Abstract
Small non-coding microRNAs (miRNAs) have been found to play critical roles in many biological processes by controlling gene expression at the post-transcriptional level. They appear to fine-tune the immune response by targeting key regulatory molecules, and their abnormal expression is associated with immune-mediated inflammatory disorders. Monocytes actively contribute to tissue homeostasis by triggering acute inflammatory reactions as well as the resolution of inflammation and tissue regeneration, in case of injury or pathogen invasion. Their contribution to tissue homeostasis can have many aspects because they are able to differentiate into different cell types including macrophages, dendritic cells, and osteoclasts, which fulfill functions as different as bone remodeling and immune response. Monocytes consist of different subsets with subset-specific expression of miRNAs linked to distinct biological processes dedicated to specific roles. Therefore, understanding the role of miRNAs in the context of monocyte heterogeneity may provide clues as to which subset gives rise to which cell type in tissues. In addition, because monocytes are involved in the pathogenesis of chronic inflammation, associated with loss of tissue homeostasis and function, identifying subset-specific miRNAs might help in developing therapeutic strategies that target one subset while sparing the others. Here, we give an overview of the state-of-the-art research regarding miRNAs that are differentially expressed between monocyte subsets and how they influence monocyte functional heterogeneity in health and disease, with descriptions of specific miRNAs. We also revisit the existing miRNome data to propose a canonical signature for each subset.
Keywords: microRNA, monocytes, Ly6Chigh, Ly6Clow, CD14+, CD16+
Introduction
MicroRNAs (miRNAs) are a class of short non-coding RNAs (18–22 nt), conserved from worms to mammals that play a regulatory role in gene expression at the posttranscriptional level (1). Since their discovery, many studies have shown that they are involved in biological processes. Quantitative and qualitative assessments of miRNA expression in various disease conditions have revealed considerable changes in their expression profiles.
The biogenesis of miRNAs occurs in the nucleus. MiRNA-encoding genes are transcribed to a primary miRNA and processed by Drosha, a class 2 RNase III enzyme, into a precursor miRNA (pre-miRNA), which is exported to the cytoplasm by exportin-5. In the cytoplasm, mature forms of miRNAs are produced after several steps involving Dicer, a RNase III type protein, and RISC, a RNA-induced silencing complex (2). MiRNA genes can be located in the context of non-coding transcription units or in the introns of protein-coding genes (3, 4). Almost half of miRNA genes are clustered and can be independently or simultaneously transcribed into single polycistronic transcripts (5, 6).
Currently, more than 2,800 and 2,100 miRNAs have been identified in human and mouse, respectively (miRBase vs22). Both in vitro and bioinformatic analyses have determined that more than 500 genes could be targeted by a single miRNA (7, 8). MiRNAs bind mRNA targets by their “seed” sequence interacting with the 3′untranslated region (UTR), and more rarely with the coding region (CDS) or 5′UTR, of the targeted mRNA (9). According to the degree of complementarity, miRNAs lead to mRNA cleavage and degradation or to the inhibition of translation, thus interfering with the downstream protein output (10). MiRNA family members can be highly conserved among vertebrates, in particular in the seed region, which corresponds to nucleotides 2 to 7/8 and is the main determinant of target specificity (11). Thus, miRNAs with similar seed sequence can target similar sets of genes and similar biological pathways.
Extensive work has been performed to identify miRNA-specific signatures in immune cells and to understand how a specific miRNA gene controls the development and function of a specific immune cell population. However, few studies have addressed the role of miRNAs in terms of subset heterogeneity of one specific immune cell type. Here we review reports of miRNAs in monocyte subsets and performed an in silico analysis that also includes new data to revisit the current knowledge of monocyte subset functions.
miRNA-Based Signatures Specific to Monocyte Subsets
Monocytes are composed of two main subsets in both mouse and human (12) that are committed to different functions (13–15): in mice, the “classical” inflammatory Ly6Chigh and the “non-classical” patrolling Ly6Clow monocyte subsets. Their human counterparts are CD14+CD16− and CD14dimCD16++, respectively (12, 13). Ly6Chigh monocytes secrete inflammatory mediators in response to bacteria and can differentiate into macrophages, inflammatory dendritic cells (DCs), and osteoclasts (OCs) (16–18). Ly6Clow monocytes survey endothelial cells and surrounding tissues to detect damage and viral threat and are involved in tissue repair (13). Although mouse and human studies have underscored the relevance of studying monocyte subsets in disease by showing differential accumulation of both subsets, factors that regulate monocyte subset fate and functional heterogeneity under pathophysiological conditions remain poorly explored.
MiRNAs play pivotal roles in regulating monocyte development and functions, including differentiation, tissue recruitment, activation, initiation, and resolution of inflammation (19); however, very little is known about their involvement when considering the heterogeneity of monocytes. An attempt to close this gap in knowledge has been addressed by miRNome analyses of monocyte subsets. Nevertheless, few miRNome analyses of monocyte subsets have been performed with human or mouse samples. To our knowledge and from free-access databases, we identified only three studies (20–22); two focused on miRNAs differently expressed between classical and non-classical monocytes, in humans and mice, without considering “intermediate” monocytes (20, 21). After showing differences in DNA methylation in the three human monocyte subsets—classical, non-classical, and intermediate (23)—Zawada et al. studied miRNA profiling for human “intermediate” monocytes (22).
Thus, in the current review, we combined all existing data with our own unpublished miRNome data for both classical and non-classical monocyte subsets isolated from human and mouse blood to provide novel insights into monocyte subset-specific miRNA signatures.
Analysis of the miRNome of Classical and Non-classical Monocyte Subsets
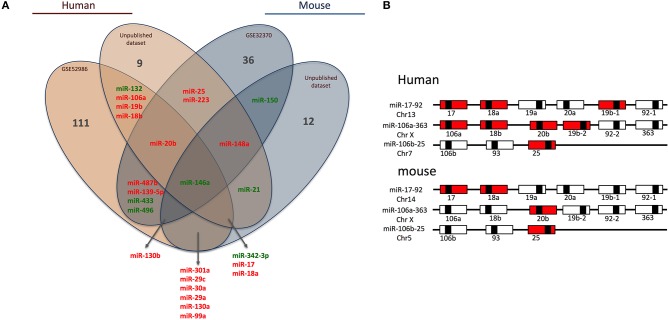
Briefly, we collected miRNome datasets (GSE52986 and GSE32370) from the GEO bank (http://www.ncbi.nlm.nih.gov/geo/). For each GEO dataset, we compared miRNA expression profiles for classical and non-classical monocyte subsets (i.e., CD14++ CD16− and CD14+ CD16++ for human blood samples, Ly6Chigh and Ly6Clow for mouse blood samples) to obtain a list of miRNAs differentially expressed between the subsets in both species. The technical platforms used in these two studies were Illumina Human v2 and Mouse v1 MicroRNA expression beadchips, respectively. Also, we performed large-scale miRNA screening using a TaqMan low-density array to identify miRNAs differentially expressed between classical and non-classical monocyte subsets in human and mouse. With false discovery rate-adjusted P ≤ 0.05, we found 25 miRNAs differentially expressed between classical and non-classical monocytes. We then used a Venn diagram to visualize common miRNAs between all four datasets (http://www.irp.nia.nih.gov/bioinformatics/vennplex.html). Only miR-146a was commonly downregulated in classical monocytes, for all available human and mouse datasets, independent of the technological platform used (Figure 1A). At the intersection of human datasets, we identified nine miRNAs (miR-132, miR-106a, miR-19b, miR-18b, miR-20b, miR-146a, miR-342-3p, miR17, and miR18a): miR-132, miR146a, and miR-342-3p showed lower expression in classical than non-classical monocytes. At the intersection of mouse datasets, we identified 4 miRNAs (miR-146a, miR-130b, miR-150, and miR-148a); miR-148a and miR-130b were upregulated in Ly6Chigh versus Ly6Clow monocytes. Only a very small number of miRNAs was specific to mouse (miR-150) or human (miR-18b, miR-19b, miR-106a, and miR-132) subsets.
Figure 1.
Human and mouse miRNome profiles identifying monocyte subset-specific miRNAs. (A) Four datasets were analyzed and clustered to obtain a Venn diagram showing miRNAs differentially regulated between classical and non-classical monocytes and common to human and mouse. Red and green represent miRNAs up- and down-regulated, respectively, in classical vs. non-classical monocytes. (B) Schematic representation of members of the miR-17/92 family of miRNA gene clusters in human and mouse. MiRNAs upregulated in classical monocytes vs. non-classical monocytes are in red.
Genomic Organization of miRNAs Specific to Monocyte Subsets
Among the 25 miRNAs identified, 16 showed sequence homology between human and mouse (Table 1) and almost 70% were organized in clusters in both species; examples are miR-17/92, miR-106a/363 and miR-106b/25 (Figure 1B). Only two miRNAs were not organized in clusters in either species: miR-342/151b and miR-150/5121 in human and mouse, respectively.
Table 1.
List of miRNAs common in human and mouse miRNome datasets.
| Name | Species | Cluster (miRNA) | Chromosome position | miRNA mature seq | miRNA* mature seq |
|---|---|---|---|---|---|
| miR-106a | hsa | Yes (miR-18b, miR-20b, miR-19b-2, miR-92a-2, miR-363) | chrX: 134170198-134170278 | AAAAGUGCUUACAGUGCAGGUAG | CUGCAAUGUAAGCACUUCUUAC |
| mmu | Yes (miR-18b, miR-20b, miR-19b-2, miR-92a-2, miR-363) | chrX: 52742503-52742567 | AAAGUGCUACAGUGCAGGUAG | CUGCAUGCAGCACUUCUUAC | |
| miR-130a | hsa | No | chr17: 59151136-59151221 | GCUCUGACUUUAUUGCACUACUCAGUGCAAUAGUAUUGUCAAAGC | |
| mmu | No | chr11: 87113004-87113089 | CAGUGCAAUAGUAUUGUCAAAGC | GCUCUGACUUUAUUGCACUACU | |
| miR-130b | hsa | yes (miR-301b) | chr22: 21653304-21653385 | CAGUGCAAUGAUGAAAGGGCAU | ACUCUUUCCCUGUUGCACUAC |
| mmu | yes (miR-301b) | chr16: 17124061-17124142 | CAGUGCAAUGAUGAAAGGGCAU | ACUCUUUCCCUGUUGCACUAC | |
| miR-132 | hsa | Yes (miR-212) | chr17: 2050271-2050380 | UAACAGUCUACAGCCAUGGUCG | ACCGUGGCUUUCGAUUGUUAC |
| mmu | Yes (miR-212) | chr11: 75173388-75173478 | UAACAGUCUACAGCCAUGGUCG | ACCGUGGCUUUCGAUUGUUAC | |
| miR-139-5p | hsa | No | chr11: 72615063-72615130 | UCUACAGUGCACGUGUCUCCAG | |
| mmu | No | chr7: 101475376-101475443 | UCUACAGUGCACGUGUCUCCAG | ||
| miR-146a | hsa | No | chr5: 160485352-160485450 | UGAGAACUGAAUUCCAUGGGUU | CCUCUGAAAUUCAGUUCUUCAG |
| mmu | No | chr11: 43374397-43374461 | UGAGAACUGAAUUCCAUGGGUU | CCUGUGAAAUUCAGUUCUUCAG | |
| miR-148a | hsa | No | chr7: 25949919-25949986 | UCAGUGCACUACAGAACUUUGU | AAAGUUCUGAGACACUCCGACU |
| mmu | No | chr6: 51269812-51269910 | UCAGUGCACUACAGAACUUUGU | AAAGUUCUGAGACACUCCGACU | |
| miR-150 | hsa | No | chr19: 49500785-49500868 | UCUCCCAACCCUUGUACCAGUG | CUGGUACAGGCCUGGGGGACAG |
| mmu | yes (miR-5121) | chr7: 45121757-45121821 | UCUCCCAACCCUUGUACCAGUG | CUGGUACAGGCCUGGGGGAAG | |
| miR-17 | hsa | Yes (miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92a-1) | chr13: 91350605-91350688 | CAAAGUGCUUACAGUGCAGGUAG | ACUGCAGUGAAGGCACUUGUAG |
| mmu | Yes (miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92a-1) | chr14: 115043671-115043754 | CAAAGUGCUUACAGUGCAGGUAG | ACUGCAGUGAGGCACUUGUAG | |
| miR-18a | hsa | Yes (miR-17, miR-19a, miR-20a, miR-19b-1, miR-92a-1) | chr13: 91350751-91350821 | UAAGGUGCAUCUAGUGCAGAUAG | ACUGCCCUAAGUGCUCCUUCUG |
| mmu | Yes (miR-17, miR-19a, miR-20a, miR-19b-1, miR-92a-1) | chr14: 115043851-115043946 | UAAGGUGCAUCUAGUGCAGAUAG | ACUGCCCUAAGUGCUCCUUCUG | |
| miR-18b | hsa | Yes (miR-106a, miR-20b, miR-19b-2, miR-92a-2, miR-363) | chrX: 134170041-134170111 | UAAGGUGCAUCUAGUGCAGUUAG | UGCCCUAAAUGCCCCUUCU |
| mmu | Yes (miR-106a, miR-20b, miR-19b-2, miR-92a-2, miR-363) | chrX: 52742331-52742413 | UAAGGUGCAUCUAGUGCGUUAG | UGCCCUAAAUGCCCCUUCU | |
| miR-19b-1 | hsa | Yes (miR-17, miR-19a, miR-18a, miR-20a, miR-92a-1) | chr13: 91351192-91351278 | UGUGCAAAUCCAUGCAAAACUGA | AGUUUUGCAGGUUUGCAUCCAGC |
| mmu | Yes (miR-17, miR-19a, miR-18a, miR-20a, miR-92a-1) | chr14: 115044305-115044391 | UGUGCAAAUCCAUGCAAAACUGA | AGUUUUGCAGGUUUGCAUCCAGC | |
| miR-19b-2 | hsa | Yes (miR-106a, miR-18b, miR-20b, miR-92a-2, miR-363) | chrX: 134169671-134169766 | UGUGCAAAUCCAUGCAAAACUGA | AGUUUUGCAGGUUUGCAUUUCA |
| mmu | Yes (miR-106a, miR-18b, miR-20b, miR-92a-2, miR-363) | chrX: 52741983-52742066 | UGUGCAAAUCCAUGCAAAACUGA | AGUUUUGCAGUUUGCAUUCA | |
| miR-20b | hsa | Yes (miR-106a, miR-18b, miR-19b-2, miR-92a-2, miR-363) | chrX: 134169809-134169877 | CAAAGUGCUCAUAGUGCAGGUAG | ACUGUAGUAUGGGCACUUCCAG |
| mmu | Yes (miR-106a, miR-18b, miR-19b-2, miR-92a-2, miR-363) | chrX: 52742113-52742192 | CAAAGUGCUCAUAGUGCAGGUAG | ACUGAGUUGGCACUUCAG | |
| miR-21 | hsa | No | chr17: 59841266-59841337 | UAGCUUAUCAGACUGAUGUUGA | CAACACCAGUCGAUGGGCUGU |
| mmu | No | chr11: 86584067-86584158 | UAGCUUAUCAGACUGAUGUUGA | CAACACAGUCGAUGGGCUGU | |
| miR-223 | hsa | No | chrX: 66018870-66018979 | UGUCAGUUUGUCAAAUACCCCA | CGUGUAUUUGACAAGCUGAGUU |
| mmu | No | chrX: 96242817-96242926 | UGUCAGUUUGUCAAAUACCCCA | CGUGUAUUUGACAAGCUGAGUU | |
| miR-25 | hsa | Yes (miR-106b, miR-93) | chr7: 100093560-100093643 | CAUUGCACUUGUCUCGGUCUGA | AGGCGGAGACUUGGGCAAUUG |
| mmu | Yes (miR-106b, miR-93) | chr5: 138165321-138165404 | CAUUGCACUUGUCUCGGUCUGA | AGGCGGAGACUUGGGCAAUUG | |
| miR-29a | hsa | Yes (miR-29b-1) | chr7: 130876747-130876810 | UAGCACCAUCUGAAAUCGGUUA | ACUGAUUUCUUUUGGUGUUCAG |
| mmu | Yes (miR-29b-1) | chr6: 31062660-3106274 | UAGCACCAUCUGAAAUCGGUUA | ACUGAUUUCUUUUGGUGUUCAG | |
| miR-29c | hsa | Yes (miR-29b-2) | chr1: 207801852-207801939 | UAGCACCAUUUGAAAUCGGUUA | UGACCGAUUUCUCCUGGUGUUC |
| mmu | Yes (miR-29b-2) | chr1: 195037547-195037634 | UAGCACCAUUUGAAAUCGGUUA | UGACCGAUUUCUCCUGGUGUUC | |
| miR-30a | hsa | No | chr6: 71403551-71403621 | UGUAAACAUCCUCGACUGGAAG | CUUUCAGUCGGAUGUUUGCAGC |
| mmu | No | chr1: 23272269-23272339 | UGUAAACAUCCUCGACUGGAAG | CUUUCAGUCGGAUGUUUGCAGC | |
| miR-342-3p | hsa | Yes (miR-151b) | chr14: 100109655-100109753 | UCUCACACAGAAAUCGCACCCGU | |
| mmu | No | chr12: 108658620-108658718 | UCUCACACAGAAAUCGCACCCGU | ||
| miR-433 | hsa | Yes (miR-337, miR-665, miR-431, miR-127, miR-432, miR-136) | chr14: 100881886-100881978 | UACGGUGAGCCUGUCAUUAUUCAUCAUGAUGGGCUCCUCGGUGU | |
| mmu | Yes (miR-337, miR-3544, miR-665, miR-3070-1, miR3070-2, miR-431, miR-127, miR-434, miR-432, miR-3071, miR-136) | chr12: 109591715-109591838 | AUCAUGAUGGGCUCCUCGGUGU | UACGGUGAGCCUGUCAUUAUUC | |
| miR-487b | hsa | Yes (miR-376c, miR-376a-2, miR-654, miR-376b, miR-300, miR-1185-1, miR-1185-2, miR-381, miR-539, miR-889, miR-544a, miR-655, miR-487a, miR-382, miR-134, miR-668, miR-485, miR-323b) | chr14: 101046455-101046538 | GUGGUUAUCCCUGUCCUGUUCGAAUCGUACAGGGUCAUCCACUU | |
| mmu | Yes (miR-495, miR-667, miR-376c, miR-654, miR-376b, miR-376a, miR-300, miR-381, miR-539, miR-889, miR-544, miR-382, miR-134, miR-668, miR-485, miR-453) | chr12: 109727333-109727414 | AAUCGUACAGGGUCAUCCACUU | UGGUUAUCCCUGUCCUCUUCG | |
| miR-496 | hsa | Yes (miR-487a, miR-382, miR-134, miR-668, miR-485, miR-323b, miR-154, miR-377, miR-541, miR-409, miR-412, miR-369, miR-410, miR-656) | chr14: 101060573-101060674 | UGAGUAUUACAUGGCCAAUCUC | |
| mmu | Yes (miR-544, miR-382, miR-134, miR-668, miR-485, miR-453, miR-154, miR-377, miR-541, miR-409, miR-412, miR-369, miR410, miR-3072) | chr12: 109739119-109739197 | UGAGUAUUACAUGGCCAAUCUC | AGGUUGCCCAUGGUGUGUUCA | |
| miR-99a | hsa | Yes (miR-let-7c) | chr21: 16539089-16539169 | AACCCGUAGAUCCGAUCUUGUG | CAAGCUCGCUUCUAUGGGUCU |
| mmu | Yes (miR-let-7c) | chr16: 77598936-77599000 | AACCCGUAGAUCCGAUCUUGUG | CAAGCUCGUUUCUAUGGGUCU |
hsa, human; mmu, mouse; chr, chromosome. Red highlights nucleotide differences between humans and mice.
Of note, miR-17, miR-18a/b, miR-19a/b, miR-20b, miR-25, and miR106a are members of the three paralog clusters: miR-17/92, miR-106a/323, and miR-106b/25. These clusters contain miRNAs that are very comparable, regulate similar sets of genes, and have overlapping functions (24). Their genomic organization is highly conserved, which suggests important functions and coordinated regulations. Overall, 7 and 4 miRNAs in human and mouse, respectively, were overexpressed in classical monocytes (Figure 1B).
The miR-17/92 cluster is a well-described cluster that plays a role in immune responses (25). In the lymphocyte lineage, this cluster is expressed in B and T precursor cells, and its expression diminishes upon differentiation (26). In the monocytic lineage, monocyte hematopoiesis is affected by loss of the miR-17/92 cluster in humans but is unaffected in mouse (27). Human CD34+ hematopoietic progenitor cells differentiate in vitro into monocytes upon exposure to macrophage-colony stimulating factor; this differentiation leads to decreased expression of the miR-17/92 cluster, which is inversely correlated with upregulation of the transcription factor acute myeloid leukemia 1, a validated human target of miR-17. In addition, overexpression of the miR-17/92 cluster delays terminal differentiation of monocytes, and its inhibition accelerates differentiation (28). Differences between human and mouse data may be caused by species-specific differences and/or the fact that experiments were performed in vitro or in vivo. Using genetic mouse models, deletion of the miR-17/92 cluster and its paralog miR-106b/25 led to severe developmental defects, so these two miRNA clusters may act synergistically on cell survival to control embryonic development (27). Moreover, miR-17, miR-20a, and miR-106a, which belong to the two cluster paralogs miR-17/92 and miR-106a/363, regulate macrophage infiltration, phagocytosis, and proinflammatory cytokine secretion via targeting signal-regulatory protein alpha expression, both in vitro and in vivo (29).
Despite rare reports describing these 25 miRNAs in the context of monocyte subsets (see below), many more exist on their role in monocyte differentiation or inflammation processes. For example, some miRNAs are involved in macrophage polarization; one is miR-148a-3p, which promotes macrophage 1 (M1) polarization and inhibits M2 polarization upon Notch activation (30). Others are involved in osteoclastogenesis [e.g., the miR-29 family regulates osteoclast commitment and migration (31)]. MiR-223 is upregulated during granulopoiesis and fine-tunes the differentiation of myeloid precursors into granulocytes or monocytes and negatively controls the activity of NLRP3 inflammasome in these cell types (32). Also, miR-433 negatively regulates the hematopoietic cell proliferation by directly targeting interferon-induced guanylate-binding protein 2 (33), and miR-130a regulates the expression of macrophage pro-fibrogenic genes in chronic inflammation (34).
Putative Function of miRNAs Specific to Human Monocyte Subsets
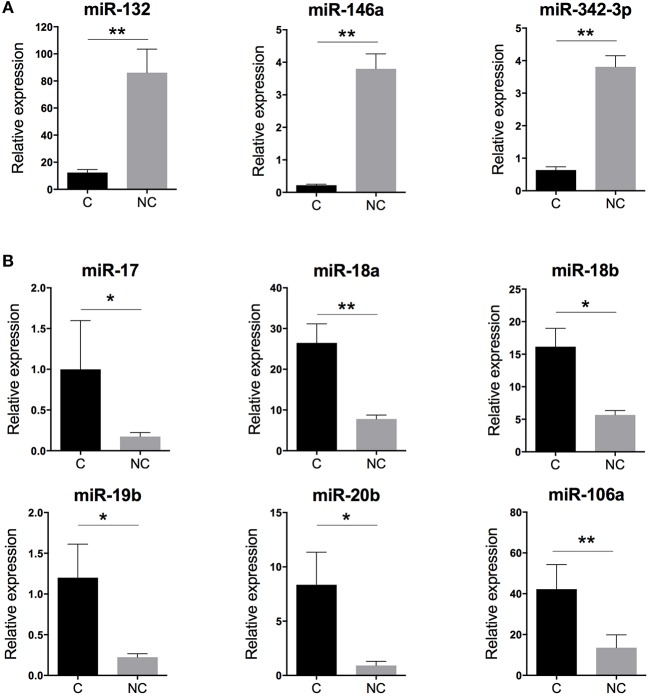
With human miRNome data from classical and non-classical monocytes (Figure 1A), we quantified the expression of the nine miRNAs commonly deregulated between both subsets by using RT-qPCR and new samples. We confirmed the overexpression of miR-132, miR-146a, and miR-342-3p in human non-classical vs. classical monocytes and the overexpression of miR-17, miR-18a, miR-18b, miR-19b, miR-20b, and miR-106a in classical vs. non-classical monocytes (Figure 2).
Figure 2.
Validation of the human monocyte subset-specific miRNA-based signature. Blood samples from healthy donors (n = 7) were collected from the French Blood Establishment (EFS). After Ficoll-Paque density gradient, classical (C) CD14++CD16− and non-classical (NC) CD14+CD16++ monocyte subsets were FACS sorted with >97% purity (Montpellier RIO Cytometry platform). Total RNA was extracted from both monocyte subsets by using a miRNeasy kit and the automatized QIAcube procedure (QIAGEN). MiRNA expression was quantified by using multiplexed TaqMan RT-qPCR (Life Technology). (A) Quantification of the three miRNAs overexpressed in non-classical vs. classical monocytes. (B) Quantification of the six miRNAs overexpressed in classical vs. non-classical monocytes. Data are mean ± SD and differences were compared by non-parametric Mann-Whitney test (*p < 0.05, **p < 0.01).
Zawada et al. hypothesized that intermediate monocytes have a distinct miRNA profile as compared with classical and non-classical monocytes and identified 38 miRNAs differentially expressed in intermediate monocytes vs. both classical and non-classical monocytes (22). Figure 3 gives a schematic representation of the miRNA expression profile patterns for the three human monocyte subsets in the Zawada et al. study. Of note, miRNAs in panel 1 with gradually increasing expression from classical to intermediate and non-classical monocyte subsets included the three miRNAs that we found upregulated in our comparative study (miR-132, miR-146a, and miR-342-3p; Figure 2A). Panels 2 and 3, showing decreasing expression from classical to non-classical monocytes, displaying (panel 2) or not (panel 3) differences between intermediate and non-classical monocytes, contained the six miRNAs that we found downregulated in our comparative study (Figure 2B). Zawada et al. identified a fourth panel, including miR-150, with downregulated miRNAs in intermediate monocytes as compared with both classical and non-classical monocytes (p < 10−10, and > 10-fold difference in expression). Our Venn diagram analysis identified miR-150 as the only miRNA with differential expression between classical and non-classical monocyte datasets in mouse but not human datasets (Figure 1A), which agrees with the Zawada et al. miRNome data (Panel 4).
Figure 3.
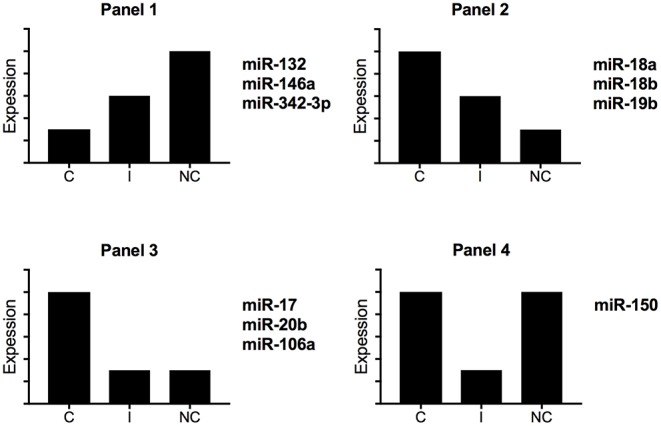
Schematic representation of miRNA expression profiles for human monocyte subsets. By using miRNome data from the study of Zawada et al. (22), we identified four different expression profiles. C, classical monocytes CD14++CD16−; I, intermediate monocytes CD14++CD16+; NC, non-classical monocytes CD14+CD16++.
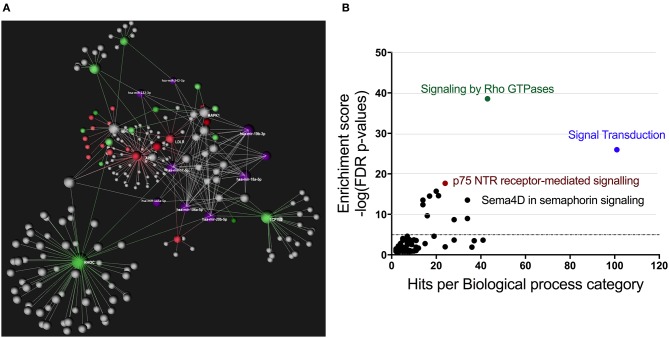
Using OmicsNet, a web-based tool for the creation and visual analysis of biological networks (35), we uploaded the list of nine monocyte subset-specific miRNAs identified in our human analysis together with the list of 182 genes found overexpressed in classical or non-classical monocytes identified in five independent transcriptomic microarray datasets (36). We aimed to create and merge different types of biological networks that could provide a clue to the pathways involved in the functional heterogeneity of monocyte subsets. Figure 4A shows the 3D OmicsNet biological networks highlighting connections between the nine miRNAs and putatively targeted genes according to the TarBase software [Table 2; (37)]. Gene Ontology analysis with the Reactome pathway database (38) showed enrichment of biological process categories such as signal transduction, small GTPases of the Rho family (Rho GTPases), p75 NTR receptor-mediated and Sema4D in semaphorin signaling (Figure 4B and Table 3). The trafficking of monocytes into tissues requires the activation of integrins via signal transduction induced by Rho GTPases such as RHOA or RAP1, which results in cell adhesion to the blood-vessel wall (39). Rho-GTPases are key regulators of cellular actomyosin dynamics and are therefore considered pharmacological targets for restricting leukocyte motility, including monocytes, in inflammatory disorders (40). A comparison of protein expression based on cell maturity (from pro-monocyte to monocyte and to macrophage lineages) suggested that Rho proteins are readily available for signaling events in response to numerous activating cues (41). Human CD100/Sema4D belongs to a large family of membrane-bound proteins named Semaphorins that are involved in numerous functions, including axon guidance, morphogenesis, carcinogenesis, and immunomodulation; Sema4D in particular influences monocyte migration (42). Resident microglia and infiltrated peripheral monocytes are two main types of immune cells in the central nervous system that control the inflammation process. Recently, the p75 neurotrophin receptor (p75NTR) was found to play a role in the peripheral expansion and central nervous system trafficking of pro-inflammatory monocytes (43).
Figure 4.
Gene ontology analysis of genes putatively targeted by monocyte subset-specific miRNAs. (A) Using OmicsNet, a force-directed sub-network was constructed for the nine miRNAs with differential expression between classical and non-classical human monocytes (color violet) and their putative target genes extracted from a list of 182 genes with differential expression in classical and non-classical monocyte subsets. Red and green represent genes up- and downregulated, respectively, in classical vs. non-classical monocytes. Genes in gray are those that link genes putatively targeted by miRNAs or are associated in the network. (B) By using Reactome pathway data, we plotted genes with differential expression between monocyte subsets as the number of genes for the respective biological function category (x-axis) against the enrichment score for log10 of p-value (y axis).
Table 2.
List of genes putatively targeted by the nine monocyte subset-specific miRNAs.
| miR-17 | miR-18a | miR-18b | miR-19b | miR-20b | miR-106a | miR-132 | miR-146a | miR-342-3p |
|---|---|---|---|---|---|---|---|---|
| AIB1 | AIB1 | AIB1 | AIB1 | AIB1 | AIB1 | AIB1 | AIB1 | |
| APP | APP | |||||||
| CCND1 | CCND1 | CCND1 | CCND1 | CCND1 | ||||
| CRK | CRK | CRK | CRK | |||||
| DCAF8 | DCAF8 | DCAF8 | DCAF8 | DCAF8 | DCAF8 | |||
| F2RL1 | F2RL1 | F2RL1 | ||||||
| FAS | FAS | FAS | FAS | FAS | ||||
| GIGYF1 | GIGYF1 | GIGYF1 | GIGYF1 | GIGYF1 | GIGYF1 | |||
| ITGA2 | ITGA2 | ITGA2 | ITGA2 | ITGA2 | ITGA2 | |||
| INSIG1 | ||||||||
| LDLR | LDLR | LDLR | LDLR | LDLR | ||||
| MAPK1 | MAPK1 | MAPK1 | MAPK1 | MAPK1 | ||||
| MAPK14 | MAPK14 | MAPK14 | NAP1L1 | |||||
| NAP1L1 | ||||||||
| PTEN | PTEN | PTEN | PTEN | PTEN | ||||
| RAC1 | ||||||||
| RHOC | RHOC | RHOC | ||||||
| RLIM | RLIM | RLIM | RLIM | RLIM | RLIM | |||
| RORA | RORA | RORA | RORA | RORA | RORA | |||
| SMAD4 | SMAD4 | SMAD4 | SMAD4 | SMAD4 | SMAD4 | |||
| TCF7L2 | TCF7L2 | TCF7L2 | ||||||
| TNRC6B | TNRC6B | TNRC6B | TNRC6B | TNRC6B | TNRC6B | |||
| UBC | UBC | UBC | ||||||
| WAC | WAC | WAC | WAC | WAC | WAC | WAC |
Table 3.
Gene ontology and functional pathway enrichment analysis.
| Pathway | Total | Expected false positives | Hits | –log (FDR p-values) |
|---|---|---|---|---|
| Sema4D induced cell migration and growth-cone collapse | 29 | 0.634 | 14 | 22.1 |
| Sema4D in semaphorin signaling | 34 | 0.743 | 14 | 18.8 |
| NRAGE signals death through JNK | 45 | 0.984 | 17 | 17.3 |
| Rho GTPase cycle | 123 | 2.69 | 43 | 16.0 |
| Signaling by Rho GTPases | 123 | 2.69 | 43 | 16.0 |
| Cell death signaling via NRAGE NRIF and NADE | 62 | 1.36 | 20 | 14.7 |
| Lipoprotein metabolism | 22 | 0.481 | 7 | 14.6 |
| p75 NTR receptor-mediated signaling | 85 | 1.86 | 24 | 12.9 |
| G alpha (12/13) signaling events | 80 | 1.75 | 21 | 12.0 |
| Semaphorin interactions | 72 | 1.57 | 16 | 10.2 |
| Signaling by NGF | 290 | 6.34 | 34 | 5.4 |
| Axon guidance | 292 | 6.38 | 28 | 4.4 |
| Platelet activation signaling and aggregation | 220 | 4.81 | 19 | 4.0 |
| Developmental Biology | 417 | 9.12 | 34 | 3.7 |
| Signal Transduction | 1,690 | 36.9 | 101 | 2.7 |
The top 10 terms were selected according to false discovery rate (FDR)-adjusted p-values.
Although these pathways and putative target genes have not yet been validated and functionally studied, further investigating their implication will increase our understanding of the functional heterogeneity of monocyte subsets.
Role of miRNAs in Monocyte Subsets
Since the first description of blood monocytes in the early 2000s as a heterogeneous population of leukocytes displaying different phenotypic markers, homing properties, and immune functions (44), the scientific community has tried to dissect the role of individual subsets by identifying protein-encoding genes that specifically control the development and function of each sub-population. For example, the lineage-defining transcription factor nuclear receptor 4a1 (Nr4a1) was found essential for Ly6Clow monocyte development because Nr4a1−/− mice lack Ly6Clow monocytes (45). Because Nr4a1 regulates inflammatory gene expression and differentiation of Ly6Clow monocytes, the functions of Ly6Chigh monocytes can be studied independently in vivo by using Nr4a1−/− mice (46). The same expectations have been expressed for miRNA-encoding genes. However, few miRNAs have been identified (see previous section), and only three have been thoroughly studied in vivo by using genetic models. The first identified and most studied is miR-146a. In 2012, the group of Mikael Pittet showed that miR-146a is the highest differentially expressed miRNA between Ly6Chigh and Ly6Clow monocytes (20). Also, until 2018, it remained the only miRNA described as regulating the functional heterogeneity of monocyte subsets.
Control of Innate Immune Response to Infections
For many years, miR-146a has beenknownas anegativeregulator of inflammation inmyeloid cells (26, 47). In-depth characterization of miR-146a−/− mice revealed decreased hematopoietic stem cell homeostasis during chronic inflammation, dysregulated hematopoietic stem cell differentiation toward myeloid cells, and abnormal myeloproliferation (48). The group of Pittet showed that miR-146a expression was inducible only in Ly6Chigh monocytes upon inflammatory stimuli, reaching levels comparable to those in Ly6Clow monocytes in basal conditions. Lack of miR-146a in mice did not alter the development of monocyte subsets but markedly amplified the inflammatory response of Ly6Chigh monocytes upon bacterial challenge by targeting RelB, a non-canonical NF-κB family member highly conserved between mice and humans. This amplification of the inflammatory response is not due to more pro-inflammatory cytokine production per cell but rather to an expansion of Ly6Chigh monocytes in the bone marrow and their increased trafficking to inflamed tissue during acute bacterial challenge because of high expression of CCR2 and responsiveness to monocyte chemoattractant protein 1-mediated chemoattraction. This interesting result parallels the fact that neither TNF receptor associated factor six nor interleukin 1 receptor associated kinase 1 expression was modified by miR-146a in Ly6Chigh monocytes (20, 49), but they were modified in monocytic cell lines (47). Overall, by maintaining a low level of miR-146a, Ly6Chigh monocytes can rapidly proliferate into the bone marrow to be the first mobilized cells to egress into the circulation and rejoin the site of bacterial attack. In contrast, Ly6Clow monocytes remain insensitive to this type of environmental danger because of constitutive high expression of miR-146a. Thus, the gradual increase of miR-146a expression in Ly6Chigh monocytes upon stimulation acts as a negative feedback loop that represses proliferation and prevents overwhelming amplification of the inflammation by so-called inflammatory Ly6Chigh monocytes within the injured tissue, which would be deleterious. However, this study does not answer the question of the role of miR-146a in Ly6Clow monocytes.
RelB can directly bind with the aryl hydrocarbon receptor (AHR) that supports the xenobiotic-detoxifying pathway, the AHR nuclear translocator like 1 (also named Bmal1) partner of Clock that regulates the circadian rhythm, and the bioenergy sensor sirtuin 1 (Sirt1) to integrate acute inflammation with changes in metabolism and mitochondrial bioenergetics. Finally, RelB is involved in chromatin modifications, and low RelB expression recapitulates the formation of silent heterochromatin upon endotoxin tolerance conditions, further halting inflammatory signaling (50). Although these functions have not all been investigated in terms of monocyte heterogeneity and/or miRNA context, the miR-146/Relb axis might be the missing link with Bmal1-dependent regulation of Ly6Chigh diurnal variations controlling their trafficking to sites of inflammation (51), AHR-dependent regulation of Ly6Chigh monocyte-derived DC differentiation (52), and Sirt1-mediated inhibition of the pro-inflammatory macrophage activation (53). These are interesting areas to be addressed.
Control of the Inflammatory Response in the Context of Atherosclerosis
In 2015, the group of Robert Raffai showed that apolipoprotein E (ApoE) expression was higher in Ly6Clow than Ly6Chigh monocytes (54). The expression of ApoE in monocytes had an anti-inflammation effect by enhancing the purine-rich PU-box binding protein 1-dependent miR-146a transcription, thereby reducing Ly6Chigh monocytosis, NF-κB–mediated inflammation, and atherosclerosis in the setting of hyperlipidemia. Thus, increasing miR-146a expression in Ly6Chigh monocytes might have therapeutic application in atherosclerosis. Also, miR-146a may play a role in controlling the proliferation of Ly6Chigh monocytes. This finding contradicts the general concept that monocytes are non-proliferating cells (55) but agrees with studies observing Ly6Chigh monocytosis in bone marrow and blood, in different pathological contexts (20, 49, 54).
Role in Bone Erosion and Formation
RelB also promotes the differentiation of myeloid precursors into DCs and OCs and activates the transcription of pro-inflammatory genes in response to immune signals and environmental stressors. Monocyte subset-specific differences in miR-146a expression, together with the well-described role of miR-146a as a negative feedback regulator of inflammation and osteoclastogenesis in myeloid cells (56, 57) and reduced level of miR-146a expression in Ly6Chigh monocytes, might explain why Ly6Chigh monocytes are prone to egress from the bone marrow upon inflammatory stimuli and differentiate into DCs and OCs upon entry into the inflamed site. Also, in contrast, this information might also explain why by maintaining constitutively high levels of miR-146a, Ly6Clow monocytes are prevented from differentiating into DCs and OCs. Indeed, monocyte subsets have a differential contribution to osteoclastogenesis (16). The capacity of the Ly6Chigh subset but not Ly6Clow subset to develop into OCs has been recently attributed to low miR-146a expression (49). Indeed, our group showed that classical monocytes display reduced miR-146a expression in both arthritic humans and mice as compared with healthy individuals; in vivo delivery of miR-146a mimics into Ly6Chigh monocytes using a specific delivery system that spares Ly6Clow monocytes (58) rescued RelB expression in Ly6Chigh monocytes, reduced their capacity to differentiate into OCs and reduced inflammatory-mediated bone erosion in an experimental model of arthritis. This is the first work to provide an in vivo proof of concept for a therapeutic strategy design targeting a subset-specific miRNA. Whether miR-146a plays other roles in Ly6Chigh monocytes and investigating its Ly6Clow-specific function(s) remains for further investigation. Our transcriptomic analyses comparing both monocyte subsets sorted from miR-146a+/+ and miR-146−/− mice showed that miR-146a modulates the expression of 1,000 genes in Ly6Chigh monocytes but only 100 genes in Ly6Clow monocytes (49), which suggests that beyond osteoclastogenesis, miR-146a may play other roles in Ly6Chigh functions but not many in Ly6Clow monocytes.
miRNA Function in Monocyte Subset Differentiation
Recently, the group of Stéphane Potteaux identified an miRNA critical for generating Ly6Clow monocytes. The authors observed increased expression of miR-21 in non-classical Ly6Clow monocytes of atherosclerotic ApoE−/− mice, which mediated their higher number and lifespan in this model (59). The frequency of Ly6Clow monocytes in blood, bone marrow and spleen was markedly reduced in ApoE−/− miR-21–deficient mice. Consequently, Ly6Clow monocyte numbers were reduced in the atherosclerotic aorta because of increased susceptibility to apoptosis. However, miR-21 deficiency did not affect trafficking of Ly6Chigh monocytes nor their number in atherosclerotic aortas or the size of lesions but was associated with the presence of more pro-inflammatory macrophages in plaque, increased necrotic core, deficient efferocytosis, and increased macrophage death. This work reveals a role for miR-21 in atherosclerosis development and reveals the proof of concept that inhibiting miR-21 in monocytes might have relevant therapeutic application in atherosclerosis. However, many questions remain, including the target gene(s) that mediates the observed phenotype in ApoE−/− mice, whether miR-21 plays a role in Ly6Clow monocytes under non- ApoE−/− conditions, and which role it plays (if any) in the biology of Ly6Chigh monocytes. In addition, because miR-21 controls macrophage polarization, apoptosis, and efferocytosis (60), determining which of these functions is affected in the context of monocyte subsets would be of interest.
In 2018, the group of Eric Solary identified miR-150 as overexpressed in Ly6Clow monocytes as compared with Ly6Chigh monocytes and critical for promoting the terminal differentiation of classical monocytes into non-classical monocytes in both humans and mice (61). The authors found a defect of Ly6Clow monocytes in miR-150–deficient mice that did not affect the total number of monocytes in peripheral blood and bone marrow and was due to the un-repressed expression of Tet methylcytosine dioxygenase 3 (Tet3) gene. Tet3 is a dioxygenase that binds DNA and mediates demethylation but also promotes open chromatin independent of its catalytic function. Overall, Tet3 enhances transcription and gene expression, especially during changes in cellular identity (62). Thus, high Tet3 expression in Ly6Chigh monocytes due to low expression of miR-150 in this subset prevented their differentiation into Ly6Clow monocytes. This finding has important clinical implications because reduced expression of miR-150 was also found in peripheral-blood CD14+ monocytes, mostly classical monocytes, of patients with chronic myelomonocytic leukemia. Thus, monitoring the repartition of monocyte subsets is now an international recommendation as a diagnostic tool for patients with monocytosis to distinguish between chronic myelomonocytic leukemia and reactive monocytosis (63).
Conclusion
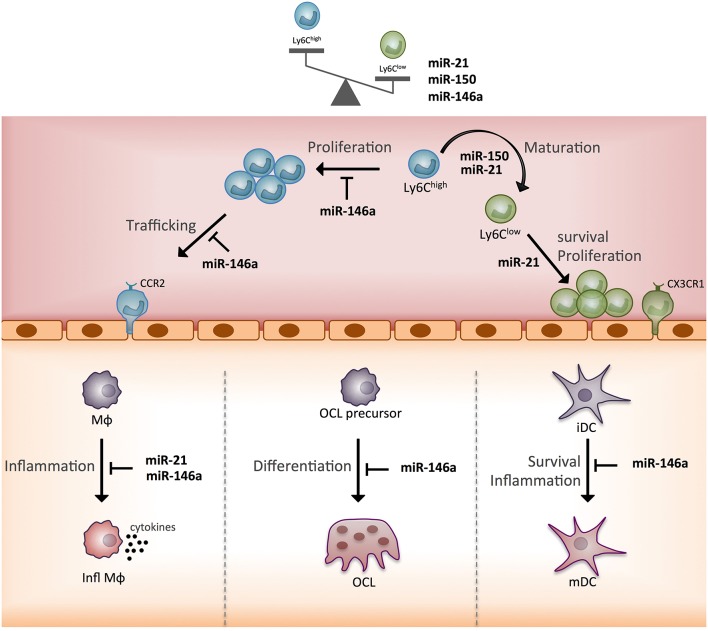
In this review, we give an overview of the state-of-the-art research of miRNAs that are differentially expressed between monocyte subsets and how they affect monocyte functional heterogeneity, with descriptions of functional and in silico studies of specific miRNAs. Three miRNAs miR-146a, miR-21, and miR-150 with differential expression in classical vs. non-classical monocyte subsets were all first identified as immune system regulators. The three miRNAs are inducible mediators of anti-inflammatory responses in the myeloid lineage acting via negative feedback loops and leading to the resolution of inflammation. Thus, they represent switches from pro- to anti-inflammatory responses of real therapeutic potential.
In terms of the functional heterogeneity of monocyte subsets, the few studies described bring new valuable insights into the role of the three miRNAs. Indeed, by identifying new target genes and functions that discriminate Ly6Clow and Ly6Chigh mouse monocytes, these studies helped specify miRNA-based mechanisms for the commitment of monocytes to a cellular subset fate and related functional specificity (Figure 5). Of note, miR-146a, miR-21, miR-150 all show greater expression in Ly6Clow rather than Ly6Chigh monocytes, and their expression is increased by inflammation. The reported role for miR-146a concerns mainly Ly6Chigh monocyte inflammatory functions, and that of miR-21 and miR-150 concerns the generation of Ly6Clow monocytes. However, so far, all studies have been performed with Ly6Chigh monocytes, thus future studies need to work with Ly6Clow monocytes to address the role of miRNAs in the function specific to Ly6Clow monocytes. In addition, the role of miRNAs in the commitment of monocyte subsets to OCs remains poorly explored and deserves further investigations (64).
Figure 5.
Schematic representation of the function of three monocyte subset-specific miRNAs. With the three miRNAs showing differential expression in monocyte subsets that were functionally studied in mouse models, we propose a scheme outlining their role. MΦ, macrophage; Infl MΦ, inflammatory macrophage; OC, osteoclast; iDC, immature dendritic cell; mDC, mature dendritic cell; CCR2, C-C chemokine receptor type 2; CX3CR1, CX3C chemokine receptor 1.
Finally, we must revisit this knowledge in light of recent works using single-cell RNA sequencing of human blood monocytes (65), high-dimensional mass cytometry (66), and ontogeny study (67), under steady state or pathological conditions (68), which broadens our perspectives by identifying new monocyte subsets and further underlines the control of monocyte plasticity by miRNAs and their target genes. Recently, 29 human immune cell types have been characterized by RNA sequencing and flow cytometry, and mRNA heterogeneity and abundance appeared to be cell type-specific (69). Most miRNAs act as rheostats, refining the expression of hundreds of genes to enhance cell differentiation. Thus, miRNA detection in single-cell monocytes is needed to understand the biology of the heterogeneity of monocytes and to propose new strategies for disease treatment and diagnosis.
Data Availability
The datasets analyzed for this study can be found in the NCBI gene expression omnibus, GSE137729 and GSE137730 for mouse and human data sets, respectively.
Ethics Statement
The studies involving human participants were reviewed and approved by the local human ethics committee (ID RCB 2008-A01087-48) and with the code of ethics of the world medical association. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
ID-R contributed to the design of the work, analysis and interpretation of the data, wrote the review, and designed the figures and tables. MR and CP contributed to the acquisition and analysis of the data and revised the manuscript. FA contributed to the conception of the work and interpretation of the data, wrote the review, and designed the figures. All authors gave approval for publication of the content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by INSERM (Institut National de la Santé et Recherche Médicale), the University of Montpellier, the Arthritis Fondation Courtin, the European Union (grant no. 115142), the French National Research Agency (ANR-16-CE14-0030-02), and the E-Rare-3 Joint Transnational Research Project on Rare Diseases INSAID that is a joint initiative between EC and ANR (ANR-15-RAR3-0013-05).
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. (2019) 20:5–20. 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. (2001) 294:853–8. 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. (2004) 14:1902–10. 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldrich P, Hsing YI, San Segundo B. Genome-wide analysis of polycistronic microRNAs in cultivated and wild rice. Genome Biol Evol. (2016) 8:1104–14. 10.1093/gbe/evw062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. (2004) 16:861–5. 10.1016/j.molcel.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 7.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. (2014) 42(Database issue):D68–73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. (2004) 32(Database issue):D109–11. 10.1093/nar/gkh023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3'UTRs. PLoS ONE. (2011) 6:e18067. 10.1371/journal.pone.0018067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. (2010) 42:1316–29. 10.1016/j.biocel.2009.09.016 [DOI] [PubMed] [Google Scholar]
- 11.Kehl T, Backes C, Kern F, Fehlmann T, Ludwig N, Meese E, et al. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget. (2017) 8:107167–75. 10.18632/oncotarget.22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. (2010) 33:375–86. 10.1016/j.immuni.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. (2009) 27:669–92. 10.1146/annurev.immunol.021908.132557 [DOI] [PubMed] [Google Scholar]
- 14.Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. Monocyte trafficking in acute and chronic inflammation. Trends Immunol. (2011) 32:470–7. 10.1016/j.it.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. (2005) 5:953–64. 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- 16.Seeling M, Hillenhoff U, David JP, Schett G, Tuckermann J, Lux A, et al. Inflammatory monocytes and Fcγ receptor IV on osteoclasts are critical for bone destruction during inflammatory arthritis in mice. Proc Natl Acad Sci USA. (2013) 110:10729–34. 10.1073/pnas.1301001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacome-Galarza CE, Lee SK, Lorenzo JA, Aguila HL. Identification, characterization, and isolation of a common progenitor for osteoclasts, macrophages, and dendritic cells from murine bone marrow and periphery. J Bone Miner Res. (2013) 2 8:1203–13. 10.1002/jbmr.1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. (2009) 325:612–6. 10.1126/science.1175202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Self-Fordham JB, Naqvi AR, Uttamani JR, Kulkarni V, Nares S. MicroRNA, dynamic regulators of macrophage polarization and plasticity. Front Immunol. (2017) 8:1062. 10.3389/fimmu.2017.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etzrodt M, Cortez-Retamozo V, Newton A, Zhao J, Ng A, Wildgruber M, et al. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. (2012) 1:317–24. 10.1016/j.celrep.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang TM, Wong WC, Ong SM, Li P, Lum J, Chen J, et al. MicroRNA expression profiling of human blood monocyte subsets highlights functional differences. Immunology. (2015) 145:404–16. 10.1111/imm.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zawada AM, Zhang L, Emrich IE, Rogacev KS, Krezdorn N, Rotter B, et al. MicroRNA profiling of human intermediate monocytes. Immunobiology. (2017) 222:587–96. 10.1016/j.imbio.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 23.Zawada AM, Schneider JS, Michel AI, Rogacev KS, Hummel B, Krezdorn N, et al. DNA methylation profiling reveals differences in the 3 human monocyte subsets and identifies uremia to induce DNA methylation changes during differentiation. Epigenetics. (2016) 11:259–72. 10.1080/15592294.2016.1158363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. (2013) 20:1603–14. 10.1038/cdd.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. (2009) 9:514–20. 10.1016/j.coph.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. (2008) 9:839–45. 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- 27.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. (2008) 132:875–86. 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. (2007) 9:775–87. 10.1038/ncb1613 [DOI] [PubMed] [Google Scholar]
- 29.Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α. J Allergy Clin Immunol. (2013) 132:426–36.e8. 10.1016/j.jaci.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang F, Zhao JL, Wang L, Gao CC, Liang SQ, An DJ, et al. miR-148a-3p mediates notch signaling to promote the differentiation and M1 activation of macrophages. Front Immunol. (2017) 8:1327. 10.3389/fimmu.2017.01327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschetti T, Kessler CB, Lee SK, Delany AM. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem. (2013) 288:33347–60. 10.1074/jbc.M113.484568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haneklaus M, Gerlic M, O'Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. (2013) 274:215–26. 10.1111/joim.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Rice KL, Buzzai M, Hexner E, Costa FF, Kilpivaara O, et al. miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia. (2013) 27:344–52. 10.1038/leu.2012.224 [DOI] [PubMed] [Google Scholar]
- 34.Su S, Zhao Q, He C, Huang D, Liu J, Chen F, et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun. (2015) 6:8523. 10.1038/ncomms9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou G, Xia J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res. (2018) 46:W514–22. 10.1093/nar/gky510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anbazhagan K, Duroux-Richard I, Jorgensen C, Apparailly F. Transcriptomic network support distinct roles of classical and non-classical monocytes in human. Int Rev Immunol. (2014) 33:470–89. 10.3109/08830185.2014.902453 [DOI] [PubMed] [Google Scholar]
- 37.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. (2018) 46:D239–45. 10.1093/nar/gkx1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. (2011) 39(Database issue):D691–7. 10.1093/nar/gkq1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imhof BA, Aurrand-Lions M. Adhesion mechanisms regulating the migration of monocytes. Nat Rev Immunol. (2004) 4:432–44. 10.1038/nri1375 [DOI] [PubMed] [Google Scholar]
- 40.Biro M, Munoz MA, Weninger W. Targeting Rho-GTPases in immune cell migration and inflammation. Br J Pharmacol. (2014) 171:5491–506. 10.1111/bph.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godek ML, Sampson JA, Duchsherer NL, McElwee Q, Grainger DW. Rho GTPase protein expression and activation in murine monocytes/macrophages is not modulated by model biomaterial surfaces in serum-containing in vitro cultures. J Biomater Sci Polym Ed. (2006) 17:1141–58. 10.1163/156856206778530731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elhabazi A, Marie-Cardine A, Chabbert-de Ponnat I, Bensussan A, Boumsell L. Structure and function of the immune semaphorin CD100/SEMA4D. Crit Rev Immunol. (2003) 23:65–81. 10.1615/CritRevImmunol.v23.i12.40 [DOI] [PubMed] [Google Scholar]
- 43.Lee S, Mattingly A, Lin A, Sacramento J, Mannent L, Castel MN, et al. A novel antagonist of p75NTR reduces peripheral expansion and CNS trafficking of pro-inflammatory monocytes and spares function after traumatic brain injury. J Neuroinflammation. (2016) 13:88. 10.1186/s12974-016-0544-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. (2003) 19:71–82. 10.1016/S1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- 45.Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. (2011) 12:778–85. 10.1038/ni.2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas GD, Hanna RN, Vasudevan NT, Hamers AA, Romanoski CE, McArdle S, et al. Deleting an Nr4a1 super-enhancer subdomain ablates Ly6C. Immunity. (2016) 45:975–87. 10.1016/j.immuni.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. (2006) 103:12481–6. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao JL, Rao DS, O'Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. Elife. (2013) 2:e00537. 10.7554/eLife.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ammari M, Presumey J, Ponsolles C, Roussignol G, Roubert C, Escriou V, et al. Delivery of miR-146a to Ly6C(high) monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics. (2018) 8:5972–85. 10.7150/thno.29313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. (2010) 285:20940–51. 10.1074/jbc.M110.115063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. (2013) 341:1483–8. 10.1126/science.1240636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S, et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity. (2017) 47:582–96.e6. 10.1016/j.immuni.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 53.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. FEBS J. (2011) 278:1167–74. 10.1111/j.1742-4658.2011.08042.x [DOI] [PubMed] [Google Scholar]
- 54.Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ Res. (2015) 117:e1–11. 10.1161/CIRCRESAHA.117.305844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. (2013) 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. (2011) 208:1189–201. 10.1084/jem.20101823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. (2011) 63:1582–90. 10.1002/art.30321 [DOI] [PubMed] [Google Scholar]
- 58.Presumey J, Courties G, Louis-Plence P, Escriou V, Scherman D, Pers YM, et al. Nicotinamide phosphoribosyltransferase/visfatin expression by inflammatory monocytes mediates arthritis pathogenesis. Ann. Rheum. Dis. (2013) 72:1717–24. 10.1136/annrheumdis-2012-202403 [DOI] [PubMed] [Google Scholar]
- 59.Chipont A, Esposito B, Challier I, Montabord M, Tedgui A, Mallat Z, et al. MicroRNA-21 deficiency alters the survival of Ly-6C. Arterioscler Thromb Vasc Biol. (2019) 39:170–7. 10.1161/ATVBAHA.118.311942 [DOI] [PubMed] [Google Scholar]
- 60.Sheedy FJ, Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. (2015) 6:19 10.3389/fimmu.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selimoglu-Buet D, Rivière J, Ghamlouch H, Bencheikh L, Lacout C, Morabito M, et al. A miR-150/TET3 pathway regulates the generation of mouse and human non-classical monocyte subset. Nat Commun. (2018) 9:5455. 10.1038/s41467-018-07801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Chen S, Zhang D, Shi Z, Li H, Zhao T, et al. Tet3-Mediated DNA demethylation contributes to the direct conversion of fibroblast to functional neuron. Cell Rep. (2016) 17:2326–39. 10.1016/j.celrep.2016.10.081 [DOI] [PubMed] [Google Scholar]
- 63.Selimoglu-Buet D, Wagner-Ballon O, Saada V, Bardet V, Itzykson R, Bencheikh L, et al. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood. (2015) 125:3618–26. 10.1182/blood-2015-01-620781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozano C, Duroux-Richard I, Firat H, Schordan E, Apparailly F. MicroRNAs, key regulators to understand osteoclast differentiation? Front Immunol. (2019) 10:375. 10.3389/fimmu.2019.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. (2017) 356:eaah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamers AAJ, Dinh HQ, Thomas GD, Marcovecchio P, Blatchley A, Nakao CS, et al. Human monocyte heterogeneity as revealed by high-dimensional mass cytometry. Arterioscler Thromb Vasc Biol. (2019) 39:25–36. 10.1161/ATVBAHA.118.311022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yáñez A, Coetzee SG, Olsson A, Muench DE, Berman BP, Hazelett DJ, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity. (2017) 47:890–902.e4. 10.1016/j.immuni.2017.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satoh T, Nakagawa K, Sugihara F, Kuwahara R, Ashihara M, Yamane F, et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature. (2017) 541:96–101. 10.1038/nature20611 [DOI] [PubMed] [Google Scholar]
- 69.Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carré C, et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. (2019) 26:1627–40.e7. 10.1016/j.celrep.2019.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed for this study can be found in the NCBI gene expression omnibus, GSE137729 and GSE137730 for mouse and human data sets, respectively.