Abstract
CRISPR-Cas genome editing technologies have revolutionized modern molecular biology by making targeted DNA edits simple and scalable. These technologies are developed by domesticating naturally occurring microbial adaptive immune systems that display wide diversity of functionality for targeted nucleic acid cleavage. Several CRISPR-Cas single effector enzymes have been characterized and engineered for use in mammalian cells. The unique properties of the single effector enzymes can make a critical difference in experimental use or targeting specificity. This review describes known single effector enzymes and discusses their use in genome engineering applications.
GRAPHICAL ABSTRACT:
PART I: CRISPR ORIGINS, FUNCTION, AND CATEGORIZATION
The evolutionary web of life represents a complex environment where organisms continuously evolve for better adaptation and survival. This competitive evolutionary process is exemplified by the relationship between hosts and their pathogens. For example, bacteriophages can outnumber bacteria 10:1,1 and parasitic mobile genetic elements such as plasmids cause unnecessary energetic or genomic burdens.2 To combat these forces, bacteria and archaea have evolved several different countermeasures to foreign genetic material, including physical blockage, restriction enzymes, abortive infection, bacteriophage exclusion, and Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-Cas systems.3 CRISPR-Cas systems provide adaptive immunity for the bacterial species by incorporating foreign DNA fragments into the host genome to create “memories” and later target the foreign nucleic acid for cleavage, based on complementarity to the encoded sequence.4
Basic Components of CRISPR-Cas Systems.
The CRISPR array and Cas proteins are the basic components in the microorganism ′s′ genomic locus necessary for the adaptive immunity process. The CRISPR array, or the repetitive targeting moiety, and its diverse cas genes together provide adaptive immunity. The CRISPR array is composed of alternating short variable spacers, which are generally identical to sequences from invading foreign genetic elements, and short direct repeat (DR) sequences.5–8 The CRISPR array is transcribed and processed into its final functional form, the CRISPR RNA (crRNA), consisting of a spacer and a DR. The partially palindromic DRs within a single CRISPR-Cas system usually have the same sequence, and when transcribed individually, or complexed to another unique trans-activating RNA (tracrRNA) sequence9 such as for Type II, V–B, and V–E systems,3 a unique hairpin-like structure forms that is recognized by the Cas protein(s).10 The cas genes are organized in an operon expression system and have diverse roles contributing to adaptive immunity.3 There are four distinct functional modules that classify Cas protein function, including spacer acquisition, crRNA processing, target cleavage, and ancillary roles.
The Three Phases of CRISPR Adaptive Immunity.
There are three phases of CRISPR adaptive immunity: spacer acquisition, crRNA processing, and target cleavage, discussed at length in other reviews.11–13 In brief, adaptation, or spacer acquisition, is the first phase of CRISPR adaptive immunity and is the process by which foreign nucleic acids are encoded into the CRISPR array4 mediated by the Cas1 and Cas2 proteins in various known bacterial species.14–16 Expression is the second phase of CRISPR adaptive immunity, during which the components for targeted nucleic acid cleavage are synthesized, processed, and complexed to form the RNA-guided endonuclease complex.9,17 Interference is the final phase of CRISPR adaptive immunity, which results in cleavage of the targeted foreign nucleic acid dependent on the proper base pairing between the crRNA and the target sequence in the protospacer and the protospacer adjacent motif (PAM).18,19 A majority of the known Cas proteins function in one of the three phases of CRISPR adaptive immunity, but some CRISPR systems have ancillary proteins that conduct various but generally less well characterized roles.20,21,22,23
CRISPR-Cas Nomenclature.
Approximately 47% of analyzed bacterial genomes and 87% of analyzed archaeal genomes have at least one CRISPR system, and many contain multiple CRISPR-Cas systems.24 These CRISPR-Cas systems in bacteria or archaea exhibit notable diversity and are organized according to a specific classification scheme. CRISPR-Cas protein classification is based on phylogenetic, comparative genomic, and protein structural analyses.24,25 Currently, there are two major classes of CRISPR-Cas systems, which are further divided into six types.24,25 The most up to date classification of CRISPR-Cas systems is in ref 3. The naming of signature proteins and CRISPR-Cas types is generally based on the timeline of characterization or experimental validation and therefore does not have a sequential naming scheme based on characterization alone.24–27
CRISPR-Cas systems are most broadly characterized as either class 1 or class 2.24 Class 1 systems require multiple Cas proteins to come together in a complex to mediate interference against foreign genetic elements. Class 1 systems are further divided into three CRISPR-Cas types based on the presence of a specific signature protein: Type I contains Cas3, Type III contains Cas10, and the putative Type IV contains Csf1, a Cas8-like protein (for more information on Class 1 systems, see Koonin et al.). In contrast to Class 1 systems, Class 2 systems use a large single Cas enzyme to mediate interference. Class 2 systems are generally less common than Class 1 systems and occur almost exclusively in the bacterial domain of life. Class 2 systems are further divided into three CRISPR-Cas types based on the presence of other specific signature proteins: Type II contains Cas9; Type V contains Cas 12a (previously known as Cpf1), Cas12b (previously known as C2c1), Cas12c (previously known as C2c3), Cas12d (previously known as CasY), and Cas12e (previously known as CasX); and Type VI contains Cas13a (previously known as C2c2), Cas13b, and Cas13c.3
PART II: GENOME ENGINEERING WITH CRISPR-CAS SYSTEMS
The ability of CRISPR-Cas systems to cleave targeted nucleic acids within bacteria or archaea may be repurposed for targeted editing of nucleic acids in heterologous contexts: harnessed CRISPR-Cas systems provide a simple platform for genome or transcriptome modulation as well as additional assays.28,29 Class 2 CRISPR-Cas systems are employed for genome engineering or assay development simply because there are fewer components to engineer compared to Class 1 CRISPR-Cas systems. The diversity of Class 2 CRISPR-Cas systems provides various opportunities for tool development, which will be elaborated on below.
CRISPR-Cas Enzyme Diversity.
There is widespread natural diversity within the Class 2 single effector Cas enzymes. This diversity is believed to be a byproduct of the competitive coevolution of CRISPR-Cas systems with different evolving viruses and associated anti-CRISPR proteins.3 Additionally, it may stem and diverge from the diverse environmental conditions from which microbes containing the Cas nucleases were derived. Such conditions may affect the temperature, pH, or ion requirements for optimal Cas nuclease activity. This diversity provides researchers with many variations of Cas proteins that may be explored for individual experiments. For example, CRISPR-Cas nucleases display a range of catalytic activity in heterologous contexts, such as in mammalian cells, as assayed by indel frequency.30–33 Orthologs that express well and display robust as well as consistent levels of targeted DNA cleavage have traditionally been selected for genome editing purposes in mammalian cells.31 Furthermore, wild-type Cas proteins have been engineered to create a number of variants with specific biochemical properties such as altered PAM specificity or reduced off-target cleavage efficiency,34–37 further adding to the diversity of these enzymes.
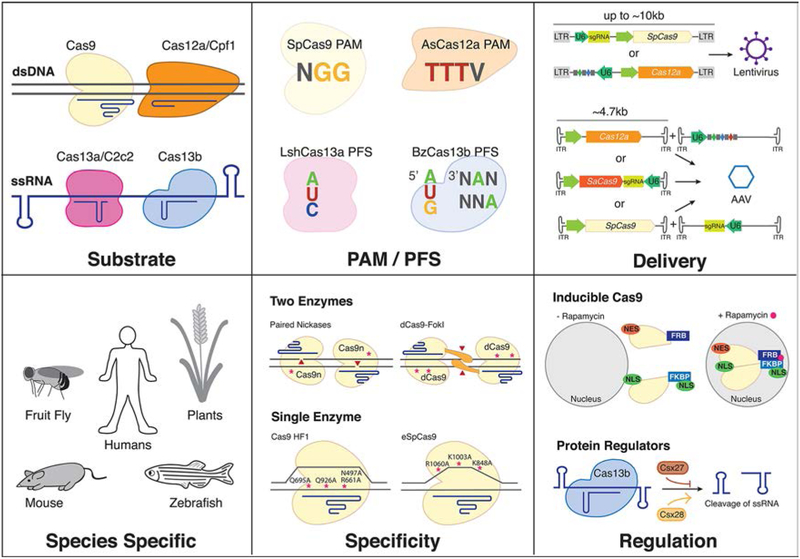
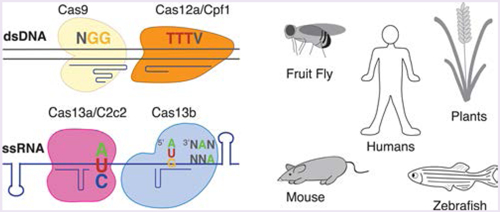
Reflecting the evolutionary diversity and unique selective pressures, CRISPR-Cas single effector enzymes exhibit a range of unique features, many of which have been leveraged for specific genome engineering contexts, see Figure 1. These features include target nucleic acid, nuclease domains, protein amino acid length, flanking sequence requirements, the number of reported orthologs, target cleavage pattern, requirement of a tracrRNA, crRNA architecture, and the ability to process its own pre-crRNA (see Table 1). Type II and Type V Class 2 CRISPR-Cas enzymes catalyze RNA-guided cleavage of dsDNA, whereas Type VI CRISPR-Cas enzymes exclusively cleave ssRNA. The DNA targeting Cas enzymes also differ with respect to their target cleavage pattern when cleaving dsDNA. Some Cas proteins even have the ability to process their own pre-crRNA.32 Some CRISPR-Cas systems are more common than others, and therefore more orthologs exist, providing opportunities to test for functionality in heterologous contexts. Different Cas orthologs may exhibit differences in protein size, which can influence the delivery method of the cas gene. Single effector Cas proteins generally range in size from ~950 to 1400 amino acids. Although different sized enzymes can be used, the smallest functional Cas proteins have been used when there are defined size limits for experiments, such as the ~4.7 kb packaging capacity of adeno-associated viruses (AAVs), a commonly used therapeutic viral vector. Protospacer adjacent sequence requirements can also vary across multiple orthologs, and this influences the available target sequence space.31 In addition to differences in protein structure, there exist specific variations with the targeting guide RNA or crRNA for each Cas protein. The architecture of the targeting RNAs may differ, and some Cas proteins require a tracrRNA, such as the commonly used Streptoccocus pyogenes Cas9. Last, some enzymes are naturally more specific than others, and this specificity can be augmented through structure-guided engineering.34,35 Specific examples of the above variations are referenced in Table 1 or in Part III.
Figure 1.
Graphic representing the diversity that enzyme choice may influence. Single effector Cas enzymes may be targeted to dsDNA or ssRNA, have different required protospacer flanking sequence requirements, may be delivered using different viral vectors, may be codon optimized and driven by promoters specific to function in different organisms, can be modified for higher specificity, and can be regulated.
Table 1.
Basic Properties for Each Unique Class 2 CRISPR-Cas Systema
| type | protein name | target nucleic acid | crystal structure(s) | nuclease domain to cleave target | array processing | PAM/PFS | # reported orthologs | target cleavage pattern | tracrRNA |
|---|---|---|---|---|---|---|---|---|---|
| II | Cas9 | dsDNA | 43–45 | RuvC and HNH | no | G-Rich PAM | 3,82225 | blunt | yes |
| V-A | Cas12a (Cpf1) | dsDNA | 73, 74, 89, 90 | RuvC | yes | T-Rich PAM32 | 7025 | staggered | no |
| V-B | Cas12b (C2c1) | dsDNA | 75, 76 | RuvC | ND | T-Rich PAM27 | 1825 | staggered | yes |
| V-C | Cas12c (C2c3) | dsDNAb | N/A | RuvCb | ND | ND | 525 | ND | nob |
| V-D | Cas12d (CasY) | dsDNA | N/A | RuvCb | ND | T-Rich77 | 677 | ND | nob |
| V-E | Cas12e (CasX) | dsDNA | N/A | RuvCb | ND | T-Rich77 | 277 | ND | yes |
| VI-A | Cas13a (C2c2) | ssRNA | 81 | HEPN | yes | H PFS (A, U, C)80 | 3025 | ssRNA regions and collateral | no |
| VI-B | Cas13b | ssRNA | N/A | HEPN | yes | 5′ D PFS (A, U, G) | 9425 | ssRNA regions and collateral | no |
| 3′ NAN or NNA23 | |||||||||
| VI-C | Cas13c | ssRNAb | N/A | HEPNb | ND | ND | 625 | ND | nob |
Properties listed have either been experimentally confirmed or marked with an asterisk if computationally predicted. ND, no data; N/A, data not available.
Computationally predicted.
CRISPR-Cas Systems in Heterologous Contexts.
To utilize CRISPR-Cas systems for targeted genome engineering in heterologous organisms such as mammals, the Cas protein as well as the guide RNA(s) have traditionally been modified for optimal expression and localization according to specific interests in cell type, species, and applications. For guide RNA expression in heterologous cells, RNA expression is generally driven by a promoter recognized by the endogenous transcription machinery. In mammalian cells, the RNA polymerase III (polIII) U6 promoter is commonly used because it is short and able to transcribe short RNAs.38 Another polIII promoter for guide RNA expression is H1, which also drives constitutive expression of short RNAs.39 Second, if a tracrRNA is required, such as for spCas9, a chimeric crRNA–tracrRNA hybrid RNA, also known as a single guide RNA (sgRNA), can be used to reduce the number of expressed RNAs required for targeted genome editing.40–42 Last, the spacer sequence of the guide RNA should be chosen in the genomic area of interest with the appropriate flanking PAM sequence for the particular Cas enzyme.
Several modifications to wild type cas genes also aid protein function and expression in heterologous contexts. The cas gene can be codon optimized for efficient translation in the specific organism. If particular organelle localization is required, such as localization of the Cas enzyme to the nucleus to edit dsDNA, localization tags can be added, such as nuclear localization sequence (NLS) tags to recruit the ribonucleoprotein complex to the nucleus.41 A promoter specific to the organism of interest can be selected for tissue specific or ubiquitous constitutive expression. The specific ortholog of the Cas enzyme will dictate the size of the Cas protein and protospacer flanking sequence requirements.
PART III: CLASS 2 CRISPR-CAS SYSTEMS
Given the broad application of Class 2 CRISPR-Cas systems as genome editing tools, there has been a recent focus on the discovery and characterization of these systems. The unique properties of these diverse systems have been used as an array of molecular tools (Figure 1). The next sections will describe specific molecular characteristics of different Cas enzymes, orthologs, or enzymes modified through targeted engineering and are summarized in Figure 2.
Figure 2.
Basic structure and functions of Class 2 CRISPR-Cas Types. Cas9 from Streptococcus pyogenes represents Type II. Cas12a from Francisella novicida U112 represents Type V, and Cas13a from Leptotrichia shahii represents Type VI.
Type II CRISPR Systems.
The first CRISPR associated single effector enzyme to be repurposed as a mammalian genome-editing tool was Cas9,41,42 which is currently the only known unique large single effector Cas protein of the Type II CRISPR-Cas systems. Cas9 is the most common large single effector Cas enzyme with 3822 reported orthologs, nearly all from the bacterial tree of life.25 Crystal structures have shown that Cas9 is a bilobed enzyme that uses two separate nuclease domains to cleave the target DNA in a blunt pattern using a RuvC-like and a HNH DNase domain (Figure 3).43–45 Each nuclease domain of Cas9 cleaves a single strand of DNA. Mutating either HNH or RuvC catalytic residues separately creates a nickase enzyme that cleaves just one of the two strands of DNA.40,46 Mutating both catalytic residues results in a catalytically inactive enzyme, which can be used as a programmable DNA binding protein. Additionally, Cas9 is not capable of cleaving its own pre-crRNA into individual crRNA units. RNase III processes the pre-crRNA.9 It appears that mammalian cells can process pre-crRNAs without expression of the bacterial RNase III, possibly due to the ability of endogenous RNase III enzymes.41 The crRNA associates with a tracrRNA that has partial complementarity to the crRNA repeat and forms a secondary RNA structure recognized by the Cas9 protein.9
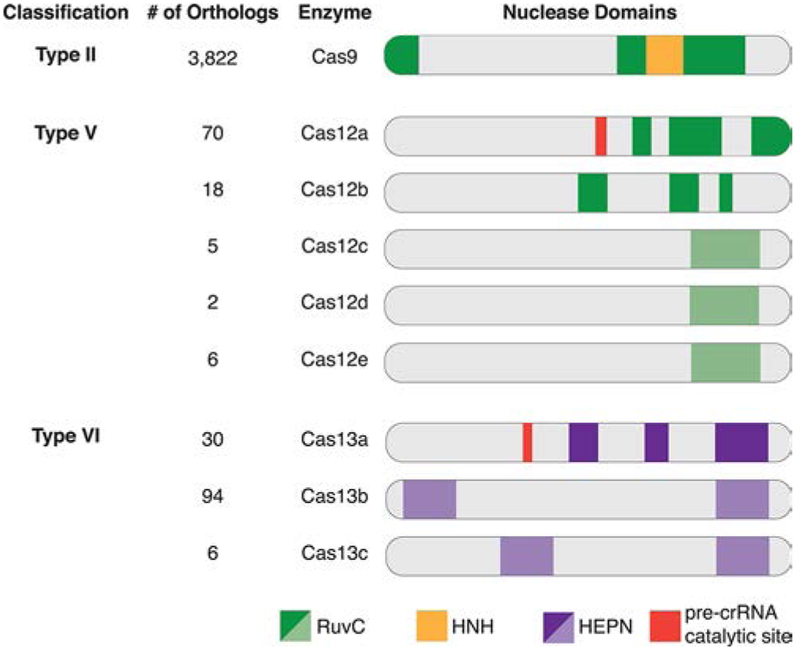
Figure 3.
Schematic representation of the unique Class 2 single effector Cas enzymes and their confirmed or predicted catalytic nuclease domains. Each Cas enzyme is grouped according to its CRISPR-Cas Type classification, and the number of orthologs is listed. The bright colors represent nuclease domains confirmed by crystal structure, and the faded colors represent computationally predicted nuclease domains. Several nuclease domains confirmed by crystal structure are not linear along the amino acid polypeptide chain and are split into sections as shown in the schematic. The split domains include RuvC-like in Cas9, Cas12a, and Cas12b and the first HEPN domain of Cas13a.
Cas9 is widely used to mediate genome editing within in vitro or in vivo environments, and several genome-wide off-target cleavage analyses have been performed in mammalian cells to better understand their on-target and off-target cleavage patterns.47,48 The most well characterized Cas9 ortholog comes from the species Streptococcus pyogenes SF370, termed SpCas9, which is a large 1368aa protein with a short, permissive NGG PAM.18 Several other orthologs of Cas9 have been harnessed for mammalian genome editing and are summarized in Table 2. Characterizing other orthologs has produced genome-engineering tools with altered PAM sequences or reduced gene sizes such as SaCas9, which is a shorter (1053aa) variant31 that may be packaged with its guide RNA into a single AAV vector. Besides the characterized orthologs, additional structure-guided engineered Cas9 variants have been produced. Cas9 engineered variants exhibit altered PAM specificity37 or enhanced cutting specificity34,35,49 as listed in Table 2. Cas9 is the subject of other relevant genome engineering reviews such as that of Karvelis et al.
Table 2.
Known Orthologs and Engineered Enzymes for Genome Engineeringa
| enzyme | abbreviation | organism/engineered alteration | PAM | size | reference using variant for genome editing |
|---|---|---|---|---|---|
| Cas9 variant orthologs | SpCas9 | Streptococcus pyogenes | 5′ SP-NGG 3′ | 1368 | 41, 42 |
| StlCas9 | Streptococcus thermophilus CRISPR1 | S′ SP-NNAGAAW 3′ | 1121 | 41 | |
| SaCas9 | Staphylococcus aureus | 5′ SP-NNGRRT 3′ | 1053 | 31 | |
| FnCas9 | Francisella novicida | 5′ SP-NGG 3′ | 1629 | 91 | |
| NmCas9 | Neisseria meningitidis | 5′ SP-NNNNGNNT 3′ | 1082 | 92 | |
| CjCas9 | Campylobacter jejuni | 5′ SP-NNNVRYM 3′ | 984 | 93, 94 | |
| BlatCas9 | Brevibacillus laterosporus | 5′ SP-NNNNCNDD 3′ | 1092 | 95 | |
| Cas9 engineered variants | SpCas9 VQR | SpCas9 variant with altered PAM recognition | 5′ SP-NGA 3′ | 1368 | 37 |
| SpCas9 VRQR | SpCas9 variant with altered PAM recognition | S′ SP-NGA 3′ | 1368 | 35 | |
| SpCas9 EQR | SpCas9 variant with altered PAM recognition | 5′ SP-NGAG 3′ | 1368 | 37 | |
| SpCas9 VRER | SpCas9 variant with altered PAM recognition | 5′ SP-NGCG 3′ | 1368 | 37 | |
| SaCas9 KKH | SaCas9 variant with altered PAM recognition | 5′ SP-NNNRRT 3′ | 1053 | 36 | |
| FnCas9 RHA | FnCas9 variant with altered PAM recognition | 5′ SP-YG 3′ | 1629 | 91 | |
| SpCas9-HFl | high-fidelity SpCas9 variant 1 | 5′ SP-NGG 3′ | 1368 | 35 | |
| eSPCas9 1.1 | enhanced fidelity SpCas9 version 1.1 | 5′ SP-NGG 3′ | 1368 | 34 | |
| HypaCas9 | high accuracy SpCas9 | 5′ SP-NGG 3′ | 1368 | 49 | |
| abbreviation | organism/engineered alteration | PAM | size | reference using variant for genome editing | |
| Casl2a/Cpfl variant orthologs | FnCpf1 | Francisella novicida U112 | 5′ TTTV-SP 3′ | 1300 | 32 |
| AsCpf1 | Acidaminococcus sp. BV3L6 | 5′ TTTV-SP 3′ | 1307 | 32 | |
| LbCpf1 | Lachnospiraceae bacterium ND2006 | 5′ TTTV-SP 3′ | 1228 | 32 | |
| TsCpf1 | Thiomicrospira sp. Xs5 | 5′ TTTV-SP 3′ | 1298 | 68 | |
| Mb2Cpf1 | Moraxella bovoculi AAX08_00205 | 5′ TTTV-SP 3′ | 1251 | 68 | |
| Mb3Cpf1 | Moraxella bovoculi AAX11 00205 | 5′ TTTV-SP 3′ S′TTV-SP 3′ | 1261 | 68 | |
| BsCpf1 | Butyrivibrio sp. NC300S | 5′ TTTV-SP 3′ | 1206 | 68 | |
| engineered variants | AsCpf1 RR | AsCpf1variant with altered PAM recognition | 5′ TYCV-SP 3′ | 1307 | 71 |
| AsCpf1 RVR | AsCpf1 variant with altered PAM recognition | S′ TATV-SP 3′ | 1307 | 71 | |
| AsCpf1 K949A | enhanced specificity AsCpf1 | S′ TTTV-SP 3′ | 1307 | 71 | |
| LbCpf1 RR | AsCpf1 variant with altered PAM recognition | S′ TYCV-SP 3′ | 1228 | 71 |
The abbreviation SP, or spacer, helps indicate the location of the PAM relative to the spacer sequence. Table adapted from ref 96.
In addition to its use as a way to edit the genome, its specific activity, ease of targeting, and modularity have enabled Cas9 to be repurposed for many other assays, summarized in refs 28 and 50. Cas9 can be used in genome-wide knockout screens in which a pooled library of guide RNAs can be delivered through lentiviral vectors to a cell population, which is then subjected to positive or negative selection.51,52 Catalytically inactive Cas9, or dCas9, has been used as a programmable DNA binding protein to direct functional effector localization.53,54 For targeted transcription initiation, either dCas9 is directly fused to a transcriptional activator such as VP6455–57 and/or the guide RNA can undergo structure-guided alterations to direct localization of VP64. An MS2 RNA stem loop, from the MS2 bacteriophage, can be added to a permissive area of the guide RNA and coexpressed with an engineered MS2 coat protein fused to the VP64 protein, in which the MS2 coat protein strongly and specifically binds to the MS2 RNA stem loop, thereby providing synergistic activation of transcription.58,59 Also, dCas9 has also been used for targeted DNA imaging60 or to alter epigenetic marks such as histone demethylation.61 More recently, Cas9n has been fused to an artificially evolved tRNA adenosine deaminase to achieve precise conversion of A·T to G·C in genomic DNA.62
Type V CRISPR Systems.
Type V CRISPR systems encompass all Cas enzymes that have a RuvC-like endonuclease domain with the RNase H fold, conserve catalytic motifs, and lack an HNH nuclease domain.3 Currently there are five such unique single large effector proteins: Cas12a (Cpf1), Cas12b (C2c1), Cas12c (C2c3), Cas12d (CasY), and Cas12e (CasX; Figure 3). Available data of the Cas12 family of proteins suggest that they exhibit several functional similarities but generally show low sequence homology.3
Cas12a/Cpf1 has been identified in 70 bacteria species25,63 and was functionally characterized as an active bacterial immune system.32 Cas12a has also been successfully harnessed for genome editing applications in several different organisms including human cells,32 mice,64 plants (tobacco and rice),65 silkworms,66 zebrafish, and Xenopus,67 illustrating its widespread applicability. Cas12a orthologs have also been examined, four of which are active in mammalian cells, as shown in Table 2.68 The wild-type Cas12a enzyme is found to have high levels of specificity.30,69 Last, Cas12a has promise for use therapeutically as illustrated by correction of muscular dystrophy in human cells and a mouse model.70
Cas12a/Cpf1 is distinct from Cas9 in several different ways. First, Cas12a enzymes have a T-rich PAM located 5′ of the guide, whereas Cas9 enzymes have G-rich PAM sequences. The T-rich PAM enables editing in AT rich genomes as well as pyrimidine-rich genomic areas such as the splice acceptor region.70 For example, the wild-type Cas12a ortholog Acidaminococcus sp. BV3L6 has a TTTV PAM 5′ of the protospacer.32 Engineered Cas12a variants exist with altered PAM specificities including 5′ TYCV and 5′ TATV, which have expanded the targeting range of Cas12a.71 Cas12a is also naturally targeted to the cognate DNA using only the single crRNA,32 which is simpler than the crRNA–tracrRNA duplex used by Cas9 and shorter than the chimeric Cas9 guide RNA. Cas12a is also unique in that the enzyme itself cleaves its own CRISPR array into individual crRNA units, making it the only known Cas nuclease with both DNase and RNase activity.72 The ability of Cas12a to process its own pre-crRNA has been leveraged for multiplexed genome editing.68 Unlike the blunt cut of Cas9, Cas12a targeted dsDNA cleavage results in a staggered cut leaving sticky overhangs after cleavage,32 potentially enhancing targeted knock-in efficiency. Crystal structures of Cas12a orthologs provide evidence that the RuvC-like domain is involved in dsDNA cleavage. Crystal structure data from the Francisella novicida U112 Cas12a suggests that the RuvC-like domain is the only domain responsible for cleaving both the target and nontarget DNA strands.73 However, in the Acidaminococcus sp. crystal structure, data show that mutation of the RuvC catalytic residues inhibits cleavage of both strands but that one mutation in the Nuc domain prevented cleavage of the target strand.74
Much less is known about Cas12b, Cas12c, Cas12d, and Cas12e, but the available data suggest they may behave similarly to Cas12a but also have some differences. For example, crystal structures show that Cas12b is structurally unique compared to Cas12a overall but shows similarity in the domain organization of the RuvC-like domain.75,76 Biochemically, Cas12b generates staggered DNA cuts distal to its T-rich PAM like Cas12a.27,75,76 Cas12b/C2c1 also has been shown to accommodate both the nontarget and target DNA strands in the RuvC-like domain. Mutagenesis of its RuvC catalytic residues prevents cleavage of both dsDNA strands, suggesting that Cas12b uses a single nuclease domain to cleave the substrate dsDNA.75 One notable difference between Cas12a and Cas12b is that Cas12b requires a tracrRNA for crRNA maturation and target cleavage.27 On the other hand, Cas12c, Cas12d, and Cas12e are less characterized. Known properties of Cas12c and Cas12d suggest they are similar to Cas12a,2777 whereas Cas12e is a small 980 amino acid protein that is predicted to use a tracrRNA.77
Type VI CRISPR Systems.
The Type VI CRISPR-Cas enzymes are RNA guided RNA nucleases. There are three unique Type VI CRISPR Cas enzymes, Cas13a (C2c2), Cas13b, and Cas13c, each of which has two Higher Eukaryotes and Prokaryotes Nucleotide-binding or HEPN RNase domains (Figure 3).78 HEPN domains generally consist of α-helical structures and have a conserved amino acid motif of E upstream of an R(X4–6)H where the amino acid after R is typically polar (often N, D, or H).79 The HEPN domains of Cas13a and Cas13c are located toward the central and C-terminal ends of the protein, whereas the HEPN domains of Cas13b are located toward the N-terminal and C-terminal ends of the protein.25 Other than the catalytic residues of the HEPN domains, there is no sequence homology between Cas13a, Cas13b, and Cas13c.
Cas13a/C2c2 has been identified in 30 bacterial species25 and specifically cleaves single stranded RNA from the two individual HEPN domains80 that come together to form the RNase active site.81 Validating in vitro cleavage data with purified protein showed cleavage of ssRNA at the RNA base uracil80 or adenine depending on the ortholog.82 ssRNA cleavage is dependent on a 3′H (A, U, or C) protospacer flanking sequence (PFS).80 Mutagenesis of arginine in either HEPN domain of Cas13a resulted in a catalytically dead enzyme.80 Cas13a also has a second, independent RNase activity that is able to process its own CRISPR array,83 which could aid in multiplexed targeting of multiple RNA sequences. Cas13a also exhibits an in vitro cleavage phenomenon called the collateral effect, i.e., promiscuous RNase cleavage activity after initial targeted RNA cleavage.80 This activity has been useful in the development of RNA detection assays83 such as SHERLOCK, which can detect specific DNA or RNA molecules with attomolar sensitivity.84 More recently, Cas13a has been engineered to achieve new functionality in mammalian cells, such as highly robust multiplexed RNA knockdown and binding,85 as well as direct adenosine to inosine editing with catalytically inactive Cas13 (dCas13) fused to DAR2.86
Cas13b functions similarly to Cas13a but with some differences, especially in terms of regulation. There are more orthologs of Cas13b,25 and they are subcategorized into two groups based on the identity of a small accessory protein, either Csx27 or Csx28, encoded in the CRISPR locus.23 These proteins differentially impact Cas13b interference activity in a bacterial host: Csx27 weakens Cas13b interference activity, whereas Csx28 increases activity.23 Cas13b also has a double-sided PFS of 5′D (A, U, or G) and 3′NAN or NNA and cleaves ssRNA flanking uracil or cytosine.23 Currently, there are no experimental data published on Cas13c. Cas13a, Cas13b, and Cas13c are the only known naturally occurring RNA targeting class 2 Cas enzymes.25 Their development as RNA-targeting tools in mammalian cells could be used for endogenous RNA knockdown, RNA editing, translation control, splicing control, localization studies, or in other applications for targeted RNA-binding assays.
Are There Undiscovered Class 2 Cas Enzymes?
The first known CRISPR-Cas systems were discovered empirically, whereas other rarer CRISPR-Cas systems were discovered through large computational searches. The first empirically discovered CRISPR-Cas systems were Types I, II, and III, which are relatively abundant.26 Next, while analyzing the genomic DNA of the intracellular pathogen Franciscella tularensis, the Type V cas12a (cpf1) gene was first discovered as an uncharacterized gene next to cas1, cas2, cas4, and a CRISPR array63 and later found in other bacterial species.24 With a growing appreciation for the diversity of CRISPR-Cas systems in combination with the successful repurposing of Cas9 as a genome-engineering tool, the first comprehensive search for uncharacterized Class 2 CRISPR proteins was conducted.27 This bioinformatics search was seeded on cas1, which encodes the most highly conserved Cas protein87 and is present in most CRISPR-Cas loci.24,26 From this search, the three large single effector Cas enzymes Cas12b, Cas13a, and Cas12c were found.27 Later bioinformatics searches for uncharacterized Class 2 CRISPR proteins were seeded on the CRISPR array and any large protein nearby.23,27 This approach identified Cas13b and Cas13c. Regardless of the starting point, large computational searches are limited by the sequenced populations available, which are generally skewed toward bacteria that can be cultured in laboratories or exhibit clinical relevance. To overcome this limitation, previously uncharacterized bacteria were sequenced, yielding the discovery of Cas12d and Cas12e.77 New data sets of bacterial or archaeal genomes are continuously being published,88 providing a rich source for future exploration of these diverse systems for discovery of novel CRISPR-Cas systems.
ACKNOWLEDGMENTS
We would like to thank R. Macrae and B. Zetsche for critical comments on the manuscript. S.C. is supported by the NIH/NCI Center for Cancer Systems Biology (1U54CA209992).
KEYWORDS
- Genome editing
a type of genetic engineering in which DNA is inserted, deleted, or replaced in the genome of a living organism using engineered nucleases, also known as genome engineering
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats, a bacterial immune system that forms the basis for CRISPR-Cas9 genome editing technology
- Immunity
biological defense of an organism to distinguish “self” from “non-self,” to fight infection, disease, or other unwanted biological invasion
- Class 2 CRISPR-Cas system
a class of CRISPR system in which the effector modules consist of single, large, multidomain proteins that might originate from mobile genetic elements
- Cas9
an RNA-guided DNA endonuclease enzyme associated with the CRISPR adaptive immunity system, particularly class 2 CRISPR
- Cas12a (Cpf1)
a key enzyme in the type V A CRISPR system, also known as CRISPR from Prevotella and Francisella 1
- Cas12b (C2c1)
a key enzyme in the type V A CRISPR system, also known as C2c1
- Cas12d (CasY)
a key enzyme in the type V D CRISPR system, also known as CasY
- Cas12e (CasX)
a key enzyme in the type V E CRISPR system, also known as CasX
- Cas13a (C2c2)
a key enzyme in the type VI A CRISPR system, also known as C2c2
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Suttle CA (2005) Viruses in the sea. Nature 437, 356–361. [DOI] [PubMed] [Google Scholar]
- (2).Doolittle WF, and Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603. [DOI] [PubMed] [Google Scholar]
- (3).Koonin EV, Makarova KS, and Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- (5).Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, and Severinov K (2010) Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol 77, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, and Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol 60, 174–182. [DOI] [PubMed] [Google Scholar]
- (7).Bolotin A, Quinquis B, Sorokin A, and Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561. [DOI] [PubMed] [Google Scholar]
- (8).Pourcel C, Salvignol G, and Vergnaud G (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663. [DOI] [PubMed] [Google Scholar]
- (9).Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, and Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Barrangou R, and Marraffini LA (2014) CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Marraffini LA (2015) CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. [DOI] [PubMed] [Google Scholar]
- (13).Sorek R, Lawrence CM, and Wiedenheft B (2013) CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem 82, 237–266. [DOI] [PubMed] [Google Scholar]
- (14).Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, and Doudna JA (2014) Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol 21, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yosef I, Goren MG, and Qimron U (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, and Semenova E (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun 3, 945. [DOI] [PubMed] [Google Scholar]
- (17).Carte J, Wang R, Li H, Terns RM, and Terns MP (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, and Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. [DOI] [PubMed] [Google Scholar]
- (19).Marraffini LA, and Sontheimer EJ (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang J, Kasciukovic T, and White MF (2012) The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One 7, e47232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Makarova KS, Anantharaman V, Grishin NV, Koonin EV, and Aravind L (2014) CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet 5, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hein S, Scholz I, Voss B, and Hess WR (2013) Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 10, 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Smargon AA, Cox DB, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, Koonin EV, and Zhang F (2017) Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 65, 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, and Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, and Koonin EV (2017) Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol 15, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, and Koonin EV (2011) Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol 9, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, and Koonin EV (2015) Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hsu PD, Lander ES, and Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Komor AC, Badran AH, and Liu DR (2017) CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 169, 559. [DOI] [PubMed] [Google Scholar]
- (30).Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, and Joung JK (2016) Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol 34, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, and Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, and Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, and Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, and Joung JK (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, and Joung JK (2015) Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol 33, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, and Joung JK (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Miyagishi M, and Taira K (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol 20, 497–500. [DOI] [PubMed] [Google Scholar]
- (39).Baer M, Nilsen TW, Costigan C, and Altman S (1990) Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 18, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, and Nureki O (2015) Crystal Structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, and Nureki O (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, and Doudna JA (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gasiunas G, Barrangou R, Horvath P, and Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A 109, E2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, and Kim JS (2015) Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243. [DOI] [PubMed] [Google Scholar]
- (48).Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, and Joung JK (2014) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, and Doudna JA (2017) Enhanced proofreading governs CRISPRCas9 targeting accuracy. Nature 550, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Schmid-Burgk JL (2017) Disruptive non-disruptive applications of CRISPR/Cas9. Curr. Opin. Biotechnol 48, 203–209. [DOI] [PubMed] [Google Scholar]
- (51).Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wang T, Wei JJ, Sabatini DM, and Lander ES (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wang F, and Qi LS (2016) Applications of CRISPR Genome Engineering in Cell Biology. Trends Cell Biol 26, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Wang H, La Russa M, and Qi LS (2016) CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem 85, 227–264. [DOI] [PubMed] [Google Scholar]
- (55).Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, and Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, and Joung JK (2013) CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, and Gersbach CA (2013) RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, and Lim WA (2015) Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, and Zhang F (2014) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, and Vale RD (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, and Maehr R (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 12, 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Gaudelli M, Komor A, Rees H, Packer M, Badran A, Bryson DL, and Liu DR (2017) Programmable base editing of A· T to G·C in genomic DNA without DNA cleavage, Nature, DOI: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Schunder E, Rydzewski K, Grunow R, and Heuner K (2013) First indication for a functional CRISPR/Cas system in Francisella tularensis. Int. J. Med. Microbiol 303, 51–60. [DOI] [PubMed] [Google Scholar]
- (64).Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, and Sung YH (2016) Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol 34, 808–810. [DOI] [PubMed] [Google Scholar]
- (65).Endo A, Masafumi M, Kaya H, and Toki S (2016) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep 6, 38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ma S, Liu Y, Liu Y, Chang J, Zhang T, Wang X, Shi R, Lu W, Xia X, Zhao P, and Xia Q (2017) An integrated CRISPR Bombyx mori genome editing system with improved efficiency and expanded target sites. Insect Biochem. Mol. Biol 83, 13–20. [DOI] [PubMed] [Google Scholar]
- (67).Moreno-Mateos MA, Fernandez JP, Rouet R, Lane MA, Vejnar CE, Mis E, and Giraldez AJ (2017) CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. bioRxiv, DOI: 10.1101/156125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, and Zhang F (2016) Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol 35, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, Federova Y, Zetsche B, Zhang F, Bienko M, and Crosetto N (2017) BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun 8, 15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, Bassel-Duby R, and Olson EN (2017) CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv 3, e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, and Zhang F (2017) Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol 35, 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Fonfara I, Richter H, Bratovic M, Le Rhun A, and Charpentier E (2016) The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. [DOI] [PubMed] [Google Scholar]
- (73).Swarts DC, van der Oost J, and Jinek M (2017) Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, Ishitani R, Zhang F, and Nureki O (2016) Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 165, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Yang H, Gao P, Rajashankar KR, and Patel DJ (2016) PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell 167, 1814–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Liu L, Chen P, Wang M, Li X, Wang J, Yin M, and Wang Y (2017) C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism. Mol. Cell 65, 310–322. [DOI] [PubMed] [Google Scholar]
- (77).Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, and Banfield JF (2016) New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Grynberg M, Erlandsen H, and Godzik A (2003) HEPN: a common domain in bacterial drug resistance and human neurodegenerative proteins. Trends Biochem. Sci 28, 224–226. [DOI] [PubMed] [Google Scholar]
- (79).Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, and Aravind L (2013) Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, and Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, and Wang Y (2017) Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 168, 121. [DOI] [PubMed] [Google Scholar]
- (82).East-Seletsky A, O′Connell MR, Burstein D, Knott GJ, and Doudna JA (2017) RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Mol. Cell 66, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).East-Seletsky A, O′Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, and Doudna JA (2016) Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, and Zhang F (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, and Zhang F (2017) RNA targeting with CRISPR-Cas13. Nature 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017) RNA editing with CRISPR-Cas13. Science, eaaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Takeuchi N, Wolf YI, Makarova KS, and Koonin EV (2012) Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol 194, 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Mukherjee S, Seshadri R, Varghese NJ, Eloe-Fadrosh EA, Meier-Kolthoff JP, Goker M, Coates RC, Hadjithomas M, Pavlopoulos GA, Paez-Espino D, Yoshikuni Y, Visel A, Whitman WB, Garrity GM, Eisen JA, Hugenholtz P, Pati A, Ivanova NN, Woyke T, Klenk HP, and Kyrpides NC (2017) 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat. Biotechnol 35, 676–683. [DOI] [PubMed] [Google Scholar]
- (89).Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, Ma C, Wang S, Wu D, Ma Y, Fan S, Wang J, Gao N, and Huang Z (2016) The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526. [DOI] [PubMed] [Google Scholar]
- (90).Gao P, Yang H, Rajashankar KR, Huang Z, and Patel DJ (2016) Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, and Nureki O (2016) Structure and Engineering of Francisella novicida Cas9. Cell 164, 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, and Church GM (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, and Kim JS (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun 8, 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, Ishitani R, Zhang F, Nishimasu H, and Nureki O (2017) Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems. Mol. Cell 65, 1109–1121. [DOI] [PubMed] [Google Scholar]
- (95).Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, and Siksnys V (2015) Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Karvelis T, Gasiunas G, and Siksnys V (2017) Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox. Curr. Opin. Microbiol 37, 88–94. [DOI] [PubMed] [Google Scholar]