Abstract
To understand fundamental mechanisms of mammalian spinal cord function, it is necessary to reveal the diverse array of constituent spinal “cell types” – populations that can be consistently identified because they share a unique and cohesive set of characteristics. Many parameters can contribute to the definition of a spinal cord cell type, including location, morphology, lineage, electrophysiological properties, circuit features, gene expression patterns, and behavioral contribution. While it is not necessary for all of these features to align completely at all times to identify an individual cell type, a correlation of these characteristics paints a rich portrait of cell identity. This review will summarize recent advances in the identification of mammalian spinal cord neuronal cell types and will highlight the power of transcriptional profiling to identify and characterize the cell types of the spinal cord.
Keywords: spinal cord neurons, spinal cord atlas, single cell sequencing
1. History
The first indication of spinal cord cellular diversity began with Ramon y Cajal’s characterization of the morphological heterogeneity within neural tissue. His detailed illustrations emphasized the staggering complexity of the nervous system and challenged the idea of continuity and uniformity within neural networks, including the spinal cord[1]. The structural organization of the spinal cord was further elaborated by Rexed, who used histological criteria to delineate spinal cells into ten discrete “laminae” along the dorso-ventral axis of the spinal cord[2]. Many elegant anatomical studies refined this system further, focusing on anatomical, morphological, and electrophysiological features of neurons and creating cell groupings or cell types such as “islet”, “radial”, and “la-inhibitory” and “Renshaw” neurons[3–5].
2.1. Single molecular markers in the adult
The advent of molecular markers – proteins that are reliably expressed in specific cellular populations – dramatically improved the ability to classify spinal cord cell types and to relate different features of cell types with a consistent reference. Within the adult spinal cord, this strategy was particularly helpful in the dorsal horn, where Neurotensin, Tachykinins, Gastrin-releasing peptide, and many other proteins are generally found in neurons that share a specific location, neurotransmitter status, morphology, or firing pattern[6,7]. The set of cell types that can be defined by molecular markers expressed in the mature spinal cord continues to expand. In a recent landmark study, Abraira and colleagues used genetic tools to label and examine eleven dorsal horn neuronal populations and described the location, morphology, connectivity, and electrophysiological properties of each[●8]. This approach provides a multifaceted understanding of these cell types by aligning multiple features of cell identity. Specific marker defined cell types in the adult spinal cord have also been probed for their contributions to behavior. Many populations are now known to be required for a wide array of functions including pain processing, itch, locomotion, and skilled reaching[9–16].
2.2. Single molecular markers during development
In parallel with studies that characterized specific molecular markers in the adult spinal cord, the embryonic origin of spinal cord cells has become an important prism though which to view spinal organization[17,18]. A transcription factor code that controls early cell fate specification of spinal cord neurons was revealed and used to define a set of cardinal classes, namely V0 – V3 and motor neurons in the ventral spinal cord and dI1 – dI6 populations in the dorsal spinal cord. Each embryonic class generally has common major characteristics such as inhibitory or excitatory neurotransmitter status and contralateral or ipsilateral neuronal targets[19,20].
A significant advantage of using embryonic lineage to define spinal cord cell types is that the progenitor domains of the known classes cover the entire dorsal-ventral axis, raising the possibility of a comprehensive classification scheme based on a complete census of the major embryonic classes. However, three major challenges have limited the utility of this approach. First, our understanding of lineage-derived cell types in the dorsal horn has lagged significantly behind our understanding of the ventral horn. Second, it has proven difficult to link each embryonic lineage-defined cell type with its mature progeny. Third, there is high degree of diversification within each embryonic class which has complicated efforts to achieve a “complete” census of spinal cord cell types. Genetic tools have revealed that each of the ventral classes are critical for particular core features of motor control, such as the rhythmic left-right and flexion-extension cycles during walking[21–24]. However, these lineage-defined populations likely include a broad range of heterogenous cell types, whose diversity we are now beginning to appreciate.
Several recent studies have greatly advanced our understanding of this diversification, most notably within the V0, V1, V2, and V3 classes. Importantly, this work aligns multiple features that contribute to the definition of cell types, including lineage, anatomical location, genetic tools, electrophysiological features, and circuit features. The V0 population provides an informative example of how a major class can be split into finer and finer sub-types. It is comprised of an excitatory V0V population and an inhibitory V0D population, as well as a small population of Pitx2-expressing neurons that itself can be further divided into V0C “C-bouton” neurons and a tiny population of V0G neurons[25–27]. Probing V1 derived cells with a set of nineteen transcription factors and with an innovative Bayesian analysis approach, Bikoff and colleagues revealed dozens of distinct V1 sub-populations that sort into four major groupings, or “clades” [●28,29]. Subsequently, Sweeney and colleagues found that these populations were found to vary between lumbar and thoracic levels of the spinal cord, adding rostro-caudal position as another dimension in cell type diversity[30]. Within V2a derived cells, Hayashi and colleagues found that there are at least two principal types that can be identified by expression levels of the marker gene Vsx2/Chx10. Overlaid onto these two major types of V2a cells are eleven cell clusters that each possess subtly distinct gene expression profiles[●31]. In a series of studies, Zhang and colleagues found that V3 cells can be divided into multiple groups based on anatomical location and that each of these display a distinct pattern of electrophysiological features and circuit integration[32,33]. Together, these meticulous dissections of broad ventral cell classes have pushed the depth of our understanding of spinal cord cell types but several key questions remain. Can each major cell type be divided further and further? How will we know when a census of spinal cord cell types is “complete”? And, more broadly, what are the organizing principles of the relationships between cell types?
3. Single cell sequencing
The answers to some of these questions are just beginning to be found in a new approach to identifying cell types. Single cell sequencing can reveal the expression of thousands of genes per cell in thousands of cells[34]. When combined with computational analysis, this data can be used to uncover common transcriptional patterns that define the cell types of a complex tissue[35,36] (Figure 1). Large scale efforts are underway to map all of the cell types of the central nervous system[34,37–39]. Three recent studies have harnessed the power of this revolution in cell type identification to focus on defining spinal cord cell types, each with a different technical approach and focus.
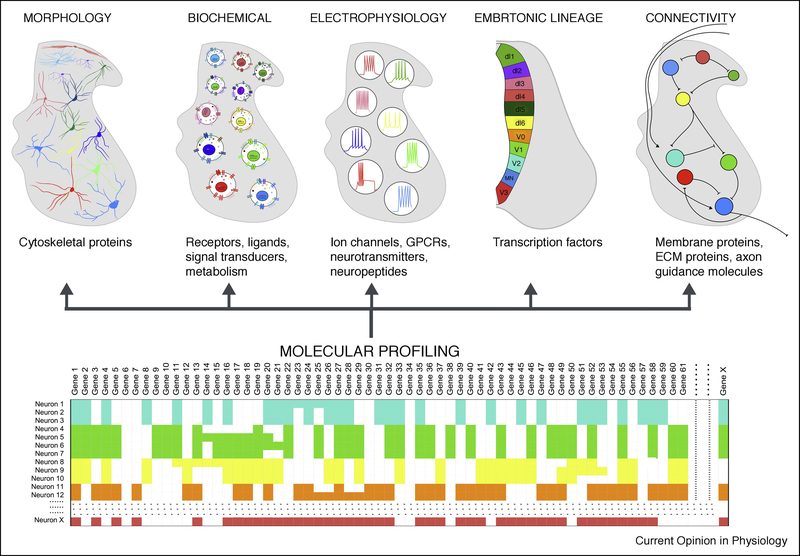
Figure 1. Towards comprehensive classification of spinal neuron features.
Schematized in the top row are current perspectives on the classification of spinal neurons. Spinal neurons have traditionally been classified based on their morphology, biochemical properties, electrophysiological properties, embryonic origin, and connectivity. Recent advances in single-cell sequencing techniques, schematized in the bottom row, have facilitated high-throughput RNA-sequencing from thousands of cells in a given tissue. Patterns of gene expression can then be used to cluster cells into types. The unique molecular repertoire of each cell type mediates its functional features.
Sathyamurthy and colleagues used single nuclei, rather than single cells, to create an atlas of the adult lumbar spinal cord because single nuclei can be easily isolated from adult tissue without experimentally-induced gene expression changes that can accompany the isolation of single cells[●●40]. They classified forty-three neuronal populations, all of which were identified according to neurotransmitter status and spatial location within the dorsal and ventral horns. To begin to link this set of spinal cord cell types to behavioral mechanisms, they used detection of immediate early genes at the single cell level to identify activated neuronal populations. As this study examined all neuronal populations throughout the dorsal-ventral axis, broad relationships amongst cell types began to emerge. Of particular interest is the separation of dorsal horn neurons into discrete, molecularly distinct cell types while ventral cells often displayed overlapping patterns of gene expression. This may indicate that dorsal horn neurons are characterized by molecular differences in the adult spinal cord while ventral horn neurons are more defined by embryonic factors and represent a continuum of transcriptionally-defined cell types in the adult.
Probing the early post-natal spinal cord, Rosenberg and colleagues developed and applied a low-cost method for transcriptome sequencing, SPLiT-seq, which harnesses the power of split pool barcoding while bypassing the requirement for custom equipment[●●41]. This study identified thirty spinal neuron populations, including two distinct types that correspond to alpha and gamma motor neurons. Enriched gene expression levels within each cluster revealed patterns of laminae specific spatial organization. Intriguingly, most of these clusters represented the dorsal horn of the spinal cord, while a very large “unresolved” cluster was enriched for many classic markers of ventral embryonic cardinal classes, including Pitx2 (V0), En1 (V1), Vsx2/Chx10 (V2a), and Sim1 (V3). This pattern may corroborate the dorsal-ventral trend observed in the work of Sathyamurthy and colleagues and, together, these studies point to a general principle of spinal cord cell type organization.
A comprehensive study by Haring and colleagues focused on the dorsal horn and revealed a highly refined, laminae specific organization of neuron types[●●42]. They classified thirty dorsal neuronal clusters and specifically presented key factors that may be driving forces behind cell type specificity: receptors, ion channels, and transcription factors. This study also included a systematic analysis of in situ gene expression patterns for multiple defining markers of each cluster to demonstrate the location and co-expression patterns of each cell type. With these in situ tools, they next probed for activated neurons following behavior to link cell types with function.
While each of these studies used different techniques, in many cases they identified the same cell types, demonstrating the robustness of these strategies. For example, an inhibitory population that is characterized by Galanin expression was found in all three studies (DI-2 in the adult spinal cord atlas, Cluster 26 in the Split-Seq atlas, and GABA3 in the dorsal horn atlas). The role of these dI4/ dILA inhibitory interneurons remains unknown. However, the single cell sequencing studies reveal that these cells are also distinguished by co-expression of the neuropeptide gene Pdyn/dynorphin, the neuropeptide receptor gene Sstr2, and the signal transducer Prkca/PKCα, which may provide clues to the functional properties of this cell type. Thus, a strength of single cell sequencing emerges: through its characterization of cell types based upon a transcriptional profile rather than a single marker, new hypotheses about cell function and mechanism can be generated.
Overall, there are three important advantages to using single cell sequencing to study spinal cord cell types. First, in addition to identifying each cell type, transcriptional profiles can be used to characterize each cell type. This includes discovering factors that mediate the cell’s physiology and function, as well as identifying new markers that may serve as genetic entry points for cell type control. Second, studies on the full spinal cord tissue provide the backbone for a comprehensive atlas and classification schema for all of the cell types of the spinal cord, including the wide array of neuronal populations as well as oligodendrocytes, astrocytes, and other cell types (Figure 2). This type of atlas will allow the relationships between cell types to be understood and will reveal broad organizing principles of spinal cord cell types. Third, a spinal cord cell atlas derived from single cell sequencing data can serve as a reference framework for aligning non-transcriptional types of data (Figures 1 and 2). While no single cellular characteristic can fully define a cell type, the integration of multiple features, such as gene expression patterns, location, and behavioral role, may identify functional cell types that have cohesive multi-dimensional identities.
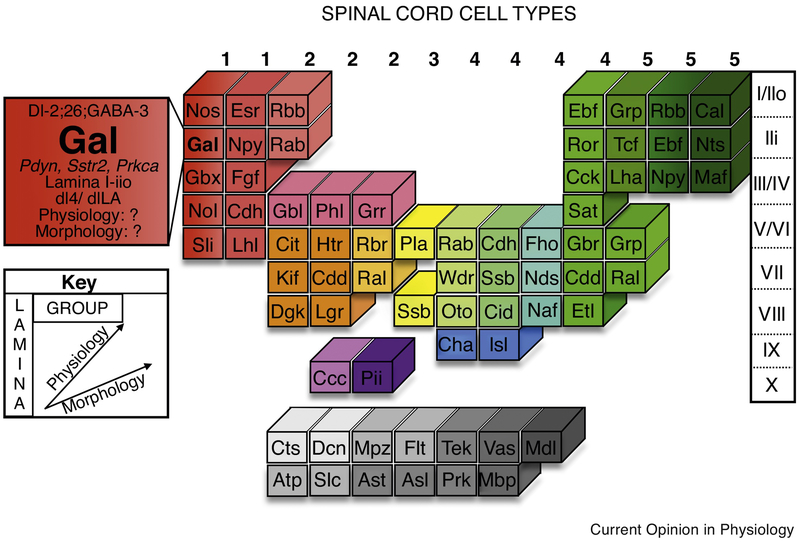
Figure 2. A model of a comprehensive census and schema for spinal cord cell types.
Advancements in single cell sequencing technologies have provided a platform by which a comprehensive spinal cord cell type atlas can be achieved. This table serves as a theoretical example of how major characteristics of spinal cord neurons can be integrated to sort cell types into a consistent schema. Similar to the patterns found within the periodic table of elements, this system can reveal organizing principles of cell type relationships and offers a multidimensional view of the complexity behind functional definitions of cell types. As possible examples, “groups” are presented that could correlate with major classes of cell types, such as a dorsal, inhibitory, peptidergic group.
4. Future Challenges and Opportunities
Still, several key challenges remain. One major technical disadvantage of using abstracted, computational, or population-based definitions of a cell type, is that it is difficult to identify an individual cell on its own. The new field of “spatial transcriptomics” provides a promising approach to this challenge by combining in situ cell location with the ability to map or sequence dozens to thousands of genes[43–46]. Another current limitation to this approach is the difficulty in aligning multiple datasets to establish a common set of cell types. Each study involves choices about analysis strategies including how supervised or unsupervised to be, whether to focus on clustering genes or cells, whether to create hierarchical branch points in cell taxonomies or cluster all cell types in parallel, when to “lump” related populations into a single cell type and when to “split” a heterogeneous cell type into refined sub-populations. Moving forward, a consensus atlas with a common cell type nomenclature could serve as a foundation for the field. With an established atlas in hand, we can realize the potential of gene expression analysis to define and decode the cell types of the spinal cord.
Highlights:
Review of approaches to define and classify spinal cord cell types
Recent studies leveraged molecular markers to identify and sub-divide cell types
Single cell sequencing studies can establish a comprehensive spinal cord “atlas”
Acknowledgements:
This work was funded by the National Institutes of Health Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
References:
- 1.Cajal SRY, Swanson N, Swanson LW: Histology of the nervous system of man and vertebrates. 1995. [Google Scholar]
- 2.REXED B: The cytoarchitectonic organization of the spinal cord in the cat. The Journal of Comparative Neurology 1952, 96:414–495. [DOI] [PubMed] [Google Scholar]
- 3.Light AR, Trevino DL, Perl ER: Morphological features of functionally defined neurons in the marginal zone and substantia gelatinosa of the spinal dorsal horn. The Journal of Comparative Neurology 1979, 186:151–171. [DOI] [PubMed] [Google Scholar]
- 4.Jankowska E: Interneuronal relay in spinal pathways from proprioceptors. Prog. Neurobiol 1992, 38:335–378. [DOI] [PubMed] [Google Scholar]
- 5.Thomas RC, Wilson VJ: Precise localization of Renshaw cells with a new marking technique. Nature 1965, 206:211–213. [DOI] [PubMed] [Google Scholar]
- 6.Todd AJ, Spike RC: The localization of classical transmitters and neuropeptides within neurons in laminae I-III of the mammalian spinal dorsal horn. Prog. Neurobiol 1993, 41:609–645. [DOI] [PubMed] [Google Scholar]
- 7.Koch SC, Acton D, Goulding M: Spinal Circuits for Touch, Pain, and Itch. Annu. Rev. Physiol 2017, 80:annurev–physiol–022516–034303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. ●.Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. : The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 2017, 168:295–310. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This comprehensive study characterizes the degree of diversity amongst eleven dorsal horn neuronal populations through differences in location, morphology, connectivity, and electrophysiological property.
- 9.Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, et al. : Identification of spinal circuits transmitting and gating mechanical pain. Cell 2014, 159:1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y-G, Zhao Z-Q, Meng X-L, Yin J, Liu X-Y, Chen Z-F: Cellular basis of itch sensation. Science 2009, 325:1531–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azim E, Jiang J, Alstermark B, Jessell TM: Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 2014, 508:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch SC, Del Barrio MG, Dalet A, Gatto G, Günther T, Zhang J, Seidler B, Saur D, Schüle R, Goulding M: RORβ Spinal Interneurons Gate Sensory Transmission during Locomotion to Secure a Fluid Walking Gait. Neuron 2017, doi: 10.1016/j.neuron.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra SK, Hoon MA: The cells and circuitry for itch responses in mice. Science 2013, 340:968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourane S, Grossmann KS, Britz O, Dalet A, Del Barrio MG, Stam FJ, Garcia-Campmany L, Koch S, Goulding M: Identification of a spinal circuit for light touch and fine motor control. Cell 2015, 160:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peirs C, Williams S-PG, Zhao X, Walsh CE, Gedeon JY, Cagle NE, Goldring AC, Hioki H, Liu Z, Marell PS, et al. : Dorsal Horn Circuits for Persistent Mechanical Pain. Neuron 2015, 87:797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh D, Pudenz C, Arber S: Context-Dependent Gait Choice Elicited by EphA4 Mutation in Lbxl Spinal Interneurons. Neuron 2016, 89:1046–1058. [DOI] [PubMed] [Google Scholar]
- 17.Alaynick WA, Jessell TM, Pfaff SL: Snapshot: spinal cord development. Cell 2011, 146:178–178. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catela C, Shin MM, Dasen JS: Assembly and function of spinal circuits for motor control. Annu. Rev. Cell Dev. Biol 2015, 31:669–698. [DOI] [PubMed] [Google Scholar]
- 19.Gosgnach S, Bikoff JB, Dougherty KJ, Manira El A, Lanuza GM, Zhang Y: Delineating the Diversity of Spinal Interneurons in Locomotor Circuits. J. Neurosci 2017, 37:10835–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu DC, Niu T, Alaynick WA: Molecular and cellular development of spinal cord locomotor circuitry. Frontiers in Molecular Neuroscience 2015, 8:3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosgnach S, Lanuza GM, Butt SJB, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M: V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature 2006, 440:215–219. [DOI] [PubMed] [Google Scholar]
- 22.Crone SA, Quinlan KA, Zagoraiou L, Droho S, Restrepo CE, Lundfald L, Endo T, Setlak J, Jessell TM, Kiehn O, et al. : Genetic ablation of V2a ipsilateral interneurons disrupts left-right locomotor coordination in mammalian spinal cord. Neuron 2008, 60:70–83. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Lanuza GM, Britz O, Wang Z, Siembab VC, Zhang Y, Velasquez T, Alvarez FJ, Frank E, Goulding M: V1 and v2b interneurons secure the alternating flexor-extensor motor activity mice require for limbed locomotion. Neuron 2014, 82:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Narayan S, Geiman E, Lanuza GM, Velasquez T, Shanks B, Akay T, Dyck J, Pearson K, Gosgnach S, et al. : V3 spinal neurons establish a robust and balanced locomotor rhythm during walking. Neuron 2008, 60:84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talpalar AE, Bouvier J, Borgius L, Fortin G, Pierani A, Kiehn O: Dual-mode operation of neuronal networks involved in left-right alternation. Nature 2013, 500:85–88. [DOI] [PubMed] [Google Scholar]
- 26.Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M: Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 2004, 42:375–386. [DOI] [PubMed] [Google Scholar]
- 27.Zagoraiou L, Akay T, Martin JF, Brownstone RM, Jessell TM, Miles GB: A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron 2009, 64:645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. ●.Bikoff JB, Gabitto MI, Rivard AF, Drobac E, Machado TA, Miri A, Brenner-Morton S, Famojure E, Diaz C, Alvarez FJ, et al. : Spinal Inhibitory Interneuron Diversity Delineates Variant Motor Microcircuits. Cell 2016, 165:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors utilized molecular genetic approaches to reveal the diversity amongst V1 spinal interneurons, which can be classified based upon the combinatorial expression pattern of nineteen unique transcription factors.
- 29.MI Gabitto, Pakman A, Bikoff JB, Abbott LF, Jessell TM, Paninski L: Bayesian Sparse Regression Analysis Documents the Diversity of Spinal Inhibitory Interneurons. Cell 2016, 165:220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney LB, Bikoff JB, Gabitto MI, Brenner-Morton S, Baek M, Yang JH, Tabak EG, Dasen JS, Kintner CR, Jessell TM: Origin and Segmental Diversity of Spinal Inhibitory Interneurons. Neuron 2018, 97:341–355. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. ●.Hayashi M, Hinckley CA, Driscoll SP, Moore NJ, Levine AJ, Hilde KL, Sharma K, Pfaff SL: Graded arrays of spinal and supraspinal V2a interneuron subtypes underlie forelimb and hindlimb motor control. Neuron 2018, 97:869–844. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors examine V2a spinal interneurons and uncover two distinct subtypes, type I and type II, that can be characterized by molecular profile, location, and functional role in forelimb and hindlimb motor outputs.
- 32.Borowska J, Jones CT, Zhang H, Blacklaws J, Goulding M, Zhang Y: Functional subpopulations of V3 interneurons in the mature mouse spinal cord. J. Neurosci 2013, 33:18553–18565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chopek JW, Nascimento F, Beato M, Brownstone RM, Zhang Y: Sub-populations of Spinal V3 Interneurons Form Focal Modules of Layered Pre-motor Microcircuits. Cell Rep 2018, 25:146–156. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell PJ, Carninci P, Clatworthy M, et al. : Science Forum: The Human Cell Atlas. Elife 2017, 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng H, Sanes JR: Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 2017, 18:530–546. [DOI] [PubMed] [Google Scholar]
- 37.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. : Molecular Architecture of the Mouse Nervous System. Cell 2018, 174:999–1014. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. : Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 2018, 174:1015–1030. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H: The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 2017, 96:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ●●.Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, Bergman TB, Kelly MC, Kelley MW, Levine AJ: Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep 2018, 22:2216–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors characterize forty-three neuronal cell types within the adult lumbar spinal cord through single nucleus transcriptional profiling. This single nucleus sequencing technique was then used to identify active neuronal populations following sensory and motor behaviors.
- 41. ●●.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, et al. : Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018, 360:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes a novel, low-cost technique for transcriptome sequencing and identifies thirty neuronal populations within the post-natal spinal cord. These clusters were found to display a laminae specific pattern of spatial organization.
- 42. ●●.Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, et al. : Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci 2018, 21:869–880. [DOI] [PubMed] [Google Scholar]; Through single-cell transcriptomics, this systematic study characterizes thirty neuronal populations within the dorsal horn of the adult spinal cord. The authors reveal a highly refined, laminae specific organization of the dorsal horn and use molecular markers to identify neuronal populations activated following sensory stimuli.
- 43.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X: RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348:aaa6090–aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lein E, Borm LE, Linnarsson S: The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 2017, 358:64–69. [DOI] [PubMed] [Google Scholar]
- 45.Wang G, Moffitt JR, Zhuang X: Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Sci Rep 2018, 8:4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. : Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361:eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]