ABSTRACT
Jasmonic acid (JA) modulates plant development, growth, and responses to stress. Previously, we showed that in Arabidopsis thaliana, JA promotes the formation of extra xylem in roots, and mutant plants unable to express PIN-FORMED 3 (PIN3) and PIN7 formed extra xylem in the absence of exogenous JA. Those results suggested that JA modulates root xylem development by controlling PIN-mediated polar auxin transport. Consistent with this, treatment with an auxin transport inhibitor induced extra xylem formation. Here, we characterized the expression of PIN3 and PIN7 in JA-treated Arabidopsis plants. PIN3 expression was not altered in response to JA; by contrast, PIN7 expression was reduced by JA, which suggested that PIN7 is involved in JA-mediated xylem development. Indeed, overexpressing PIN7 suppressed the formation of extra xylem in response to JA. Based on these results, we propose that JA mediates xylem development by controlling polar auxin transport with PIN7 critically involved in this process.
KEYWORDS: Jasmonic acid, xylem, cytokinin, auxin, polar auxin transport, PIN-formed proteins
Text
Auxin is a key phytohormone regulating various aspects of plant development including xylem development. The identification of the auxin carrier proteins including PINs and AUXs revealed that polar auxin transport provides crucial positional information for the specification of cells and tissues.1–3 Xylem cells in vascular tissues show high auxin responses compared with other cells, and the auxin signaling-defective mutant axr3-1 produces a no-xylem phenotype, demonstrating the essential role of auxin in xylem development.4,5 This suggests that xylem-specific auxin accumulation produces the responses in xylem, consistent with the recent finding that inhibition of polar auxin transport affects xylem-specific auxin accumulation and xylem development.6
JA regulates plant defenses against biotic and abiotic stresses.7 JA is synthesized from linolenic acid via the octadecanoid pathway, after which it is metabolized into an isoleucine conjugate (JA-Ile).8,9 JA-signaling pathways are activated by the interaction between JA-Ile and the CORONATINE INSENSITIVE1 (COI1) receptor, which prompts proteolysis of transcriptional repressor JASMONATE ZIM-DOMAIN proteins.10,11 JA vitally underpins many plant responses to biotic and abiotic stresses and also modulates plant growth and development, strongly suggesting that JA is essential for coordinating development and stress responses in plants.12–14
Our prior work revealed that JA promotes the formation of extra xylem in Arabidopsis15,16 (Figure 1). Wild-type plants form xylem cells in a single axis in the middle of the vasculature; by contrast, JA-treated wild-type plants produce extra xylem cells adjacent to the axis. Recently, we observed the extra-xylem phenotype in mutant plants that lacked expression of PIN3 and PIN7.6 PIN3 and PIN7 are responsible for polar auxin transport from the procambium to the xylem;5 this suggested that JA modulates xylem development by controlling auxin movement. Consistent with this, the PIN3 and PIN7 expression responsible for xylem patterning is activated by cytokinin (CK), yet JA antagonistically interacts with CK to modulate xylem development.5,6,15 Therefore, CK-responsive PIN3 and PIN7 expression could be involved in JA-mediated xylem development in plants.
Figure 1.
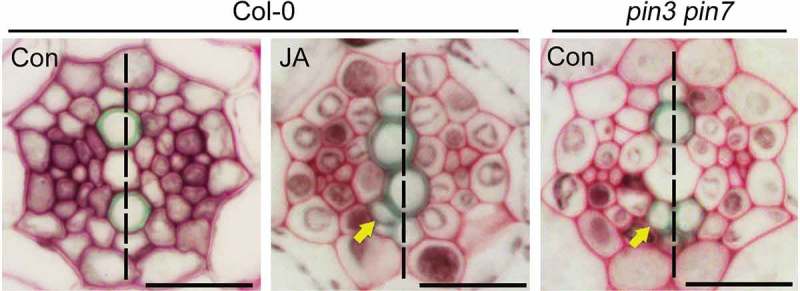
JA and knockout-out mutation of PIN3 and PIN7 promoted the formation of extra xylem cells.
Cross-sectional images showing the extra xylem formed in the roots of Col-0 plants grown in 10 μM of methyl jasmonate (JA) for 7 days unlike those of JA-untreated Col-0 plants (Con). The mutant plants lacking expression of PIN3 and PIN7 (pin3 pin7) formed extra xylem even under JA-untreated conditions. Arrows and dashed lines respectively indicate the formation of extra xylem and the xylem axis. Scale bar = 20 μm.
To investigate the involvement of PIN3 and PIN7 in JA-mediated xylem development, we analyzed changes in CK responses and PIN3 and PIN7 expression in response to JA (Figure 2). When the CK response was visualized in Arabidopsis plants expressing the CK-responsive marker ARR5::GFP, JA reduced the CK response in the vasculature and columella (Figure 2a). To analyze changes in PIN3 and PIN7 expression in response to JA, we performed qRT-PCR using total RNA extracted from these plants. The PIN7 expression level in JA-treated plants was approximately 3-fold lower than that in untreated plants (Figure 2b). In contrast to PIN7, expression levels of PIN3 were similar between JA-untreated and -treated plants, indicating that JA has a negligible effect on PIN3 expression. The JA effect on PIN7 expression was also observed in wild-type plants (Figure 2c). JA-treated wild type exhibited reduced expression of PIN7 compared to the JA-untreated wild type, suggesting that PIN7 is involved in JA-mediated xylem development.
Figure 2.
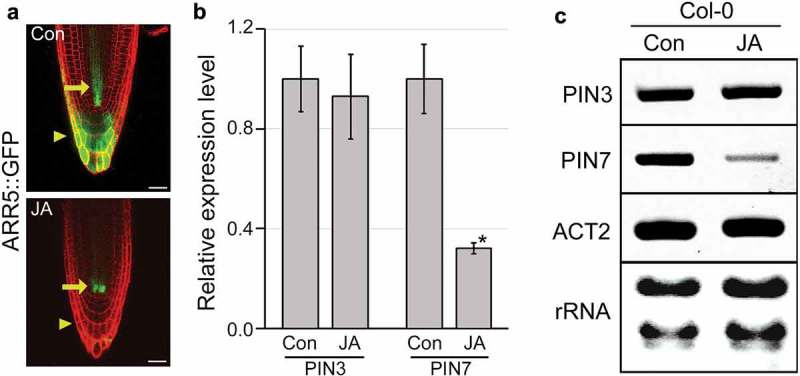
Suppression of PIN7 expression by JA.
(a) Images showing the distribution and intensity of GFP signals in ARR5::GFP transgenic plants grown in JA-untreated (Con) and -treated conditions (JA) for 5 days (JA, 10 μM of methyl jasmonate). (b) Expression levels of PIN3 and PIN7 genes in the ARR5::GFP transgenic plants. Data represent mean values of three technical replicates, and error bars indicate SD. Asterisks show statistically significant differences between JA-untreated and -treated samples (p-value < 0.01, Student’s t-test). GAPDH (At1G13440) was used as an internal control to normalize gene expression. (c) Semi-quantitative RT-PCR results showing that expression levels of PIN3 and PIN7 in wild-type plants (Col-0) grown in JA-untreated and -treated conditions (10 μM of methyl jasmonate) for 9 days. ACT2 (At3G18780) was used as an internal control. Scale bar = 20 μm.
Based on the evidence that JA reduces expression of PIN7, we hypothesized that plants overexpressing PIN7 would not show JA-induced development of extra xylem. To test this, we quantified formation of extra xylem in PIN7-overexpressing plants (35S::PIN7) grown in JA-untreated and -treated conditions (Figure 3). Approximately 15% of these plants we tested displayed the extra-xylem phenotype, a proportion approximately 4-fold lower than that of wild-type plants grown under the same conditions (n > 20). This indicates that JA regulates xylem development by controlling polar auxin transport, in a process mediated by PIN7. This crucial role of PIN7 in xylem development is consistent with the findings of Murano et al. (2014), whose computational study showed that PIN7 activity was capable of establishing the xylem-specific auxin accumulation responsible for xylem development, by directing polar auxin transport from the procambium to xylem cells.17 Taken together, these findings suggest that PIN7 mediates JA-dependent xylem development by controlling polar auxin transport to xylem structures.
Figure 3.
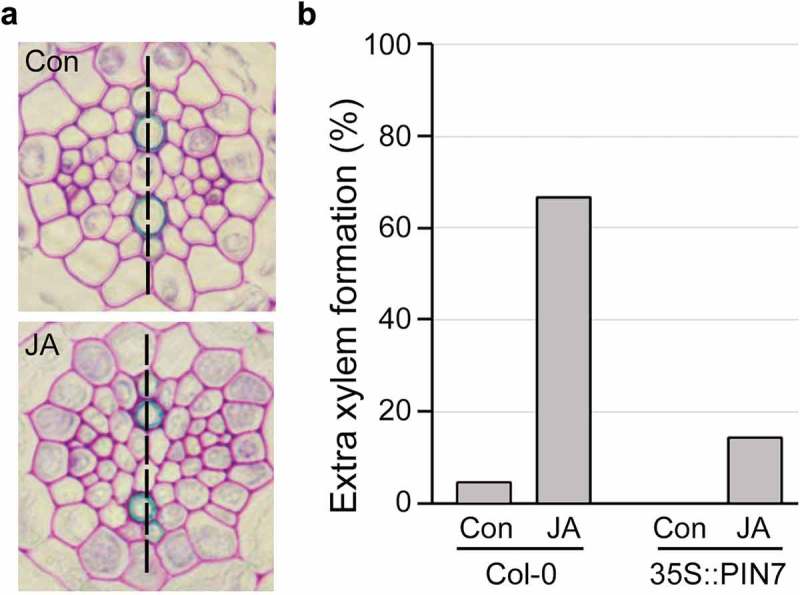
Overexpression of PIN7 suppressed the JA-mediated extra-xylem phenotype.
Cross-sectional images of the roots of 35S::PIN7 grown in JA-untreated (Con) and -treated conditions (JA) for 7 days (JA, 10 μM methyl jasmonate). (b) Quantification of extra xylem formation in these plants. Percentages were calculated by dividing the number of plants with extra xylem by total number of plants observed (n > 20). Scale bar = 20 μm.
Modulation of plant development under stress conditions largely occurs through the interaction between hormones that mediate plant developmental process and stress response. Growing numbers of studies have proposed that JA extensively interacts with other phytohormones to regulate plant development under stress conditions. For example, JA interacts with gibberellic acid in the coordination of plant development and stress responses.18–20 Based on our results presented here, we propose that JA interacts with auxin and CK to modulate xylem development. Further molecular and genetic studies will enhance our understanding of the molecular mechanisms underlying JA-mediated xylem development and the regulatory interactions between JA and other key phytohormones.
Funding Statement
This work was supported by the Rural Development Administration (ROK) [PJ01364301]; National Research Foundation (ROK) [NRF-2019R1A2C1007103]; Rural Development Administration (ROK) [PJ01323901].
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01364301 and PJ01323901)” Rural Development Administration, Republic of Korea and the National Research Foundation of Korea grant funded by the Korean Government (MOE) [NRF-2019R1A2C1007103].
Abbreviations
| JA | jasmonic acid |
| CK | cytokinin, PIN, pin-formed protein |
| GFP | green fluorescent protein |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G.. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:1–4. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 2.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. [DOI] [PubMed] [Google Scholar]
- 3.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 4.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Jang G, Lee S, Chang SH, Kim J-K, Do Choi Y. Jasmonic acid modulates xylem development by controlling polar auxin transport in vascular tissues. Plant Biotechnol Rep. 2018;12:265–271. doi: 10.1007/s11816-018-0491-x. [DOI] [Google Scholar]
- 7.Creelman RA, Mullet JE. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Zitzmann N, Deane C, Ohkura H, Wakefield JG. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chini A, Fonseca S, Fernandez G, Adie B, Chico J, Lorenzo O, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011;23:3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, Xiao L-T, Sun T-P, Li J, Deng X-W, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang G, Chang SH, Um TY, Lee S, Kim J-K, Do Choi Y. Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Sci Rep. 2017;7:10212. doi: 10.1038/s41598-017-10634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang G, Choi YD. Drought stress promotes xylem differentiation by modulating the interaction between cytokinin and jasmonic acid. Plant Signal Behav. 2018;13:e1451707. doi: 10.1080/15592324.2018.1451707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muraro D, Mellor N, Pound MP, Lucas M, Chopard J, Byrne HM, Byrne HM, Godin C, Hodgman TC, King JR, et al. Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc Natl Acad Sci USA. 2014;111:857–862. doi: 10.1073/pnas.1221766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J, Copenhaver GP. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Um TY, Lee HY, Lee S, Chang SH, Chung PJ, Oh K-B, Kim J-K, Jang G, Choi YD. JASMONATE ZIM-DOMAIN PROTEIN 9 interacts with SLENDER RICE 1 to mediate the antagonistic interaction between jasmonic and gibberellic acid signals in rice. Front Plant Sci. 2018;9:1866. doi: 10.3389/fpls.2018.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]


