Abstract
Atherosclerosis is a progressive vascular disease triggered by interplay between abnormal shear stress and endothelial lipid retention. A combination of these and, potentially, other factors leads to a chronic inflammatory response in the vessel wall, which is thought to be responsible for disease progression characterized by a buildup of atherosclerotic plaques. Yet molecular events responsible for maintenance of plaque inflammation and plaque growth have not been fully defined. Here we show that endothelial TGFβ signaling is one of the primary drivers of atherosclerosis-associated vascular inflammation. Inhibition of endothelial TGFβ signaling in hyperlipidemic mice reduces vessel wall inflammation and vascular permeability and leads to arrest of disease progression and regression of established lesions. These pro-inflammatory effects of endothelial TGFβ signaling are in stark contrast with its effects in other cell types and identify it as an important driver of atherosclerotic plaque growth and show the potential of cell-type specific therapeutic intervention aimed at control of this disease.
Introduction
Atherosclerotic vascular disease is the major cause of myocardial infarctions, strokes, peripheral vascular disease and other illnesses that collectively account for the most deaths in the world. Whilst its initiating factors are well understood, the cause of the relentless progression of the disease, once it is established, is less certain. Chronic vascular inflammation and continuous abnormal mechanotransduction are thought to play a role, but how they affect the atherosclerosis progression has not been established 1-3. Recent studies have suggested that activation of endothelial TGFβ signaling may be an important driver of atherogenesis 4,5. Yet this has been difficult to reconcile with a general perception of anti-inflammatory effects TGFβ signaling.
Results
Prop-inflammatory effects of endothelial TGFβ signaling
To explore the effect of TGFβ signaling activation in endothelial vs. other cell types thought to be involved in atherosclerosis, we used bulk RNA-seq to examine transcriptional changes induced by TGFβ treatment in endothelial (EC), smooth muscle (SMC), CD4+ effector memory T-cells (T) and macrophages (M). As expected, the growth factor treatment resulted in both activation and suppression of expression of various genes in all cell types. Surprisingly, however, TGFβ-regulated molecular programs appeared very cell type-specific: there was little overlap between gene expression signatures in the four cell types examined (Fig 1A). A closer examination of TGFβ-induced gene expression in ECs revealed induction of expression of a number of pro-inflammatory chemokines and cytokines and their receptors (including CCL2), leucocyte adhesion molecules (such as ICAM-1 and VCAM-1), matrix metalloproteinases (MMP2) as well fibronectin, a pro-inflammatory extracellular matrix (ECM) component long linked to inflammation6,7 (Fig 1B). In contrast, TGFβ signature in SMCs was largely anti-inflammatory (Fig 1B). In particular, CCL2, ICAM-1 and VACAM-1 expression was reduced, unlike the increased observed in ECs.
Figure 1. TGFβ endothelial-specific induction of inflammatory response.
(A, B) Bulk RNA-seq analysis of HUVEC (EC), HASMC (SMC), CD4+ effector memory T cells (T) and macrophages (M) gene expression before and after stimulation with TGFβ1 (A) and inflammatory gene expression (B). Each lane is an average of two biologically independent samples. (C) ChIP-seq analysis of canonical TGFβ signaling in EC and SMCs. Note increased SMAD2/3 binding to regulatory elements of CCL2, CLDN5 and SERPINE1 in ECs but not SMCs. Pol II S2 and Pol II S5 ChIP in combination with bulk RNA-seq indicate increased expression of CCL2 and SERPINE1 and decreased expression of CLDN5 after TGFβ1 stimulation. N=1 per experimental condition. (D) Relative CLDN5 expression in primary ECs isolated from control (Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-mT/mG) and TGFβRiEC mice treated with corn oil. Bars: mean ± SEM. 0 hr P=0.004, 6 hr P=0.001. Two-tailed Student’s t tests (N=3 animals per group for 0 hr, N=6 animals per group for 6 hr). (E) i.v. Evans blue dye administration following a subcutaneous TNFα injection into mouse ear. Scale bar: 2 mm; mean ± SEM. 0 min P=0.56, 15 min P=0.000066, Two-tailed Student’s t tests (N=6 animals per group for 0 min, N=7 animals per group for 15 min). (F,G) Mouse ear sections were stained with anti-ICAM, VCAM-1, CD45 and Ly6G antibodies 6 hrs and 3 days following a s.c. TNFα injection. Note a strong reduction in expression of both adhesion molecules (F) and reduced presence of inflammatory cells (G) in TGFβRiEC mice. Scale bar: 16 μm. N=7 animals per group. Bars: mean ± SEM. ICAM1+ area 6 hrs P=0.001, ICAM1+ area 3 days P=0.0002, VCAM1+ area 6 hrs P=0.0001, VCAM1+ area 3 days P=0.00001, Ly6G+ cells 6 hrs P=0.0499, Ly6G+ cells a 3 days P=0.0004, CD45+ cells 6 hrs P=0.051, CD45+ cells a 3 days P=0.00004. Two-tailed Student’s t tests.
To get further insights into the observed differences, we carried out ChIP-seq using SMAD2/3 to assess canonical TGFβ signaling-dependent regulation of transcription in combination with Pol II S2 and Pol II S5 to assess the status of target gene’s transcription in ECs and SMCs. In agreement with bulk RNA-seq results, scanning of genomic SMAD2/3 binding sites revealed several EC-specific regulatory elements among inflammation-related genes. In particular, there was a prominent binding to CCL2 and SERPINE1 regulatory elements that was associated with increased transcription of these genes (Fig 1C). At the same time, SMAD2/3 binding to Claudin-5 promoter resulted in suppression of its expression. In marked contrast, no SMAD2/3 enrichment was detected in these regulatory elements of these genes in SMCs and there were no changes in gene expression. These results further emphasize cell-type specificity of TGFβ signaling that is pro-inflammatory in ECs but not in SMCs.
To test functional significance of these findings, we generated mice with an inducible endothelial specific knockout of TGFβ receptors 1 and 2 (Cdh5CreERT2;Tgfbr1l/l;Tgfbr2l/l hereafter referred to as TGFβRiEC; Fig S1A,B). qPCR analysis of primary ECs isolated from TGFβRiEC showed 50% higher levels of Claudin-5 compared to controls (Fig 1D). In agreement with this finding, intravenous TNFα administration resulted in a significantly smaller vascular leak in TGFβRiEC animals (Fig 1E). An intradermal injection of TNFα into the mouse ear induced lower ICAM-1and VCAM-1 expression in ear blood vessels 6 hours and 3 days later (Fig. 1F) and a significant reduction in leukocyte infiltration in TGFβRiEC mice compared to control mice (Fig. 1G). Together, these results indicate that suppression of EC TGFβ signaling has a profound anti-inflammatory effect including reduced vascular permeability.
Endothelial TGFβ receptors knockout reduces atherosclerotic plaque growth.
To test if this would result in reduced vascular inflammation in atherosclerosis settings, TGFβRiEC mice were crossed onto the Apoe−/− background to induce atherosclerotic susceptibility, and the mT/mG strain to fate-map ECs (Fig S1A, B). We chose to delete both TGFβ receptors to avoid possible aberrant signaling after a single receptor deletion8,9. Furthermore, a knockdown of both TGFβR1 and TGFβR2 more effectively abolished TGFβ-driven Smad2 and Smad3 phosphorylation than either receptor alone (Fig S1C). The resultant mutant mice (Cdh5CreERT2;Tgfbr1fl/fl;Tgfbr2fl/fl;Apoe−/−;mT/mGfl/fl), hereby referred to as TGFβRiEC-Apoe, with controls (absent Cdh5CreERT2, mice without Tgfbr1 and Tgfbr2 loci or non-induced mice) were used for subsequent experiments. Activation of the Cdh5CreERT2 gene at six weeks of age led to a complete deletion of both targeted Tgfbr genes (Fig S1D). This fully blocked TGFβ signaling while preserving BMP signaling (Fig S1D, E).
Two-month-old TGFβRiEC-Apoe, and control mice were placed on high cholesterol, high fat diet (HCHFD) two weeks after induction of Tgfbr1/2 excision (Fig S2A). The dietary intervention resulted in an increase in body weight, total plasma cholesterol, and triglycerides which was similar in both groups (Fig S2B-D). Serial analysis of whole aortas opened en face and aortic arch using Oil-Red-O staining, demonstrated a significantly delayed onset and reduced extent of lipid deposition in TGFβRiEC-Apoe mice (Fig. 2A-C). Quantitative assessment showed a 61–79% reduction in Oil-Red-O staining in aortas of TGFβRiEC-Apoe mice over this time course (Fig, 2B).
Figure 2: Endothelial cell Tgfbr1/Tgfbr2 knockout inhibits atherosclerosis plaque development.
(A) Microphotographs of en face aortas from Apoe−/− and TGFβRiEC-Apoe mice treated with corn oil or Tamoxifen after 0, 0.5, 1, 2, 3, and 4 months of HCHFD stained with Oil-Red-O. (B) Lesion area quantification: % lesion area refers to ORO stained as a % of the total aortic surface. All data shown as mean ± SEM. Ø: not detected. P=0.000000007 for Apoe−/− 0.5 month of HCHFD compared to TGFβRiEC-Apoe 0.5 month of HCHFD, P=0.0000058 for Apoe−/− 1 month of HCHFD compared to TGFβRiEC-Apoe 1 month of HCHFD, P=0.00000000002 for Apoe−/− 2 month of HCHFD compared to TGFβRiEC-Apoe 2 month of HCHFD, P=0.0000051 for Apoe−/− 3 month of HCHFD compared to TGFβRiEC-Apoe 3 month of HCHFD, P=0.0000000000094 for Apoe−/− 4 month of HCHFD compared to TGFβRiEC-Apoe 4 month of HCHFD.Two-tailed Student’s t tests. 0M N=3 animals per group, 0.5M-4M N=11 animals per group. (C) Representative photomicrographs of Oil-Red-O stained atherosclerotic lesions in aortic arches of Apoe−/− and TGFβRiEC-Apoe mice treated with corn oil or Tamoxifen after 0, 0.5, 1, 2, 3, and 4 months of HCHFD. Scale bar: 5 mm. (N=3 animals per group) (D) (Left) Representative examples of cross-sections from the aortic root after 4 months of HCHFD stained with Oil-Red-O. Scale bar: 200 μm. (Right) Quantification of aortic root lesion areas. Bars indicate the mean ± SEM. P=0.000011, Two-tailed Student’s t tests (N=11 animals per group). (E) Representative images of brachiocephalic artery from Apoe−/− and TGFβRiEC-Apoe mice treated with corn oil or tamoxifen (Movat staining). Scale bar: 100 μm. (F) Measurement of lesion area. Bars: mean ± SEM. P=0.000000136 for for Apoe−/− 1 month of HCHFD compared to TGFβRiEC-Apoe 1 month of HCHFD, P=0.0000006 for Apoe−/− 2 month of HCHFD compared to TGFβRiEC-Apoe 2 month of HCHFD, P=0.0000000019 for Apoe−/− 3 month of HCHFD compared to TGFβRiEC-Apoe 3 month of HCHFD, P=0.000000022 for Apoe−/− 4 month of HCHFD compared to TGFβRiEC-Apoe 4 month of HCHFD. Two-tailed Student’s t tests. (N=11 animals per group). (G) Quantifications of the extent of necrotic areas in brachiocephalic artery of Apoe−/− and TGFβRiEC-Apoe mice. Bars: mean ± SEM. P=0.000000023 for Apoe−/− 1 month of HCHFD compared to TGFβRiEC-Apoe 1 month of HCHFD, P=0.000000000014 for Apoe−/− 2 month of HCHFD compared to TGFβRiEC-Apoe 2 month of HCHFD, P=0.000000000029 for Apoe−/− 3 month of HCHFD compared to TGFβRiEC-Apoe 3 month of HCHFD, P=0.00000000000013 for Apoe−/− 4 month of HCHFD compared to TGFβRiEC-Apoe 4 month of HCHFD. Two-tailed Student’s t tests. (N=11 animals per group). (H) Morphologic classification of brachiocephalic artery lesion severity in Apoe−/− and TGFβRiEC-Apoe mice.
To study the effect of TGFβ receptors deletion on the composition and size of atherosclerotic plaques, the aortic root and brachiocephalic artery from both groups of mice were serially sectioned. Histological examination of Oil-Red-O-stained aortic root (Fig 2D) and Movat-stained brachiocephalic artery (Fig 2E) demonstrated a marked reduction in the plaque size (60% and 68%, respectively) in TGFβRiEC-Apoe mice fed HCHF diet for 4 months (Fig. 2D, F). The differences were most pronounced early in the time course: histological analysis of Movat-stained brachiocephalic sections, using the Virmani classification10,11, showed that after 1 month of HCHFD, 45% of the Apoe−/− mice exhibited pathological intimal thickening (PIT) and 22% had evidence of intimal xanthomas (IX) while 90% TGFβRiEC-Apoe mice had no signs of atherosclerosis. Fibrous cap atheromas (FCA) were evident after 2 months of HCHFD in the majority of Apoe−/− mice but did not appear in TGFβRiEC-Apoe mice until a month later. Even after 4 months of HCHFD, FCA were present in only a subset of the TGFβRiEC-Apoe mice (Fig 2E-H). This attenuation of plaque progression observed in TGFβRiEC-Apoe mice was also reflected by a significant reduction in the necrotic core area, which is a prominent feature of advance atherosclerotic lesions (Fig 2G).
Serial evaluation of plaque cellularity (H&E staining) showed a significant decrease in total cell numbers in lesions from TGFβRiEC-Apoe compared to Apoe−/− mice at all time points (Fig S3A-C). Immunostaining demonstrated a decrease in the number of neointimal αSMA+ cells and a reduction in plaque collagen content (Fig S3D, E). In addition, fibronectin deposition and endothelial VCAM-1 expression decreased (Fig S3F, G), indicating a reduction in the “inflammatory” state of the vessel wall.
Endothelial TGFβ receptors knockout induces regression of established atherosclerotic lesions.
Given profound effects of suppression of EC TGFβ activation during atherogenesis, we next addressed whether the same approach would reduce progression and induce regression of established atherosclerotic lesions. To this end, Cdh5CreERT2;Tgfbr1fl/fl;Tgfbr2fl/fl;Apoe−/−;mT/mGfl/fl mice were placed on HCHFD and, after two months, were randomized to tamoxifen-driven Cre activation (generating TGFβRiEC-Apoe mice) or sham treatment while continuing on the HCHF diet (Fig S4A). Eight weeks later (16 weeks of HCHFD), both groups were sacrificed, and the extent of atherosclerotic burden was determined using whole en face aorta Oil-Red-O staining (Fig S4B). As expected, the control mice demonstrated extensive progression of disease with the total aortic lesion area increasing from 6.8% to 17%. At the same time, mice with the induced EC TGFβR1/R2 deletion showed no significant disease progression (6.6% to 8.5%, p=NS) (Fig S4C). Thus, EC deletion of TGFβR1 and TGFβR2 prevented progression of atherosclerosis under hypercholesterolemic conditions.
To test how the absence of TGFβR1 and TGFβR2 in ECs influences the regression of atherosclerosis, Cdh5CreERT2;Tgfbr1fl/fl;Tgfbr2fl/fl;Apoe−/−;mT/mGfl/fl mice were kept on the HCHFD for 3 months. At that time, they were switched to the normal chow diet (which still results in elevated cholesterol levels on the Apoe−/− background) and randomized to Cdh5CreERT2 activation, inducing endothelial-specific Tgfbr1 and Tgfbr2 deletions (Fig S4D). One month later, while both Apoe−/− and TGFβRiEC-Apoe mice showed an expected decrease in the aortic wall neutral lipid accumulation, it was far more profound in the latter (Fig S4E,F).
Histological sections of the aortic root were used to analyze atherosclerotic plaque size and composition after 1 or 2 months of normal chow diet (ND, Fig 3A,B). There was no significant reduction in the aortic root plaque size in Apoe−/− mice either after 1 or 2 months of ND (Fig 3B,C). However, TGFβRiEC-Apoe mice showed a 47% decrease in plaque size after 1 month and a 71% decrease after 2 months on ND (Fig 3B,C). Morphological analysis showed the expected plaque progression from initial lesions (IX, PIT) towards established fibrous cap atheromata in Apoe−/− mice. In contrast, deletion of Tgfbr1/Tgfbr2 abolished plaque progression and the fraction of IX and PIT even increased after 1 and 2 months of ND. These observations, together with the decrease in plaque area, suggest true plaque regression in mice lacking endothelial TGFβR1 and TGFβR2 (Fig 3D).
Figure 3: Endothelial cell Tgfbr1/Tgfbr2 knockout facilitates regression of advanced murine atherosclerotic plaques.
(A) Experiment timeline: mice were fed HCHFD for 3 months to induce advanced atherosclerotic lesions. At that point, tamoxifen or vehicle control were administered and the diet was changed to ND for additional 1–2 months. (B) Representative images of aortic root cross-sections stained with Movat (scale bar: 200 μm). (C) Root lesion area quantification. Data are means ± SEM. NS: not significant, P=0.53 for Apoe−/− 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.00000097 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.125 for Apoe−/− 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.000000079 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.00033 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD + 1M ND, P=0.0000079 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD + 2M ND. Two-tailed Student’s t tests. 3M Apoe−/− baseline N=12 animals per group, 3M HFD + 1M ND Apoe−/− N=6 animals per group, 3M HFD + 2M ND Apoe−/− N=6 animals per group, 3M HFD + 1M ND TGFβRiEC-Apoe N=5 animals per group, 3M HFD + 2M ND TGFβRiEC-Apoe N=4 animals per group. (D) Morphologic classification of aortic root lesions severity in Apoe−/− and TGFβRiEC-Apoe mice. (3M Apoe−/− baseline N=10 animals per group, 3M HFD + 1M ND Apoe−/− N=10 animals per group, 3M HFD + 2M ND Apoe−/− N=10 animals per group, 3M HFD + 1M ND TGFβRiEC-Apoe N=10 animals per group, 3M HFD + 2M ND TGFβRiEC-Apoe N=10 animals per group) (E) Representative images of cross-sections of brachiocephalic artery stained with Movat (scale bar: 200 μm). (F) Lesion area quantification. All data shown as mean ± SEM. NS: not significant, P=0.58 for Apoe−/− 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.00086 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.047 for Apoe−/− 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.00039 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.021 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD + 1M ND, P=0.0000075 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD + 2M ND. Two-tailed Student’s t tests. 3M Apoe−/− baseline N=4 animals per group, 3M HFD + 1M ND Apoe−/− N=5 animals per group , 3M HFD + 2M ND Apoe−/− N=8 animals per group, 3M HFD + 1M ND TGFβRiEC-Apoe N=7 animals per group, 3M HFD + 2M ND TGFβRiEC-Apoe N=7 animals per group. Two-tailed Student’s t tests. (G) Morphologic classification of brachiocephalic artery lesion severity in Apoe−/− and TGFβRiEC-Apoe mice. (3M Apoe−/− baseline N=4 animals per group, 3M HFD + 1M ND Apoe−/− N=5 animals per group, 3M HFD + 2M ND Apoe−/− N=7 animals per group, 3M HFD + 1M ND TGFβRiEC-Apoe N=8 animals per group, 3M HFD + 2M ND TGFβRiEC-Apoe N=7 animals per group).
Analysis of serial brachiocephalic sections confirmed these findings; while there was no change in plaque size in Apoe−/− mice, atherosclerotic lesion size in TGFβRiEC-Apoe mice decreased 58.1% after 1 month and 67.4% after 2 months of ND following disruption of TGFβ signaling (Fig 3E,F). Analysis of plaque morphology showed an even more pronounced plaque regression (Fig 3G).
Endothelial single cell RNA-seq analysis identifies molecular signature of atherosclerosis.
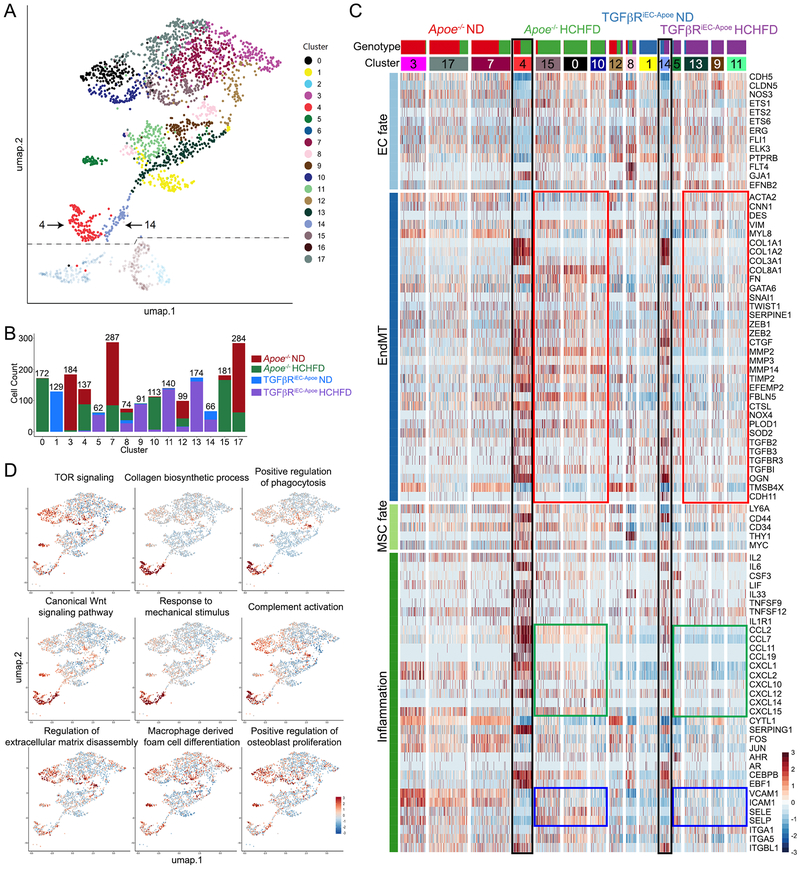
To understand the contribution of endothelial TGFβ signaling to atherosclerosis, we carried out a single cell RNA-seq (scRNA-seq) analysis of aortic ECs (CD31+CD45- population) from Apoe−/− and in TGFβRiEC-Apoe mice on ND and after 4 months of HCHFD (Fig S5A). Our generative deep neural network strategy (Fig S6A) originally identified 18 distinct EC clusters (Fig 4A, Fig S6B). The distribution of the 4 groups is represented across the aggregated data matrix (Fig S6C, Supplementary Table 3). Cluster 8 contains cells from all 4 groups of mice while cluster 12 contains cells mainly from 3 of the 4 groups of mice (TGFβRiEC-Apoe mice on ND is about 1% of these cells). The 15 remaining EC clusters largely segregated by genotype (Fig 4B and S6D) displaying distinct genotype- and cluster-specific expression profiles (Fig 4C). Three clusters were determined to be SMC contamination based on their high expression of MYH11 and MYOCD in comparison to all other cell populations (Fig S6B,E) and were subsequently removed from further analysis.
Figure 4. Single cell RNA-sequencing (scRNA-seq) analysis of endothelial gene expression in atherosclerosis.
(A) tSNE representation of single-cell transcriptomes of ECs from Apoe−/− (Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice treated with corn oil) and TGFβRiEC-Apoe (Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice treated with Tamoxifen) mice on ND or HCHFD. (B) Bar graph showing the cell distributions by genotype across cell clusters. Total number of cells in the cluster is at the top of the bar. (C) Heat map representation of gene expression from scRNA-seq data arranged by mouse genotype. (D) The z-score of the first principal component of the nGO pathway
Exposure of Apoe−/− mice to HCHFD resulted in a strong increase in endothelial to mesenchymal transition (EndMT) gene expression signature (Fig 4C, red boxes, Supplementary Data 1). This was accompanied by a broad increase in expression of chemokines (CCLs) and chemokine receptors (CXCLs, Fig 4C green boxes) and EC adhesion molecules (Fig 4C blue boxes). Increased EndMT, inflammation and cell adhesion signature were dramatically reduced in HCHFD-fed TGFβRiEC-Apoe mice. Two clusters (4 and 14), characterized by an almost complete loss of EC fate gene expression, are of particular interest (Fig 4A,C). Cluster 4 is largely composed of ECs derived from Apoe−/− mice on HCHFD and account for 11% of all EC of that genotype. These cells express mesenchymal stem cell (MSC) marker and show a strong increase in EndMT and inflammation-related genes signatures and likely represent endothelial-derived MSC-like cells, the ultimate product of EndMT. Cluster 14 is largely composed of EC derived from TGFβRiEC-Apoe mice on HCHFD and account for 5.8% of that genotype. While EC fate gene expression is still absent, there is a reduction in expression of MSC markers as well as EndMT and inflammation-related genes. We interpret this group as a “recovering” EC-derived MSC-like population.
Cluster-specific nested functional enrichment of gene ontology terms12 further illustrates key differences between the genotypes (Fig 4C,D, Fig S6F, and Supplementary Data 2). The two MSC-like clusters (4 and 14) display increased expression of genes related to complement activation, collagen synthesis, response to mechanical stimuli, Wnt signaling and phagocytosis. Clusters derived from Apoe−/− mice on HCHFD show a distinct increase in expression of the extracellular matrix, osteoblast proliferation, macrophage-derived foam cell differentiation and TOR signaling genes. Overall, these data are consistent with a significant reduction in inflammation following endothelial TGFβ receptors knockout.
To verify these insights, we studied the presence of inflammatory cells in plaques of Apoe−/− and TGFβRiEC-Apoe mice. The number of Mac-3+ macrophages and CD3+ T-cells were profoundly reduced in TGFβRiEC-Apoe mice (Fig 5A). Since the plaque size in these mice is smaller than in Apoe−/− mice and since the number of inflammatory cells is related to the plaque size, we compared the presence of Mac-2+ macrophages, CD45+ leukocytes and CD3+ T cells in size-matched plaques from TGFβRiEC-Apoe mice after four months and Apoe−/− mice after two months of HCHFD. Once again, there was a significant reduction in the number of inflammatory cells as well as a significant decrease in endothelial ICAM-1 expression (Fig 5B). We also evaluated the number of Mac-3+ macrophages in the aortic root plaques of Apoe−/− and TGFβRiEC-Apoe mice during regression, one or two months after the switch to ND. While there were no changes in macrophage plaque content in Apoe−/− mice, their numbers were reduced 57% (one months) and 99% (two months) after deletion of TGFβ receptors (Fig S7A,B). Analysis of brachiocephalic arteries showed similar trends: the presence of Mac-3+ macrophages and CD3+ T-cells and was profoundly reduced after TGFβ receptors deletion (Fig S7C-F).
Figure 5: Endothelial cell Tgfbr1/Tgfbr2 deletion decreases inflammation.
(A,B) Brachiocephalic artery atherosclerotic plaques after 4 months (A) and 2 months (B) of HCHFD in Apoe−/− and TGFβRiEC-Apoe mice. (A) Top: immunocytochemistry. Nuclei are counterstained with DAPI (blue). L: lumen. Scale bar: 62 μm. (4M Apoe−/− N=7 animals per group and 4M TGFβRiEC-Apoe N=8 animals per group). Bottom: Image quantification. Bars: mean ± SEM. P=0.000016 for Mac3, P=0.0083 for CD3 Two-tailed Student’s t tests. (B) Top: immunocytochemistry. Nuclei are counterstained with DAPI (blue). L: lumen. Scale bar: 62 μm. (4M Apoe−/− N=6 animals per group and 4M TGFβRiEC-Apoe N=6 animals per group). Bottom: Image quantification. Bars: mean ± SEM. P=0.000011 for ICAM-1, P=0.025 for Mac-2, P=0.02 for CD45, P=0.044 for CD3. Two-tailed Student’s t tests. (C) qPCR analysis of cytokines and cytokine receptors in Apoe−/− and TGFβRiEC-Apoe mice heart endothelial cells. β-actin was used for sample loading normalization. Bars: mean ± SEM. P=0.000036 for CCL2, P=0.00023 for CCL7, P=0.000058 for TNFRSFLA, P=0.00000021 for TNFRSFLB, P=0.0000042 for TLR4, P=0.000032 for MyD88. Two-tailed Student’s t test. Histogram is representative of six biologically independent samples. (D) (Left) THP-1 cell attachment to TNFα-treated (10 ng/ml for 5hrs) HUVEC monolayer in the presence or absence of TGFβR1/2 knockdown. (Right) Quantification of attached THP-1 cells per field. Bars: mean ± SEM. P=0.00053 for PBS treated groups, P=0.0000000033 for TNFα-treated groups. Two-tailed Student’s t test. N=8 biologically independent samples for PBS treated groups, N=7 for TNFα-treated groups. (E-F) Mice were fed the HCHFD for 3 months to induce advanced atherosclerotic lesions then treated with tamoxifen or vehicle control and switched to ND for additional 2 weeks. Top: Immunocytochemistry of brachiocephalic artery plaques from Apoe−/− and TGFβRiEC-Apoe mice stained with indicated antibodies. Nuclei counterstained with DAPI (blue). L: lumen. Bottom: Quantification of ICAM-1 area, Mac-2+/Ki67+, Mac-2+/TUNEL+, and VCAM-1+ EC. Bars: mean ± SEM. P=0.03 for Mac2+/Ki67+ cells, P=0.002 for Mac2+/TUNEL+ cells, P=0.0019 for ICAM-1 area, P=0.0002 for VCAM-1+ EC. Two-tailed Student’s t test. Scale bar: 16 μm. (Apoe−/− N=4 animals per group, TGFβRiEC-Apoe N=7 animals per group).
To further assess the mechanism by which the absence of TGFβ receptors in ECs significantly reduce the number of macrophages during the progression and regression of atherosclerosis, we examined whether TGFβ receptors expression in ECs have influence on the responsiveness to inflammatory cytokines. To this end, we studied endothelial cell inflammatory cytokine, inflammatory cytokine receptor expression and leukocyte adhesion isolated from Apoe−/− and TGFβRiEC-Apoe mice. There was a marked reduction in inflammatory cytokine and inflammatory cytokine receptor expression (Fig 5C) and less leukocyte adhesion after TNFα treatment (Fig 5D). In addition to decreased leukocyte adhesion, we also observed a decreased macrophage proliferation and vascular inflammation and increased macrophage apoptosis 2 weeks after the switch to ND (Fig 5E-F). While TGFβRiEC-Apoe mice had fewer Ly6Chi monocytes compared to controls, there was no difference in the size of Ly6Chi population taking up the beads between Apoe−/− and TGFβRiEC-Apoe mice (Fig S8).
Nanoparticle-targeted suppression of endothelial TGFβ signaling reduces plaque growth and induces regression.
Given this effect of endothelial TGFβ receptors knockout on atherosclerosis progression, we next tested whether a therapeutic intervention can be designed on this basis. Since the adverse effect of TGFβ signaling is cell-type specific, we utilized a nanoformulation previously demonstrated to deliver RNA to the endothelium, termed “7C1” 13. To test the ability of these nanoparticles to deliver their payload to the atherosclerotic aortic endothelium, endothelial fate-mapped Apoe−/− mice kept for 2 months on the HCHF diet or on regular chow diet were injected with 7C1 nanoparticles loaded with Alexa647-conjugated Luciferase siRNA as indicated (Fig S9A-D). FACS analysis of endothelial cells isolated from the aorta, heart, lung, and blood two hours after intravenous injection of 7C1-siLuciferase Alexa647 showed that over 85% of ECs (defined as GFP+ cells) in the aorta, heart, lung and took up 7C1 nanoparticles while no signal was detected in the peripheral blood (Fig S9E.) To confirm and expand these findings, we carried out a detailed assessment of various aortic segments (Fig S9F). In all segments examined, virtually all luminal endothelial cells demonstrated 7C1 uptake. The presence of atherosclerosis had no observable effect on the efficiency of 7C1 delivery to the aortic endothelium (Fig S9G).
Ten different TGFβR1 and TGFβR2 siRNAs were tested in vitro for their ability to suppress the target receptor expression (Fig S10A). The two siRNA with the best activity were chosen for testing, chemically modified to increase stability, packaged into 7C1 nanoparticles13,14 and used for in vivo delivery in Apoe−/− mice. Various amounts of the siRNAs packaged into 7C1 nanoparticles were injected into mice and qPCR assessment of TGFβ receptors expression in lung and heart endothelial cells was then used to determine an effective dose for subsequent studies (Fig S10B-C).
Two-months old Apoe−/− mice were placed on HCHFD and 7C1-packaged Tgfbr1/2 siRNAs were administered as indicated (Fig S10D). PBS and 7C1-formulated luciferase were used as controls. siRNAs alone had no effect on normal weight gain associated with HCHFD (Fig S10E). Evaluation of Oil-Red-O stained aortas after 4 months of therapy showed a ~50% reduction of total aorta lipid deposition and a significant reduction in brachiocephalic plaque sizes in Tgfbr1/2 siRNAs -treated group (Fig 6A,B). To test the effect of therapy on plaque regression, Apoe−/− mice subjected to 3 months of HCHFD were switched to ND and 7C1-packaged Tgfbr1/2 siRNAs were administered as indicated (Fig 6C). Oil-Red-O analysis of aortic lipid deposition showed a significant reduction in the total aortic lipid burden 1 and 2 months later (Fig 6D), size of brachiocephalic plaques and regression of key histological features (Fig 6E-G). Finally, we used ICH to assay macrophage burden in aortic root (Fig S11A,B) and brachiocephalic artery (Fig S11C,D). In both cases, Tgfbr1/2 siRNAs treatment was associated with profound reduction in the number of Mac3+ plaque macrophages.
Figure 6: 7C1-siTgfbr1/2 therapy suppress atherosclerosis lesion development and facilitates regression of advanced atherosclerotic plaques.
Apoe−/− mice treated with PBS, 7C1-siLuc or 7C1-siTgfbr1/2. (A) Representative images of the Oil-Red-O (ORO)-stained atherosclerotic lesions in the aorta (left) and their quantification (right). All data: mean ± SEM. P=0.00018 for siTgfbr12 compared to Luc. Two-tailed Student’s t tests. (PBS Apoe−/− N=4 animals per group, Luc Apoe−/− N=4 animals per group, siTgfbr1/2 Apoe−/− N=8 animals per group). (B). Movat-stained brachiocephalic artery plaques Scale bar: 200 μm. Lesion area quantification (bottom). Bars: mean ± SEM. Lesion area: P=0.065 for siTgfbr12 compared to Luc. Necrotic core area: P=0. 00015 for siTgfbr12 compared to Luc. Two-tailed Student’s t tests. PBS Apoe−/− N=4 animals per group, Luc Apoe−/− N=4 animals per group, siTgfbr1/2 Apoe−/− N=8 animals per group for lesion area; PBS Apoe−/− N=4 animals per group, Luc Apoe−/− N=4 animals per group, siTgfbr1/2 Apoe−/− N=5 animals per group for necrotic core area. (C) Experiment timeline. (D) (Left) Representative images of the ORO-stained atherosclerotic aorta lesions in Apoe−/− mice treated with 7C1-siLuciferase or 7C1-siTgfbr1/2. (Right) Lesion area quantification; data: mean ± SEM. NS: not significant, P=0.41 for Apoe−/− 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.0015 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.17 for Apoe−/− 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.000019 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.0015 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD + 1M ND, P=0.00022 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD + 2M ND. Two-tailed Student’s t tests. 3M Apoe−/− baseline N=7 animals per group, 3M HCHFD + 1M ND 7C1-Luciferase Apoe−/− N=6 animals per group, 3M HCHFD + 1M ND 7C1-siTgfbr1/2 Apoe−/− N=6 animals per group, 3M HCHFD + 2M ND 7C1-Luciferase Apoe−/− N=6 animals per group, 3M HCHFD + 2M ND 7C1-siTgfbr1/2 Apoe−/− N=6 animals per group). (E-G) Representative images of brachiocephalic artery from 7C1-siLuciferase or 7C1-siTgfbr1/2 treated mice stained with H&E (scale bar: 200 μm) (E) and their histological (F) and morphological (G) quantification. NS: not significant, P=0.57 for Apoe−/− 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.0027 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD, P=0.036 for Apoe−/− 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.00065 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD, P=0.0012 for TGFβRiEC-Apoe 3M HCHFD + 1M ND compared to Apoe−/− 3M HCHFD + 1M ND, P=0.00038 for TGFβRiEC-Apoe 3M HCHFD + 2M ND compared to Apoe−/− 3M HCHFD + 2M ND. Two-tailed Student’s t tests. 3M Apoe−/− baseline N=6, 3M HCHFD + 1M ND 7C1-Luciferase Apoe−/− N=6 animals per group, 3M HCHFD + 1M ND 7C1-siTgfbr1/2 Apoe−/− N=6 animals per group, 3M HCHFD + 2M ND 7C1-Luciferase Apoe−/− N=6 animals per group, 3M HCHFD + 2M ND 7C1-siTgfbr1/2 Apoe−/− N=6 animals per group).
Discussion
The results of this study show that activation of endothelial TGFβ signaling plays a key role in induction of vessel wall inflammation and development and progression of atherosclerosis. The recruitment of leukocytes into the vessel wall is a key event in atherogenesis. This is typically viewed as a form of chronic inflammation, but some critical processes, such as activation of endothelial cells, may be shared with acute inflammatory processes where microvascular endothelial cells recruit circulating leukocytes. Strikingly, the ablation of TGFβ signaling genetically confined to the endothelium, reduced both expression of ICAM-1 and VCAM-1 on endothelial cells and the extent of leukocyte recruitment following injection of TNFα. These findings were extended to chronic inflammation of the arterial wall in Apoe−/− mice fed a western diet. Selective genetic inactivation or pharmacological inhibition of endothelial TGFβ signaling, using either Tgfbr1/Tgfbr2 deletions or nanoparticle-based TGFβR1/R2 siRNA delivery, delayed the onset of atheroma formation, reduced the rate of its progression in the settings of hypercholesterolemia, and induced regression. Taken together, these data implicate endothelial TGFβ signaling as a key factor responsible for atherosclerotic plaque growth and maintenance.
TGFβ involvement has long been recognized in a variety of biological processes including cell proliferation, differentiation, migration, adhesion, and extracellular matrix production 15,16 and TGFβ signaling signature and expression of TGFβ ligands, receptors and various Smad proteins has been reported in atherosclerotic plaques 15,17,18. Yet its role has been controversial, with both pro-and anti-atherosclerotic effects reported2,19. In particular, systemic inhibition of TGFβ signaling, using a neutralizing anti-TGFβ1, -β2, and -β3 antibody, was shown to accelerate the development of atherosclerosis in Apoe−/− mice 20, while treatment with anti-TGFβR2 antibody decreased plaque size of advanced lesions, but increased plaque vulnerability 21. Mice with TGFβR2 knockout in CD11c+ dendritic cells exhibited an increase in plaque area 22 as did the Apoe−/− mice with disrupted TGF-beta signaling in T cells 23,24 while activation of TGFβ signaling in SMCs reduced atherosclerosis 25. These results are consistent with the general belief in anti-inflammatory activity of TGFβ.
Yet recent studies suggested that TGFβ signaling in the endothelium accelerates disease progression and leads to formation of vulnerable plaque 4,5. Among the deleterious effects of activated EC TGFβ signaling is the induction of EndMT 26. EndMT is frequently observed in human atherosclerotic lesions 4,5 and its extent strongly correlates with the severity of disease4. EndMT contributes directly to atherosclerotic plaque growth by increasing neointima cellularity due to ECs acquiring mesenchymal properties and extensive deposition of the extracellular matrix. It is also an important driver of inflammation due to increased EC expression of leukocyte adhesion molecules and deposition of pro-inflammatory fibronectin-rich matrix 4.
While all endothelial cells undergoing EndMT displayed increased inflammatory signature, one unique cell population, characterized by a complete loss of EC fate marker expression and high-level expression of MSC markers, stands out. These cells are particular pro-inflammatory and may well represent the key cell population driving the disease progression. This conjecture is supported by a significantly reduced frequency of this cell population in TGFβRiEC-Apoe mice. Thus, activation of endothelial TGFβ signaling in atherosclerosis settings drives vascular inflammation and EndMT, establishing a vicious circuit.
The data presented in this study provides support and explanation for the pro-inflammatory effects of endothelial TGFβ signaling. Both bulk RNA-seq and ChIP-seq analysis point to the ability of TGFβ to induce inflammation of key inflammatory pathways in ECs and not in other cell types. These observations are supported by functional studies that point to decreased TNFα-induced permeability and inflammation in TGFβRiEC mice and reduced atherosclerosis (and all its inflammatory components) in TGFβRiEC-Apoe mice. Furthermore, the anti-inflammatory TGFβ effects in SMCs observed here in both RNA-seq and ChIP-seq studies, are in agreement with our recent study demonstrating reduced atherosclerosis in mice with activated SMC TGFβ signaling25. Taken together, these considerations highlight inadvisability of systemic suppression of TGFβ signaling: any beneficial effects seen in the endothelium will be overridden by negative effects in smooth muscle and inflammatory cells.
Therefore, we specifically targeted EC TGFβ signaling cascade using genetic and molecular techniques. Both approaches were effective in reducing the total lesion burden and plaque size. Plaque morphology was favorably affected with a decrease in the necrotic core size implying increased plaque stability. Importantly, favorable changes were seen in multiple vascular locations including the whole aortic endothelium, aortic root, and brachiocephalic artery.
Associated with these changes was a reduction in EndMT as suggested by reduced expression of EndMT markers. One important consequence of less extensive EndMT was a decrease in expression of leukocyte adhesion molecules by luminal endothelial cells and reduced presence of inflammatory cells in the plaque. A combination of these factors led to a decrease in plaque cellularity and reduced matrix deposition resulting in a large decrease in plaque inflammation. Indeed, as expected with such a profound reduction in plaque inflammation following EC TGFβR1/R2 deletion, plaques displayed a true regression, with a decrease in plaque area, as well as a profound change in plaque composition, revealing that inhibition of EC TGFβR1/2 is one of the most powerful tools to regress plaques in a laboratory setting.
In addition to its anti-inflammatory effects, suppression of EC TGFβ signaling increased expression of key genes involved in the regulation of vascular permeability, leading to a reduction in increased permeability normally observed in inflammatory and atherosclerotic settings. A combination of reduced inflammation and reduced sensitivity to inflammation (e.g. decrease in vascular permeability) likely explains decreased atherosclerosis observed in this study. In summary, this study establishes endothelial TGFβ signaling as an important driver of atherosclerotic plaque growth and demonstrates a potential utility of a therapeutic intervention aimed at suppression of this process.
Methods
Generation of mice.
Cdh5-CreERT2 mice were obtained from Ralf Adams (Max Planck Institute for Molecular Biomedicine, Münster, Germany). Tgfbr2fl/fl mice were obtained from Harold L. Moses (Vanderbilt University), and Tgfbr1fl/fl mice were obtained from Martin M. Matzuk (Baylor College). To generate Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl mice, we mated Cdh5-CreERT2; Tgfbr2fl/f mice with Tgfbr1fl/fl mice. To generate Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-mT/mG mice, we mated Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl mice with mT/mG mice (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, Stock No: 007676, The Jackson Laboratory). To generate Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice, we mated Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-mT/mG mice with Apoe−/− mice (B6.129P2-Apoetm1Unc/J, Stock No: 002052). This strain had been back-crossed more than ten times to C57BL/6 background. We have generated and analyzed both female and male mice. The results are similar. The data presented in the manuscript are all from male mice to focus on a single experimental cohort with similar body size and hormonal milieu as has been traditional in the field. In addition, we are fully aware of the strain effects on our atherosclerosis models and have carefully examined all possible controls in our experiments. For the experiments required lineage tracing (Fig 1F&G, Fig S3A&B, Fig S9), we used Cdh5-CreERT2; mT/mG or Cdh5-CreERT2; Apoe−/−mT/mG mice as our controls. For progression and regression experiments (Fig 2-5, Fig S1-S2, Fig S4, Fig S7, Fig S8, Fig S11), we used Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG without Tamoxifen treatment as our controls. For nanoparticle experiments (Fig 6, Fig S9, Fig S10), we used Apoe−/− mice (purchased from JAX lab). Our mouse breeding pairs are all Cdh5-CreERT2 positive, but occasionally, we get Cdh5-CreERT2 negative offspring. We treated Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice with tamoxifen and used them as additional controls for our progression experiment in Fig 2. All animal procedures were performed under protocols approved by Yale University Institutional Animal Care and Use Committee.
Mouse genotyping.
Genotyping was performed by mouse ear DNA PCR analysis. Mouse ear DNA was isolated using the DNeasy Blood & Tissue kit (QIAGEN #69506). Genotyping was performed using the following PCR primers: Cdh5-CreERT2 (5’-GCC TGC ATT ACC GGT CGA TGC AAC GA-3’, and 5’-GTG GCA GAT GGC GCG GCA ACA CCA TT-3’), Tgfbr1fl/fl (5’-ACT CAC ATG TTG GCT CTC ACT GTC-3’, and 5’-AGT CAT AGA GCA TGT GTT AGA GTC-3’), Tgfbr2fl/fl (5’-TAA ACA AGG TCC GGA GCC CA-3’, and 5’-ACT TCT GCA AGA GGT CCC CT-3’), Apoe (5’-GCC TAG CCG AGG GAG AGC CG-3’, 5’-TGT GAC TTG GGA GCT CTG CAG C-3’, and 5’-GCC GCC CCG ACT GCA TCT-3’), mT/mG (5’-CTC TGC TGC CTC CTG GCT TCT-3’, 5’-CGA GGC GGA TCA CAA GCA ATA-3’, and 5’-TCA ATG GGC GGG GGT CGT T-3’).
Synthesis of siTgfbr1, siTgfbr2.
Chemically-modified siRNA were synthesized at Alnylam Pharmaceuticals (Cambridge, MA). 2-O-methyl-nucleotide modifications (indicated in lower case) were introduced to both strands to decrease the likelihood of triggering an innate immune response27. The siRNA sequence for Tgfbr1 sense strand (UGUCAAGGAGAUGCUUCAAuAdTsdT) and antisense (UAUUGAAGCAUCUCCUUGACAUAdTsdT); for Tgfbr2 is sense (GGCUCGCUGAACACUACCAAAdTsdT) antisense (UUUGGUAGUGUUCAGCGAGCCAUdTsdT). LNPs formulated with siRNA targeting luciferase (siLuc), were used as the control. The siLuc, which was also incorporated 2-O-methyl-nucleotide modifications, is commonly used as a control for in vivo siRNA studies13,14.
siRNA formulation in lipid nanoparticles (LNPs).
siLuc, siTGFβR1, and siTGFβR2 were encapsulated in LNPs formulated with the lipid 7C1. More specifically, 7C1 was synthesized and purified13 and it was then combined with C14PEG2000 in a glass syringe (Hamilton Company), and diluted with 100% Ethanol. siLuc, siTGFβR1, and siTGFβR2 were diluted in 10 mM citrate buffer and loaded into a separate syringe. In the siTGFβR1 / siTGFβR2 in vivo experiments, siTGFβR1 and siTGFβR2 were added at a 1:1 mass ratio in the syringe. The two syringes were connected to a microfluidic mixing device, before the 7C1 and RNA solutions were mixed together at a flow rate of 300 and 900 μL/min, respectively. The resulting nanoparticles were dialyzed into 1X PBS, before being sterile filtered using a 0.22 μm filter. The amount of 7C1 used in in vivo formulations was kept constant.
Animal treatment.
Cre-Lox recombination was induced by tamoxifen (Sigma T5648) at 1 mg/day i.p. for 5 days versus vehicle (corn oil, Sigma C8267) alone. For PBS, siLuc, and siTGFβR1 and siTGFβR2 delivery in vivo, 8 to 10-week-old mice were placed on a high cholesterol high fat diet (HCHFD; 40% kcal% Fat, 1.25% Cholesterol, 0% Cholic Acid) for 16 weeks (Research Diets, product #D12108). During this period there were injected intravenously (every 10 days) with one of the of the following: sterile PBS (100 μl/mouse), siLuc (1 mg/kg), siTgfbr1/2 (1 mg/kg). For TNFα administration, mice were given TNFα (Peprotech 300–01A) prepared in 0.1 ml of sterile saline and administered i.v. by single injection at a dose of 1 μg/mouse. Control groups received 0.1 ml of saline i.p. Animals were studied 2 h after the injections.
7C1-siLuciferase Alexa647 intravenous injection, tissue dissociation, and FACS analysis.
Apoe−/− (Cdh5-CreERT2;Apoe−/−mT/mG mice treated with Tamoxifen) mice were injected with 1 mg/kg 7C1 formulated with Alexa647-tagged siLuciferase (VasoRx, Inc.). Two hours after intravenous injection with 1 mg/kg 7C1-siLuciferase Alexa647, the aorta, heart, and lungs were dissected, rinsed in cold PBS, and sliced into small pieces. The finely minced tissue was transferred to a digestion mix consisting of Hank’s balanced salt solution (Gibco) + 1 mg/ml collagenase type A (Sigma 10103578001) + 0.5 mg/ml elastase (Worthington LS006365) for 3 hr at 37°C and pipetting every 30 min. The cell suspension was passed through a 40 μm filter. DAPI (Sigma D9542) was used to detect dead cells. A FACS machine (BD FACSAria) was used to analyze GFP+7C1-siLuciferase Alexa647+ cells.
In vivo mRNA measurements.
Endothelial cells were isolated from the lung and heart using the same technique as isolation and culture of mouse endothelial cells described below.
Cells.
Mouse bEnd.3 cells (ATCC CRL-2299) were maintained in Dulbecco’s modified Eagle’s medium (ATCC 30–2002) with 10% fetal bovine serum (Life Technologies 16000–044) and penicillin-streptomycin (Gibco 15140–122). Cells were grown at 37°C, 5% CO2.
Primary mouse endothelial cells were isolated from the lung or heart using rat anti-mouse CD31 antibody (BD #553370) and Dynabeads (Invitrogen 110.35). Briefly, minced lung or heart were digested with Type I collagenase (2 mg/ml; Sigma C0130) at 37°C for 45 min with agitation. The cells were then filtered through a 70 μm disposable cell strainer (BD Falcon 352350) and centrifuged at 1300 rpm for 10 min at 4°C, and then resuspended in 2 ml of EC medium DMEM (LONZA 12–709F), 20% FBS (Sigma 26140–079), 10 units/ml Penicillin/ 10 μg/ml Strep (Gibco 15140–122), 1X non-essential amino acid (Gibco 11140–050), 2 mM L-glutamine (Gibco 25030–081), 1.2 μg/ml Amphotericin B (Fisher Scientific BP2645–50), 60 μg/ml Gentamicin sulfate (Gibco 15750–060). The cells were then incubated with anti-mouse CD31 Dynabeads on a rotator at room temperature for 15 min. After several washes, the cells were plated on gelatin-coated 10 cm dishes. The cells were fed with EC medium containing 100 mg/ml heparin (Sigma H-3933), 100 mg/ml ECGS (Alfa Aesar J64516). Primary mouse endothelial cells between passages 3 and 4 were used in all experiments.
PBMCs were collected by leukapheresis from anonymous volunteer donors and isolated using Ficoll Lymphocyte Separation Medium (MP Biomedicals). Purified PBMCs were cryopreserved in 10% DMSO and 90% FBS in liquid nitrogen before use. For CD4+ effector memory T cells (TEM) isolation and stimulation, total CD4+ T lymphocytes were purified from PBMCs using Dynabeads CD4 Positive Isolation Kit (Invitrogen 11331D) per the manufacturer’s protocol. To obtain the resting TEM subset, naive T cells, central memory T cells, and activated TEM were depleted from the purified total CD4+ populations using antibodies against human CD45RA (eBioscience, HI100), CCR7 (Biolegend, clone G043H7), CD62L (eBiosicence, clone DREG56), and HLA-DR (clone LB3.1), followed by removal of antibody-bound cells by magnetic separation using Dynabeads Pan Mouse IgG (Invitrogen 11041). The isolated populations were routinely greater than 95% CD45RA–CCR7–CD62L–HLA-DR–CD45RO+CD4+ TEM lymphocytes. TEM were then cultured in RPMI 1640 media (Gibco) with 10% FBS, 2% L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml), thereafter referred to as complete RPMI media. Freshly isolated CD4+ TEM were resuspended in complete RPMI media at the concentration of 1 million cells/mL, and were activated with Dynabeads Human T-Activator CD3/CD28 (Gibco 11131D) per manufacturer’s instructions. Fresh TGFβ1 (5 ng/mL, Biolegend 580702) or PBS control were added to the CD4+ TEM on Day 0, Day 1, Day 2, Day 3, and Day 4 of culture. On Day 4, CD4+ TEM were collected for RNA isolation. For macrophage isolation and stimulation, PBMCs were plated in tissue culture dishes at a density of 3×106 cells/mL and monocytes were obtained by adhesion for 24 hours in DMEM supplemented with 2 mM L-glutamine (Gibco), 100 U/ml penicillin (Gibco), 100 μg/ml streptomycin (Gibco), 0.1 mM sodium pyruvate (Gibco), 1% non-essential amino acids (Gibco), 50 μM 2-mercaptoethanol (Gibco), and 10% fetal bovine serum (Gibco). After 24 hours cells were washed twice with DMEM to remove the non-adherent cells and cultured for additional five days in the culture media described above, supplemented with 20 ng/mL M-CSF (R&D 216-MC-025). After 5 days, cells were washed with DMEM to remove non-adherent or dead cells, stimulated with TGFβ1 (5 ng/mL, Biolegend 580702) or PBS for 48 hrs followed by RNA isolation.
Growth factors.
Recombinant human BMP9 (BioLegend 553104), recombinant human TGFβ1 (BioLegend 580702), recombinant human TNFα (Peprotech 300–01A), and recombinant human M-CSF (R&D 216-MC-025) were reconstituted in 0.1% BSA/PBS.
Generation of lentiviruses.
Mouse TGFβR1 and TGFβR2 shRNA lentiviral constructs were purchased from Sigma. For the production of shRNA lentivirus, 3.7 μg of ∆8.2, 0.2 μg of VSVG, and 2.1 μg of pLKO.1 carrying the control, TGFβR1 or TGFβR2 shRNA were co-transfected into 293T cells using X-tremeGENE 9 DNA Transfection Reagent (Roche 06365787001). Forty-eight hrs later the medium was harvested, cleared by 0.45 μm filter (PALL Life Sciences PN4184), mixed with polybrene (5 μg/ml) (Sigma H9268), and applied to cells. After a 6 hr incubation, the virus-containing medium was replaced by the fresh medium.
RNA isolation and qRT-PCR.
Cells were suspended in TRIzol Reagent (Invitrogen #15596018) and the total RNA (QIAGEN #74134) were isolated according to the manufacturer’s instructions. Reverse transcriptions were performed by using iScript cDNA synthesis kit (Bio-Rad 170–8891). qRT-PCR was performed using Bio-Rad CFX94 (Bio-Rad) by mixing equal amounts of cDNAs, iQ SYBR Green Supermix (Bio-Rad 170–8882), and gene specific primers SA Biosciences (a QIAGEN company). All reactions were done in a 20 μl reaction volume in duplicate. Individual mRNA expression was normalized in relation to expression of endogenous β-actin. PCR amplification consisted of 5 min of an initial denaturation step at 95°C, followed by 46 cycles of PCR at 95°C for 15 s, 60°C for 30 s.
Western Blot Analysis.
Cells were lysed with HNTG lysis buffer (20 mM HEPES, pH 7.4/ 150 mM NaCl/ 10% glycerol/ 1% Triton-X 100/ 1.5 mM MgCl2/ 1.0 mM EGTA) containing complete mini EDTA-free protease inhibitors (Roche #11836170001) and phosphatase inhibitors (Roche #04906837001). 20 μg of total protein from each sample was resolved on Criterion TGX Precast Gels (Bio-Rad #567–1084) with Tris/Glycine/SDS Running Buffer (Bio-Rad #161–0772), transferred to nitrocellulose membranes (Bio-Rad #162–0094) and then probed with various antibodies. Chemiluminescence measurements were performed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific Prod #34080).
Cell adhesion assay.
HUVECs were seeded into 6-cm plates (must be 100% confluent the next day). Next day, HUVECs were treated with or without TNF-α (10 ng/mL) for 5 hr. In the last hour of the HUVEC TNF-α incubation, THP-1 cells were spun down and resuspended at 1 × 106/mL in serum-free RPMI-1640. HUVECs were washed twice with RPMI 1640, and 2ml of THP-1 cells were added to each well at 37°C. After 1 hr incubation, non-adherent cells were carefully removed and the endothelial monolayers with adherent THP-1 cells were gently washed 10 times. Adherent THP-1 cells were visualized under a microscope.
Antibodies.
We used the following antibodies for flow cytometry (flow cyto), immunoblotting (IB), or immunohistochemistry (IHC): ALK1 (Fitzgerald 70R-49334; IB 1:1000), BMPR2 (Cell Signaling 6979; IB 1:1000), CD115 (eBioscience 12–1152-81; flow cyto 1:100), CD3 (Dako; IHC 1:50), CD45 (BD 550539; IHC 1:100) (abcam ab10558; flow cyto 1:100), Collagen 1 (Novus Biologicals NB600–408; IHC 1:200), CD45 (BioLegend 103112; flow cyto 1:100), Fibronectin (Sigma F3648; IHC 1:100), GAPDH (Cell Signaling 2118; IB 1:1000), Gr-1 (BioLegend 108426), ICAM-1 (BioLegend 116102; IHC 1:100), Ki-67 (abcam ab66155; IHC 1:100), Ly6G (abcam ab25377; IHC 1:100), Mac-2 (CEDARLANE CL8942AP; IHC 1:100), Mac-3 (BD 550292; IHC 1:100), RNA pol II CTD phospho Ser2 (Active Motif 61083; ChIP:1:100), RNA pol II CTD phospho Ser5 (Active Motif 39749; ChIP:1:100), phospho-Smad1/5/8 (Ser465/467) (Cell Signaling 9511; IB 1:1000), Smad1/5/8 (Santa Cruz sc-6031; IB 1:1000), phosphorylated Smad2 (Ser465/467) (Cell Signaling 3108; IB 1:1000), phospho-Smad3 (Ser465/467) (R&D AB3226; IB 1:1000), Smad2/3 (BD 610843; IB 1:1000), Smad2/3 (Cell Signaling 8685; ChIP 1:100), Smad2/3 (R&D AF3797; ChIP 1:100), smooth muscle α-actin-APC (allophycocyanin) (R&D IC1420A; IHC 1:10), TGFβR1 (Santa Cruz sc-398; IB 1:1000), TGFβR2 (Santa Cruz sc-400; IB 1:1000), In Situ Cell Death Detection Kit, TMR red (Sigma 12156792910), VCAM-1 (abcam ab19569; IHC 1:100), and VE-cadherin (Santa Cruz sc-6458; IB 1:100).
Lipid measurements.
Mice were fasted for 12–16 hrs overnight before blood samples were collected by retro-orbital venous plexus puncture, and plasma was separated by centrifugation. Total plasma cholesterol and triglycerides were enzymatically measured (Wako Pure Chemicals Tokyo, Japan) according to the manufacturer’s instructions.
Monocyte Labeling.
1-μm Fluoresbrite fluorescein isothiocyanate (FITC)-dyed (YG) plain microspheres (Polysciences) were diluted 1∶4 in sterile PBS, and 250 μl of the solution was intravenously injected into mice to selectively label Gr1lo monocytes. The labeling efficiency of a representative mouse from each genotype was verified by flow cytometry one day after the beads injection (Fig S8). After red blood cell lysis (100 μl of blood), monocytes were identified by flow cytometry using fluorochrome-conjugated combinations of the following mAbs: CD115 (PE conjugated anti-mouse CD115, clone AFS98; eBioscience), CD45 (APC-Cy7 anti-mouse CD45, clone 30-F11; BioLegend) and Gr-1 (PerCP/Cy5.5 anti-mouse Ly-6G/Ly-6C (Gr1); BioLegend). Beads were counted in plaques using a fluorescent microscope (light range, 450–490 nm).
Permeability assay.
Animals were anesthetized with ketamine/xylazine and TNFα (10 ng) or saline solution was injected intradermally (10 μl total) into the dorsal ear skin before Evans blue dye (30 mg/kg) injection and circulation for 30 min. Animals were then killed and perfused with 4% paraformaldehyde before tissues were excised and photographed. The areas of blue skin (vascular leak) were removed and placed into formamide for 5 days. The intensity of the reaction was quantified by reading the samples at a wavelength of 620 nm on a SpectraMax plate reader.
Histological analysis of atherosclerotic lesions.
The animals were euthanized and perfusion-fixed with 4% paraformaldehyde (Polysciences, Inc. Cat #18814) via the left ventricle for 5 min. The lesions located in the aorta and aortic roots were analyzed using Oil Red O staining. To measure lesions in the aorta, the whole aorta, including the ascending arch, thoracic and abdominal segments, was dissected, gently cleaned of adventitial tissue and stained with Oil Red O (Sigma O0625)4. The surface lesion area was quantified with ImageJ software (NIH). To measure lesions in the aortic root, the heart and proximal aorta were excised and the apex and lower half of the ventricles were removed.
Immunohistochemical staining.
Blocks were sectioned at 5 μm intervals using a Microm cryostat (for frozen blocks) or a Paraffin Microtome (for paraffin blocks). For frozen tissue sections, slides were fixed in acetone for 10 min at −20ºC. For paraffin sections, slides were dewaxed in xylene, boiled for 20 min in citrate buffer (10 mM, pH 6.0) for antigen retrieval, and rehydrated. After washing three times with phosphate-buffered saline, tissue sections were incubated with primary antibodies diluted in blocking solution (10% BSA and horse serum in PBS) overnight at 4ºC in a humidified chamber.
Sections were washed three times with tris-buffered saline, incubated with appropriate Alexa Fluor 488-, Alexa Fluor 594-, or Alexa Fluor 647-conjugated secondary antibodies diluted 1:1000 in blocking solution for 1 hr at room temperature, washed again 3 times, and mounted on slides with ProLong Gold mounting reagent with DAPI (Life Technologies P36935). All immunofluorescence micrographs were acquired using Zeiss and Leica SP8 microscopes.
For CD3 and Mac-3 staining (Figure 5A,B), after incubation with the primary anti-CD3 and anti-Mac-3 antibodies, sections were incubated with the appropriate biotinylated secondary antibodies (1:1000), followed by washing and incubation with the ABCHRP kit, and chromogenic staining with diaminobenzidine (DAB). Images were captured using Velocity software and quantifications performed using ImageJ software (NIH), or counted manually by 2 independent observers.
Mouse aorta digestion and single cell FACS sorting.
The whole aorta of Apoe−/− (Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice treated with corn oil) and TGFβRiEC-Apoe (Cdh5-CreERT2; Tgfbr1fl/fl-Tgfbr2fl/fl-Apoe−/−mT/mG mice treated with Tamoxifen) mice were dissected from the mice and rinsed in cold PBS. The tissue was opened longitudinally and sliced into small fragments roughly 2 mm in length. The finely minced tissue was transferred to a digestion mix consisting of Hank’s balanced salt solution (Gibco) + 1 mg/ml collagenase type A (Sigma 10103578001) + 0.5 mg/ml elastase (Worthington LS006365) for 3 hrs at 37°C and pipetting every 30 min. DAPI (Sigma D9542) was used to detect dead cells. The cell suspension was passed through a 40 μm filter before sorting. A FACS machine (BD FACSAria) was used to sort CD31+CD45- live cells. Single cells were sorted into 0.4% BSA-PBS.
Chromatin immunoprecipitation (ChIP).
10×106 cells were first cross-linked with 1.5 mM EGS in PBS for 30 mins, followed by 1% paraformaldehyde for 10 minutes at room temperature. Glycine was added at a final concentration of 125 mM for 5 minutes at room temperature in order to quench the crosslinking reaction. The cross-linked material was washed once each with Buffer 1 (0.25% Triton, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH7.5) and Buffer 2 (200 mM NaCl, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH7.5) before resuspension in Shearing Buffer (0.1% SDS, 1 mM EDTA, 1 mM Tris-HCl pH7.6) with 1X Complete Protease Inhibitor Cocktail (Roche). Cell pellet was lysed in 130 μl SDS lysis buffer (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS, 50 mM PMSF) and sonicated using an S220 Focused-ultrasonicator (Covaris) to generate 200 to 600 bp fragments. Fragmented chromatin was centrifuged at 10,000 g for 5 minutes and the supernatants were diluted in ChIP dilution buffer (1% Triton, 150 mM NaCl, 20 mM Tris pH 8.0). Immunoprecipition was performed by rotating samples at 4°C with magnetic beads (Dynabeads Protein A or G, Invitrogen) pre-bound with 5 μg of antibody. The beads were washed once each with low salt buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Deoxycholate, 1% NP40, 1 mM EDTA), high salt buffer (50 mM Tris pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% Deoxycholate, 1% NP40, 1 mM EDTA), LiCl wash buffer (50 mM Tris pH 8.0, 250 mM LiCl, 0.5% Deoxycholate, 1% NP40, 1 mM EDTA), Morohashi RIPA buffer (50 mM Tris pH7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP40, 0.1% SDS), DOC/Triton Buffer (25 mM Tris pH7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton-X-100, 0.5% DOC), and twice with Tris-EDTA buffer (10 mM Tris pH 8.0, 1 mM EDTA). All washes took place on a rotator for 10 minutes at 4°C. Beads were treated twice with 100μl elution buffer for 15 minutes on a shaker at room temperature, to obtain a total of 200 μl eluate. 8 μl of 5M NaCl was added to the eluate and the sample was reverse cross-linked overnight at 55°C. DNA was then purified using the QIAquick PCR Purification Kit (Qiagen).
ChIP-Seq analyses.
ChIP DNA samples was used in a 23.3 μl combined end repair and A-tailing reaction using a KAPA Hyper Prep Kit (Kapa Biosciences) for 30 minutes at 20°C followed by 30 minutes at 65°C. 10 μl of ligase buffer, 3.7 μl of Adapters and 3.3 μl ligase (KAPA Hyper Prep Kit) were added and incubated at 20°C for 4 hours. Double-stranded DNA fragments were purified from this reaction using KAPA Pure Beads (Kapa Biosciences) and eluted in 22 μl 10 mM Tris pH8.0. Libraries were generated by PCR and size-selected on a 2% E-Gel EX agarose gel (Invitrogen) and fragments between 150 and 400 bp were extracted using a QIAEX II Gel Extraction Kit (Qiagen) performed at room temperature. Libraries were submitted to the Yale Center for Genome Analysis for quality control analyses and next-generation sequencing 28. Short reads from ChIP-Seq data sets were aligned to human reference genome GRCh38 (build 38) using BWA-MEM for PBS and TGFβ1 conditions. DeepTools (version 3.0.0) was then used to convert bam files to bigwig files using default options 29. The matrices were then computed and used for creating average profiles and signal visualization around promoter region of −5/+5 kb and bin size of 25.
RNA-seq, Bulk RNA-Seq data processing and differential analysis:
For population (bulk) RNA-seq, cells were lysed with RLT Plus buffer and RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Next-generation whole-transcriptome sequencing of unstimulated or TGFβ treated endothelial cells (EC), smooth muscle cells (SMC), CD4+ effector memory T-cells and macrophages was performed at the Shanghai Sequencing Lab of WuXiNextCODE (n=2 for each cell type and condition). RNA-Seq reads were aligned to the human reference genome (human genome build 38 GRCh38) using short reads aligner STAR. Quantification of gene expression was performed using RSEM with GENCODE annotation (human release 27, http://www.gencodegenes.org)30,31. Genes with 1 count per million in less than 10% of samples were removed. Read counts were normalized using the trimmed mean of M-values method (TMM) and differentially expressed genes were identified (edgeR R package) 32,33. Macrophage comparisons were batch corrected using ComBat (sva R package) to remove the effects of sequencing at different times. Differentially expressed genes with a false discovery corrected p-value ≤ 0.05 were used for further analysis (heatmaps and functional enrichment).
Droplet-based scRNA-seq library preparation and sequencing.
Sorted CD31+CD45- cells were processed for scRNA library preparation using the Chromium™ Single Cell Platform (10x Genomics) as per the manufacturer’s protocol. Briefly, single cell suspensions were partitioned into Gel Beads in Emulsion in the Chromium™ system (10x Genomics) at Yale Center for Genome Analysis, followed by cell lysis and barcoded reverse transcription of RNA, cDNA amplification and shearing, and 5’ adaptor and sample index attachment. Final scRNA-seq libraries were sequenced on an Illumina NextSeq 500 (WuXi AppTec, China).
Single cell RNA-Seq data processing:
Single cell RNA sequencing (scRNA-Seq) was performed at the Shanghai Sequencing Lab of WuXiNextCODE (Illumina). We processed RNA-Seq short reads from each cell group using UMI-tools34. Briefly, we first identified cell barcodes from R1 reads, extracted cell barcodes and UMIs from transcript sequences, and added these to the read names for R2 reads (Supplementary Data 3). We aligned R2 reads to the reference mouse genome build 38 (GRCm38) with short read aligner STAR. Quantification was performed using UMI-tools count function to find the number of unique UMIs mapping to each gene (GENCODE mouse annotation, release M15). Count data was filtered by: (1) removing the cells without detected expression of CD31; (2) requiring cell’s total number of detected genes to be greater than or equal to 1,500 and cell’s library UMI counts to be greater than or equal to 3,000; (3) requiring genes to be expressed in more than 5% of cells in at least one genotype group (Fig S5B-D). We then checked the fraction of mitochondrial genes within the filtered gene matrix and found minimal expression of mitochondrial genes (minimum: 0.2%, median: 0.6%, maximum: 2.2%), so no additional filtering was necessary. In addition, we removed the TGFβRiEC-Apoe and Apoe−/− UMI counts from the processed data matrix to prevent downstream analysis from segregating cells solely based on genotypes. From initial 3,298 cells and 27,498 genes we included in our analysis 2,547 cells and 11,326 genes (Fig S5E). The percentage of cells considered in our analysis is consistent across the four mouse genotypes (Fig S5F). Finalized UMI counts were normalized using the trimmed mean of M-values method (TMM) and used for downstream clustering and modeling.
Deep learning framework for single-cell RNA-seq analysis:
We built a custom generative deep neural network strategy in TensorFlow to carry out unsupervised analysis and clustering of single cell RNA-seq data using a variational autoencoder (VAE) framework (Fig S6A) 34,35.
1.1. Variational Autoencoder Model
Briefly, suppose x is a vector of D variables, z is a vector of stochastic latent variables of dimension M, and pθ(x∣z) is a parametric model of the joint distribution. When this model is parametrized by a neural network (discussed below), optimization becomes difficult due to the computational intractability of calculating the marginal likelihood. The most common method to avoid this is to apply variational inference, and optimize the estimated lower bound (ELBO):
| (1) |
Where q(x) is the empirical distribution, qφ(z∣x) is the variational posterior (i.e. encoder), pθ(x∣z) is the generative model (i.e. the decoder), pλ(z) is the prior, and φ, θ, λ are their parameters. We re-formulated Eq. (1) by assuming z is composed of continuous and independent Gaussian distributions, using the re-parameterization trick for qφ(z∣x), and using a Monte Carlo estimate of the second expectation over L sample points:
| (2) |
The first term in Eq. (2) is known as the reconstruction error and the second and third terms function similar to a regularization term that keeps the distributions of the encoder matching the prior. An alternative formulation of Eq. (2) is the InfoVAE prior,
| (3) |
which instead uses the maximum mean discrepancy (MMD) and Gaussian kernels as the regularization term.
| (4) |
| (5) |
Eq. (3) maximizes the mutual information between x and z, which can lead to a richer latent space36. We set β = 50 as given in the original MMD manuscript 36. qφ(z∣x), the encoder, is modeled using a typical feed-forward deep neural network structure (section 1.3), and similarly we let pθ(x∣z), the decoder, be a zero-inflated negative binomial distribution (section 1.2) with its parameters computed from z with a feed-forward deep neural network structure (section 1.3).
1.2. Zero-Inflated Negative Binomial Distribution (ZINB)
We model the posterior distribution of the decoder with a ZINB distribution in order to address the issue of ‘dropout’, where genes can be observed in one cell but not detected in another cell 37. The probability mass function for the ZINB distribution (fZINB) is parameterized by inverse dispersion θ ≥ 0, mean μ ≥ 0, dropout probability π ∈ [0, 1], and Dirac function δ(⋅).
| (6) |
The dropout probability π represents the likelihood of observing a zero in the data (i.e. observing ‘dropout’) instead of an actual numerical read count, thus inflating the observation of zeros within the distribution. We restrict the ZINB inverse dispersion parameter θ to be feature-dependent (i.e. the inverse dispersion is modeled across a gene and not on a per gene per sample basis) as it is both computationally easier and has shown to be sufficient for both single-cell RNA-seq and bulk RNA-seq analysis. 38,39
1.3. Feed-Forward Neural Networks
The basic formulation of a simple fully-connected feed-forward frameworks where each layer (l) uses a specific activation function (ρ), is defined as:
| (7) |
Where the dimensions of W and b are determined by the number of neurons in each layer (d1, d2, … dm). Each layer used rectified linear units (ReLUs) as activation functions:
| (8) |
In addition, we utilized the concept of ‘dropout’ which randomly sets a portion of input values (η) to the layer to zero during the training phase 40. This has a strong regularization effect, essentially by injecting random noise, that helps prevent models from over-fitting. Layers that included ‘dropout’ were formulated as:
| (9) |
When evaluating models on test datasets the ‘dropout’ mask is not used.
1.3. VAE Model Training
We used the InfoVAE formulation and the log-likelihood of the ZINB distribution as the three terms in the loss function (section 1.1). For learning, we utilized the RMSProp gradient descent algorithm with mini-batch size of 256. We trained models for a minimum of 250 epochs and utilized early stopping with a look ahead (i.e. patience) of 50 epochs (on a validation dataset consisting of a random 20% of the total data). In addition, the regularization term of the loss function was annealed (i.e. ‘warmed-up’) for the first 50 epochs of training. The per sample loss was weighted by the frequency of samples in the experimental design to help account for unbalanced datasets (i.e. total number of cells per experimental condition). The final log-likelihoods were generated by sampling 100 times from the final model and calculating the mean value.
1.4. VAE Hyper-Parameter Search
The following hyper-parameters for were optimized using a random search strategy over the parameter grid: layer-dimension, number of layers, dropout-rate, learning rate, and latent-dimension. Detailed lists of model hyper-parameters and ranges used for evaluating hyper-parameters are given in Supplementary Table 1 and Supplementary Table 2. We selected the model with the lowest validation loss.
1.5. PhenoGraph Clustering
We fixed the hyper-parameters and structure of the zero-inflated VAE and ran 100 replicate model runs. We applied the PhenoGraph algorithm, using k=30 nearest-neighbors and the Jaccard similarity metric, to cluster the latent space of each run41;42. We determined the number of clusters within the dataset by selecting the most frequent across the 100 runs (in case of ties we selected the lower cluster number). Lastly, we calculated a consensus clustering across all runs using the using the Cluster-based Similarity Partitioning Algorithm, spectral-clustering, and the specified cluster number (as determined above) 43,44
Nested GOSeq (nGOseq) Functional Enrichment:
GOseq analysis was performed, using all annotated genes as the background distribution, on either the differentially expressed genes from bulk RNA-seq comparisons (false discovery corrected p-value ≤ 0.01) or the most informative genes from single cell RNA-seq to identify enriched gene ontology (GO) terms 45. Briefly, nested GOseq (nGOseq), a modified version of the nested Expression Analysis Systematic Explorer (nEASE) algorithm, was then used to identify enriched nested GO terms driving the statistical enrichment of upper-level GOseqterms12,46. We performed nGOseq enrichment using the top 10% of genes identified by sensitivity mapping (1133 genes) with a background distribution of all measured genes that were functionally annotated (11,326 genes).
ChIP-seq Analysis:
Next-generation ChIP-Sequencing and quality control analyses were performed at Yale Center for Genome Analysis and Shanghai Sequencing Lab of WuXi NextCODE for human smooth muscle cells and human umbilical vein endothelial cells stimulated with either PBS or TGFβ1, respectively. Short reads from Smad2/3, Pol II S2, and Pol II S5 ChIP-Seq data sets were aligned to human reference genome GRCh38 (build 38) using bwa-mem. deepTools (version 3.0.0) was used to convert bam files to bigwig files (default options). The matrices were then computed and used for creating average profiles and signal visualization around promoter region of −5/+5kb and bin size of 25.
Statistics.
Statistical analyses were performed with GraphPad Prism version 8 (GraphPad Software). Parameters such as sample size, the number of replicates, the number of independent experiments, measures of center, dispersion, and precision (mean ± SEM), and statistical significance are reported in Figures and Figure Legends. Results were considered statistically significant when P < 0.05 (*), P < 0.01 (**), P < 0.001 (***).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Software Availability.
The code and version of the dependencies are available in https://github.com/wxncailab/NatureMetabolism_Endothelial.
Data Availability Statement.
High-throughput sequencing data associated with this study (scRNA-seq, ChIP-seq and bulk RNA-seq) have been deposited Gene Expression Omnibus (GEO). Awiting GEO accession codes AAA, BBB, CCC.”
Supplementary Material
Acknowledgments
We are grateful to J. Fang for aortic endothelial cell isolation used in scRNA-seq. We would like to thank K. Hirschi for helpful discussion and R. Webber and L. Coon for maintaining mice colonies used in this study and N. Cilfone for assistance with RNA-seq data analysis. This work was supported, in part, by National Nature Science Foundation of China Grant 81600365 (G. L.) and NIH Grant R01 HL135582 (M.S. and MAS).
Footnotes
Competing Interests Statement
M Simons and P-Y Chen are holders of a provisional US Patent Application 62/311,086 and 62/406,732 dealing with endothelial-specific treatment of atherosclerosis. M Simons, P-Y Chen and D.G. Anderson are scientific founders of VasoRx, Inc. M Simons and D.G. Anderson are members of VasoRx, Inc. Scientific Advisory Board. The other authors declare no competing interests.
Bibliography
- 1.Baeyens N & Schwartz MA Biomechanics of vascular mechanosensation and remodeling. Mol Biol Cell 27, 7–11, doi: 10.1091/mbc.E14-11-1522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabas I, Garcia-Cardena G & Owens GK Recent insights into the cellular biology of atherosclerosis. J Cell Biol 209, 13–22, doi: 10.1083/jcb.201412052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz MA, Vestweber D & Simons M A unifying concept in vascular health and disease. Science 360, 270–271, doi: 10.1126/science.aat3470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P-Y et al. Endothelial-to-mesenchymal transition drives atherosclerosis progression. The Journal of clinical investigation 125, 4529–4543, doi: 10.1172/JCI82719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrard SM et al. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nature communications 7, 11853, doi: 10.1038/ncomms11853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feaver RE, Gelfand BD, Wang C, Schwartz MA & Blackman BR Atheroprone hemodynamics regulate fibronectin deposition to create positive feedback that sustains endothelial inflammation. Circulation research 106, 1703–1711, doi: 10.1161/CIRCRESAHA.109.216283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohwedder I et al. Plasma fibronectin deficiency impedes atherosclerosis progression and fibrous cap formation. EMBO molecular medicine 4, 564–576, doi: 10.1002/emmm.201200237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwata J et al. Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. The Journal of clinical investigation 122, 873–885, doi: 10.1172/JCI61498 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P et al. Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events. Scientific reports 6, 35444, doi: 10.1038/srep35444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutgens E et al. Deficient CD40-TRAF6 signaling in leukocytes prevents atherosclerosis by skewing the immune response toward an antiinflammatory profile. The Journal of experimental medicine 207, 391–404, doi: 10.1084/jem.20091293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virmani R, Kolodgie FD, Burke AP, Farb A & Schwartz SM Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol 20, 1262–1275 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Yu P et al. FGF-dependent metabolic control of vascular development. Nature 545, 224–228, doi: 10.1038/nature22322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlman JE et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol 9, 648–655, doi: 10.1038/nnano.2014.84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sager HB et al. RNAi targeting multiple cell adhesion molecules reduces immune cell recruitment and vascular inflammation after myocardial infarction. Science translational medicine 8, 342ra380, doi: 10.1126/scitranslmed.aaf1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardali E & Ten Dijke P TGFbeta signaling and cardiovascular diseases. International journal of biological sciences 8, 195–213, doi: 10.7150/ijbs.3805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhurst RJ & Hata A Targeting the TGFbeta signalling pathway in disease. Nature reviews. Drug discovery 11, 790–811, doi: 10.1038/nrd3810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCaffrey TA TGF-beta signaling in atherosclerosis and restenosis. Frontiers in bioscience 1, 236–245 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Schunkert H et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics 43, 333–338, doi: 10.1038/ng.784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toma I & McCaffrey TA Transforming growth factor-beta and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell and tissue research 347, 155–175, doi: 10.1007/s00441-011-1189-3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallat Z et al. Inhibition of transforming growth factor-beta signaling accelerates atherosclerosis and induces an unstable plaque phenotype in mice. Circulation research 89, 930–934 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Lutgens E et al. Transforming growth factor-beta mediates balance between inflammation and fibrosis during plaque progression. Arterioscler Thromb Vasc Biol 22, 975–982 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Lievens D et al. Abrogated transforming growth factor beta receptor II (TGFbetaRII) signalling in dendritic cells promotes immune reactivity of T cells resulting in enhanced atherosclerosis. European heart journal 34, 3717–3727, doi: 10.1093/eurheartj/ehs106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gistera A et al. Transforming growth factor-beta signaling in T cells promotes stabilization of atherosclerotic plaques through an interleukin-17-dependent pathway. Science translational medicine 5, 196ra100, doi: 10.1126/scitranslmed.3006133 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Robertson AK et al. Disruption of TGF-beta signaling in T cells accelerates atherosclerosis. The Journal of clinical investigation 112, 1342–1350, doi: 10.1172/JCI18607 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PY, Qin L, Li G, Tellides G & Simons M Smooth muscle FGF/TGFbeta cross talk regulates atherosclerosis progression. EMBO molecular medicine, doi: 10.15252/emmm.201506181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Meeteren LA & ten Dijke P Regulation of endothelial cell plasticity by TGF-beta. Cell and tissue research 347, 177–186, doi: 10.1007/s00441-011-1222-6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deleavey GF & Damha MJ Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 19, 937–954, doi: 10.1016/j.chembiol.2012.07.011 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Wang Z et al. A Non-canonical BCOR-PRC1.1 Complex Represses Differentiation Programs in Human ESCs. Cell stem cell 22, 235–251 e239, doi: 10.1016/j.stem.2017.12.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic acids research 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323, doi: 10.1186/1471-2105-12-323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD & Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology 11, R25, doi: 10.1186/gb-2010-11-3-r25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomczak JM & Welling M VAE with a VampPrior. arXiv 1705.07120 (2017). [Google Scholar]
- 35.Doersch C Tutorial on Variational Autoencoders. arXiv 1606.05908v2 (2016). [Google Scholar]
- 36.Zhao S, Song J & Ermon S InfoVAE: Information Maximizing Variational Autoencoders,. arXiv 1706.02262 (2017). [Google Scholar]
- 37.Kharchenko PV, Silberstein L & Scadden DT Bayesian approach to single-cell differential expression analysis. Nature methods 11, 740–742, doi: 10.1038/nmeth.2967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risso D, Perraudeau F, Gribkova S, Dudoit S & Vert JP A general and flexible method for signal extraction from single-cell RNA-seq data. Nature communications 9, 284, doi: 10.1038/s41467-017-02554-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD & Smyth GK Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332, doi: 10.1093/biostatistics/kxm030 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Srivastava N, Hinton G, Krizhevsky A, Sutskever I & Salakhutdinov R Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Journal of Machine Learning Research 15, 1929–1958 (2014). [Google Scholar]
- 41.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197, doi: 10.1016/j.cell.2015.05.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shekhar K et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323 e1330, doi: 10.1016/j.cell.2016.07.054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strehl A & Ghosh J Cluster Ensembles – A Knowledge Reuse Framework for Combining Multiple Partitions. Journal of Machine Learning Research 3, 583–617 (2002). [Google Scholar]
- 44.Shi J & Malik J Normalized Cuts and Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22, 888–905 (2000). [Google Scholar]
- 45.Young MD, Wakefield MJ, Smyth GK & Oshlack A Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 11, R14, doi: 10.1186/gb-2010-11-2-r14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chittenden TW et al. nEASE: a method for gene ontology subclassification of high-throughput gene expression data. Bioinformatics 28, 726–728, doi: 10.1093/bioinformatics/bts011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data associated with this study (scRNA-seq, ChIP-seq and bulk RNA-seq) have been deposited Gene Expression Omnibus (GEO). Awiting GEO accession codes AAA, BBB, CCC.”