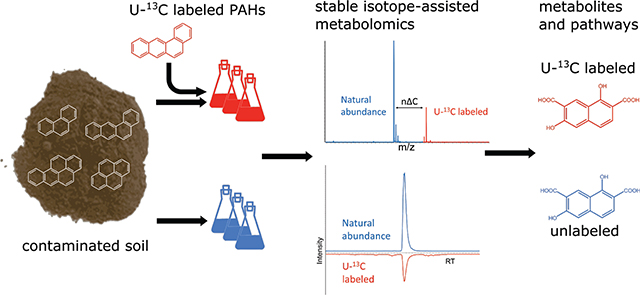
Abstract
Biotransformation of organic pollutants may result in the formation of oxidation products more toxic than the parent contaminants. However, to trace and identify those products, and the metabolic pathways involved in their formation, is still challenging within complex environmental samples. We applied stable isotope-assisted metabolomics (SIAM) to PAH-contaminated soil collected from a wood treatment facility. Soil samples were separately spiked with uniformly 13C-labeled fluoranthene, pyrene, or benzo[a]anthracene at a level below that of the native contaminant, and incubated for 1 or 2 weeks under aerobic biostimulated conditions. Combining high-resolution mass spectrometry and automated SIAM workflows, chemical structures of metabolites and metabolic pathways in the soil were proposed. Ring-cleavage products, including previously unreported intermediates such as C11H10O6 and C15H12O5, were detected originating from fluoranthene and benzo[a]anthracene, respectively. Sulfate conjugates of dihydroxy compounds were found as major metabolites of pyrene and benzo[a]anthracene, suggesting the potential role of fungi in their biotransformation in soils. A series of unknown N-containing metabolites were identified from pyrene, but their structural elucidation requires further investigation. Our results suggest that SIAM can be successfully applied to understand the fate of organic pollutants in environmental samples, opening lines of evidence for novel mechanisms of microbial transformation within such complex matrices.
Graphical Abstract
INTRODUCTION
Bioremediation is a well-accepted technology for the treatment of PAH-contaminated soil, but it might not be sufficient to reduce soil genotoxicity due to the formation of oxidation products.1–3 In a recent study we identified a previously unknown bacterial metabolite of pyrene in contaminated soil after bioremediation,4 illustrating that current knowledge on PAH biodegradation pathways gathered from the study of isolated strains on single compounds can be limited. Interactions among organisms in soil microbial communities or between components of complex PAH-mixtures5,6 can result in cometabolism,7 competitive metabolism,8–10 or cooperation; 11,12 such phenomena can enhance, inhibit, or modify the extent and/or profiles of metabolite formation.
An effective strategy to trace the biotransformation of specific pollutants within environmental samples is using stable or radioactive isotope-labeled compounds.13–16 For radioactive isotopes, their special safety requirements can limit wider application. Conversely, 13C-labeled substrates could be a suitable tool to shed light on the biodegradation pathways of organic pollutants.17,18 However, the identification of labeled metabolites has been restricted so far by the resolution of mass spectrometry and by data analysis methods. Recently, isotope labeling combined with high-resolution mass spectrometry (HRMS) has been applied to track the abiotic transformation of pollutants in aqueous mixtures.19,20 Adapting this combination of techniques to understanding biotransformation of the components of complex environmental mixtures, such as coal or oil derivatives, requires more powerful analysis of the HRMS data.
Stable isotope-assisted metabolomics (SIAM) could provide new insights on the biotransformation of pollutants and unleash the inherent power of isotope labeling. Such methods have already been established in the field of metabolomics, 21–24 and various automatic workflows are available.25–28 The basic principle of SIAM is that when 13C-labeled substrates are added to mixtures containing natural-abundance (unlabeled) counterparts and are biotransformed into labeled metabolites, they can be detected in groups with unlabeled metabolites by high performance liquid chromatography (HPLC) coupled with HRMS. Algorithms enable such group selection by picking up features with (1) the same chromatographic retention time; (2) certain exact mass differences; and (3) diagnostic intensity ratios.29,30 The SIAM approach differs in this respect from previous applications of HRMS with isotope-labeled compounds in environmental science cited above. Among other differences, SIAM cannot be used with deuterated compounds because they elute at different retention times than their unlabeled counterparts. Here, we applied SIAM to trace the biotransformation of PAHs in contaminated soil samples. Our objectives were (1) to check the feasibility of this method for organic pollutants in complex mixtures using uniformly 13C (U-13C) labeled PAHs as a model, and (2) to apply this method to identify the compound-specific metabolite profiles and degradation pathways for three four-ring PAHs in real contaminated soil samples during an active, laboratory-scale bioremediation process.
METHODS
Chemicals.
Anhydrous sodium sulfate and HPLC-grade solvents were purchased from Fisher Scientific (Pittsburgh, PA, U.S.A.). U-13C labeled fluoranthene, pyrene, and benzo[a]anthracene (B[a]A) were synthesized according to previous publications.31,32
Contaminated soil and spiking.
Creosote-contaminated soil was collected from a wood-treatment facility in Andalucía, Spain with a 100-year history of pollution.33 PAH concentrations are listed in Table S1. For the preliminary method assessment, treated soil from a different site was collected from a lab-scale aerobic bioreactor.4
For each U13C-labeled PAH (fluoranthene, pyrene, or B[a]A), the desired mass in acetone solution was spiked into 0.4 g (20% of a 2 g sample) of dry contaminated soil. After solvent evaporation overnight, the spiked soil was mixed with the remaining 1.6 g of unspiked soil in successive 0.4 g increments.34 The amount of spiked U-13C PAH was adjusted to maintain a 1:3 (mass:mass) proportion with respect to the corresponding native PAH concentration in the soil. Unspiked samples were amended with the same level of acetone as the spiked samples.
Microcosm incubations.
Aliquots of 1 g of soil (dry wt) were placed in sterile 125 mL Erlenmeyer flasks containing 30 mL of 10 mM phosphate buffer (pH 7.5) supplemented with urea (1 mM) as nitrogen source.4 All flasks were incubated at 25 ºC under agitation at 150 rpm. Triplicate unspiked flasks were sacrificed initially and after 7, 14, and 21 days of incubation. Triplicate killed controls were prepared by acidifying (pH 2) using phosphoric acid, and sacrificed after 21 days. Duplicate flasks containing U-13C fluoranthene, pyrene or B[a]A were sacrificed after 7 or 14 d. Optimal incubation times for each target PAH were determined in a preliminary incubation using unspiked soil as described in SI (Figure S1).
After incubation, the flask contents were centrifuged to recover the solid and aqueous phases. The solid phase was processed as described previously.1 The aqueous phase was acidified (pH 2) and extracted with ethyl acetate. Further details on sample extraction and processing are provided in SI.
Instrumental analysis.
Extracts were analyzed using an Agilent 1200 series HPLC interfaced with a 6520 accurate mass quadrupole time-of-flight mass spectrometer (qTOF-MS, Agilent Technologies, Santa Clara, CA). Full scan (m/z 100–1000) with electrospray ionization (ESI) was used for SIAM analysis, and product ion scanning (MS/MS) was applied to elucidate the putative structures of metabolites. Details and parameters are provided in SI.
Data analysis.
Raw data from qTOF-MS were converted into. mzData or. mzXML format for in-depth analysis. Peak detection and alignment were achieved using the R package XCMS35 and the publicly available Scripps XCMS Online platform (https://xcmsonline.scripps.edu/).36 Isotope group picking was conducted in R using the package X13CMS.25 Detailed processing methods and parameter settings are provided in SI.
RESULTS AND DISCUSSION
Assessing performance of the method.
Considering the previous identification of a genotoxic metabolite (2H-naphtho[2,1,8-def]chromen-2-one; NCO) during the incubation of a bioreactor-treated soil with pyrene,4 we first assayed three different ratios of U-13C to natural abundance (native) pyrene in the bioreactor slurry to test the feasibility of the method. Ratios of 1:1 and 1:3 produced 13C-NCO in sufficient quantity for its analytical detection, while 1:9 could not. Considering the cost and scarcity of U-13C compounds, and to minimize their impact on the microbial community as an enrichment substrate, we chose the 1:3 ratio to perform our experiments on the creosote-contaminated soil.
To detect the natural abundance and 13C-enriched pairs of metabolites, we applied the “getIsoDiffReport” function of the X13CMS package to select the peak groups that shared the same retention time but presented m/z values that differed in N × 1.00335, where N is the number of 13C atoms (N varied depending on the metabolite from a given PAH).25 However, with this approach most of the selected groups were false positives that only contained ions corresponding to [M-H]- and [M-H+1]-, assigned to the background of natural abundance 13C.
To restrict the number of selected groups, we first restricted the ratio of labeled and unlabeled metabolites according to the 1:3 ratio between the spiked 13C PAHs and the native compounds, using the “enrichmentLvsU” parameter in the X13CMS package. We then compared spiked and unspiked samples obtained from the same incubation time using XCMS Online. These two samples differed only in the presence of isotope labeling, and the features significantly higher in the spiked samples should correspond to labeled metabolites. By integrating the results from both approaches we generated the final list of prioritized features (Table 1).
Table 1.
Detected metabolites from the four-ring PAHs
| RTa | Phaseb | Ionization | m/z>12C | m/z13C | Formula | Confidence levelc | Tentative identification | PubChem Identifier |
|---|---|---|---|---|---|---|---|---|
| Fluoranthene | ||||||||
| 23.1 | W | ESI- | 217.0652 | 233.1192 | C16H10O | 3 | Hydroxy-fluoranthene | 3014645 |
| 20.1 | S | ESI+ | 233.0567 | 249.1094 | C16H8O2 | 3 | Fluoranthene o-quinone | 21481 |
| 14.2 | W | ESI- | 249.0562 | 265.1068 | C16H10O3 | 4 | 9-fluorenone-1-(2-propene)-carboxylic acid | |
| 14.1 | W | ESI- | 195.0459 | 208.0874 | C13H8O2 | 3 | Hydroxy-fluorenone | 80663 |
| 7.3 | W | ESI- | 237.0398 | 248.0695 | C11H10O6 | 4 | (2-Carboxybenzyl) malonic acid | |
| Pyrene | ||||||||
| 20.0 | S | ESI+ | 233.0567 | 249.1094 | C16H8O2 | 3 | Pyrene o-quinone | 160814 |
| 18.1 | W | ESI- | 313.0179 | 329.0716 | C16H10O2·SO3 | 3 | Dihydroxy-pyrene sulfate conjugate | |
| 20.4 | W | ESI- | 358.0043 | 374.0572 | C16H9NO4·SO3 | 4 | ||
| 21.9, 22.3 | W | ESI- | 369.9945 | 385.0452 | C15H5N3O9 | 4 | ||
| 21.2 | W | ESI- | 342.0005 | 356.0448 | C14H5N3O8 | 4 | ||
| 21.4 | W | ESI- | 386.9854 | 401.0332 | C14H4O4·2N2O3 | 4 | ||
| Benzo[a]anthracene | ||||||||
| 19.9 | W | ESI- | 339.0331 | 357.0932 | C18H12O2·SO3 | 3 | Dihidroxy benzo(a)anthracene | |
| 15.9 | W | ESI- | 289.0178 | 303.0645 | C14H10O2·SO3 | 3 | Dihydroxy-anthracene; dihydroxy-phenanthrene | |
| 11.1 | W | ESI- | 271.0614 | 286.1104 | C15H12O5 | 4 | 1-Hydroxy-2-carboxy-anthracene dihydrodiol | |
| 2.6, 5.9 | W | ESI- | 247.0246 | 259.0640 | C12H8O6 | 3 | Dihydroxynaphthalene-dicarboxylic acid (2 isomers) | 69414683 |
RT, retention time
W, aqueous (water) phase; S, solid phase
Confidence levels for metabolite identification were assigned according to the criteria established by Schymanski et al.53
Metabolite identification.
Because we tested only ESI in positive and negative ionization modes, it is possible that some metabolites escaped analysis. Metabolites present in low abundance, with very low ionization potential, or which would be better detected with a different ionization source could have been missed. However, our primary objective was to establish proof-of-concept of SIAM for applications in complex environmental systems, rather than an exhaustive search for all possible metabolites.
For each target PAH, several labeled features were detected. Tentative structures of metabolites (Table 1) were based on their exact mass, MS/MS fragmentation patterns, deduced number of labeled carbons, and further comparison with data available in the literature and pathway prediction systems such as EAWAG-BBD37 and enviPath38. Standards were not available for any of the proposed metabolites. With a formula and MS/MS pattern, each structure was processed with the MetFrag39 platform and searched in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and five of them matched the record (Table 1).
In the solid phase, labeled metabolites were detected only for fluoranthene and pyrene. The identified products with the formula C16H8O2 corresponded to the respective o-quinones (see Figure S2 for the fluoranthene quinone), which were previously found to accumulate during incubation with pure bacterial strains 40,41. The low number of products detected in the solid phase suggested that most of the metabolites were formed in or transferred to the aqueous phase due to their increased polarity relative to the parent compounds.
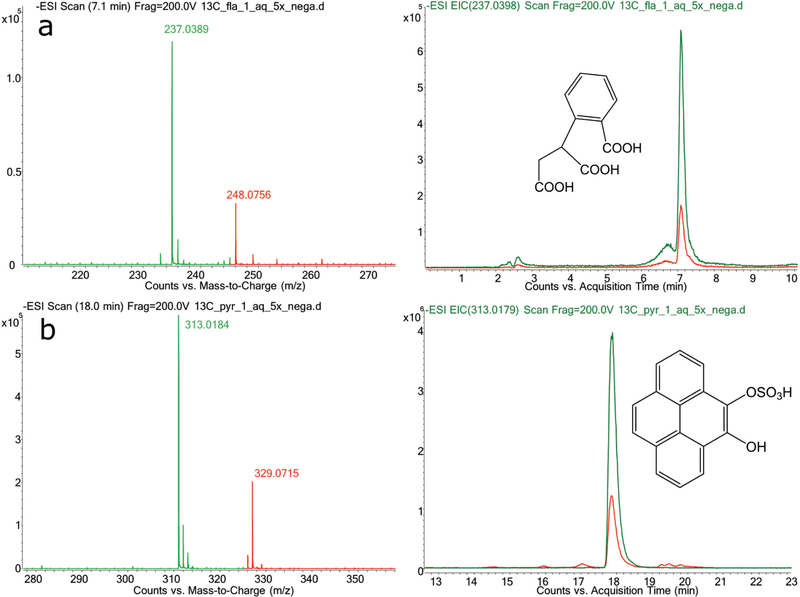
Two major groups of compounds were detected in the liquid phase: highly oxygenated ring-cleavage products and PAH conjugates. Within the first group, we detected several formulas originating from fluoranthene (C11H10O6; Figure 1) and B[a]A (C15H12O5, Figure S3; and C12H8O6) corresponding to ring-cleavage products containing one or more carboxyl group (mass spectra consistent with loss of at least one fragment of m/z 44). The fluoranthene product C11H10O6 did not correspond to any previously identified metabolite, and is proposed as (2-carboxybenzyl) malonic acid (Figure 1a and Figure S6d). The B[a]A metabolite C15H12O5 has not been previously described, and is proposed as a dihydrodiol of the 1-hydroxy-2-anthracene carboxylic acid. For the formula C12H8O6, the MS/MS spectral characteristics (Figure S6e) are identical to those of 1,6-dihydroxynaphthalene-2,7-dicarboxylic acid, a known ring-fission product of B[a]A from Sphingobium sp. strain KK22.42
Figure 1.
Mass spectra, extracted ion chromatograms (EIC), and proposed structures of two representative metabolites. a: C11H10O6 from fluoranthene; b: C16H10O2·SO3 from pyrene. The green lines represent 12C metabolites, and the red lines represent U-13C labeled metabolites.
A second group of products were conjugates of PAH metabolites, mainly sulfate conjugates of dihydroxylated PAHs. We detected several peaks corresponding to the sulfate conjugates of dihydroxy-pyrene (Figure 1b), dihydroxy-B[a]A, and either dihydroxy-anthracene or -phenanthrene derived from B[a]A (Figure S3). Each of these dihydroxy-compounds was always associated with a co-eluting peak with m/z corresponding to [M-H+80]- (Figure S5). According to the exact mass (m/z 79.9570), this fragment was assigned to an SO3 group, indicative of a sulfate conjugate.43 To confirm this observation, we repeated the analysis decreasing the fragmentor voltage to 100V, which increased the sulfate conjugate signal while that of the dihydroxy compounds disappeared, confirming the sulfate conjugates as the actual metabolites. Such sulfate conjugation of dihydroxy-PAHs is common in fungi,44,45 but has never been observed in bacteria. These results suggest that fungi might have played a significant role in PAH biotransformation and detoxification in the contaminated soil.
Besides the sulfate conjugates, we observed a number of other candidate conjugation products in the incubations containing U-13C pyrene (m/z 342.0005, 358.0003, 369.9945, and 386.9854, Figure S4). Although their formulas could not be unambiguously assigned, all of these features had an m/z over 300, and some of the deprotonated ions presented nominal masses in even numbers, suggesting the existence of nitrogen atoms. Their MS/MS spectra revealed fragments with exact masses compatible with the loss of different nitrogen oxides (NO or NO2) but especially of m/z 75.99, corresponding to N2O3 (Figure S6). The formulas of the conjugates were therefore assigned according to their fragmentation patterns. PAH metabolites with such fragments, as well as conjugation reactions with nitrogen oxides, have not previously been described. Based on current evidence, it is difficult to elucidate the identities of these unknown pyrene metabolites. Considering their novelty and diversity, these nitrogen-containing metabolites should be interesting targets for future research.
Proposed compound specific degradation pathways in soil.
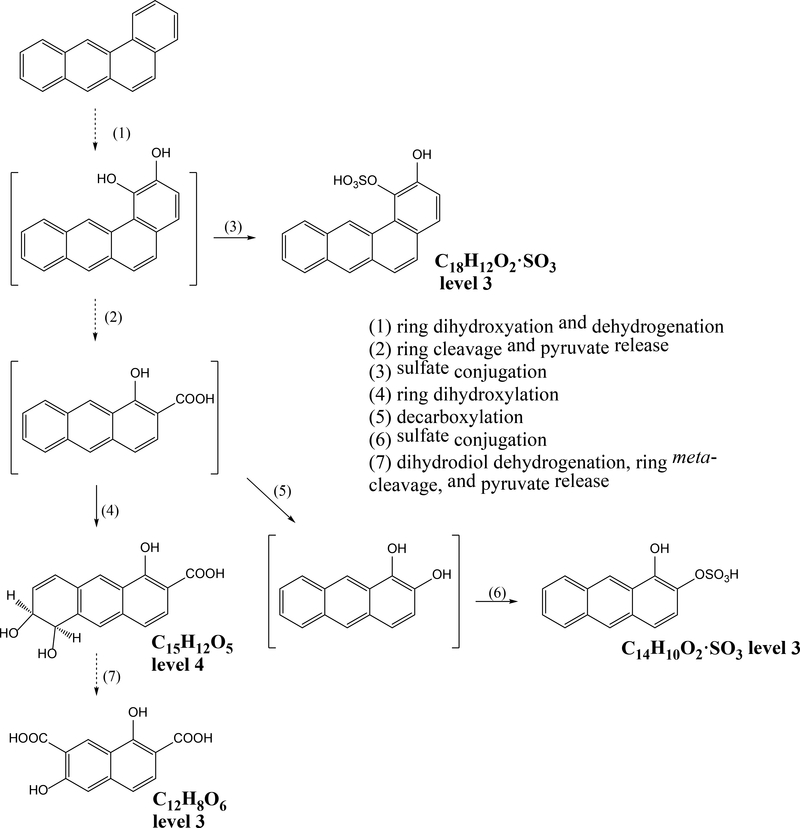
Based on the detected 13C-labeled metabolites, pathways for B[a]A and fluoranthene degradation in the PAH-contaminated soil are proposed (Figure 2 and Figure S7, respectively). It is important to note that these pathways do not necessarily result from metabolism of the parent compound by an individual organism, and therefore the specific metabolites and their accumulation over time are less meaningful than for incubation of a single substrate with a pure culture. B[a]A biodegradation mainly followed the preferential degradation route previously described for Sphingobium yanoikuyae B1, (formerly Beijerinckia),46,47 which is initiated by dioxygenation of the molecule in positions 1,2. Further meta-cleavage and pyruvate release would lead to the formation of 1-hydroxy-2-anthracenoic acid, an intermediate also observed in other Sphingobium strains,42 followed by oxidation to a dihydrodiol (C15H12O5) that is reported here for the first time. The identification of two isomers of dihydroxynaphthalene dicarboxylic acid (Table 1) indicated the dehydrogenation and ring cleavage of the latter.42 Several fluoranthene metabolites are not consistent with established pathways, but the identification of hydroxy-fluorenone suggests the processing of one of the two fused aromatic rings.48,49 We observed evidence for both initial mono- (C16H10O) and dioxygenation (C16H8O2) of the fluoranthene molecule (Figure S2), but none of the previously described ring-cleavage products was detected. The MS/MS analysis of the formula C16H10O3, corresponding to a metabolite not previously identified, presented a fragment of m/z 44 indicative of a carboxyl group (Figure S6), suggesting its formation as a product from the initial ring fission. Because this metabolite contained 16 C atoms and only three oxygen atoms, instead of the expected four for a common meta- or ortho-cleavage product, it could not be assigned to any previously described ring fission metabolites.48,50 Finally, the product with formula C11H10O6, identified here for the first time, could be consistent with a product from the cleavage and further degradation of the biphenylene derivative originating from angular attack on fluorenone.48
Figure 2.
Proposed degradation pathway for benzo[a]anthracene based on detected metabolites and published literature.42 Note that substituent positions are either inferred from known mass spectra or are hypothesized. Dashed arrows indicate more than one successive reaction. Structures in brackets are putative and were not observed directly. Reactions follow the canonical reactions in aerobic fungal and bacterial metabolism of PAHs summarized in Cerniglia51 and Kweon et al.,52 respectively. The levels indicated next to structures are the level of confidence as defined in Schymanski et al.53 Level 3: tentative candidates (compounds identified by molecular formula and substructures), based on accurate mass, MS/MS pattern, and database search. Level 4: unequivocal molecular formula based on accurate mass, but no other supporting data.
Time-course evolution of the identified metabolites.
We traced the detected metabolites through the course of the incubations according to their m/z and retention time (Figure S1). Due to the lack of standards, metabolites were quantified relative to their peak areas. Consistent with PAH removal (Figure S1), fluoranthene metabolites peaked at 7 d (except C16H8O2, which peaked at 14 d), then decreased in abundance, clearly indicative of their subsequent metabolism. B[a]A metabolites, including two sulfate conjugates, appeared at 14 d and increased in abundance after 21 d incubation. Sulfate conjugates from pyrene also accumulated over the course of incubation, suggesting the potential persistence of such conjugates even in biologically active soil. The recalcitrance of analogous PAH conjugates accumulated by the fungus Cunninghamella elegans to soil microbial communities was previously demonstrated.45
Environmental implications and further applications.
In this study, we demonstrated that SIAM can be successfully applied to the biotransformation of pollutants. This method allowed us to trace the biotransformation of individual pollutants in the context of a real contaminated sample. SIAM in combination with MS/MS enabled the detection of novel metabolites and degradation pathways that could result from interactions within complex microbial communities and contaminant mixtures. Despite the analytical limitations associated with the use of a single ionization source, our method allowed us to highlight major mechanisms implicated in PAH biodegradation during biostimulation of a contaminated soil. The SIAM workflow could be applied to a wider range of studies in environmental sciences, such as the biotransformation of micropollutants in aquatic systems and in water treatment processes. In addition, the combination of SIAM with compound specific isotope analysis (CSIA) may become a powerful tool for field-based bioremediation studies.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) under its Superfund Research Program, grant number P42ES005948, and the UNC Center for Environmental Health and Susceptibility, grant number P30ES010126. JV was supported by a Marie Skłodowska Curie Individual Fellowship from the European Commission (Grant Agreement 661361 – NETPAC – H2020-MSCA-IF-2014). We thank Prof. Avram Gold and Prof. Zhenfa Zhang for providing U-13C labeled PAHs, and Mr. Leonard Collins for his help in instrumental analysis.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information Available
Experimental details; parameter values for data analysis; time course of biodegradation for target PAHs; mass spectra and extracted ion chromatograms (EIC) of major metabolites; evolution of metabolites over the course of incubation.
REFERENCES
- (1).Hu J; Nakamura J; Richardson SD; Aitken MD Evaluating the effects of bioremediation on genotoxicity of polycyclic aromatic hydrocarbon-contaminated soil using genetically engineered, higher eukaryotic cell lines. Environ. Sci. Technol 2012, 46, 4607–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Lemieux CL; Long AS; Lambert IB; Lundstedt S; Tysklind M; White PA In vitro mammalian mutagenicity of complex polycyclic aromatic hydrocarbon mixtures in contaminated soils. Environ. Sci. Technol 2015, 49, 1787–1796. [DOI] [PubMed] [Google Scholar]
- 3).Chibwe L; Geier MC; Nakamura J; Tanguay RL; Aitken MD; Simonich SLM Aerobic bioremediation of PAH contaminated soil results in increased genotoxicity and developmental toxicity. Environ. Sci. Technol 2015, 49, 13889–13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Tian Z; Gold A; Nakamura J; Zhang Z; Vila J; Singleton DR; Collins LB; Aitken MD Nontarget analysis reveals a bacterial metabolite of pyrene implicated in the genotoxicity of contaminated soil after bioremediation. Environ. Sci. Technol 2017, 51, 7091–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Grifoll M; Selifonov SA; Gatlin CV; Chapman PJ Actions of a versatile fluorene-degrading bacterial isolate on polycyclic aromatic compounds. Appl. Environ. Microbiol 1995, 61, 3711–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Zhong Y; Luan T; Zhou H; Lan C; Tam NFY Metabolite production in degradation of pyrene alone or in a mixture with another polycyclic aromatic hydrocarbon by Mycobacterium sp. Environ. Toxicol. Chem 2006, 25, 2853–2859. [DOI] [PubMed] [Google Scholar]
- 7).Bouchez M; Blanchet D; Vandecasteele J-P Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol 1995, 43, 156–164. [DOI] [PubMed] [Google Scholar]
- 8).Stringfellow WT; Aitken MD Competitive metabolism of naphthalene, methylnaphthalenes, and fluorene by phenanthrene-degrading pseudomonads. Appl. Environ. Microbiol 1995, 61, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Knightes CD; Peters CA Multisubstrate biodegradation kinetics for binary and complex mixtures of polycyclic aromatic hydrocarbons. Environ. Toxicol. Chem 2006, 25, 1746–1756. [DOI] [PubMed] [Google Scholar]
- 10).Hennessee CT; Li QX Effects of polycyclic aromatic hydrocarbon mixtures on degradation, gene expression, and metabolite production in four mycobacterium species. Appl. Environ. Microbiol 2016, 82, 3357–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Zhong Y; Luan T; Lin L; Liu H; Tam NFY Production of metabolites in the biodegradation of phenanthrene, fluoranthene and pyrene by the mixed culture of Mycobacterium sp. and Sphingomonas sp. Bioresour. Technol 2011, 102, 2965–2972. [DOI] [PubMed] [Google Scholar]
- 12).Luo S; Chen B; Lin L; Wang X; Tam NF-Y; Luan T Pyrene degradation accelerated by constructed consortium of bacterium and microalga: effects of degradation products on the microalgal growth. Environ. Sci. Technol 2014, 48, 13917–13924. [DOI] [PubMed] [Google Scholar]
- 13).Richnow HH; Eschenbach A; Mahro B; Seifert R; Wehrung P; Albrecht P; Michaelis W The use of 13C-labelled polycyclic aromatic hydrocarbons for the analysis of their transformation in soil. Chemosphere 1998, 36, 2211–2224. [DOI] [PubMed] [Google Scholar]
- 14).Richnow HH; Annweiler E; Koning M; Lüth J-C; Stegmann R; Garms C; Francke W; Michaelis W Tracing the transformation of labelled [1-13C]phenanthrene in a soil bioreactor. Environ. Pollut 2000, 108, 91–101. [DOI] [PubMed] [Google Scholar]
- 15).Liu J; Wang Y; Jiang B; Wang L; Chen J; Guo H; Ji R Degradation, metabolism, and bound-residue formation and release of tetrabromobisphenol A in soil during sequential anoxic–oxic incubation. Environ. Sci. Technol 2013, 47, 8348–8354. [DOI] [PubMed] [Google Scholar]
- 16).Wang Y; Xu J; Shan J; Ma Y; Ji R Fate of phenanthrene and mineralization of its non-extractable residues in an oxic soil. Environ. Pollut 2017, 224, 377–383. [DOI] [PubMed] [Google Scholar]
- 17).Morasch B; Hunkeler D; Zopfi J; Temime B; Höhener P Intrinsic biodegradation potential of aromatic hydrocarbons in an alluvial aquifer – potentials and limits of signature metabolite analysis and two stable isotope-based techniques. Water Res. 2011, 45, 4459–4469. [DOI] [PubMed] [Google Scholar]
- 18).Fischer A; Manefield M; Bombach P Application of stable isotope tools for evaluating natural and stimulated biodegradation of organic pollutants in field studies. Curr. Opin. Biotechnol 2016, 41, 99–107. [DOI] [PubMed] [Google Scholar]
- 19).Kolkman A; Martijn BJ; Vughs D; Baken KA; van Wezel AP Tracing nitrogenous disinfection byproducts after medium pressure UV water treatment by stable isotope labeling and high resolution mass spectrometry. Environ. Sci. Technol. 2015, 49, 4458–4465. [DOI] [PubMed] [Google Scholar]
- 20).Sun K; Liang S; Kang F; Gao Y; Huang Q Transformation of 17β-estradiol in humic acid solution by ο-MnO2 nanorods as probed by high-resolution mass spectrometry combined with 13C labeling. Environ. Pollut 2016, 214, 211–218. [DOI] [PubMed] [Google Scholar]
- 21).Hegeman AD; Schulte CF; Cui Q; Lewis IA; Huttlin EL; Eghbalnia H; Harms AC; Ulrich EL; Markley JL; Sussman MR Stable isotope assisted assignment of elemental compositions for metabolomics. Anal. Chem 2007, 79, 6912–6921. [DOI] [PubMed] [Google Scholar]
- 22).Giavalisco P; Hummel J; Lisec J; Inostroza AC; Catchpole G; Willmitzer L High-resolution direct infusion-based mass spectrometry in combination with whole 13C metabolome isotope labeling allows unambiguous assignment of chemical sum formulas. Anal. Chem 2008, 80, 9417–9425. [DOI] [PubMed] [Google Scholar]
- 23).Guo K; Li L Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal. Chem 2009, 81, 3919–3932. [DOI] [PubMed] [Google Scholar]
- 24).Hiller K; Metallo CM; Kelleher JK; Stephanopoulos G Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal. Chem 2010, 82, 6621–6628. [DOI] [PubMed] [Google Scholar]
- 25).Huang X; Chen Y-J; Cho K; Nikolskiy I; Crawford PA; Patti GJ X13CMS: Global tracking of isotopic labels in untargeted metabolomics. Anal. Chem 2014, 86, 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Zhou R; Tseng C-L; Huan T; Li L IsoMS: Automated processing of LC-MS data generated by a chemical isotope labeling metabolomics platform. Anal. Chem 2014, 86, 4675–4679. [DOI] [PubMed] [Google Scholar]
- 27).Leeming MG; Isaac AP; Pope BJ; Cranswick N; Wright CE; Ziogas J; O’Hair RAJ; Donald WA High-resolution twin-ion metabolite extraction (HiTIME) mass spectrometry: Nontargeted detection of unknown drug metabolites by isotope labeling, liquid chromatography mass spectrometry, and automated high-performance computing. Anal. Chem 2015, 87, 4104–4109. [DOI] [PubMed] [Google Scholar]
- 28).Bueschl C; Kluger B; Neumann NKN; Doppler M; Maschietto V; Thallinger GG; Meng-Reiterer J; Krska R; Schuhmacher R MetExtract II: A software suite for stable Isotope-assisted untargeted metabolomics. Anal. Chem 2017, 89, 9518–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Kluger B; Bueschl C; Neumann N; Stückler R; Doppler M; Chassy AW; Waterhouse AL; Rechthaler J; Kampleitner N; Thallinger GG; et al. Untargeted profiling of tracer-derived metabolites using stable isotopic labeling and fast polarity-switching LC–ESI-HRMS. Anal. Chem 2014, 86, 11533–11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Neumann NKN; Lehner SM; Kluger B; Bueschl C; Sedelmaier K; Lemmens M; Krska R; Schuhmacher R Automated LC-HRMS(/MS) approach for the annotation of fragment ions derived from stable isotope labeling-assisted untargeted metabolomics. Anal. Chem 2014, 86, 7320–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Singleton DR; Sangaiah R; Gold A; Ball LM; Aitken MD Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol 2006, 8, 1736–1745. [DOI] [PubMed] [Google Scholar]
- 32).Zhang Z; Sangaiah R; Gold A; M. Ball L. Synthesis of uniformly 13C-labeled polycyclic aromatic hydrocarbons. Org. Biomol. Chem 2011, 9, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Tejeda-Agredano MC; Gallego S; Vila J; Grifoll M; Ortega-Calvo JJ; Cantos M Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol. Biochem 2013, 57, 830–840. [Google Scholar]
- 34).Jones MD; Singleton DR; Carstensen DP; Powell SN; Swanson JS; Pfaender FK; Aitken MD Effect of incubation conditions on the enrichment of pyrene-degrading bacteria identified by stable-isotope probing in an aged, PAH-contaminated soil. Microb. Ecol 2008, 56, 341–349. [DOI] [PubMed] [Google Scholar]
- 35).Patti GJ; Tautenhahn R; Siuzdak G Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat. Protoc 2012, 7, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Tautenhahn R; Patti GJ; Rinehart D; Siuzdak G XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem 2012, 84, 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Wicker J; Fenner K; Ellis L; Wackett L; Kramer S Predicting biodegradation products and pathways: a hybrid knowledge- and machine learning-based approach. Bioinformatics 2010, 26, 814–821. [DOI] [PubMed] [Google Scholar]
- 38).Latino DARS; Wicker J; Gütlein M; Schmid E; Kramer S; Fenner K Eawag-Soil in enviPath: a new resource for exploring regulatory pesticide soil biodegradation pathways and half-life data. Environ. Sci. Process. Impacts 2017, 19, 449–464. [DOI] [PubMed] [Google Scholar]
- 39).Ruttkies C; Schymanski EL; Wolf S; Hollender J; Neumann S MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminformatics 2016, 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Kazunga C; Aitken MD; Gold A; Sangaiah R Fluoranthene-2,3- and −1,5-diones are novel products from the bacterial transformation of fluoranthene. Environ. Sci. Technol 2001, 35, 917–922. [DOI] [PubMed] [Google Scholar]
- 41).Kazunga C; Aitken MD Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl. Environ. Microbiol 2000, 66, 1917–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Kunihiro M; Ozeki Y; Nogi Y; Hamamura N; Kanaly RA Benz[a]anthracene biotransformation and production of ring fission products by sphingobium sp. strain KK22. Appl. Environ. Microbiol 2013, 79, 4410–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Riva M; Tomaz S; Cui T; Lin Y-H; Perraudin E; Gold A; Stone EA; Villenave E; Surratt JD Evidence for an unrecognized secondary anthropogenic source of organosulfates and sulfonates: gas-phase oxidation of polycyclic aromatic hydrocarbons in the presence of sulfate aerosol. Environ. Sci. Technol 2015, 49, 6654–6664. [DOI] [PubMed] [Google Scholar]
- 44).Pothuluri JV; Evans FE; Heinze TM; Cerniglia CE Formation of sulfate and glucoside conjugates of benzo[e]pyrene by Cunninghamella elegans. Appl. Microbiol. Biotechnol 1996, 45, 677–683. [Google Scholar]
- 45).Schmidt SN; Christensen JH; Johnsen AR Fungal PAH-metabolites resist mineralization by soil microorganisms. Environ. Sci. Technol 2010, 44, 1677–1682. [DOI] [PubMed] [Google Scholar]
- 46).Jerina DM; Van Bladeren PJ; Yagi H; Gibson DT; Mahadevan V; Neese AS; Koreeda M; Sharma ND; Boyd DR Synthesis and absolute configuration of the bacterial cis-1,2-, cis-8,9-, and cis-10,11-dihydrodiol metabolites of benz[a]anthracene formed by strain of Beijerinckia. J. Org. Chem 1984, 49, 3621–3628. [Google Scholar]
- 47).Mahaffey WR; Gibson DT; Cerniglia CE Bacterial oxidation of chemical carcinogens: formation of polycyclic aromatic acids from benz[a]anthracene. Appl. Environ. Microbiol 1988, 54, 2415–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Kweon O; Kim S-J; Jones RC; Freeman JP; Adjei MD; Edmondson RD; Cerniglia CE A polyomic approach to elucidate the fluoranthene-degradative pathway in Mycobacterium vanbaalenii PYR-1. J. Bacteriol 2007, 189, 4635–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Rehmann K; Hertkorn N; Kettrup AA Fluoranthene metabolism in Mycobacterium sp. strain KR20: identity of pathway intermediates during degradation and growth. Microbiology 2001, 147, 2783–2794. [DOI] [PubMed] [Google Scholar]
- 50).van Herwijnen R; Wattiau P; Bastiaens L; Daal L; Jonker L; Springael D; Govers HAJ; Parsons JR Elucidation of the metabolic pathway of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res. Microbiol 2003, 154, 199–206. [DOI] [PubMed] [Google Scholar]
- 51).Cerniglia CE Fungal metabolism of polycyclic aromatic hydrocarbons: past, present and future applications in bioremediation. J. Ind. Microbiol. Biotechnol 1997, 19, 324–333. [DOI] [PubMed] [Google Scholar]
- 52).Kweon O; Kim S-J; Holland RD; Chen H; Kim D-W; Gao Y; Yu L-R; Baek S; Baek D-H; Ahn H; et al. Polycyclic aromatic hydrocarbon metabolic network in Mycobacterium vanbaalenii PYR-1. J. Bacteriol 2011, 193, 4326–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol 2014, 48, 2097–2098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.