ABSTRACT
Plant surface-localized pattern recognition receptors (PRRs) recognize pathogen- or damage-associated molecular patterns (PAMP/DAMPs) and activate pattern-triggered immunity (PTI). PRRs recruit receptor-like cytoplasmic kinases (RLCKs) to transduce the perceived signal to downstream signaling components. Brassinosteroid-signaling kinase 5 (BSK5) is a member of the RLCK XII subfamily and mutational analysis revealed its involvement in plant immunity. Here, we provide evidence that overexpression of BSK5 in transgenic Arabidopsis plants enhanced disease resistance to the bacterial pathogen Pseudomonas syringae and to the fungus Botrytis cinerea. Remarkably, upon treatment with the flg22, elf18 and pep1 PAMP/DAMPs, BSK5-overexpressing plants displayed higher levels of immune responses, including production of reactive oxygen species, callose deposition at the cell wall, and PATHOGENESIS-RELATED1 (PR1) gene expression. Together, these findings further substantiate the role of BSK5 in plant immunity and illustrate its potential use for improving plant disease resistance.
KEYWORDS: Arabidopsis thaliana, Botrytis cinerea, Pseudomonas syringae, BSK5, PAMP/DAMPs, PTI
Text
Plants possess a multilayered recognition system that detect invading pathogens.1 Early pathogen detection is performed by membrane-localized pattern recognition receptors (PRRs) that recognize pathogen- or damage-associated molecular patterns (PAMPs or DAMPs).1 Major examples of PRRs that recognize bacterial PAMPs are the Arabidopsis thaliana FLAGELLIN SENSITIVE2 (FLS2) receptor and the ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR), which bind the flg22 epitope of flagellin and the elf18 epitope of EF-Tu, respectively.2,3 Most extensively studied PRRs that perceive DAMPs are the PEP1 RECEPTOR1 (PEPR1) and PEPR2, which recognize the endogenous peptide pep1.4 PAMP/DAMP recognition by PRRs promptly triggers the activation of pattern-triggered immunity (PTI),5 which represents the first line of plant defense.5 Early PTI responses include production of reactive oxygen species (ROS), activation of mitogen-activated protein kinases (MAPKs), and deposition of callose at the plant cell wall.1,6 Late PTI responses include production of ethylene and salicylic acid and transcriptional reprogramming of a large number of defense-related genes.7–9 Collectively, these PTI responses defend plants against the invading pathogen. PRRs recruit receptor-like cytoplasmic kinases (RLCKs) for linking ligand perception and downstream signaling.6,10
Arabidopsis brassinosteroid signaling kinases (BSKs) belong to the RLCK subfamily XII that includes 12 members (BSK1–BSK12).11,12 BSKs contain an N-terminal kinase domain and a C-terminal tetratricopeptide repeats domain.11 Besides their established role in brassinosteroid signaling and growth,11,12 recent investigation indicates that BSKs are also involved in plant immunity. BSK1 interacts with FLS2 and is required for flg22-induced ROS burst and MAPK signaling.13,14 BSK3 and BSK8 have been detected in PRR protein complexes.15,16 In a recent study, we investigated the role of Arabidopsis BSK5 in PTI.17 BSK5 interacted with multiple RLKs including the EFR and PEPR1 PRRs in a direct and specific manner. PEPR1 and EFR phosphorylated BSK5 suggesting that BSK5 acts as a signaling component downstream of these PRRs. Phenotypic and genetic analyses revealed that a bsk5 loss-of-function mutation causes increased susceptibility to both the biotrophic bacterium Pseudomonas syringae and the necrotrophic fungus Botrytis cinerea. Consistently, bsk5 mutant plants displayed reduced accumulation of ROS, callose, and PATHOGENESIS-RELATED1 transcripts. Phosphorylation by PRRs, kinase activity, and localization to the cell periphery were shown to be important for BSK5 immune function.17
To expand the investigation on the function of BSK5 in plant immunity, we used Agrobacterium-mediated transformation protocols to generate Arabidopsis transgenic plants of the Columbia-0 (Col-0) line that express BSK5 fused to a C-terminal HA tag and driven by the CaMV 35S promoter.18 A total of 11 independent transgenic lines were obtained and five transgenic homozygous lines were selected for further analysis. The accumulation of the BSK5-HA fusion protein was detected in all five overexpression lines (OE1-OE5) using anti-HA antibodies (Figure 1A). We then tested the susceptibility of transgenic Arabidopsis lines overexpressing BSK5 (BSK5 OE1-OE5) to the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 (Pst) and to the fungus B. cinerea. Five-week-old Arabidopsis Col-0 wild-type and BSK5 OE1-OE5 plants were infected either by infiltrating leaves with a Pst bacterial suspension (1 × 105 CFU/mL) or by placing a droplet of B. cinerea spore suspension (5 × 105 conidia/mL) on the leaves. Pst bacterial populations were determined in leaf tissues sampled at 0 and 4 days post-inoculation (dpi). In plants inoculated with B. cinerea, lesion size was measured at 3 dpi. All five BSK5 OE lines displayed a significantly lower growth of Pst bacteria and smaller B. cinerea-induced lesions as compared to wild-type plants (Figure 1B, C). These results indicate that overexpression of BSK5 in Arabidopsis enhanced immunity to both pathogens.
Figure 1.
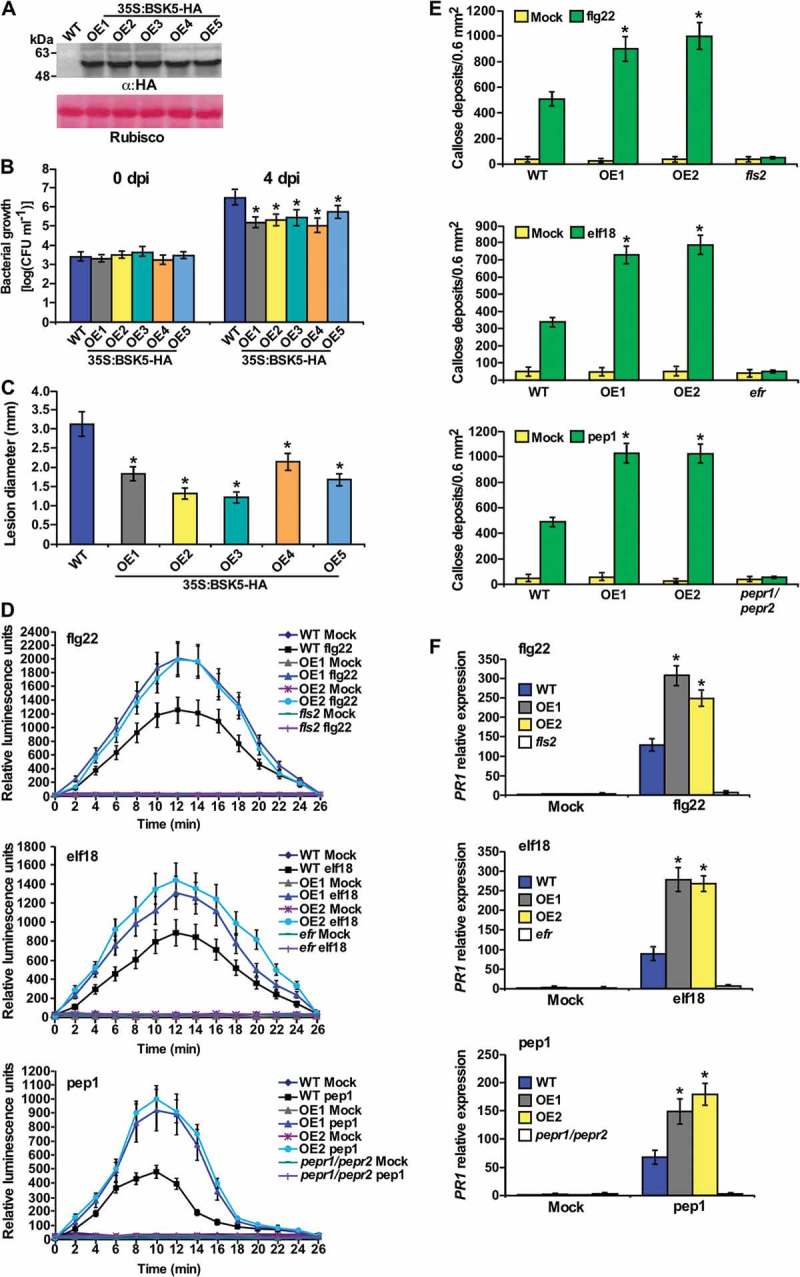
Enhanced resistance and PTI responses in plants overexpressing BSK5.
(A) Western blot analysis of BSK5-HA in total protein extracts of five BSK5 overexpression lines (OE1-OE5) performed with anti-HA antibodies (α:HA). Ponceau S staining of Rubisco is shown as a loading control. (B) Leaves of wild-type, and OE1-OE5 plants were inoculated by infiltration with a suspension of Pst (1x105 CFU/mL). Bacterial growth was measured at 0 and 4 dpi. Data are means ± SE of three biological replicates each including five plants. (C) Leaves of wild-type and OE1-OE5 plants were droplet-inoculated with a suspension of B. cinerea spores (5 x 105 conidia/mL). The size of disease lesions was measured at 3 dpi. Data are means ± SE of three biological replicates each including five plants. (D) ROS production. Leaf disks from plants of the indicated genotypes were treated with flg22 (100 nM), elf18 (100 nM), pep1 (1 μM) or water, and incubated with luminol and horseradish peroxidase. Luminescence was measured as relative luminescence unit for 26 min after treatment every 2 min. Data are means ± SE of three biological repeats each including ten samples. (E) Callose deposition. Leaves were treated with 1 μM flg22, elf18 and pep1 or water, and samples were collected 16 h later. Callose deposits were visualized by fluorescence microscopy and counted. Data are means ± SE of four biological replicates each with five leaves. (F) PR1 mRNA expression. Leaves were sprayed with 100 nM flg22, elf18 and pep1 or water. After 12 h, PR1 mRNA levels were measured by RT-qPCR analysis relative to wild-type mock-inoculated plants. ACTIN2 was used as normalizer. Data are means ± SE of three biological repeats. In B-F, asterisks indicate a significant difference (Student’s t test, P value < .05) compared to wild-type plants.
Furthermore, we analyzed PTI responses in the BSK5 overexpression lines OE1 and OE2. Interestingly, both lines did not exhibit constitutive ROS accumulation nor callose deposition (Figure 1D, E), while higher ROS accumulation and callose deposits were observed in OE1 and OE2 compared to the wild-type plants upon treatment with the PAMPs flg22 and elf18, or the DAMP pep1 (Figure 1D, E). The fls2, efr, and pepr1/pepr2 mutants were used as controls in these experiments. As expected, the fls2, efr, and pepr1/pepr2 mutants were unable to respond to flg22, elf18, and pep1, respectively, and did not accumulate ROS or callose (Figure 1D, E). We then tested the expression pattern of the defense related gene PR119 in BSK5 overexpression lines treated with PAMP/DAMPs. Constitutive upregulation of PR1 was not observed in OE1 and OE2 lines, but PR1 expression levels were significantly enhanced in these lines upon flg22, elf18 or pep1 treatment (Figure 1F). These results suggest that BSK5 positively regulates PTI responses.
In conclusion, our study demonstrates that BSK5 acts as a positive regulator of immunity against Pst and B. cinerea in Arabidopsis through the activation of PTI responses upon PAMP/DAMP perception. Future studies should determine how BSK5 regulates immune signal transduction. We hypothesize that this can be achieved by promoting either phosphorylation of the NADPH oxidase RESPIRATORY BURST HOMOLOG PROTEIN D (RBOHD) or the activity of calcium-dependent protein kinases that directly influence ROS production. Future identification of BSK5 interacting partners and substrates will shed light on PTI signaling pathways and enhance our understanding of their activation mechanisms. Potentially, the BSK5 and its homologs from other plant species may serve as tools to enhance plant disease resistance to bacterial and fungal pathogens.
Funding Statement
This work was supported by the Israel Science Foundation (ISF) under grant 309/15, the United States-Israel Binational Agricultural Research and Development Fund (BARD) under grant no. IS-4931-16C, and the European Cooperation in Science and Technology (COST) under grant EuroXanth CA16107 from the European Union.
References
- 1.Boller T, Felix G.. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T.. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02437. [DOI] [PubMed] [Google Scholar]
- 3.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 4.Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KAS, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285:13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05376. [DOI] [PubMed] [Google Scholar]
- 6.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 7.van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 9.Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant. 2015;8:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Liang X, Zhou J-M. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol. 2018;69:267–299. doi: 10.1146/annurev-arplant-042817-040540. [DOI] [PubMed] [Google Scholar]
- 11.Tang W, Kim T-W, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang Z-Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, et al. BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 2013;74:905–919. doi: 10.1111/tpj.12175. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25:1143–1157. doi: 10.1105/tpc.113.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan H, Zhao Y, Shi H, Li J, Wang Y, Tang D. BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 2018;176:2991–3002. doi: 10.1104/pp.17.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P, Xu S-L, Li Z-J, Tang W, Burlingame AL, Wang Z-Y. A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol Plant. 2014;7:441–444. doi: 10.1093/mp/sst105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Tsuda K, Glazebrook J, Katagiri F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol. 2011;12:702–708. doi: 10.1111/j.1364-3703.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majhi BB, Sreeramulu S, Sessa G. BRASSINOSTEROID-SIGNALING KINASE5 associates with immune receptors and is required for immune responses. Plant Physiol. 2019;180:1166–1184. doi: 10.1104/pp.18.01492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Henriques R, Lin -S-S, Niu Q-W, Chua N-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 19.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]


