ABSTRACT
Cytokinin Response Factor (CRF) genes are a subgroup of AP2/ERF domain-containing transcription factors that are defined by the CRF domain, from which five clades of CRF genes have been identified. Clade III CRFs are strongly induced by cytokinin, as well as other abiotic stress factors, such as oxidative stress. While this appears well studied for the Clade III CRFs in Arabidopsis and tomato, there have been almost no studies done outside of these model systems. This study expands upon that and represents the first CRF research in the Sunflower family, Asteraceae. Fifty Asterid Clade III CRF protein sequences were examined, and novel Clade III CRF C-terminus motifs were identified. Clade III CRF genes of Marshallia mohrii and M. caespitosa were assembled from genome-skimming and transcriptomic data. Expression experiments were conducted on M. caespitosa to test responsiveness to both cytokinin and oxidative stress. Low levels of basal expression for the McCRF1 were found to be strongly induced in both treatment groups. These are the first experiments to show regulation of a nuclear gene in a Marshallia species, and these results suggest there is broad conservation in the sequence, form, and regulation of Clade III CRF genes and proteins.
KEYWORDS: Cytokinin response, Marshallia, CRF, oxidative stress, gene expression
Introduction
Cytokinin Response Factors (CRFs) are a subfamily of AP2/ERF domain-containing transcription factors with strong connections to cytokinin, as several members were originally identified as being highly induced in Arabidopsis thaliana (L.) Heynh by cytokinin.1,2 CRFs proteins have a family specific AP2/ERF DNA-binding domain, a subfamily-specific CRF domain involved in protein–protein interactions as well as a predicted mitogen-activated protein kinase (MAPk) site.2,3 Phylogenetic analyses using the CRF and AP2/ERF domains of CRF proteins across a wide range of flowering plant lineages have indicated that there are at least five major clades of CRFs, each containing a unique C-terminus motif that is well conserved.2 While CRFs do have related expression patterns, such as preferential localization to vascular tissue, individual CRF clades appear to also have distinct regulation, such as induction by cytokinin.2,3
Perhaps the best-studied CRF clade is Clade III, yet, surprisingly, this really consists of an examination of only three genes: AtCRF5 and 6 from Arabidopsis and SlCRF5, a tomato (Solanum lycopersicum L.) CRF.2,4–7 While there has been a greater examination of AtCRF6 than the others, genes showing direct connections to leaf senescence, it appears that features common to all Clade III CRF members are strong induction by cytokinin and oxidative stress, with possible connections to other abiotic stress responses.2,4–6
This study aimed to further our knowledge of Clade III CRFs beyond Arabidopsis and tomato. To do this, we looked for conserved motifs unique to Clade III CRF proteins and characterized the Clade III CRF gene for the genus Marshallia Shreb. (Asteraceae) for a response to cytokinin and oxidative stress to test if broad regulation of Clade III exists. This also represents the first research to identify and characterize regulatory mechanisms of any nuclear genes in Marshallia.
Materials and methods
Asterid clade III CRF data
Asterid Clade III CRF protein sequences were collected from public databases (e.g., NCBI and OneKP (One Thousand Plant Project)) and aligned manually in SeaView v. 4.4.2.8,9 Sequences of low quality (e.g., missing large regions or short sequences) were excluded from further analysis. ClustalO v. 1.2.0 was used to align the remaining sequences to compare to manual alignment.10 Fifty unaligned sequences were submitted to MEME motif analysis (See Supplemental Table 1 for species and accession information).11 Parameters for MEME analysis were occurrence of motif = zero to one, number of motifs = 10, minimum number sites = 35, maximum number sites = 50, minimum width = five, and maximum width = 100.
Extraction and sequencing
DNA from all species of Marshallia were extracted from fresh leaf tissue using a modified 2X CTAB protocol as described by Doyle and Doyle (1987) or E.Z.N.A. kits (Omega Bio-tek, Inc., Norcross, GA) per manufacturer protocol.12 RNA was extracted from fresh leaf material of five individuals of Marshallia Schreb. (two M. mohrii Beadle and F.E. Boynt., one M. caespitosa Nutt. Ex D.C., one M. obovata (Walt.) Beadle and F.E. Boynt., and one M. trinervia (Walt.) Trel.) and from fresh shoots and roots of M. caespitosa using Plant RNA extraction kit (Qiagen, Hilden, Germany) per manufacturer protocol. DNA samples were submitted to HudsonAlpha Institute for Biotechnology (Huntsville, AL) for paired-end library prep and 100 bp sequencing via an ILLUMINA (ILLUMINA Inc., San Diego, CA) HiSEQ 2000 platform. RNA samples were submitted to the Auburn University Genomics and Sequencing Laboratory (Auburn, AL) where cDNA libraries were prepared using an Illumina mRNA TruSeq kit and sequenced on an ILLUMINA HiSEQ 1500 platform.
Assembly and description
Reads from unassembled genome-skimming readsets were identified using BLAST and assembled using CAP3 for the coding region of Clade III CRF gene.13,14 RNA reads from Marshallia accessions M2.9 (M. trinervia), M3.9 (M. obovata), M10.1.1 (M. caespitosa), M20r, and M21r (M. mohrii) were assembled using Trinity v20131110.15 Both DNA and RNA reads were mapped to the resulting DNA contig and RNA transcript using Bowtie2 v. 2.1.0 with the flags – local, – qc-filter, and – no-unal.16 Maps were visualized using Tablet 1.14.04.10 to assess the quality of the mapping and to assess the accuracy of the assemblies.17
5ʹ UTR sequence assembly was performed by identifying appropriate reads from all data sets via BLAST followed by hand assembly in SeaView. Bowtie2 was used to map read pairs from all data sets to further identify the reads of the 5ʹ UTR. Within the mapping, reads were selected that were properly mapped, along with their pair, and that exhibited soft clipping of the 5ʹ region. These reads were then extracted from the database and added to the assembly in an iterative process. The cis-element analysis was examined using the New PLACE cis-element web tool and manual examination of the sequence.18
Characterization of expression
Individuals of Marshallia caespitosa were grown in the Auburn University Plant Research Center greenhouses. Cypselae were planted in Sunshine (Sun Gro Horticulture, Agawam, MA) mix #8 potting soil just below the surface of the soil and covered with a thin layer peat moss. A weekly watering/fertilizing regime and natural light and photoperiod were utilized. Plants were grown for approximately one year and then harvested while in the rosette growth stage. Whole harvested plants were used for oxidative stress and cytokinin induction tests. Three plants were sprayed with a control spray, three plants were sprayed with 0.5 μM solution of benzyladenine (BA), and three were sprayed with a 41.65mM or 0.5% solution of hydrogen peroxide. Plants were given a 6-h period to allow for uptake of the solutions and for changes in expression to occur. Each plant was separated into shoots and roots for RNA extractions using a Qiagen Plant RNA extraction kit (Qiagen, Hilden, Germany) per manufacturer protocol. cDNA was prepared using Quanta qScript cDNA supermix (Quanta BioSciences, Inc., Gaithersburg, MD, USA) per manufacturer protocol with an Eppendorf Thermal Cycler (Eppendorf, Hamburg, Germany). q-PCR was performed using two technical replicates of three biological replicates per experimental group in an Eppendorf Realplex with Quanta Sybr Taq mix per a modified protocol (10μl G-Bio taq, 0.4 μl primer mix, 9.6 μl template) using the following program: one cycle at 95.0°C, followed by 40 cycles of Tm = 95.0 for 30s, Ta = 55.0°C for 20s, and Te = 68.0°C for 30s; melting curve program was set to 95.0°C for 15 s, 60.0°C for 15 s, a 20-m temperature increase period followed by 95.0°C for 15 s. Primers were designed using NCBI BLAST Primer and were prepared by Eurofins Scientific (McCRF1 Forward = GCTTCTGGTTCTGTGTCCCGA, McCRF1 Reverse = CCAAACCGTAACACGGAGGT, EF1A Forward = GATGATTCCCACCAAGCCCA, EF1A Reverse = CAAACAACCGACGAACCCAC). Mean Ct was calculated for each biological replicate. Mean Ct of EF1A was subtracted from mean Ct of the respective experimental replicate to calculate ΔCt. ΔΔCt was calculated by subtracting ΔCt of the experimental group from the ΔCt of the control group. Fold change (FC) was calculated by the equation 2^ΔΔCt.
Results and discussion
Clade III CRF genes have been found to be strongly induced by both the plant hormone cytokinin and oxidative stress, yet there has been little to no experimental study of these responses outside of Arabidopsis and tomato. Here, we aligned and analyzed 50 Asterid Clade III CRFs to provided valuable insight into the structure of these genes across a broader phylogenetic context and identified several conserved motifs. MEME analysis identified motifs common to all CRF proteins, including the CRF domain motif (VRI[SY]VTD[CG]DATD; e-value = 7.3e-066; Figure 1(a)) and the putative MAPk motif (SPTSVLRFD; e-value of 6.1e-054; Figure 1(b)). The most strongly supported motifs in the MEME analysis were those of the C-terminus. The first C-terminus motif was found to be [LY]D[QS]CFL[NK][DE][FY]FDFRSPSP[LI][IM]Y[ED]E (e-value of 2.2e-586; Figure 1(c)). The second was found at the end of the C-terminus and featured a WDV[DN]DF[FL] sequence (e-value = 9.3e-06; Figure 1(d)). These motifs are unique to Clade III CRFs and likely play an important role in the activation of the gene by acting as a trans-activation site.2,19,20
Figure 1.
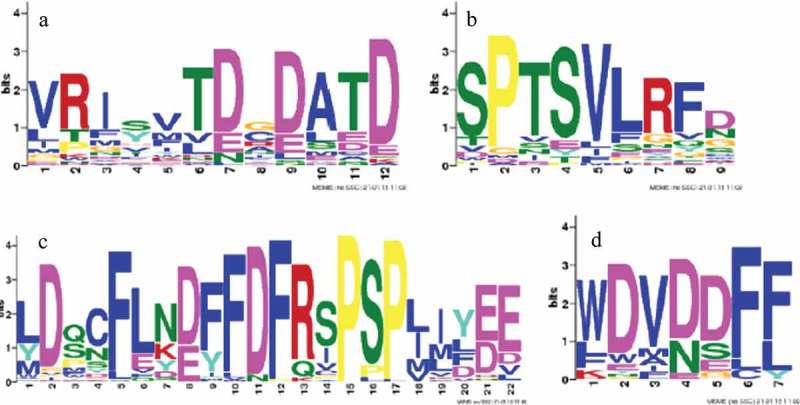
MEME analysis output for the CRF (a), putative MAPk (b), and putative 3ʹ trans-activation domain (c and d) motifs. The CRF domain motif (a) is the first of the motifs identified by the MEME analysis to occur in the amino acid sequence and is supported by an e-value of 7.3e-066. The putative MAPk site motif (b) occurs downstream of the AP2/ERF domain and was supported by an e-value of 6.1e-054. The first putative 3ʹ trans-activation domain (c) motif occurs downstream of the MAPk site and was supported by an e-value of 2.2e-586. The second putative 3ʹ trans-activation domain (d) typically occurs just upstream of the stop codon and was supported by an e-value of 9.3e-0.6.
Assembly of Clade III CRF genes from non-model plants will help increase our understanding of conserved structure within their sequences and help determine if work done in model systems truly serves as a model for other plants. Here, we used novel sequence analysis from the non-model genus Marshallia to fully examine this premise. Assembly of a Marshallia Clade III CRF from genome-skimming data was difficult due to low coverage. However, using Trinity, a Clade III CRF transcript from start to stop codons successfully assembled, with some up- and downstream elements, from the M21r RNA read set (henceforth referred to as MmCRF1, GenBank accession MF687408; refer to Table 1 for coding sequence). The Trinity assembly of the M10.1.1 (M. caespitosa) RNA read set provided a near-complete transcript, from just downstream of the CRF domain to just downstream of the stop codon. The top BLAST hit from the Trinity output provided a transcript that contained all elements of Clade III CRFs. To extend the M. caespitosa transcript, M. caespitosa DNA reads were mapped to the M21r transcript via previously described methods and a majority-rule consensus was called (henceforth referred to as McCRF1, GenBank accession MF687407; refer to Table 1 for coding sequence). The resulting consensus matched the Trinity transcript where overlap occurred and extended the sequence to the start codon. Other accessions provided only a truncated transcript (M20r) or no assembly at all (M2.9 and M3.9). Mappings of DNA readsets offered low and incomplete coverage. When compared to other genes, such as the tubulin genes, the read depth was quite low, reaching only ~50% for some readsets. Attempts to assemble 5ʹ UTR produced a contig approximately 500 bp in length (Table 3). A region either lacking read data or containing highly repetitive sequence was encountered in each data set that precluded assembly. Approximately 500 nucleotides were assembled upstream of the start codon, from which an analysis of cis-promoter elements (New PLACE) revealed a number of potentially conserved cis-element promoter element motifs.18 Importantly, several of these can be generally linked to various stresses, as well as four ARR1AT-motifs connected to cytokinin regulation and two motifs (TCTCT/AGAGA) found in other CRFs linked to vascular localization (Table 3).2,18
Table 1.
Sequences of the McCRF1 and MmCRF1 gene reading frames. SNPs between the McCRF1 and MmCRF1 coding regions are highlighted in yellow.
 |
Table 3.
Sequence of 5ʹ UTR assembled from all Marshallia spp. readsets. This region extended approximately 500 bps upstream of the start codon and includes both a known vascular-localized motif (TCTCT/AGAGA) in yellow and a known cytokinin-regulated motif ARR1AT (NGATT/NCTAA) in green.
 |
Both McCRF1 and MmCRF1 protein sequences were highly similar to each other, yet exhibit some variation in the conserved domains of the protein (Figure 2). When compared to SlCRF5, the CRF domain of McCRF1 varied in 18 positions while MmCRF1 varied in 15 positions, with strong conservation of the core of the motif, D[CG]DATDDD. Within the most conserved region of the C-terminus motif, the MmCRF1 protein was found to differ in three positions and the McCRF1 protein varied in four. The core of this motif was a strongly conserved DF[FL]DFR[SI]PSP[LI]M pattern. The MAPk site showed no variation within its sequence, exhibiting the typical SP[TS]SVL motif, with a T in the third position. The small motif at the end of the C-terminus varied in three of five positions, from KWAND in SlCRF5 to VWDVD in both Marshallia Clade III CRFs (Figure 3). McCRF1 and MmCRF1 featured 16 SNPs relative to each other, with 10 resulting in amino acid changes (Tables 1 and 2). NCBI BLASTn was used to calculate the percent similarity between McCRF1, MmCRF1, and SlCRF5. The McCRF1 and MmCRF1 proteins had a 96.5% similarity with each other and a 36.55% and 37.94% similarity with SlCRF5, respectively.
Figure 2.

Relative position map of conserved domains/motifs, highlighted in red, of the Marshallia Clade III CRF proteins.
Figure 3.

(a) Comparison of the CRF domains of SlCRF5 (top), McCRF1, and MmCRF1 (bottom) Clade III CRF proteins. This motif occurred in amino acid positions 26–47 in SlCRF5 and 27–48 in McCRF1 and MmCRF1. (b) Comparison of the putative MAPk site motif in SlCRF5 (top), McCRF1, and MmCRF1 (bottom) Clade III CRF proteins. This motif occurred in amino acid positions 180 – 186 in SlCRF5 and 188–195 in McCRF1 and MmCRF1. (c) Comparison of strongly conserved motif in putative trans-activation domain in SlCRF5 (top), McCRF1, and MmCRF1 Clade III CRF proteins. This motif occurred in amino acid positions 237–253 in SlCRF5 and 230–247 in McCRF1 and MmCRF1. (d) Comparison of 3ʹ motif of trans-activation domain in SlCRF5 (top), McCRF1, and MmCRF1 Clade III CRF proteins. This motif occurred in amino acid positions 283–287 in SlCRF5 and 263–267 in McCRF1 and MmCRF1.
Table 2.
Amino acid sequences for the McCRF1 and MmCRF1 proteins. Amino acid differences between McCRF1 and MmCRF1 proteins are highlighted in yellow.
 |
Examination of expression levels by qPCR analysis found while McCRF1 has a low basal level it is very strongly induced by both cytokinin (0.5 µM Benzyl Adenine) and oxidative stress (41.65 mM H2O2). Average Fold Change (FC) was calculated for samples achieving exponential increase during amplification and showed that McCRF1 was strongly induced by both oxidative stress and cytokinin treatment in roots and shoot tissues. The largest increase of 259.31 FC expression was found in shoot tissue from cytokinin treatment. This strong increase is consistent with previous work on Clade III CRFs, which showed the strongest induction in aerial organs in cytokinin treatment groups, as well as the identification of the ARR1AT cytokinin responsive cis-elements in the promoter (Table 3).2,3 There was also very strong 72.11 FC increase in shoots after oxidative stress treatment, which would be similar to previous findings for AtCRF6.7 While the average FC increases in the roots were lower than for the shoots they were still strong; 11.13 and 65.61 for oxidative stress and cytokinin treatment, respectively. The results of this analysis show that there is conservation in Marshallia for, at least, cytokinin and oxidative stress regulation found in the other examined Clade III CRFs, AtCRF5, AtCRF6, and SlCRF5.
This work has increased our understanding of the structure and conserved C-terminus motifs that are unique to the Clade III CRFs. It has also demonstrated that these genes have conservation in regulatory mechanisms across a broader phylogenetic breadth than previously known. Furthermore, we have investigated genetic processes in Marshallia for the first time, increasing our understanding of this clade and expanding analyses into CRF genes across a greater phylogenetic context.
Funding Statement
This work was supported by the Auburn University Graduate School.
Acknowledgments
Funding was provided by the Auburn University Graduate School via a Thesis Grant and the Auburn University Cellular and Molecular Biology Program via a Summer Biosciences Peaks of Excellence Research Fellowship.
Disclosure of Potential Conflicts of Interest
The authors have no interests to disclose.
Supplementary data
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Rashotte AM, Goertzen LR.. The CRF domain defines cytokinin response factor proteins in plants. BMC Plant Biol. 2010;10:1. doi: 10.1186/1471-2229-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwack PJ, Shi X, Robinson BR, Gupta S, Compton MA, Gerken DM, Goertzen LR, Rashotte AM. Vascular expression and C-terminal sequence divergence of cytokinin response factors in flowering plants. Plant Cell Physiol. 2012;53:1683–6. doi: 10.1093/pcp/pcs110. [DOI] [PubMed] [Google Scholar]
- 3.Rashotte AM, Mason MG, Hutchinson CE, Ferreira FJ, Schaller GE, Kieber JJ. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. PNAS. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zwack PJ, Robinson BR, Risley MG, Rashotte AM. Cytokinin response factor 6 negatively regulates leaf senescence and is induced in response to cytokinin and numerous abiotic stresses. Plant Cell Physiol. 2013;54:971–981. doi: 10.1093/pcp/pct049. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, Gupta S, Rashotte AM. Solanum lycopersicum cytokinin response factor (SlCRF) genes: characterization of CRF domain-containing ERF genes in tomato. J Exp Bot. 2012;63:973–982. doi: 10.1093/jxb/err325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S. Characterization of CRF domain containing ERF genes- Solanum lycopersicum cytokinin response factors SlCRF3 and SlCRF5 in tomato development [dissertation]. Auburn (AL): Auburn University; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwack PJ, De Clercq I, Howton TC, Hallmark HT, Hurny A, Keshishian EA, Parish AM, Benkoca E, Mukhtar MS, Van Breusegem F, et al. Cytokinin response factor 6 represses cytokinin-associated genes during oxidative stress. Plant Physiol. 2016. doi: 10.1104/pp.16.00415171:1249-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 10.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal Omega. Mol Syst Biol. 2011. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Stanford (California. Menlo Park (CA)): AAAI Press;1994. August 14–17. doi: 10.3168/jds.S0022-0302(94)77044-2 [DOI] [PubMed] [Google Scholar]
- 12.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bulletin. 1987;19:11–15. [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 18.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ketelsen B. Characterization of a Cytokinin Response Factor in Arabidopsis thaliana. [dissertation]. Tromsø (Norway): University of Tromsø; 2012. doi: 10.1094/PDIS-11-11-0999-PDN [DOI] [Google Scholar]
- 20.Striberny B, Melton AE, Schwacke R, Krause K, Fischer K, Goertzen LR, Rashotte AM. Cytokinin response factor 5 has transcriptional activity governed by its C-terminal domain. Plant Signal Behav. 2017;12:e1276684. doi: 10.1080/15592324.2016.1276684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


