diversity with overall genome integrity. This constraint provides the impetus for the rational engineering of the enzyme responsible for RNA virus replication, the RNA-dependent RNA polymerase (RdRp)10, 18, 19, 25, 26. Targeting amino acid residues involved in key structural changes necessary for RdRp function can result in RdRp variants with altered nucleotide incorporation rates and error frequencies10, 18, 19; viruses with altered mutation frequencies are attenuated in the host and can provide immunoprotection against lethal challenges from ‘wild-type’ virus15, 25–27. A better understanding of the molecular determinants of RdRp fidelity can thus potentially aid in the generation of live-attenuated vaccine strains. Here, we have rationally generated poliovirus (PV) RdRp variants with higher and lower fidelity by engineering the structural dynamics of an important active site loop, known as the motif-D loop.
The RdRp is structurally similar to DNA polymerases and reverse transcriptases (RTs), which have been described as having a cupped right hand structure with fingers, palm and thumb subdomains28–31. Most of the catalytic machinery lies in the palm subdomain that contains seven conserved structural motifs (A-G)32–35. The motif-D loop has been of special interest, because it contains the conserved catalytic lysine that has been proposed to act as a general acid to protonate the pyrophosphate leaving group and facilitate bond cleavage36–39. The motif-D lysine is unique to RdRps and RTs as this function is carried out by other structural elements in DNA polymerases37, 40–44. Modification of the motif-D lysine slows nucleotide incorporation and increases polymerase fidelity37, 38. In PV RdRp, the motif-D lysine is Lys359. Virus encoding the K359R variant are attenuated, but mice preinnoculated with this strain are immunoprotected from wild-type challenge, almost to the same extent as the clinically-used Sabin vaccine26. Considering the conservation of the motif-D lysine, it was proposed that modification of this residue may offer a universal, mechanism-based strategy for the development of vaccine strains for other RNA viruses26, 45.
Other amino acid substitutions on motif D have also been shown to change RdRp nucleotide incorporation rates and fidelity19, 46, 47. For example, the T362I substitution is one of the four RdRp amino acid substitutions encoded by the Sabin strain48, 49, and we have previously shown that the T362I substitution lowers RdRp fidelity46. PV encoding the T362I substitution also has reduced virulence in a transgenic PV-receptor mouse model46. Molecular dynamics (MD) simulations of wild-type (WT) and T362I RdRp suggested that the motif-D loop exchanges between an “open” conformation in which Lys359 is not in a catalytic position, and a “closed” conformation in which Lys359 can perform its catalytic function46. The T362I variant favors the “closed” conformation, consistent with solution-state NMR studies that suggested that T362I more readily fluctuates into the active conformation even in the presence of incorrect nucleotide46. Studies on the Coxsackievirus B3 (CVB3) RdRp have likewise indicated that modifications in motif D that affect active site closure can change RdRp fidelity and processivity, leading to attenuated virus growth and pathogenesis19.
The PV RdRp MD simulations indicated interesting sets of interactions between the motif-D residue Glu364, Lys228 and Asn37046, 50. According to the MD simulations, Glu364 swivels between making interactions with Lys228 in the “open” state and with Asn370 in the “closed” state (Figure 1). Based on our MD-derived model, we proposed that substitutions at these residues would result in a change in the dynamic equilibrium between the “open” and “closed” conformations of the motif-D loop, which would then affect RdRp function. For example, Lys228 makes an interaction with motif-D residue Glu364 in the “open” state, so we predicted that the K228A variant would disfavor the “open” state, even in the presence of incorrect nucleotide. In the “closed” state, the interaction between Lys228 and Glu364 is broken, and Glu364 makes a new interaction with Asn370, such that we predicted that the N370A substitution would disfavor the “closed” state, even in the presence of the correct nucleotide. Based on previous studies with other RdRp variants38, 46, 51, 52, we expected that the K228A and N370A would have lower and higher nucleotide selection fidelities than WT RdRp, respectively. Indeed, we show in this manuscript that amino acid substitutions at positions 228 and 370 change the relative populations of the “open” and “closed” states to affect RdRp nucleotide incorporation rates and fidelity. Similar structural elements could likewise impact the conformational dynamics of the motif-D loop to affect function in other viral RdRps.
Figure 1.
Motif-D loop interactions in the poliovirus RNA-dependent RNA polymerase. (A) The X-ray crystal structure of PV RdRp53 (PDB 1RA6) showing conserved structural motifs colored as followed: A, red; B, green; C, yellow; D, blue; E, purple; F, pink. Key residues on and making interactions with motif D are indicated. (B) Chemical mechanism of PV RdRp indicating the general acid role of Lys359 to protonate the leaving pyrophosphate group. The “?” represents a proposed general base that has yet to be identified. (C) Simplified model showing the interactions made by loop residue Glu364 in the “open” and “closed” states of PV RdRp. In the “open” state, Glu364 makes an interaction with Lys228 and Lys359 is positioned away from the active site, unable to serve as the general acid. In the “closed” state, Glu364 makes an interaction with Asn370 and Lys359 is repositioned such that it can act as the general acid.
MATERIALS AND METHODS
Materials.
[γ−32P]ATP and [α−32P]UTP (>7000Ci/mmol) were from VWR-MP Biomedical. Nucleoside 5’-triphosphates and 2’-deoxynucleoside 5’-triphosphates (all nucleotides were ultrapure solutions) were from GE Healthcare. 3’-Deoxyadenosine 5’-triphosphate (cordycepin) was from Trilink Biotechnologies. All RNA oligonucleotides were from Dharmacon Research, Inc. (Boulder, CO). T4 polynulceotide kinase was from New England Biolabs, Inc. [Methyl-13C] Methionine was from Cambridge Isotope Laboratories. HisPur Ni-NTA resin was from Thermo Scientific. Q-Sepharose fast flow was from Amersham Pharmacia Biothce, Inc. The QuikChange site-directed mutagenesis kit was from Stratagene. The plasmid DNA isolation Miniprep kit was from Omega Bio-Tek. All other reagents were of the highest grade available from Sigma or Fisher.
Plasmid construction.
The K228A and N370A variants were generated using the QuikChange method with appropriate primers. Mutations were confirmed by DNA sequencing (Nucleic Acid Facility, The Pennsylvania State University). It should be noted that all RdRp proteins, including WT protein, have two interface amino acid substitutions (L446D and R455D) to prevent polymerase polymerization.
Heterologous expression of PV RdRp in Escherichia coli.
PV RdRp (UniProtKB P03300(1748–2209)) was expressed in Escherichia coli B834(DE3) pRARE cells using autoinduction as previously described54–56. Protein samples used in NMR experiments were 13C-labeled by the addition of [methyl-13C]Met, while RdRps involved in kinetic studies did not incorporate this isotope.
Purification of PV RdRp.
Protein purification of PV WT and N370A variant was performed as described54–56. For the K228A RdRp variant, cell lysates were centrifuged for 30 min at 5,000 rpm at 4°C. The supernatant was then loaded onto a pre-equilibrated Ni-NTA column with buffer (20% glycerol, 100 mM potassium phosphate pH 8.0, 1 μg/mL pepstatin, 1 μg/ml leupeptin, 500 μM ethylenediaminetetraacetate (EDTA), 60 μM ZnCl2, 5mM β-mercaptoethanol (BME), 500 mM NaCl, 1mM phenylmethylsulfonyl fluoride (PMSF), 0.1% NP40 and 5 mM imidazole) at a flow rate of 1 mL/min. This Ni-NTA column with protein was incubated for 30 min at 4°C. The remaining purification procedures were the same as previously described54.
Enzyme kinetic assays.
Kinetic assays, including activesite titration assays, RdRp-RNA-nucleotide complex assembly assays, RdRp-RNA dissociation assays and nucleotide incorporation assays, for the K228A and N370A RdRp variants were conducted as previously described46, 57. All assays contained 50 mM HEPES pH 7.5, 10 mM BME, 5 mM MgCl2, and 60 μM ZnCl2. Experiments were conducted at 30°C unless otherwise noted.
NMR Sample Preparation and Spectroscopy.
NMR samples for the K228A and N370A RdRp variants were prepared as previously described38, 54. The NMR buffer consisted of 10 mM HEPES pH 8.0, 200 mM NaCl, 0.02% NaN3, 5 mM MgCl2 and 10 μM ZnCl2. NMR data collection followed previous procedures38, 54, using a Bruker Avance III 600 MHz spectrometer equipped with a 5-mm “inverse detection” triple-resonance (13C,1H,15N) single axis gradient TCI probe.
RESULTS
The K228A and N370A variants have lower and higher nucleotide selection fidelity, respectively.
Our MD-derived model suggested that the interactions between Glu364 on the motif-D loop and Lys228 and Asn370 on other surrounding structural elements are important in determining the motif-D loop conformation, which we have suggested is important for determining RdRp nucleotide incorporation rates and fidelity38, 46, 50. To test this model, we generated the K228A and N370A PV RdRp variants. To make sure these variants were suitable for further kinetic and NMR experiments, we first compared their abilities to form and maintain the pre-chemistry RdRp-RNA reaction complexes through enzyme assembly and dissociation assays, respectively, as we have done previously46, 57 (Figure 2). In the kinetic and NMR studies, we used the 10-mer primer/template substrate (ssU) RNA template that contains a six-base pair duplex flanked by four nucleotide 5’ overhangs (i.e. 5’-GCAUGGGCCC)57. The rate and yield of competent RdRp-RNA complexes and the stability of the assembled RdRp-RNA complexes were very similar with WT RdRp, indicating that these amino acid substitutions did not substantially affect RNA affinity or complex stability. These results indicated that these variants were suitable for further kinetic and structural investigations.
Figure 2.
Formation and stability of RdRp-RNA complexes is not affected by the K228A and N370A substitutions. The data for WT (black), K228A (blue) and N370A (red) RdRp are shown. (A) Assembly assay for the RdRp-RNA-NTP ternary complex. RNA (0.5 μM duplex) and 500 μM ATP (i.e. which templates against U) were pre-incubated for 5 min at 30°C before the addition of 1 μM RdRp. Reactions were quenched at the indicated times by adding 25 mM EDTA. (B) RdRp-RNA dissociation assay. RdRp (1 μM) and RNA (0.1 μM) were pre-incubated at 30°C for 90 s before the addition of 100 μM unlabeled RNA (i.e. “trap”). After the indicated times, the reaction buffer was mixed with 500 μM ATP and then quenched after 30 s by the addition of 25 mM EDTA. The dissociation rate constants of WT, K228A and N370A for ssU RNA were determined to be 3.04±0.19×10−4 s−1, 2.82±0.24×10−4 s−1 and 3.19±0.20×10−4 s−1, respectively.
We next performed single nucleotide incorporation kinetic experiments to determine the maximal rate constant for nucleotide incorporation, kpol, and the apparent dissociation constant for the incoming nucleoside triphosphate (NTP), Kd,app, where the correct and incorrect NTPs were considered to be ATP and GTP, respectively (Figure 3). The K228A and N370A variants had slightly elevated and diminished kpol values for correct AMP incorporation, respectively, but very similar Kd,app for ATP, compared to WT RdRp (Table 1). The K228A variant had a lower Kd,app for GTP, and the N370A variant had a lower kpol, resulting in increased and decreased second-order rate constants (i.e. kpol/Kd,app), respectively, for GMP misincorporation compared to WT RdRp. One measure of RdRp fidelity is the ratio of the second-order rate constants for correct and incorrect nucleotide (i.e. (kpol/Kd,app)correct/(kpol/Kd,app)incorrect). Based on these kinetic parameters, the K228A and N370A variants had lower and higher nucleotide incorporation fidelity, respectively, compared to WT RdRp (Table 1).
Figure 3.
The K228A and N370A substitutions lead to lower and higher fidelity polymerases respectively. (A) Experimental design for single nucleotide incorporation assays using ATP and 2’-dATP. RdRp (1 μM) was pre-incubated with ssU RNA (1 μM) before being quickly mixed with equal volume of ATP or 2’-dATP with different concentrations. These reactions were monitored by fluorescence changes over time using a stopped-flow apparatus. (B) Kinetic data for correct AMP incorporation (templated against U) for WT (black), K228A (cyan) and N370A (red) PV RdRp. The lines represent data fit to a hyperbola function to give an apparent dissociation constant (Kd,app) and a maximal rate constant for nucleotide incorporation (kpol). Kinetic parameters were determined at least three times; data shown represents one set of experiments. (C) Experimental design for single nucleotide incorporation assays using GTP. RdRp (1 μM) was pre-incubated with ssU RNA (1 μM) at room temperature for 3 min and then at the assay temperature of 30 °C for 2 min, before being quickly mixed with equal volume of GTP at different concentrations. In this case, RNA was 32P labeled on the 5′-end. (D) Kinetic data for incorrect GMP incorporation (templated against U) for WT (black), K228A (cyan) and N370A (red) PV RdRp. The lines represent data fit to a hyperbola function to give an apparent dissociation constant (Kd,app) and a maximal rate constant for nucleotide incorporation (kpol). Kinetic parameters were determined at least three times; data shown represents one set of experiments.
Table 1.
Comparison of the kinetic constants of nucleotide incorporation for variant and WT RdRp enzymes1
| Variant | NTP | kpol (s−1) | Kd,app (μM) | kpol/Kd,app (M−1s−1) | kpol,correct/ kpol,incorrect |
(kpol/Kd,app)correct/ (kpol/Kd,app)incorrect |
|---|---|---|---|---|---|---|
| WT | ATP | 68.4±1.5 | 44.1±3.5 | (1.55±0.13) ×106 | - | - |
| 2’-dATP | 0.94±0.05 | 98.3±16.2 | (9.56±1.66) ×103 | 73±4 | 160±30 | |
| GTP | (1.40±0.05) × 10−2 | 180±12 | (7.77±0.58) ×101 | (5.0±0.2) ×103 | 19900±2200 | |
| K228A | ATP | 74.3±1.0 | 44.8±2.2 | (1.66±0.08) ×106 | - | - |
| 2’-dATP | 0.99±0.03 | 77.5±9.3 | (1.27±0.15) ×104 | 75±3 | 130±17 | |
| GTP | (1.40±0.05) ×10−2 | 120±7 | (1.16±0.07) ×102 | (5.3±0.1) ×103 | 14300±1100 | |
| N370A | ATP | 58.9±2.8 | 40.6±3.2 | (1.45±0.13) ×106 | - | - |
| GTP | (7.1±0.03) ×10−3 | 170±20 | (4.17±0.49) ×101 | (8.3±0.4) ×103 | 34800±5100 |
Comparison of the K228A and N370A variants to other PV RdRp fidelity-variants.
We have previously analyzed a number of PV RdRp variants through kinetics, NMR analyses, computer simulations and biological assays38, 46, 49, 51, 52, 54, 58. It has also been shown that PV encoding lower- and higher-fidelity RdRps have reduced virulence in mice transgenic for the PV receptor15, 20, 21, 26, 46, 59. Within this context, it is worth noting then that the K228A variant had nucleotide incorporation fidelity (defined as (kpol/Kd,app(ATP) /kpol/Kd,app (GTP)) similar to the lower-fidelity T362I variant (Figure 4); we have shown that PV encoding the T362I substitution has reduced virulence in the PV-receptor mouse model46. Likewise, the N370A variant has nucleotide selection fidelity similar to the higher-fidelity G64S variant (Figure 4); PV carrying the G64S substitution has reduced virulence in the PV-receptor mouse model20, 21. These results indicate that the K228A and N370A variants had altered single nucleotide incorporate rates and fidelities on the same order of magnitude as previously identified RdRp variants.
Figure 4.
Comparison of the PV RdRp kinetic fidelity variants. (A) In most cases, nucleotide incorporation fidelity was defined as the second-order rate constant for correct nucleotide incorporation divided by the second-order rate constant for incorrect nucleotide incorporation (i.e. kpol/Kd,app (ATP) /kpol/Kd,app (GTP)). The low catalytic activities for K359R and R174K precluded accurate rate measurements at lower nucleotide concentrations (especially for incorrect nucleotide), and so nucleotide incorporation fidelities for these variants were based on estimates of kpol using a higher nucleotide concentration38, 52. In the K359R and R174K variants, fidelity is defined as the kpol for correct nucleotide incorporation divided by the estimated kpol for incorrect nucleotide incorporation (i.e. kpol (ATP) / kpol (GTP)). Viruses encoding the H273R15, T362I46, G64S20, 21 and K359R26 variants all have reduced virulence in a PV mouse model. Asterisks indicate that the kinetic parameters for the variant were statistically different from WT values, according to p value (0.01<*≤0.05; 0.001<**≤0.01; ***≤0.001; n=3) (B) Structure of PV RdRp53 (PDB 1RA6), showing the locations of amino acid substitutions used to generate lower and higher fidelity variants. Conserved structural motifs are also colored: A, red; B, green; C, yellow; D, blue; E, purple; F, pink.
Structural dynamics changes induced by the K228A and N370A substitutions as monitored by solution-state NMR.
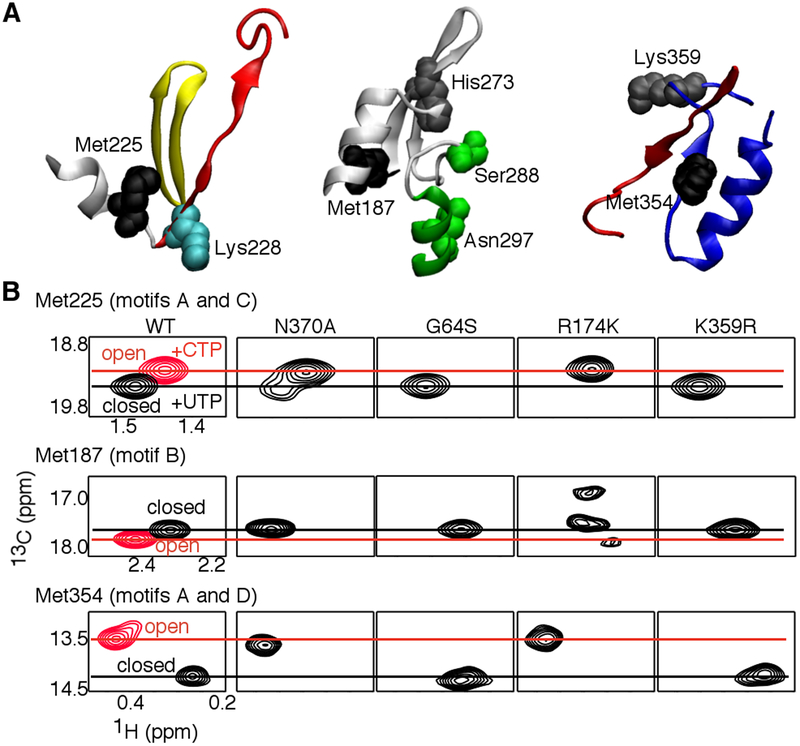
Our model (Figure 1) predicted that the K228A and N370A substitutions would disfavor the “open” and “closed” conformations of the motif-D loop respectively, which would also likely lead to structural changes in connected regions. To test this proposal, we collected [methyl-13C]Met 1H-13C HSQC spectra for RdRp:RNA:NTP ternary complexes with “correct” nucleotide (in this case, UTP) and “incorrect” nucleotide (either 2’-dUTP or CTP). We have previously established NMR “fingerprints” for when the enzyme is in the “open” and “closed” states38, 46, 54. For example, the chemical shift position for Met354 (in motif D) in the WT RdRp:RNA:UTP ternary complex (“closed”) is distinct from that observed in the RdRp:RNA:CTP ternary complex (“open”) (see Figure 5). Chemical shift changes in Met354 generally correlate to chemical shift changes associated with other Met residues, including Met225 that makes contact with residues in motifs A and C (Figure 5). X-ray crystal structures had indicated that active site closure involves a realignment of the β-strands in motifs A and C to form a proper three strand β-sheet60; chemical shift changes in Met225 are likely a response to these structural changes. We note here that Lys359 did not appear to be positioned in these X-ray crystal structures to act as a general acid, although MD simulations indicated that dynamic motions could appropriately reposition Lys35939, 51, 58. We suspect that relocation of Lys359 is among the last events before catalysis, which may not be absolutely required for nucleotide addition performed in crystallo given that Lys359 substitutions only decrease nucleotide incorporation rates by 10–50-fold37, 38. Here, we define the “closed” conformation as encompassing those structural changes identified by both X-ray crystallography and MD simulations, and we do not attempt to parse out the realignment of motifs A and C from structural changes in the motif-D loop.
Figure 5.
NMR chemical shift analyses to identify structural dynamic differences between WT and higher fidelity variants of PV RdRp. (A) There are Met residues in key positions to report on structural dynamic differences in conserved motifs between WT and fidelity variant RdRps. Met225 is in contact with residues from both motifs A (red) and C (yellow). Met187 is in contact with residues in motif B (green), and likely reports on conformational changes in motif B and nearby residues necessary for the interactions with the incoming nucleotide. In particular, Ser288 and Asn297 have been shown to make critical interactions with the ribose moiety of the incoming nucleotide and are important for correct sugar selection60–62. Met354 is present in motif D (blue), and likely reports on structural changes important in active site closure involving motifs A (red) and D. These structural changes may include conformational changes in the motif-D loop to reposition Lys359 for catalysis38. (B) Comparison of the chemical shift positions of Met225, Met187 and Met354 in RdRp-RNA-nucleotide ternary complexes based on [methyl-13C]Met 1H-13C HSQC spectra. Chemical shift positions characteristic of the “open” and “closed” conformations are highlighted in red and black, respectively. For these ternary complexes, RdRp was pre-incubated with ssU RNA (5’-GCAUGGGCCC-3’) and 3’-dATP to terminate RNA synthesis, buffer exchanged to remove excess 3’-dATP, and then UTP (black spectra) or CTP (red spectra) was added (i.e. the next templating base is A). Spectra were collected at 293 K with 250 μM RdRp, 500 μM duplex ssU RNA and 4 μM UTP or 12 μM CTP. Aspects of the NMR spectra for WT, G64S, R174K and K359R have been previously reported38, 52, 54.
Another region that was of particular interest was motif B; residues in this motif, including Ser288 and Asn297, have been shown to be important for ribose sugar recognition60–65. We have previously identified Met187 as an informative probe for conformational changes involving motif B (Figure 5). Arg188 has also been shown to be important in interacting with the RNA template strand60, 63, such that the chemical shift position of Met187 may also be sensitive to the binding and positioning of the RNA. We have noted that some fidelity variants (e.g. K359R and H273R) differentially affect Met354/Met225 and Met187, suggesting that these variants might decouple structural changes occurring in motifs A, B and D52.
We thought it helpful to compare the N370A variant to other higher fidelity variants (i.e. relative to WT RdRp) (Figure 5) and compare the K228A variant to other lower fidelity variants (i.e. relative to WT RdRp) (Figure 6). It was interesting to note that that the Met354 resonance for the N370:RNA:UTP ternary complex was at nearly the same chemical shift position as the WT:RNA:CTP ternary complex, suggesting that the motif-D loop and surrounding areas do not “close” in the N370A variant, even in the presence of the correct NTP. We have previously made a similar observation for the R174K variant52. However, the R174K variant has lower catalytic activity and higher fidelity than the N370A variant, likely because of the other functional roles for Arg174; Arg174 contacts the α-phosphate of the incoming NTP and is likely important for proper alignment of the α-phosphate for reaction with the primer 3’-OH52, 60, 61. The N370A variant may also more readily fluctuate into the “closed” conformation compared to the R174K variant. In the N370A:RNA:UTP complex, there were two resonances attributed to Met225, which appeared to correlate to the resonances observed in the WT:RNA:UTP and WT:RNA:CTP complexes. This finding suggested that Met225 and the surrounding region fluctuates between “open” and “closed”-like conformations. This conformational exchange was not observed in the R174K variant.
Figure 6.
NMR chemical shift analyses to identify structural dynamic differences between WT and lower fidelity variants of PV RdRp. Comparison of the chemical shift positions for Met225, Met187 and Met354, which report on structural dynamic changes in important structural motifs (see Figure 4), based on [methyl-13C]Met 1H-13C HSQC spectra of (A) RdRp:RNA:UTP, (B) RdRp:RNA:2’-dUTP and (C) RdRp:RNA:CTP ternary complexes. In this case, UTP is considered to be the “correct” nucleotide (i.e. templated against A), so that 2’-dUTP has the incorrect sugar and CTP has the incorrect nucleobase. Chemical shift positions characteristic of the “open” and “closed” conformations are indicated. The change in the “open-closed” conformational equilibrium is most apparent in the K228A and T362I RNA:2’-dUTP complexes. For these ternary complexes, RdRp was pre-incubated with ssU RNA (5’-GCAUGGGCCC-3’) and 3’-dATP to terminate RNA synthesis, buffer exchanged to remove excess 3’-dATP, and then (A) UTP, (B) 2’-dUTP or (C) CTP was added (i.e. the next templating base is A). Spectra were collected at 293 K with 250 μM RdRp, 500 μM duplex ssU RNA and 4 μM UTP, 8 μM 2’-dUTP or 12 μM CTP. Aspects of the NMR spectra for WT, H273R and T362I have been previously reported38, 46, 51, 52, 54.
The chemical shift position for Met187 in the N370A:RNA:UTP complex was also unique compared to that observed in WT or other high fidelity variant complexes, suggesting that Met187 and/or surrounding areas may access a different conformation in the N370A variant. We note that the G64S and K359R variants have very similar NMR spectra as WT RdRp, suggesting that increased nucleotide incorporation fidelity for these variants arise because of factors outside a change to the “open-closed” conformational equilibrium. As previously discussed, the G64S variant has altered nanosecond structural dynamics in many regions across the protein58, 66, and altered acid-catalysis in the K359R variant is likely responsible for its increased fidelity37, 38.
Comparisons of the K228A NMR spectra to WT RdRp and the lower fidelity variants were also illuminating (Figure 6). The Met354 chemical shift position was nearly identical between K228A and WT RdRp (and the other low fidelity variants) for the RNA:UTP complexes, and there were only small chemical shift differences for the Met225 and Met187 resonances. The small chemical shift change in the Met225 resonance was not surprising considering the close proximity of the K228A substitution. More substantial differences for the K228A RdRp were observed for the RNA:2’-dUTP and RNA:CTP complexes. In the K228A:RNA:2’-dUTP spectra, there were two resonances attributed to both Met225 and Met354, with chemical shift positions similar to those observed in the RNA:UTP and RNA:CTP spectra. A similar observation was previously made for the T362I variant (see also Figure 6). Similar to what we have suggested for the T362I variant46, we propose that the K228A variant can more readily fluctuate into the “closed” conformation even in the presence of incorrect NTP (i.e. 2’-dUTP), helping to explain the K228A variant’s reduced ability to discriminate NTPs. It should be noted that there were only single resonances for Met225 and Met354 for the K228A:RNA:CTP complex; the K228A variant may still affect the “closed-open” conformational equilibrium for this complex, but not sufficiently enough to observe changes in the NMR spectrum.
There were also interesting spectral differences for Met187 observed for the K228A and the other low fidelity variants. It was interesting to note that Met187 in the WT:RNA:2’-dUTP complex was associated with three or more overlapping resonances, suggesting Met187 and/or surrounding regions may access multiple conformations when binding 2’-dUTP. We observed a subset of these resonances for K228A and the other low fidelity variants, perhaps indicating that these substitutions also affect structural dynamics in motif B and related regions.
DISCUSSION
We had previously identified the structural dynamics of the motif-D loop as being important for RdRp nucleotide incorporation rates and fidelity38, 46, 50. Our previous MD simulations suggested that the motif-D loop toggles between less active “open” and more active “closed” conformations, which changes the position of the catalytic residue Lys35946. These conformations are partially stabilized through interactions between Glu364 on the motif-D loop and Lys228 in the “open” state and Asn370 in the “closed” state. The NMR studies were consistent with the MD-derived model; the K228A and N370A variants less favor the “open” and “closed” states, respectively, compared to WT RdRp. The changes to the “open-closed” conformational equilibrium likely explain changes to the single nucleotide incorporation rates and fidelity. The K228A variant favors the active “closed” state more, leading to a slightly elevated kpol for correct nucleotide incorporation, and a greater ability to incorporate incorrect nucleotide. The N370A variant favors the “closed” state less, leading to reduced rate constants, and a decreased ability to incorporate incorrect nucleotide. Although the kinetic changes are modest, they are similar to previously characterized RdRp variants. Similar interactions that likely help to modulate the motif-D loop structure and dynamics have been previously identified in other RdRps46.
It is interesting to note previous studies on the CVB3 RdRp, which studied the effects of substituting Phe364 (equivalent to Phe363 in PV RdRp) and interacting residues19. The “open” conformation is also stabilized through a stacking interaction between the motif-D residues Phe364 and Pro357 (Pro356 in PV RdRp)19, 60, 63, 67. Amino acid substitutions at position 364 appear to interfere with the “open-closed” conformational change19. Unlike previous studies67, there did not appear to be a correlation between 2’-OH discrimination based on single nucleotide incorporation assays (similar to performed here) and in vivo virus mutation frequencies for the F364-modified viruses19. Perhaps the Phe364 substitutions decouple structural events important for ribose discrimination from other fidelity-governing conformational changes, as we have observed for some PV RdRp variants52. The NMR studies presented here also suggest that the K228A and N370A substitutions differently affect motif B, as reported by the Met187 resonance(s), compared to motifs A and D, as reported by the Met225 and Met354 resonance(s). The impact on motif B and/or surrounding regions is especially relevant considering that the motif-B loop is also conformationally dynamic and Asn370 amino acid substitutions also affect RdRp fidelity29, 62, 65, 68; these studies further highlight the multifactorial nature of replication fidelity.
CVB3 encoding the F364W RdRp also had significantly reduced pathogenesis in mice, possibly suggesting a pathway to rationally engineer CVB3 vaccine strains19. This finding was especially significant considering that the analogous G64S, R174K and K359R substitutions in CVB3 do not lead to functional virus18, 67. Altogether the previous19, 38, 46 and current studies identify structural changes involving motif D as an important checkpoint in nucleotide incorporation fidelity, and suggest that modifying the interactions that govern these structural changes affects RdRp function and fidelity. These findings are important considering that it has been suggested that viruses encoding RdRps with altered replication fidelities may serve as live, attenuated vaccine strains25.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grants AI104878 (to D.D.B.) and AI45818 (to C.E.C.).
ABBREVIATIONS
- BME
β-mercaptoethanol
- CVB3
coxsackievirus CVB3
- EDTA
ethylenediaminetetraacetate
- HEPES
4-{2-hydroxyethyl)-1-piperazineethane sulfonic acid
- HSQC
heteronuclear single quantum coherence
- MD
molecular dynamics
- Ni-NTA
nickelnitrilotriacetic acid
- NMR
nuclear magnetic resonance
- NTP
nucleoside 5’-triphosphate
- PMSF
phenylmethylsulfonyl fluoride
- PV
poliovirus
- RdRp
RNA-dependent RNA polymerase
- RT
reverse transcriptase
- ssU
symmetrical primer/template substrate
- WT
wild-type
Footnotes
Accession Codes
UniProtKB P03300(1748–2209)
The authors declare no competing financial interest.
REFERENCES
- [1].Yun SI, and Lee YM (2017) Zika virus: An emerging flavivirus, J Microbiol 55, 204–219. [DOI] [PubMed] [Google Scholar]
- [2].Barr KL, and Vaidhyanathan V (2019) Chikungunya in Infants and Children: Is Pathogenesis Increasing?, Viruses 11, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Castro-Jorge LA, Siconelli MJL, Ribeiro BDS, Moraes FM, Moraes JB, Agostinho MR, Klein TM, Floriano VG, and Fonseca B (2019) West Nile virus infections are here! Are we prepared to face another flavivirus epidemic?, Rev Soc Bras Med Trop 52, e20190089. [DOI] [PubMed] [Google Scholar]
- [4].Holm-Hansen CC, Midgley SE, and Fischer TK (2016) Global emergence of enterovirus D68: a systematic review, Lancet Infect Dis 16, e64–e75. [DOI] [PubMed] [Google Scholar]
- [5].Baggen J, Thibaut HJ, Strating J, and van Kuppeveld FJM (2018) The life cycle of non-polio enteroviruses and how to target it, Nat Rev Microbiol 16, 368–381. [DOI] [PubMed] [Google Scholar]
- [6].Cassidy H, Poelman R, Knoester M, Van Leer-Buter CC, and Niesters HGM (2018) Enterovirus D68 - The New Polio?, Front Microbiol 9, 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Warmbrod KL, Patterson EI, Kautz TF, Stanton A, Rockx-Brouwer D, Kalveram BK, Khanipov K, Thangamani S, Fofanov Y, and Forrester NL (2019) Viral RNA-dependent RNA polymerase mutants display an altered mutation spectrum resulting in attenuation in both mosquito and vertebrate hosts, PLoS Pathog 15, e1007610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kautz TF, and Forrester NL (2018) RNA Virus Fidelity Mutants: A Useful Tool for Evolutionary Biology or a Complex Challenge?, Viruses 10, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li C, Wang H, Yuan T, Woodman A, Yang D, Zhou G, Cameron CE, and Yu L (2018) Foot-and-mouth disease virus type O specific mutations determine RNA-dependent RNA polymerase fidelity and virus attenuation, Virology 518, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rai DK, Diaz-San Segundo F, Campagnola G, Keith A, Schafer EA, Kloc A, de Los Santos T, Peersen O, and Rieder E (2017) Attenuation of Foot-and-Mouth Disease Virus by Engineered Viral Polymerase Fidelity, J Virol 91, e00081–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Naito T, Mori K, Ushirogawa H, Takizawa N, Nobusawa E, Odagiri T, Tashiro M, Ohniwa RL, Nagata K, and Saito M (2017) Generation of a Genetically Stable High-Fidelity Influenza Vaccine Strain, J Virol 91, e01073–01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Arias A, Thorne L, Ghurburrun E, Bailey D, and Goodfellow I (2016) Norovirus Polymerase Fidelity Contributes to Viral Transmission In Vivo, mSphere 1, e00279–00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Riemersma KK, Steiner C, Singapuri A, and Coffey LL (2019) Chikungunya Virus Fidelity Variants Exhibit Differential Attenuation and Population Diversity in Cell Culture and Adult Mice, J Virol 93, e01606–01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lloyd SB, Lichtfuss M, Amarasena TH, Alcantara S, De Rose R, Tachedjian G, Alinejad-Rokny H, Venturi V, Davenport MP, Winnall WR, and Kent SJ (2016) High fidelity simian immunodeficiency virus reverse transcriptase mutants have impaired replication in vitro and in vivo, Virology 492, 1–10. [DOI] [PubMed] [Google Scholar]
- [15].Korboukh VK, Lee CA, Acevedo A, Vignuzzi M, Xiao Y, Arnold JJ, Hemperly S, Graci JD, August A, Andino R, and Cameron CE (2014) RNA virus population diversity, an optimum for maximal fitness and virulence, J Biol Chem 289, 29531–29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheung PP, Watson SJ, Choy KT, Fun Sia S, Wong DD, Poon LL, Kellam P, Guan Y, Malik Peiris JS, and Yen HL (2014) Generation and characterization of influenza A viruses with altered polymerase fidelity, Nat Commun 5, 4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meng T, and Kwang J (2014) Attenuation of human enterovirus 71 high-replication-fidelity variants in AG129 mice, J Virol 88, 5803–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gnadig NF, Beaucourt S, Campagnola G, Borderia AV, Sanz-Ramos M, Gong P, Blanc H, Peersen OB, and Vignuzzi M (2012) Coxsackievirus B3 mutator strains are attenuated in vivo, Proc Natl Acad Sci U S A 109, E2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McDonald S, Block A, Beaucourt S, Moratorio G, Vignuzzi M, and Peersen OB (2016) Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo, J Biol Chem 291, 13999–14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, and Andino R (2006) Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population, Nature 439, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pfeiffer JK, and Kirkegaard K (2005) Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice, PLoS Pathog 1, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Domingo E, and Perales C (2018) Quasispecies and virus, Eur Biophys J 47, 443–457. [DOI] [PubMed] [Google Scholar]
- [23].Dolan PT, Whitfield ZJ, and Andino R (2018) Mechanisms and Concepts in RNA Virus Population Dynamics and Evolution, Annu Rev Virol 5, 69–92. [DOI] [PubMed] [Google Scholar]
- [24].Shirogane Y, Watanabe S, and Yanagi Y (2019) Cooperation between different variants: A unique potential for virus evolution, Virus Res 264, 68–73. [DOI] [PubMed] [Google Scholar]
- [25].Vignuzzi M, Wendt E, and Andino R (2008) Engineering attenuated virus vaccines by controlling replication fidelity, Nat Med 14, 154–161. [DOI] [PubMed] [Google Scholar]
- [26].Weeks SA, Lee CA, Zhao Y, Smidansky ED, August A, Arnold JJ, and Cameron CE (2012) A polymerase mechanism-based strategy for viral attenuation and vaccine development, J Biol Chem 287, 31618–31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsai YH, Huang SW, Hsieh WS, Cheng CK, Chang CF, Wang YF, and Wang JR (2019) Enterovirus A71 Containing Codon-Deoptimized VP1 and High-Fidelity Polymerase as Next Generation Vaccine Candidate, J Virol 93, e02308–02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Trakselis MA, and Murakami KS (2014) Introduction to nucleic acid polymerases: families, themes and mechanisms, Nucleic Acids and Molecular Biology 30, 1–15. [Google Scholar]
- [29].Ferrero D, Ferrer-Orta C, and Verdaguer N (2018) Viral RNA-Dependent RNA Polymerases: A Structural Overview, Subcell Biochem 88, 39–71. [DOI] [PubMed] [Google Scholar]
- [30].Peersen OB (2017) Picornaviral polymerase structure, function, and fidelity modulation, Virus Res 234, 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hansen JL, Long AM, and Schultz SC (1997) Structure of the RNA-dependent RNA polymerase of poliovirus, Structure 5, 1109–1122. [DOI] [PubMed] [Google Scholar]
- [32].Poch O, Sauvaget I, Delarue M, and Tordo N (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements, EMBO J 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].O’Reilly EK, and Kao CC (1998) Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure, Virology 252, 287–303. [DOI] [PubMed] [Google Scholar]
- [34].Lang DM, Zemla AT, and Zhou CL (2013) Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases, Nucleic Acids Res 41, 1464–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zemla AT, Lang DM, Kostova T, Andino R, and Ecale Zhou CL (2011) StralSV: assessment of sequence variability within similar 3D structures and application to polio RNA-dependent RNA polymerase, BMC Bioinformatics 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Gotte M, Konigsberg W, and Cameron CE (2007) Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases, Proc Natl Acad Sci U S A 104, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, and Cameron CE (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer, Nat Struct Mol Biol 16, 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang X, Smidansky ED, Maksimchuk KR, Lum D, Welch JL, Arnold JJ, Cameron CE, and Boehr DD (2012) Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition, Structure 20, 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shen H, Sun H, and Li G (2012) What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus?, PLoS Comput Biol 8, e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Franklin MC, Wang J, and Steitz TA (2001) Structure of the replicating complex of a pol alpha family DNA polymerase, Cell 105, 657–667. [DOI] [PubMed] [Google Scholar]
- [41].Kaushik N, Pandey VN, and Modak MJ (1996) Significance of the O-helix residues of Escherichia coli DNA polymerase I in DNA synthesis: dynamics of the dNTP binding pocket, Biochemistry 35, 7256–7266. [DOI] [PubMed] [Google Scholar]
- [42].Johnson SJ, and Beese LS (2004) Structures of mismatch replication errors observed in a DNA polymerase, Cell 116, 803–816. [DOI] [PubMed] [Google Scholar]
- [43].Kiefer JR, Mao C, Braman JC, and Beese LS (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal, Nature 391, 304–307. [DOI] [PubMed] [Google Scholar]
- [44].Wu EY, and Beese LS (2011) The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism, J Biol Chem 286, 19758–19767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee CA, August A, Arnold JJ, and Cameron CE (2016) Polymerase Mechanism-Based Method of Viral Attenuation, Methods Mol Biol 1349, 83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu X, Yang X, Lee CA, Moustafa IM, Smidansky ED, Lum D, Arnold JJ, Cameron CE, and Boehr DD (2013) Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity, J Biol Chem 288, 32753–32765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shen H, Deng M, and Zhang Y (2017) Effects of mutations on active site conformation and dynamics of RNA-dependent RNA polymerase from Coxsackievirus B3, J Mol Graph Model 77, 330–337. [DOI] [PubMed] [Google Scholar]
- [48].Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, and Imura N (1982) Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome, Proc Natl Acad Sci U S A 79, 5793–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu X, Musser DM, Lee CA, Yang X, Arnold JJ, Cameron CE, and Boehr DD (2015) Nucleobase but not Sugar Fidelity is Maintained in the Sabin I RNA-Dependent RNA Polymerase, Viruses 7, 5571–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Boehr DD, Liu X, and Yang X (2014) Targeting structural dynamics of the RNA-dependent RNA polymerase for anti-viral strategies, Curr Opin Virol 9, 194–200. [DOI] [PubMed] [Google Scholar]
- [51].Moustafa IM, Korboukh VK, Arnold JJ, Smidansky ED, Marcotte LL, Gohara DW, Yang X, Sanchez-Farran MA, Filman D, Maranas JK, Boehr DD, Hogle JM, Colina CM, and Cameron CE (2014) Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase, J Biol Chem 289, 36229–36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yang X, Liu X, Musser DM, Moustafa IM, Arnold JJ, Cameron CE, and Boehr DD (2017) Triphosphate Reorientation of the Incoming Nucleotide as a Fidelity Checkpoint in Viral RNA-dependent RNA Polymerases, J Biol Chem 292, 3810–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Thompson AA, and Peersen OB (2004) Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase, EMBO J 23, 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yang X, Welch JL, Arnold JJ, and Boehr DD (2010) Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance, Biochemistry 49, 9361–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Arnold JJ, Bernal A, Uche U, Sterner DE, Butt TR, Cameron CE, and Mattern MR (2006) Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity, Anal Biochem 350, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, and Cameron CE (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli, Protein Expr Purif 17, 128–138. [DOI] [PubMed] [Google Scholar]
- [57].Arnold JJ, and Cameron CE (2000) Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub), J Biol Chem 275, 5329–5336. [DOI] [PubMed] [Google Scholar]
- [58].Moustafa IM, Shen H, Morton B, Colina CM, and Cameron CE (2011) Molecular dynamics simulations of viral RNA-dependent RNA polymerases link conserved and correlated motions of functional elements to fidelity, J. Mol. Biol 410, 159–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Arnold JJ, Vignuzzi M, Stone JK, Andino R, and Cameron CE (2005) Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase, J Biol Chem 280, 25706–25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gong P, and Peersen OB (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase, Proc Natl Acad Sci U S A 107, 22505–22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, and Cameron CE (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B, J Biol Chem 275, 25523–25532. [DOI] [PubMed] [Google Scholar]
- [62].Korneeva VS, and Cameron CE (2007) Structure-function relationships of the viral RNA-dependent RNA polymerase: fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket, J Biol Chem 282, 16135–16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gong P, Kortus MG, Nix JC, Davis RE, and Peersen OB (2013) Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts, PLoS One 8, e60272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gohara DW, Arnold JJ, and Cameron CE (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection, Biochemistry 43, 5149–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Garriga D, Ferrer-Orta C, Querol-Audi J, Oliva B, and Verdaguer N (2013) Role of Motif B Loop in Allosteric Regulation of RNA-Dependent RNA Polymerization Activity, J Mol Biol 425, 2279–2287. [DOI] [PubMed] [Google Scholar]
- [66].Cameron CE, Moustafa IM, and Arnold JJ (2009) Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase?, Curr Opin Struct Biol 19, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Campagnola G, McDonald S, Beaucourt S, Vignuzzi M, and Peersen OB (2015) Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases, J Virol 89, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, and Verdaguer N (2007) Sequential structures provide insights into the fidelity of RNA replication, Proc Natl Acad Sci U S A 104, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]