ABSTRACT
Piriformospora indica, an endophytic fungus of Sebacinales, has a wide host range and promotes the performance of mono- and eudicot plants. Here, we compare the interaction of P. indica with the roots of seven host plants (Anthurium andraeanum, Arabidopsis thaliana, Brassica campestris, Lycopersicon esculentum, Oncidium orchid, Oryza sativa, and Zea mays). Microscopical analyses showed that the colonization time and the mode of hyphal invasion into the roots differ in the symbiotic interactions. Substantial differences between the species were also observed for the levels and accumulation of jasmonate (JA) and gibberellin (GA) and the transcript levels for genes involved in their syntheses. No obvious correlation could be detected between the endogenous JA and/or GA levels and the time point of root colonization in a given plant species. Our results suggest that root colonization strategies and changes in the two phytohormone levels are highly host-specific.
KEYWORDS: Piriformospora indica, jasmonate, gibberellin, ethylene, phytohormone, symbiosis
Introduction
Piriformospora indica, an endophytic fungus of Sebacinales, was first isolated from the rhizosphere of two woody shrubs in the Indian Thar Desert.1,2 The root-colonizing fungus has a broad host range, which includes bryophytes, pteridophytes, gymnosperms, and all tested monocot and eudicot plants.3–13 P. indica colonizes the roots, grows inter- and intracellularly, forms pear-shaped and auto-fluorescent spores in the cortex and rhizosphere zone, but does not enter the endodermis and aerial parts of the plants.2 Root colonization does not cause pathogenic symptoms in the roots and aerial parts of the plants. Conversely, P. indica improves nutrition uptake into the host plant,14–17 promotes biomass production,7,18–20 induces early flowering19,20 and stimulates the accumulation of secondary metabolites.17,21 Furthermore, the endophyte also enhances the plant´s resistance to biotic and abiotic stresses such as pathogens, insects, nematodes, high-temperature stress, salinity, drought, and radiation.4,19,22–24
Because of these benefits, many researchers use P. indica as a model system to understand the molecular, biochemical and cellular mechanisms leading to a beneficial symbiotic interaction.25–31 However, huge differences in the responses to P. indica are described for different plant species, such as Chinese cabbage,4 maize,17 orchid,31 tomato,3 Arabidopsis7 or barley.32 Furthermore, the interaction of P. indica with A. thaliana or barley roots results in a quite different regulation of genes involved in phytohormone metabolism,33,34 and the role of phytohormones in the symbiotic interaction is a matter of debate.4,34–36 The broad host range of P. indica suggests that the mutualist has evolved highly effective interaction and colonization strategies. Therefore, it is conceivable that the fungus recruits similar or identical hormone and signaling pathways from the plants for the establishment of a successful symbiosis. Successful root colonization requires also the repression of the host innate immunity.26 Beneficial microbes manipulate the levels of the plant hormones jasmonic acid (JA), abscisic acid (ABA), salicylic acid (SA), gibberellic acid (GA) and ethylene (ET) in order to balance innate immune responses and the progression of root colonization. The quite different reports on the involvement of hormones in the colonization process in different host plants make it difficult to understand which primary targets are used by P. indica. Furthermore, the fungus protects Arabidopsis seedlings against pests, such as powdery mildew via JA, but not SA signaling.37 Zarea et al.,38 as well as Molitor et al.5 reported that P. indica induces different biotic and abiotic stress tolerance and hormone responses in different host species. Barley plants impaired in GA synthesis and perception showed reduced mutualistic colonization, which was associated with an elevated expression of defense-related genes. GA and ET are also recruited by P. indica for correct root colonization and growth promotion in Arabidopsis.26,33,34,36,39,40 These often conflicting reports raise the question whether the described differences are caused by different experimental conditions or reflect differences in the host response to the endophyte. The goal of this study was to compare the interaction of P. indica with different host species under comparable growth conditions. We focus on root colonization, changes in the phytohormone levels and a few phytohormone-related mRNA levels to understand principles in the symbiotic interaction of the fungus with different host species.
Results
P. indica colonizes the roots of different plant species at different time points and with different colonization strategies
In order to investigate how P. indica colonizes the roots of different plant species, the efficiency and velocity of root colonization of seven different plant species were compared. The roots were exposed to the same amount of fungal hyphae, and the distance between the hyphal plaque and the roots of the maturation zone was the same for all plant species.41 Progression of colonization was detected microscopically (Figure 1) and by qPCR analyses (Figure 2). Microscopic observations were used to monitor the invasion and propagation of hyphae in the roots, and to detect the earliest root colonization time point and number of hyphae that enters the root cells. Criteria for a successful interaction were the initiation of root hair development after hyphae were detected around the primary and lateral roots. Following this, we monitored the progression of hyphal growth that started to invade between individual epidermal cells. Finally, root cells were scored based on the enlarged cortex and thick epidermal and cortex layers. According to Figure 1 and Table 1, extracellular colonization of A. thaliana roots is already visible 2 days after infection (dai). At this time point, hyphae can be detected around the primary and lateral roots and started to invade into the root tissue. The hyphae can be detected between individual epidermal cells but did not invade the root cells yet. Only 4 dai, we observe a few roots cells in which hyphae are invaded. In contrast to Arabidopsis, only 3 dai, the first hyphae are visible around the epidermal cells of B. campetris and Oncidium orchid, and single rhizodermal cells have already penetrated into the cells of the host roots without the formation of specific penetration organs (Figure 1 and Table 1). Therefore, while root colonization and the rarely occurring invasion of the hyphae into the root cells can be separated temporarily in Arabidopsis, it appears that in B. campetris and Oncidium orchid roots, hyphal invasion into root cells occurs almost immediately after the contact is established. Lycopersicon esculentum and Oryza sativa cv. Japonica appear to follow the Arabidopsis colonization strategy, and we observed first the formation of a hyphal network on the root surface and between the epidermal cells. The first hyphae which invaded the root cells were only detected one day later. The establishment of the physical contact between hyphae and roots of L. esculentum and O. sativa cv. Japonica is slower, and the first hyphae attached to their root surfaces can only be detected 4 dai. Immediately after the attachment, the hyphae start to form intra-cellular hyphae in L. esculentum roots, whereas in case of O. sativa cv. Japonica, the vast majority of the P. indica mycelium remains on the root surface (Figure 1 and Table 1). Entry into the root cells is rare. Thus, the time points of extracellular root colonization, invasion into the root tissue and entry into root cells appear to be quite different for the different plant species: e.g., 4 dai, when Arabidopsis, B. campestris and Oncidium orchid roots are already massively colonized, the colonization of L. esculentum and O. sativa cv. Japonica roots is still developing. The colonization process for Zea mays plants is even slower. Three dai, no physical contact between the hyphae and roots can be detected yet. Having established the contact, a rapid progression of the symbiotic interaction is visible. Between the 5th and the 6th day, a massive propagation of hyphae in the intracellular space of the roots is visible, which is also accompanied by the entry of hyphal inside of the Z. mays root cells (Figure 1 and Table 1). Thus, compared to the other described plant species, the establishment of the symbiotic interaction starts later but progresses fast once the contact is established. At the 6th day, the massive growth of the fungus results in a hyphal network that is found throughout the analyzed root sections, and such a massive hyphal growth within 2 days after the establishment of the first contact was never observed for the other plant species. Figure 1(f) demonstrates that the colonization pattern of the maize roots is quite different from the patterns observed for the other plant species, while hyphae can be detected almost everywhere in the maize roots, the fungal propagation in the other species is more focused on specific root sections which are highly colonized while other sections remain initially uncolonized.
Figure 1.
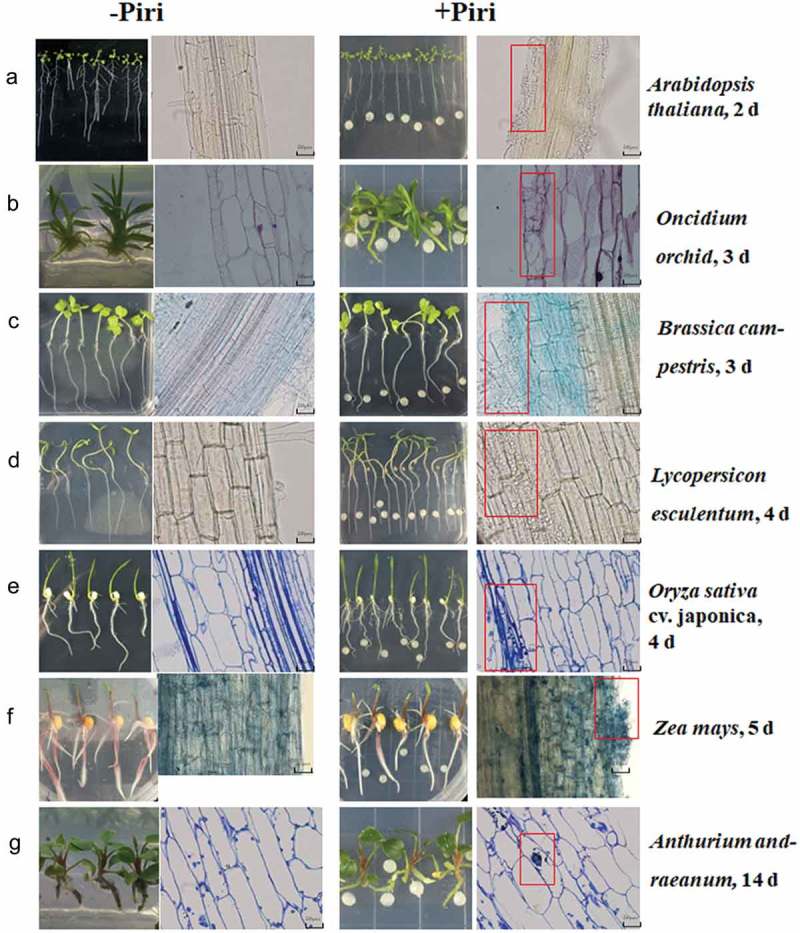
Microscopic detection of root colonization of different host species by P. indica. Root segments, starting 3 cm away from the root tip, were cut into 2 cm sections for microscopic analyses. Left panel: mock treatment, right panel: co-cultivation with P. indica. The pictures show root section from the seven investigated plant species stained with lactophenol or trypan blue between 2 and 14 dai. (a) A. thaliana, 2 dai; (b) Oncidium orchid, 3 dai; (c) Brassica campestris, 3 dai; (d) Lycopersicon esculentum, 4 dai; (e) Oryza sativa cv. japonica, 4 dai; (f) Zea mays, 5 dai; (g) A. andraeanum, 14 dai. Bar = 20 um. Red boxes highlight sections colonized by P indica in root tissues.
Figure 2.
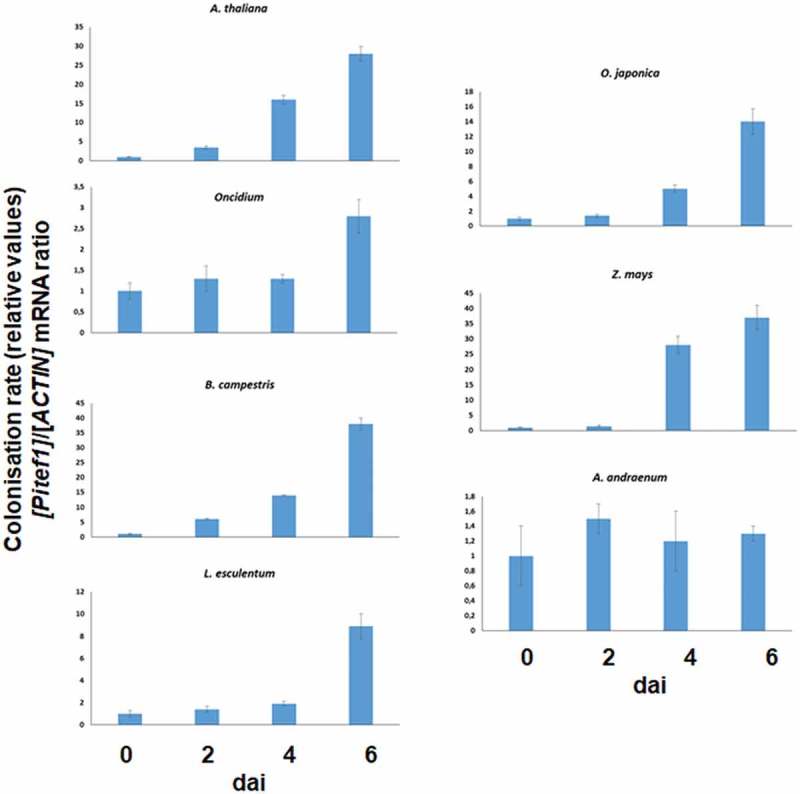
qPCR analyses for root colonization of different host species by P. indica. The plants were co-cultivated with P. indica for 0, 2, 4 and 6 days and the relative colonization rate determined as described in the Methods and Material chapter. The data are based on three independent experiments, and bar represents SEs. The values for the time points 0 were set as 1.0, and the other values were expressed relative to them.
Table 1.
Earliest time point in days at which a successful colonization of the roots of various plant species by P. indica can be detected.
| Plant type | Species | Successful colonization |
|---|---|---|
| Eudicot | Arabidopsis thaliana | 2 dai |
| Monocot | Oncidium orchid | 3 dai |
| Eudicot | Brassica campestris | 3 dai |
| Eudicot | Lycopersicon esculentum | 4 dai |
| Monocot | Oryza sativa cv. Japonica | 4 dai |
| Monocot | Zea mays | 5 dai |
| Monocot | Anthurium andraeanum | 14 dai |
Within the first 6 dai with P. indica, fungal sporulation could not be seen in any of the six host plants described so far. Furthermore, during the entire period (6 days), we did not observe significant changes in growth rates, leaf sizes, or morphology of the areal parts of the investigated plant species between P. indica-colonized and mock-treated plants. The first morphological changes, that can be detected, occur between 5 and 7dai, and the roots become visibly bushier. This phenotypic development is again very host-specific. While the bushy phenotype is most obvious for Chinese cabbage, it is less developed in Arabidopsis, followed by maize, rice, tomato and finally Oncidium. Apparently, the velocity of root colonization does not translate into corresponding alterations in root development. Finally, the symbiotic interaction between P. indica and Anthurium andraeanum starts only 14 dai. Once the hyphae reach the roots, a hyphal network around the roots and between the epidermal and subepidermal root cells become rapidly established, and the first few hyphae can be detected in the root epidermis of the host, almost immediately after the colonization occurred (Figure 1 and Table 1). Within the experimental period of time, we did not observe any visible alterations of the root phenotype by the fungus. These results demonstrate that the onset of root colonization (hyphae found on the surface of the primary and secondary roots as well as root hairs, entry of the hyphae into the root tissue, and occasional entry of the hyphae into the root cells) and reprogramming of root development by the fungus are quite different among the seven plant species.
Quantification of root colonization using the fungal RNA for the translation elongation factor 1 (Pitef1) relative to the plant RNA for actin basically confirms these results, although the differences observed by microscopy cannot be resolved by this technique (Figure 2). Overall, except for A. andraeanum, we observed an increase in the fungal RNA within the first 6 dai. As expected the kinetics are different for the seven plant species tested.
The JA and GA levels in the roots of the hosts respond differently to root colonization by P. indica
Previous studies have shown that symbiotic interactions are often associated with changes in the host’s phytohormone levels, including JA and GA. In order to test whether co-cultivation of the roots of the different plant species affects the host´s phytohormone levels, the JA and GA concentrations were determined in the roots of P. indica-exposed plants 0, 2, 4, and 6 dai.
Interestingly, we observed remarkable differences among the seven plant species (Figure 3). In A. thaliana, the JA levels remained low, while the GA level strongly increased 2 dai and dropped again until the end of experimentation. In Chinese cabbage roots, we observed an increase in the JA level which reached its maximum 6 dai, while the GA level remained more or less constant over the period of experimentation. In Anthurium roots, the JA level did not respond to P. indica, while a strong, more than 100-fold increase was observed for the GA level 6 dai. The JA level in rice roots increased 3–5 times 2 and 4 dai and dropped again, while the GA level did not change at all. In Oncidium roots, the JA level initially dropped, but the hormone started to re-accumulate at the end of the experiment, while the GA level is initially high and decreased dramatically 4 and 6 dai. In tomato roots, both phytohormones started to accumulate at the end of the experiment, while in maize roots, only the GA, but not the JA level showed a transient, but significant increase 2 and 4 dai. In addition, linear regression analysis showed that there is a negative correlation between P. indica colonization period and the JA hormone level for Chinese cabbage (R2 = 0.21, p> .05). However, the relationship between P. indica colonization and JA was positively correlated for Arabidopsis (R2 = 0.024, p > .05) and tomato (R2 = 0.21, p> .05). Similarly, for monocots, only maize (R2 = 0.025) showed a positive correlation for P. indica colonization (Figure 4). Interestingly, a positive correlation was noted for the GA levels and root colonization for dicots (Arabidopsis (R2= 0.001, p > .05), Chinese cabbage (R2= 0.046, p > .05) and tomato (R2= 0.16, p > .05), although the correlation was weak in significance. For the monocots Oncidium (R2= 0.051, p > .05), rice (R2= 0.038, p < .05), maize (R2= 0.075, p < .05) and Anthurium (R2= 0.071, p < .05)), the correlation was negative for P. indica colonization, but the analyses showed a strong significance (Figure 4). Based on these results, it is obvious that different plants show quite different responses in the regulation of their phytohormone levels in response to P. indica colonization. An obvious correlation between root colonization and changes in the phytohormone levels has been detected for the individual hosts, and it appears that the fungus has to cope with quite different JA and GA levels in the roots of the different host species.
Figure 3.
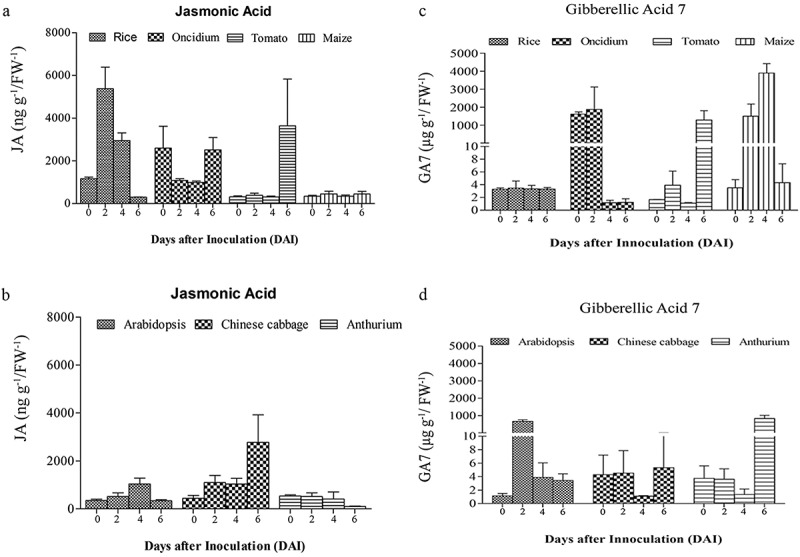
JA and GA7 levels in the roots of seven plant species after infection with P. indica. The roots of A. thaliana, Oncidium orchid, Brassica campestris, Lycopersicon esculentum, Oryza sativa cv. Japonica, Zea mays and Anthurium andraeanum were harvested 0, 2, 4, and 6 dai for the determination of the JA (a and b) and GA7 levels (c and d). For details, see Methods and Materials. The values are means ± SDs of three independent biological replications.
Figure 4.
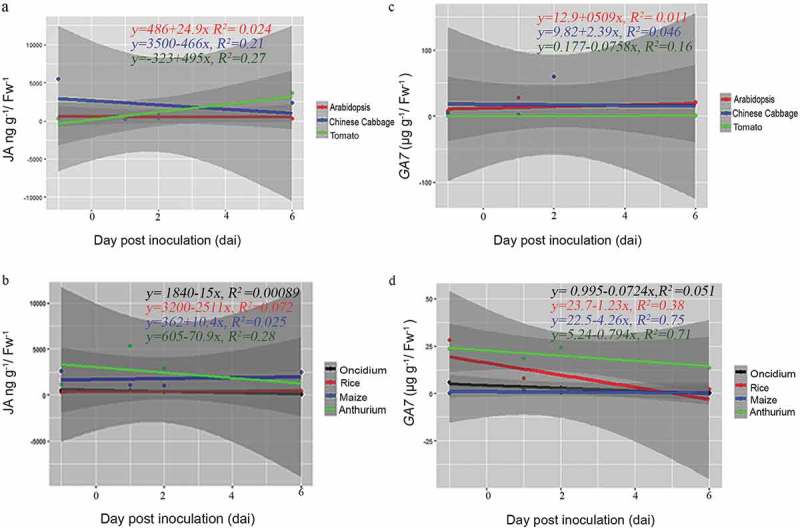
Scatter plots showing linear relationship between P. indica colonization and phytohormones (JA-, GA-) levels across plant species. Correlation of each dicot and monocot plants, P indica colonization period in days and JA (a and b) and GA (c and d) levels by rejecting `null’ hypothesis (equals to zero) are shown by linear regression analysis. Summary of regression coefficient values and R2 values are represented. Testing the significance of the differences by the comparison of dependent (y) and independent variables (x) (p<=0.05 or >=0.05) was performed using R-packages. The graphs are for statistical purpose only.
P. indica induces host genes involved in the biosynthesis of JA, GA, and ethylene (ET)
To further investigate the role of phytohormones during early phases of root colonization, the expression of marker genes for JA and GA synthesis (LOX1, AOC1, OPR3, and GA20-oxidase) were analyzed by qRT-PCR. The ET-responsive ERF1 gene was also included in our study.
Again, the gene expression pattern differed for different hosts (Figure 5). For instance, the LOX1, AOC1 and OPR3 mRNA levels representing JA biosynthesis remained relatively constant for A. thaliana, while an increase can be detected for Chinese cabbage within the first 4 dai. Furthermore, while the GA20 RNA level increased in both species, the AOC1 RNA level increased in Chinese cabbage roots, but not in A. thaliana roots. In tomato, only the OPR3 RNA level appears to increase initially. In rice and maize, the AOC1, OPR3 and GA20 RNA levels appear to increase slightly during early colonization stages. Finally, in orchid and A. andraeanum roots, all transcript levels decrease after the 2 dai. The ERF1 mRNA level was initially high in the seven species (Figure 5). In Chinese cabbage, it increased significantly until the 4th dai and decreased thereafter. In Arabidopsis, tomato, rice, and A. draeanum, the level decreased in response to P. indica infection. In Oncidium orchid, the level increased until 2nd dai and decreased dramatically during the later co-cultivation stages. In contrast, the ERF1 mRNA level in maize decreased initially until 2nd dai but increased strongly again. We also analyzed the expression levels of these genes at 8 and 10 dai in A. andraenum roots, because they became colonized much later than the other species. The mRNA levels for the JA- and GA-related genes still declined, and the AOC1 expression level was too low to be detected by our means (Supplementary Figure 1). Interestingly, the ET-related ERF1 mRNA level increased and was higher 8 and 10 dai when compared to 6 dai suggesting that the ERF1 mRNA level appears to respond to root colonization (Figure 5). Linear regression analyses for P. indica colonization and the expression of the tested genes often showed a good relationship across the plant species (Figure 6). For instance, LOX1 expression in Arabidopsis (R2 = 0.067, p < .05) and Chinese cabbage (R2 = 0.32, p > .05) and AOC1 expression in Arabidopsis (R2 = 0.51, p < .05) and Chinese cabbage (R2 = 0.08232, p > .05) showed a positive correlation, whereas LOX1 (R2 = 0.22, p > .05) and AOC1 (R2 = 0.64, p < .05) expression were negatively correlated to root colonization in tomato. Also, GA20 expression in Arabidopsis (R2 = 0.83, p > .05), Chinese cabbage (R2 = 0.15, p > .05) and tomato (R2 = 0.083, p > .05) as well as OPR3 expression in Arabidopsis (R2 = 0.043, p > .05), Chinese cabbage (R2 = 0.15, p > .05) and tomato (R2 = 0.033, p > .05) showed a positive relationship to root colonization (Figure 6), while ERF1 expression was only positively related to root colonization in Chinese cabbage (R2 = 0.24, p > .05) (Figure 6). Interestingly, while the expressions of LOX1, AOC1, OPR3, GA20, and ERF3 exhibited a strong positive linear relationship to P. indica colonization in maize and rice (Figure 6), this was not the case for Oncidium and Anthurium. Taken together, our results demonstrate that various hormone-related mRNA levels respond quite differently to the presence of P. indica.
Figure 5.
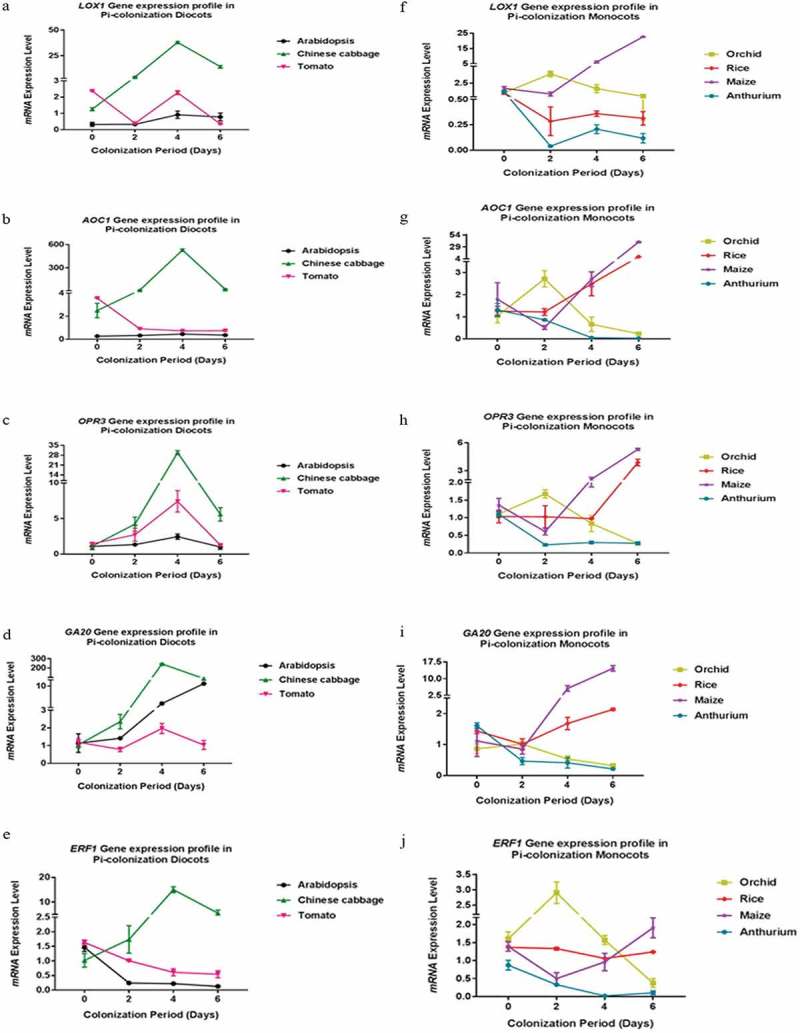
Relative mRNA levels for JA-, GA-, and ET-related genes in the roots of seven plant species after infection with P. indica for 0, 2, 4, and 6 dai. (a–j) The RNA levels of the JA synthesis-related LOX1, AOC1, and OPR3, the GA-responsive GA20-oxidase and the ET-related ERF1 genes were analyzed in the roots of A. thaliana, Oneidium orchid, Brassica campestris, Lycopersicon esculentum, Oryza sativa cv. Japonica, Zea mays and A. andraeanum at 0, 2, 4, and 6 dai by qRT-PCT. For the latter species, additional data are also presented for later time points. The mRNA levels were normalized to those of the ACTIN mRNA level. The values are means ± SDs of three independent biological replications.
Figure 6.
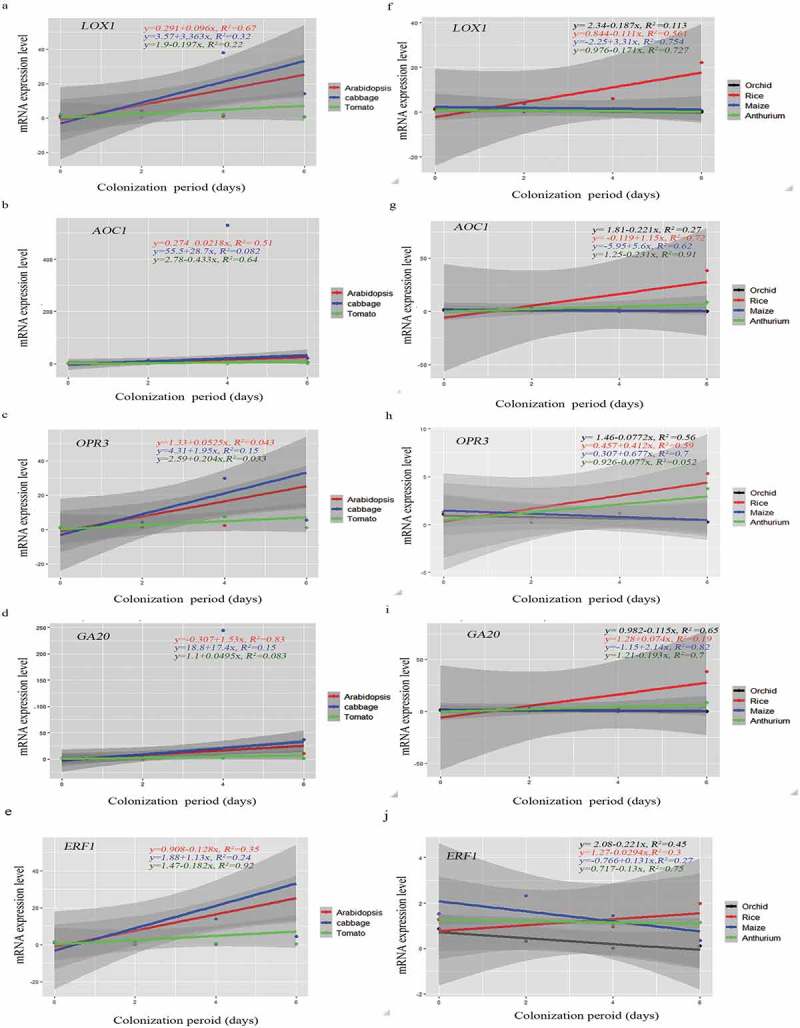
Scatter plots showing linear relationship between P. indica colonization and gene expression across plant species. (a–J) Correlation of each dicot and monocot plants, P. indica colonization period in days and gene expression levels by rejecting ‘null’ hypothesis (equals to zero) are shown by linear regression analysis. Summary of regression coefficient values and R2 values are represented. Testing the significance of the differences by the comparison of dependent (y) and independent variables (x) (p<=0.5 or >=0,05) was performed using R-packages. The graphs are for statistical purpose only.
Discussion
We demonstrate that the time point of root colonization by P. indica is highly specific for each host (Figure 1) and cannot be explained by changes in the endogenous JA and GA levels in the roots which are induced by P. indica (Figures 1–3). However, for an individual host, we often observed a correlation between the expression of the investigated phytohormone-related genes and the root colonization strategy (Figures 3 and 4), but these correlations were found for specific gene/host combinations and cannot be generalized. It appears that enormous differences in the endogenous phytohormone levels in the roots before the presence of the fungus might also be important for colonization, since the hormone balance is critical for overcoming the plant innate immunity. In some species, such as Chinese cabbage, the innate immunity appears to be immediately activated after exposure of the roots to P. indica, as reflected by the stimulation of JA and GA accumulation to orchestrate a complicated and interactive network of defense pathways. In other cases, these phytohormone levels do not respond strongly to the presence of the fungus (orchid and tomato) or occur only after colonization, such as in rice (Figures 1–3). The different responses of the JA levels in the host´s roots investigated in this study indicate that the fungus induces different threats either because the hosts recognize them differently, endogenous hormone levels restrict the fungal entry into the host or the fungus has different access to the root cells, e.g. due to different physical barriers. As long as the plant does not know whether it is approached by a beneficial or pathogenic fungus, it activates a mild JA response to restrict fungal growth in the roots and its rapid distribution. JA signaling affects other phytohormone functions including GA and ET signaling to prevent overcolonization and consequently loss of the benefits for the plants, since the plant resources have to be redirected from growth to defense when the roots become dominated by the overgrowing fungal hyphae. It appears that different species have established different mechanisms to cope with P. indica. It would be interesting to know whether the same or different fungal compound(s) induce the different responses in the host plants, and whether the fungus utilizes different strategies for a successful root colonization in different host species.
GA has been investigated in innate immune responses and early flowering induced by P. indica in various plant species.20,42 Cosme et al.43 showed that P. indica helps rice plants to tolerate root herbivory, and GA functions as a signal component of inducible plant tolerance against the biotic stress. Barley mutants impaired in GA synthesis, as well as perception, showed reduced colonization by P. indica which implicates that GA functions as a modulator of the root’s basal defense.34 A quintuple-DELLA mutant displaying constitutive gibberellin (GA) responses and the GA-biosynthesis mutant ga1-6 (for GA requiring 1) showed higher and lower degrees of colonization in Arabidopsis roots, respectively, suggesting that P. indica recruits GA signaling to help establish root cell colonization.26 Ent-kaurene synthases and ent-kaurene-like synthases are involved in the biosynthesis of phytoalexins and/or GAs. Li et al.44 demonstrated that kaurene synthase activity is required for successful root colonization of P. indica in barley. Finally, P. indica-induced growth promotion of Chinese cabbage and barley seedlings correlated to an increased GA level in the colonized roots,4,34 and the GA2ox gene involved in the inactivation of GA was down-regulated in P. indica-colonized barley roots.34,45 How these two phytohormones interact in order to control root colonization and entry of fungal hyphae into the root cell is a matter of intensive studies. However, our study clearly demonstrates that generalization over plant species is not possible.
The morphological observation also revealed that the growth pattern of root tissue was varied among the seven plant species. Especially, Chinese cabbage showed an obvious bushy root phenotype and Arabidopsis produces more branches. These observations are consistent with previous reports.4 However, no changes were observed in Oncidium and Anthurium. The results demonstrated that plant has established a different mechanism to reprogram the host responses after P. indica colonization.
In this study, four monocots (orchid, rice, maize, and Anthurium) and three eudicots (Arabidopsis, cabbage and tomato) were cultivated with P. indica. Previous studies showed that P. indica preferentially colonizes the maturation zone of roots, and that fungal hyphae appeared later in the meristematic and elongation zones.26,41,46 Therefore, microscopic analyses of colonization were performed with samples taken from the maturation zones of the roots, i.e. 3 cm away from the root tip. We show that the time period that is required for the appearance of P. indica in the maturation zone varied substantially among the species (Table 1). However, from the investigated plant species, rice, maize, and Anthurium were domesticated for the longest period of time.47–49 Therefore, one might speculate that longer domestication of a species results in the retardation of root colonization because these plants were bred to develop an optimal intrinsic adaptability to harm and resistance against pathogen attack. A stronger innate immunity in these species does not only prevent the invasion of pathogens but also of beneficial microbes which have to overcome the plant defense responses. A testable hypothesis might be that reprogramming of the phytohormone balance and signaling network to the demands of the beneficial endophyte P. indica might thus be more difficult for domesticated plants compared to those not exposed to a selection pressure for optimizing the immune system.
Multiple reports have shown that effective innate immune systems of the hosts suppress successful root colonization by P. indica in Arabidopsis and barley.26,34 The JA and GA levels in Arabidopsis increased after co-cultivation with P. indica, which is consistent with recent observations.34,50,51,52 However, the stimulatory effect on JA accumulation is much stronger in rice and reverse in Oncidium (Figure 3). This suggests that P. indica might use JA to stimulate the host´s innate immunity in rice and Arabidopsis, whereas in Oncidium, the fungus already represses JA accumulation to promote root colonization. Tomato has been reported as a pathogenically vulnerable species due to its low endogenous JA level.53,56 The strong increase during the later phases of P. indica colonization might indicate that the root utilizes GA to restrict root colonization.
The regulation of JA- and GA-related genes in response to P. indica colonization is also species-specific, which is not surprising considering the numerous processes which are regulated by these phytohormones. LOX, AOC, OPR3, and GA20-oxidase activate JA and GA biosynthesis and play important roles in senescence, wound and stress responses, pathogen attack and growth promotion.39,40,57–61 Stimulation of LOX1, AOC1 and OPR3 expression by P. indica might be a crucial regulatory step for JA biosynthesis to balance root colonization and to stimulate growth.50,57,58,62 GA20-oxidase produce active GA and the enzyme might be recruited by P. indica for correct colonization26,34 Interestingly, the observed strong changes in the mRNA levels of these four genes during co-cultivation with P. indica occur during early phases of root colonization (summarized in Table 1). The only exception is the tomato AOC1 mRNA level. A possible explanation could be that AOC1 is expressed mainly in vascular bundles61,63 which are not reached by the fungal hyphae. Furthermore, constitutive overexpression of AOC1 in tomato resulted in elevated JA levels in flower organs but not in leaves.61
Ethylene (ET) is another important hormone that participates in the activation and regulation of plant innate immunity. It is a negative regulator in mutualistic root symbioses, but a positive modulator of mutualistic root – P. indica symbiosis.33,64,65 ERF1 is a target of ET and participates in defense-related gene expression, but also to P. indica.33,66 Khatabi et al.64 showed that overexpressors of ERF1 in Arabidopsis plants displayed constitutive ET-associated defense which also improved root colonization by P. indica. Similar to JA- and GA-biosynthesis genes, the ERF1 mRNA levels were elevated in orchid, Chinese cabbage and maize roots 2, 4, and 6 dai with P. indica, while the mRNA level is downregulated in the other four hosts following the interaction with the fungus. Thus, ERF1 represents another example of a phytohormone-related gene that is quite differently responding to P. indica in different host species.
In conclusion, the huge differences observed in root colonization and phytohormone actions in the symbioses between P. indica and different host species suggest a high degree of species-specificity and warn of generalizations during early phases of the establishment of the symbiosis.
The onset of root colonization and reprogramming of root development by the fungus are quite different among the seven plant species. This might be the result of a combination of different factors: The hyphal growth towards the roots may occur with different velocities because chemical mediators either promote or inhibit the contact between the two symbionts. Once a physical contact is established, propagation of the hyphae on the root surface and between the epidermal and subepidermal cell layers might be controlled by different plant-specific factors, such as the features of the cell walls. Finally, the plant machinery controlling the invasion of hyphae into the plant cell as well as the signaling events leading to the reprogramming of the root cells appears to be highly species-specific.
In conclusion, based on these results, it is difficult to come to a general conclusion by analyzing the symbiotic interaction of P. indica with just one host plant.
Materials and methods
Plant growth and fungal culture conditions
Rice (Oryza sativa L.) and A. thaliana seeds were surface-sterilized for 30 and 10 min, respectively, in a NaClO solution (1%), and maize (Zea mays L.), tomato (Lycopersicon esculentum) and Chinese cabbage (Brassica campestris) seeds for 10, 5 and 3 min, respectively, in a NaClO solution (5%). All seeds were then washed five times with sterile water and finally placed on a petri dish containing 1/2 MS nutrient medium.67 Plates with rice, tomato, maize, and Chinese cabbage were incubated at 22°C under continuous illumination (100 mol m−2 s−1) for seed germination. A. thaliana seeds were incubated at 4°C in the dark for 48 h before transfer to 22°C and continuous illumination (100 µmol m−2 s−1) for 10 days. Anthurium andraeanum and Oncidium orchid plants were cultured for two weeks until rooting, and then transferred to 1/2 MS medium for five days. After seed germination, the growing seedlings with healthy roots were transferred to fresh plates containing 1/2 MS medium. P. indica was cultured on Kaefer medium for 6 days.68 One to six seedlings were used per petri dish (depending on the size of the seedlings) and one fungal plague of 5 mm in diameter per seedling was placed at a distance of 1 cm from the roots.4 After exposure to the fungus, all co-cultures were grown under identical conditions to allow a comparative analysis. Mock-treatments were simultaneously performed with media without the fungus.
Microscopic detection of P. indica in roots
Seedlings of the seven species listed above were co-cultivated with P. indica for 0, 2, 4 or 6 days to detect the establishment of the symbiotic interaction and root colonization. For microscopical observations, the plant material was washed in running tap water to remove the planting medium, and the primary roots, 3 cm away from the root tip, were cut off and dissected into 2 cm sections. For the thin and soft root tissue of A. thaliana, tomato, and Chinese cabbage, cut roots were dissected into pieces of 1 cm each and boiled in 2% KOH for 2 min, then washed in sterile water and stained with 0.5% lactophenol blue solution (Fluka, USA) for 10 min. Subsequently, it was squeezed on a glass slide for microscopical observation. For the thick and soft root tissue of maize, orchid, and Anthurium, each root section was sliced into thin pieces before staining with the same method. For the harder root tissue of rice, the roots were first fixed in formalin-acetic acid solution (formalin: acetic acid: 50% ethanol; 1:1:18; v/w) for 6 h and immersed in 2.5% sucrose solution overnight, and finally embedded in a frozen gel, followed by longitudinal microdissection into 25–30 μm slices. The frozen gel-embedded slices were stained with lactophenol solution as described above, before analyses with a bright field microscope. In pilot experiments with different staining procedures, we found that this analysis allows the best comparison of the roots from the different plant species.
Phytohormone analyses
Sample preparation
The seedlings were co-cultured with P. indica for 0, 2, 4 and 6 days, then their roots were washed under running tap water for 5 min and dried gently. Roots and cotyledons (excised from the hypocotyls) were harvested separately and quick-frozen in liquid N2. Frozen samples were then ground under liquid N2 with mortar and pestle. Samples (250 mg) were extracted with 750 μl MeOH–H2O–HOAc (90:9:1, v/v/v) for 2 h in 4°C and centrifuged for 1 min at 10,000 rpm. The supernatant was collected, and the extraction was repeated. Pooled supernatants were dried under N2, resuspended in 200 μl of 0.05% HOAc in H2O–MeCN (85:15, v/v), and finally filtered through a Millex-HV 0.45 μm filter from Millipore (Bedford, USA). Samples were dried, resuspended in 80% HPLC-grade methanol (MeOH) and filtrated before use. In parallel, HPLC-grade SA and JA (100 ng) was treated in the same way and used as internal standards.
JA analysis
Analyses were carried out using a Dionex Ultimate 3000 RSLC at ambient temperature, and the injected volume was 10 μl. The elution gradient was carried out with binary solvent system consisting of 0.05% HOAc in H2O (solvent A) and MeCN (solvent B) at a constant flow rate of 600 μl min−1 and a split 1/3. A linear gradient profile with the following proportions (v/v) of solvent B was applied (t (min), %B): (0, 15), (3, 15), (5, 100), (6, 100), (7, 15), (8, 15) with 5 min for re-equilibration. MS and MS/MS experiments were performed on a Bruker maXis UHR-QTOF mass spectrometer. All analyses were performed using the Turbo Ionspray source in negative ion mode with the following settings: capillary voltage −3500 V, nebulizer gas (N2) 10 (arbitrary units), curtain gas (N2) 12 (arbitrary units), collision gas (N2) 4 (arbitrary units).
GA analysis
Analyses were carried out using a Dionex Ultimate 3000 RSLC at ambient temperature, and the injected volume was 10 μl. The elution gradient was a binary solvent system consisting of 0.05% HOAc in H2O (solvent A) and MeCN (solvent B) at a constant flow rate of 600 μl min−1 and a split 1/3. A linear gradient profile with the following proportions (v/v) of solvent B was applied (t (min), %B): (0, 15), (3, 15), (5, 100), (6, 100), (7, 15), (8, 15) with 5 min for re-equilibration. MS and MS/MS experiments were performed on a Pegasus 4D GC×GC-TOFMS mass spectrometer. Column flow was set to 1 ml/min helium, injector temperature was set to 250°C, GC temperature program was set to 40°C at 1 min, 10°C/min to 300°C and held constant for 8 min, solvent delay was set to 470 s, transfer line temperature was set at 280°C, mass range saved was set to m/z 50–600, and ion source temperature was set to 200°C.
RNA preparation and analysis of gene expression by qRT-PCR
Roots of the seven species exposed to P. indica for 0, 2, 4 or 6 days were harvested. RNA was isolated from roots with an RNA isolation kit (RNA Plus Mini Kit, LabPrepTM). Total RNA was isolated from three independent replicates of the roots. After DNAse treatment, the cDNA was synthesized with the RevertAidTM first strand cDNA synthesis kit (Fermentas, Waltham, MA, USA) using 2 μg of RNA. The quantitative reverse transcription-PCR (qRT-PCR) was performed with synthesized cDNA using the KAPA SYBR FAST qPCR kit (KAPA, Wilmington, MA, USA) and gene-specific primers listed in the following Table 2 in ABI 7500 (Applied Biosystems, USA) according to the manufacturer’s instructions. To normalize the gene expression, the plant ACTIN gene was co-amplified as an internal control. The PCR cycle condition for qRT-PCR was set as default (40 cycles): 95.0°C for 3 min, 95.0°C for 3 s, 60.0°C for 30 s (instruction manual, ABI Biosystems 7500 RT-PCR machine). The mRNA levels for each cDNA probe were normalized with respect to the plant ACTIN mRNA level. The relative gene expression was analyzed by the 2–ΔΔCt method.69 For optimizing the annealing temperatures, the PCR products were first analyzed on agarose gels to confirm that a single band with the expected length is generated.
Table 2.
The following primer sequences were used in this study.
| Primer names | Primer sequences (5ʹ-3ʹ) |
|---|---|
| Actin-F | TTTGCTGGTGATGATGC |
| Actin-R | GCAACATACATGGCAGG |
| LOX1-F | TTGACCCAACCAAGCGGATA |
| LOX1-R | TTACCGGACGAGCAAGTTCA |
| AOC1-F | CGCATTCAGCTTTAGGTTTGG |
| AOC1-R | GTGATTCCCACGCGTTTCTT |
| OPR3-F | AAGATTCGATCTCTCTCATCGAGT |
| OPR3-R | GGAGTGGTCCGTTGAGCA |
| GA20-oxidase-F | CTCACCCAAACTCGTTCAACA |
| GA20-oxidase-R | TCTGCTTACTCCAAGGCTCAT |
| ERF1-F | GGCCATGGGGGAAATTCGC |
| ERF1-R | GCTTGATCGTAGGCTAAAGC |
| PiTef-F | ACCGTCTTGGGGTTGTATCC |
| PiTef-R | TCGTCGCTGTCAACAAGATG |
Statistical analyses
Data display means with standard errors of three independent biological repeats. Statistical significance of the treatments was determined using Student’s t-test in EXCEL 2010 at a significance level of P < .05. GraphPad Prism 5 software was used for statistical analysis. In all graphs, the error bars indicate the standard deviation. Linear regression analysis was performed using R-packages (http://www.R-project.org/). The coefficient of determination (R2) was analyzed, and test for the significance of the difference was performed.
Acknowledgments
We thank the Zhejiang Academy of Agricultural Sciences of the Republic of China for their financial support for the international cooperative project to K.-W. Yeh. Thanks are also to the National Taiwan University for partial financial support of international collaboration funding to K.-W. Yeh. R. Oelmüller was supported by the CRC1127.
Author contribution
HL, GA, QZ, XS, DT, FS, and MSH performed the experiments and analyses of the data, RSK performed statistical analysis. RO and KWY developed and supervised the experiments and wrote the manuscript.
Disclosure of potential conflicts of interest
There is no conflict of interests.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Varma A, Verma S, Sahay N, Büttehorn B, Franken P.. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol. 1999;65:2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma S, Varma A, Rexer K, Hassel A, Kost G, Sarbhoy A, Bisen P, Bütehorn B, Franken P.. Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia. 1998;90:896–903. doi: 10.1080/00275514.1998.12026983. [DOI] [Google Scholar]
- 3.Fakhro A, Andrade-Linares DA, von Bargen S, Bandte M, Büttner C, Grosch R, Schwarz D, Franken P. Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza. 2010;20:191–200. doi: 10.1007/s00572-009-0279-5. [DOI] [PubMed] [Google Scholar]
- 4.Lee YC, Johnson JM, Chien CT, Sun C, Cai D, Lou B, Oelmüller R, Yeh K-W. Growth promotion of Chinese cabbage and Arabidopsis by Piriformospora indica is not stimulated by mycelium-synthesized auxin. Mol Plant Microbe Interact. 2011;24:421–431. doi: 10.1094/MPMI-05-10-0110. [DOI] [PubMed] [Google Scholar]
- 5.Molitor A, Zajic D, Voll L, Pons-Kuehnemann J, Samans B, Kogel KH, Waller F. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Mol Plant Microbe Interact. 2011;24:1427–1439. doi: 10.1094/MPMI-06-11-0177. [DOI] [PubMed] [Google Scholar]
- 6.Oelmüller R, Sherameti I, Tripathi S, Varma A. Piriformospora indica, a cultivable root endophyte with multiple biotechnological applications. Symbiosis. 2009;49:1–17. doi: 10.1007/s13199-009-0009-y. [DOI] [Google Scholar]
- 7.Peškan-Berghöfer T, Shahollari B, Giong PH, Hehl S, Markert C, Blanked V, Kost G, Varma A, Oelmüller R. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant–microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plant. 2004;122:465–477. doi: 10.1111/j.1399-3054.2004.00424.x. [DOI] [Google Scholar]
- 8.Pham GH, Kumari R, Singh A, Sachdev M, Prasad R, Kaldorf M, Buscot F, Oelmüller R, Peskan T, Weiss M, et al Axenic cultures of Piriformospora indica In: Varma A, Abbott L, Werner D, Hampp R, editors. Plant surface microbiology. Berlin Heidelberg New York: Springer; 2004b. p. 593–616. [Google Scholar]
- 9.Prasad RP. Studies on interaction between symbiotic fungus (Piriformospora indica), rhizobacteria and selected plants [PhD Thesis]. Merrut, India: Merrut University; 2008. [Google Scholar]
- 10.Qiang X, Weiss M, Kogel KH, Schäfer P. Piriformospora indica-a mutualistic basidiomucete with an exceptionally large plant host range. Mol Plant Pathol. 2012;13(5):508–518. doi: 10.1111/j.1364-3703.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahollari B, Varma A, Oelmüller R. Expression of a receptor kinase in Arabidopsis roots is stimulated by the basidiomycete Piriformospora indica and the protein accumulates in Triton X-100 insoluble plasma membrane microdomains. Plant Physiol. 2005;162:945–958. doi: 10.1016/j.jplph.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Hückelhoven R, Neumann C, von Wettstein D, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci U S A. 2005. Sep 20;102(38):13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiß M, Waller F, Zuccaro A, Selosse MA. Sebacinales - one thousand and one interactions with land plants. New Phytol. 2016;211:20–40. doi: 10.1111/nph.13977. [DOI] [PubMed] [Google Scholar]
- 14.Kumar M, Yadav V, Tuteja N, Johri AK. Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Supporting Inf Microbiol. 2009;155:780–790. [DOI] [PubMed] [Google Scholar]
- 15.Johri AK, Oelmüller R, Dua M, Yadav V, Kumar M, Tuteja N, Varma A, Bonfante P, Persson BL, Stroud R. Fungal association and utilization of phosphate by plants: success, limitations, and future prospects. Front Microbiol. 2015;6:984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherameti I, Tripathi S, Varma A, Oelmüller R. The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol Plant Microbe Interact. 2008;21:799–807. doi: 10.1094/MPMI-21-6-0799. [DOI] [PubMed] [Google Scholar]
- 17.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–26544. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmüller R. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 2011;7:e1002051. doi: 10.1371/journal.ppat.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das A, Kamal S, Shakil Najam A, Sherameti I, Oelmüller R, Dua M, Tuteja N, Johri Atul K, Varma A. The root endophyte fungus Piriformospora indica leads to early flowering, higher biomass and altered secondary metabolites of the medicinal plant, Coleus forskohlii. Plant Sign Behav. 2012;7:1–10. doi: 10.4161/psb.7.1.18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan R, Xu L, Wei Q, Wu C, Tang W, Oelmüller R, Zhang W. Piriformospora indica promotes early flowering in Arabidopsis through regulation of the photoperiod and gibberellin pathways. PLoS One. 2017;12:e0189791. doi: 10.1371/journal.pone.0189791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua MS, Senthil R, Shyur LF, Cheng YB, Tian ZH, Oelmuller R, Yeh KW. Metabolomic compounds identified in Piriformospora indica-colonized Chinese cabbage roots delineate symbiotic functions of the interaction. Sci Rep. 2017;7(1). doi: 10.1038/s41598-017-08715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baltruschat H, Fodor J, Harrach BD, Niemczyk E, Barna B, Gullner G, Janeczko A, Kogel K-H, Schäfer P, Schwarczinger I, et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 2008;180:501–510. doi: 10.1111/j.1469-8137.2008.02583.x. [DOI] [PubMed] [Google Scholar]
- 23.Daneshkhah R, Cabello S, Rozanska E, Sobczak M, Grundler FM, Wieczorek K, Hofmann J. Piriformospora indica antagonizes cyst nematode infection and development in Arabidopsis roots. J Exp Bot. 2013;64(12):3763–3774. doi: 10.1093/jxb/ert213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knecht K, Seyffarth M, Desel C, Thurau T, Sherameti I, Lou B, Oelmüller R, Cai D. Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol Plant Microbe Interact. 2010;23(4):446–457. doi: 10.1094/MPMI-23-4-0446. [DOI] [PubMed] [Google Scholar]
- 25.Franken P. The plant strengthening root endophyte Piriformospora indica: potential application and the biology behind. Appl Microbiol Biotechnol. 2012;96:1455–1464. doi: 10.1007/s00253-012-4506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel K-H, Schafer P. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011;156:726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Döhlemann S, von Wirén N, Parniske M, Zuccaro A. Host-related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci U S A. 2013;110:13965–13970. doi: 10.1073/pnas.1301653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahrmann U, Zuccaro A. Opprimo ergo sum–evasion and suppression in the root endophytic fungus Piriformospora indica. Mol Plant Microbe Interact. 2012;25:727–737. doi: 10.1094/MPMI-11-11-0291. [DOI] [PubMed] [Google Scholar]
- 29.Nongbri PL, Johnson JM, Sherameti I, Glawischnig E, Halkier BA, Oelmüller R. Indole-3-acetaldoxime-derived compounds restrict root colonization in the beneficial interaction between Arabidopsis roots and the endophyte Piriformospora indica. Mol Plant Microbe Interact. 2012;25:1186–1197. doi: 10.1094/MPMI-03-12-0071-R. [DOI] [PubMed] [Google Scholar]
- 30.Vadassery J, Oelmüller R. Calcium signaling in pathogenic and beneficial plant microbe interactions: what can we learn from the interaction between Piriformospora indica and Arabidopsis thaliana. Plant Signal Behav. 2009;4:1024–1027. doi: 10.4161/psb.4.11.9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye W, Shen C-H, Lin Y, Chen P-C, Xu X, Oelmüller R, Yeh K-W LZ. Growth promotion-related miRNAs in Oncidium orchid roots colonized by the endophytic fungus Piriformospora indica. PLoS One. 2014;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achatz B, Kogel KH, Franken P, Waller F. Piriformospora indica mycorrhization increases grain yield by accelerating early development of barley plants. Plant Signal Behav. 2010;5(12):1685–1687. doi: 10.4161/psb.5.12.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, Oelmüller R. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 2010;185:1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x. [DOI] [PubMed] [Google Scholar]
- 34.Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, Scholz U, Pons-Kühnemann J, Sonnewald S, Sonnewald U, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J. 2009;59:461–474. [DOI] [PubMed] [Google Scholar]
- 35.Hilbert M, Voll LM, Ding Y, Hofmann J, Sharma M, Zuccaro A. Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 2012;196(2):520–534. doi: 10.1111/j.1469-8137.2012.04275.x. [DOI] [PubMed] [Google Scholar]
- 36.Vadassery J, Ritter C, Venus Y, Camehl I, Varma A, Shahollari B, Novák O, Strnad M, Ludwig-Müller J, Oelmüller R. The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica. Mol Plant Microbe Interact. 2008;21:1371–1383. doi: 10.1094/MPMI-21-10-1371. [DOI] [PubMed] [Google Scholar]
- 37.Stein E, Molitor A, Kogel KH, Waller F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 2008;49:1747–1751. doi: 10.1093/pcp/pcn147. [DOI] [PubMed] [Google Scholar]
- 38.Zarea MJ, Hajinia S, Karimi N, Goltapeh EM, Rejali F, Varma A. Effect of Piriformospora indica and Azospirillum strains from saline or non-saline soil on mitigation of the effects of NaCl. Soil Biol Biochem. 2012;45:139–146. doi: 10.1016/j.soilbio.2011.11.006. [DOI] [Google Scholar]
- 39.Chandler PM, Marion-Poll A, Ellis M, Gubler F. Mutants at the Slender1 locus of barley cv. Himalaya. Molecular and physiological characterization. Plant Physiol. 2002;129:181–190. doi: 10.1104/pp.010917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun TP. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book. 6:e0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong SQ, Tian ZH, Chen PJ, Senthilkumar R, Shen CH, Cai D, Oelmüller R, Yeh KW. The maturation zone is an important target of Piriformospora indica in Chinese cabbage roots. J Exp Bot. 2013;64:4529–4540. doi: 10.1093/jxb/ert265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Abdelaziz ME, Ntui VO, Guo X, Al-Babili S. Colonization by the endophyte Piriformospora indica leads to early flowering in Arabidopsis thaliana likely by triggering gibberellin biosynthesis. Biochem Biophys Res Commun. 2017;490:1162–1167. doi: 10.1016/j.bbrc.2017.06.169. [DOI] [PubMed] [Google Scholar]
- 43.Cosme M, Lu J, Erb M, Stout MJ, Franken P, Wurst S. A fungal endophyte helps plants to tolerate root herbivory through changes in gibberellin and jasmonate signaling. New Phytol. 2016;211:1065–1076. doi: 10.1111/nph.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Chen X, Ma C, Wu H, Qi S. Piriformospora indica requires kaurene synthase activity for successful plant colonization. Plant Physiol Biochem. 2016;102:151–160. doi: 10.1016/j.plaphy.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Hedden P. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;20:317–318. [DOI] [PubMed] [Google Scholar]
- 46.Deshmukh S, Hückelhoven R, Schäfer P, Imani J, Sharma M, Weiss M, Waller F, Kogel KH. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA. 2006;103:18450–18457. doi: 10.1073/pnas.0605697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX. Genetic control of rice plant architecture under domestication. Nat Gen. 2008;40:1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 48.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Li CB, Zhou AL, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 50.Vahabi K, Sherameti I, Bakshi M, Mrozinska A, Ludwig A, Reichelt M, Oelmüller R. The interaction of Arabidopsis with Piriformospora indica shifts from initial transient stress induced by fungus-released chemical mediators to a mutualistic interaction after physical contact of the two symbionts. BMC Plant Biol. 2015;15:58. doi: 10.1186/s12870-015-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vahabi K, Camehl I, Sherameti I, Oelmüller R. Growth of Arabidopsis seedlings on high fungal doses of Piriformospora indica has little effect on plant performance, stress, and defense gene expression in spite of elevated jasmonic acid and jasmonic acid-isoleucine levels in the roots. Plant Signal Behav. 2013;8:e26301. doi: 10.4161/psb.26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahrmann U, Strehmel N, Langen G, Frerigmann H, Leson L, Ding Y, Scheel D, Herklotz S, Hilbert M, Zuccaro A. Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol. 2015;207:841–857. doi: 10.1111/nph.13411. [DOI] [PubMed] [Google Scholar]
- 53.Vahabi K, Dorcheh SK, Monajembashi S, Westermann M, Reichelt M, Falkenberg D, Hemmerich P, Sherameti I, Oelmüller R. Stress promotes Arabidopsis - Piriformospora indica interaction. Plant Signal Behav. 2016;11:e1136763. doi: 10.1080/15592324.2015.1136763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 55.Chen H, Daniel JA, Howe GA. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett. 2006;580:2540–2546. doi: 10.1016/j.febslet.2006.03.070. [DOI] [PubMed] [Google Scholar]
- 56.Pedranzani H, Racagni G, Alemano S, Miersch O, Ram´ırez I, Pena-Cortes H, Taleisnik E, Machado-Domenech E, Abdala G. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003;41:149–158. doi: 10.1023/A:1027311319940. [DOI] [Google Scholar]
- 57.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vicentea J, Cascona T, Vicedob B, Garcı´a-Agustı´nb P, Hambergc M, Castresanaa C. Role of 9-lipoxygenase and a-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol Plant. 2012;5(4):914–928. doi: 10.1093/mp/ssr105. [DOI] [PubMed] [Google Scholar]
- 60.Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C. Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato – amplification in wound signaling. Plant J. 2003;33:577–589. [DOI] [PubMed] [Google Scholar]
- 61.Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol. 2003;51:895–911. [DOI] [PubMed] [Google Scholar]
- 62.Melan MA, Enriquez AD, Peterman TK. The LOX1 gene of Arabidopsis is temporally and spatially regulated in germinating seedlings. Plant Physiol. 1993;105:385–393. doi: 10.1104/pp.105.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mierscha O, Weichert H, Stenzel I, Hause B, Maucher H, Feussner I, Wasternack C. Constitutive overexpression of allene oxide cyclase in tomato (Lycopersicon esculentum cv. Lukullus) elevates levels of some jasmonates and octadecanoids in flower organs but not in leaves. Phytochemistry. 2004;65:847–856. doi: 10.1016/j.phytochem.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 64.Khatabi B, Molitor A, Lindermayr C, Pfiffi S, Durner J, von Wettstein D, Kogel KH, Schäfer P. Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0035502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khatabi B, Schäfer P. Ethylene in mutualistic symbioses. Plant Sign Behav. 2012;7:1634–1638. doi: 10.4161/psb.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. [DOI] [PubMed] [Google Scholar]
- 67.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 68.Hill TW, Kaefer E. Improved protocols for Aspergillus medium: trace elements and minimum medium salt stock solutions. Fungal Genet Newsl. 2001;48:20–21. [Google Scholar]
- 69.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


