Abstract
Recent advances highlight a pivotal role for cellular metabolism in programming immune responses. Here, we demonstrate that cell-autonomous generation of nicotinamide adenine dinucleotide (NAD+) via the kynurenine pathway (KP) regulates macrophage immune function in aging and inflammation. Isotope tracer studies revealed that macrophage NAD+ derives substantially from KP metabolism of tryptophan. Genetic or pharmacological blockade of de novo NAD+ synthesis depleted NAD+, suppressed mitochondrial NAD+-dependent signaling and respiration, and impaired phagocytosis and resolution of inflammation. Innate immune challenge triggered upstream KP activation but paradoxically suppressed cell-autonomous NAD+ synthesis by limiting the conversion of downstream quinolinate to NAD+, a profile recapitulated in aging macrophages. Increasing de novo NAD+ generation in immune-challenged or aged macrophages restored oxidative phosphorylation and homeostatic immune responses. Thus, KP-derived NAD+ operates as a metabolic switch to specify macrophage effector responses. Breakdown of de novo NAD+ synthesis may underlie declining NAD+ levels and rising innate immune dysfunction in aging and age-associated diseases.
Activated innate immune cells, in particular circulating mono-cytes and tissue macrophages, initiate a range of actions as first responders in pathogen clearance, including chemotaxis, generation of respiratory bursts, phagocytosis, elaboration of pro-inflammatory factors, and ultimately resolution of inflammation. Initiation and maintenance of these immune cell responses requires upregulation of glycolytic and mitochondrial metabolism to meet intensie demand for energy and biosynthetic precursors1,2. The cofactor nicotinamide adenine dinucleotide (NAD+) plays a central role in both glycolysis and oxidative phosphorylation, accepting high-energy electrons from glycolytic and tricarboxylic acid (TCA) intermediates and eventually feeding electrons into complex I of the electron transport chain (ETC) to drive oxidative phosphorylation, the major source of adenosine triphosphate.
NAD+ is also a co-substrate consumed by covalent modification of proteins and by signaling enzymes, including the poly(ADP-ribose) polymerases (PARPs), the cyclic ADP-ribose synthases, and the sirtuin deacetylases (SIRTs)3–6. NAD+-consuming enzymes release nicotinamide, which can then be reconverted into NAD+ by the salvage pathway initiated by the enzyme nicotinamide phospho-ribosyltransferase (NAMPT). NAD+ concentration reflects the balance between NAD+ consumption and synthesis from the salvage pathway, the Preiss-Handler pathway, which makes NAD+ from dietary nicotinic acid and nicotinic acid mononucleotide (NaMN), and the de novo pathway, where NAD+ synthesis is derived from tryptophan (Trp).
NAD+ homeostasis is frequently disrupted in aging and metabolic disease, where increased NAD+ consumption or decreased synthesis deplete NAD+ stores3,7,8. NAD+ homeostasis may also be disrupted in immune cells, where activation of PARPs or the cADPR CD38 consume NAD+9. Therefore, it is interesting to note that the enzyme indole-2,3-dioxygenase 1 (IDO1), which catalyzes the first committed step of the kynurenine pathway (KP), is rapidly and highly induced in the setting of immune challenge10,11. IDO1 metabolizes the essential amino acid Trp to the immunosuppressive metabolite kynurenine12,13; immunostimulatory IDO1 inhibitors have been under intensive investigation in clinical trials for their potential to augment cancer immunotherapy14. However, kynurenine (KYN) is not the final product of the KP. Instead, it is further metabolized through the KP to quinolinic acid (QA), which is converted by the enzyme quinolate phosphoribosyltransferase (QPRT) to NaMN and ultimately to NAD+ via the Preiss-Handler pathway.
Although the KP is the principal source of de novo NAD+ synthesis15,16, the focus on kynurenine and neuroactive intermediates has resulted in relatively little attention being paid to the potential function of KP-derived NAD+. Evidence to date argues for de novo NAD+ synthesis being localized to the liver17,18, with peripheral tissues believed to depend on liver-derived nicotinamide to meet NAD+ demand. In this study, using a combination of genetic and pharmacological approaches, coupled with isotope labeling and metabolic profiling, we demonstrate the activity of the complete KP from Trp to NAD+ in macrophages both in vitro and in vivo, where the KP is a biologically significant source of NAD+ and critically regulates macrophage immune function in resting, immune-challenged, and aged macrophages.
Results
The KP contributes to macrophage de novo NAD+ synthesis.
The nicotinamide ring of NAD+ is synthesized from the vitamin nicotinic acid or de novo via the KP from the amino acid Trp (Fig. 1a). Although liver-based synthesis is assumed to supply the NAD+ precursor nicotinamide to peripheral cells, we investigated the possibility that decreased cellular NAD+ might be replenished in macrophages by endogenous synthesis from Trp via the KP. Accordingly, in human monocyte-derived macrophages (MDMs), we disrupted the salvage pathway using the NAMPT inhibitor FK866 and examined the effects on KP enzyme and metabolite levels (Fig. 1a–d). Quantitative immunoblot analysis demonstrated marked increases of the KP enzymes IDO1, kynurenine-3-monooxygenase (KMO), kynureninase (KYNU), and QPRT (Fig. 1b,c and Supplementary Fig. 1a) and parallel increases in KP metabolites (Fig. 1d). The abundance of QA was not increased, which is suggestive of efficient metabolism of QA to NaMN via QPRT and subsequent generation of de novo NAD+. KYN supplementation restored basal amounts of the QA and NAD+ precursors NaMN and nicotinic acid adenine dinucleotide (NaAD) (Supplementary Fig. 1b,c) and prevented the FK866-induced depletion of NAD+ (Fig. 1e).
Fig. 1|. The KP contributes to de novo NAD+ synthesis.
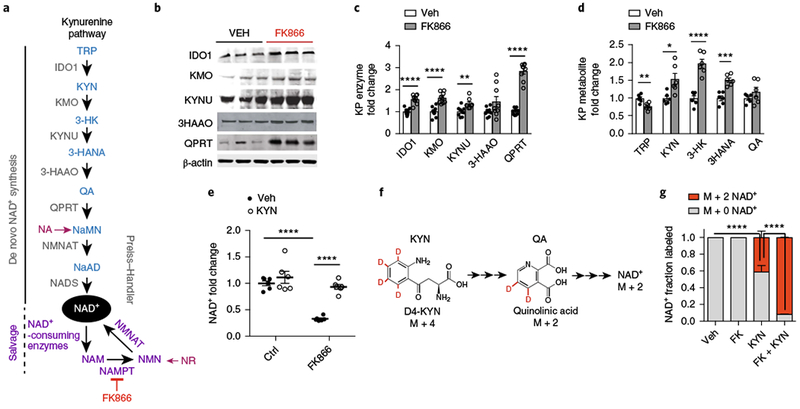
a, Diagram of the KP and formation of QA, the precursor for de novo NAD+ synthesis, which is then metabolized by the rate-limiting QPRT to NaMN. NMNAT, nicotinamide mononucleotide adenylyltransferase; NADS, NAD+ synthase. b-d, Human MDMs were treated with vehicle or the NAMPT inhibitor FK866 (10 μM for 20 h). b, Representative immunoblot of three independent experiments measuring KP enzymes ± FK866 (10 μM for 20 h). c, Quantification of KP enzyme levels normalized to β-actin; n = 9 biologically independent samples per group, represented as the mean ± s.e.m.; **P < 0.01 and ****P < 0.0001, two-tailed Student’s t-test). d, LC-MS of KP metabolites from human MDM cell lysates; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, two-tailed Student’s t-test. See also Supplementary Fig. 1a. e, Human MDMs ± FK866 (10 μM, 20 h) were supplemented with KYN (25 μM) or vehicle for 20 h. LC-MS measurements of NAD+; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-way ANOVA with Tukey’s post hoc test. See also Supplementary Fig. 1b,c. f, Experiments in e were repeated using mass-labeled D4-KYN. See Supplementary Fig. 1d. g, Human MDM cell lysates were assayed by LC-MS 20 h after the administration of FK866 and/or KYN and assayed for M + 2 NAD+. Data are represented as the mean ± s.e.m.; n = 3 biologically independent samples per group; ****P < 0.0001, two-tailed Student’s t-test.
To further confirm that de novo NAD+ is generated from the KP, we performed isotope labeling. Human MDMs were grown in medium supplemented with KYN labeled with four deuteriums (D4-KYN) (Fig. 1f and Supplementary Fig. 1d). Measurement of D2-labeled NAD+ by liquid chromatography-mass spectrometry (LC-MS) revealed substantial basal de novo NAD+ production of 40% of total NAD+. With FK866 blockade of the salvage pathway, de novo NAD+ production accounted for almost all NAD+ (Fig. 1g). Thus, in macrophages at rest, the KP functions as a de novo source of NAD+.
Loss of the KP disrupts mitochondrial respiration.
To understand the contribution of the KP to de novo NAD+ synthesis, we investigated the impact of blocking the pathway genetically using Ido1−/− mice19 or pharmacologically with 1-methyl-L-tryptophan (1MT)20 (Fig. 2 and Supplementary Figs. 2 and 3). Since reduced NAD+ (NADH) is the main electron donor for the ETC, we tested whether inhibition of IDO1 activity might alter mitochondrial oxidative phosphorylation. Genetic loss of Ido1 in primary mouse peritoneal macrophages reduced cellular NAD+ amounts compared to wild-type (WT) controls (Fig. 2a), and suppressed oxygen consumption while increasing glycolysis, as measured by the extracellular acidification rate (ECAR) (Fig. 2b–d). Pharmacological inhibition of IDO1 in human MDMs similarly decreased NAD+ and mitochondrial respiration (Supplementary Fig. 2d–g). Supplementation of Ido1−/− macrophages with KYN restored NAD+ levels, and normalized oxygen consumption and ECAR (Fig. 2a–d and Supplementary Fig. 2h). Thus, disruption of the KP decreases cellular NAD+ abundance and mitochondrial respiration.
Fig. 2|. Loss of ID01 lowers cellular NAD+ and disrupts macrophage mitochondrial respiration and dynamics.
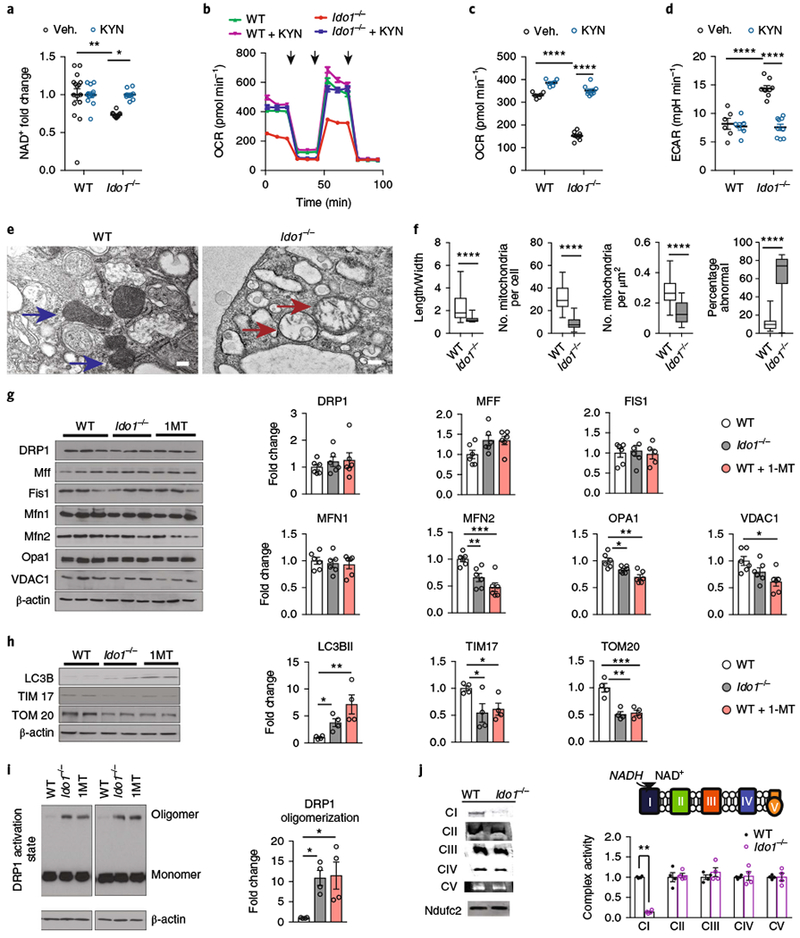
a-d, Primary peritoneal macrophages from WT and Ido1−/− mice were supplemented with vehicle or 25 μM KYN for 20 h. See also Supplementary Fig. 2. a, Measurement of NAD+ levels; n = 16 biologically independent samples per WT group and n = 10 biologically independent samples per Ido1−/− group, represented as the mean ± s.e.m.; **P = 0.0093 for WT vehicle versus Ido1−/− vehicle *P = 0.0220 for Ido1−/− vehicle versus Ido1−/− KYN, two-way ANOVA with Tukey’s post hoc test. b, Representative trace from two independent experiments of real-time changes in oxygen-consumption rate (OCR) after treatment with oligomycin (1 μM), FCCP (2 μM), and rotenone (0.5 μM), respectively, represented by three vertical black arrows, using the Seahorse extracellular flux assay. Data are represented as the mean ± s.e.m. with n = 6 biologically independent samples for the WT group and n = 9 biologically independent samples for the Ido1−/− group. c, Quantification of basal respiration; n = 6 biologically independent samples for the WT group and n = 9 biologically independent samples for the Ido1−/− group, data are represented as the mean ± s.e.m.; ****P < 0.0001 for WT vehicle versus Ido1−/− vehicle, ****P < 0.0001 for Ido1−/− vehicle versus Ido1−/− KYN, two-way ANOVA with Tukey’s post hoc test. d, Quantification of ECAR; n = 6 biologically independent samples for the WT group and n = 9 biologically independent samples for the Ido1−/− group, represented as the mean ± s.e.m.; ****P < 0.0001 for WT vehicle versus Ido1−/− vehicle, ****P < 0.0001 for Ido1−/− vehicle versus Ido1−/− KYN by two-way ANOVA with Tukey’s post hoc test. See also Supplementary Fig. 2d–h. e, Representative TEM images from two independent experiments of WT (blue arrows) and Ido1−/− (red arrows) macrophages (scale bar, 2 μm). f, Quantification of length-to-width ratio, numbers, density, and percentage abnormal mitochondria in Ido1−/− macrophages; n = 257 biologically independent samples for the WT group and n = 269 biologically independent samples for the Ido1−/− group, represented as a box and whisker plot (5–95 percentile); ****P < 0.0001, two-tailed Student’s t-test. g, Peritoneal macrophages from WT, Ido1−/−, and WT + 1MT (200 μM, 20 h) animals were assayed for mitochondrial fission, fusion, and mass proteins using quantitative immunoblot analysis. Data are represented as the mean ± s.e.m. (n = 6 biologically independent samples per group; *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA with Tukey’s multiple comparisons test). h, Representative immunoblot for markers of autophagy and mitochondrial mass (n = 9 biologically independent samples per group; *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA with Tukey’s multiple comparisons test). i, Representative immunoblot and quantification of DRP1 oligomerization in Ido1−/− and 1MT-treated macrophages (n = 4 biologically independent samples per group, *P < 0.05, one-way ANOVA with Tukey’s multiple comparisons test). j, Left: representative BN-PAGE from two independent experiments of complexes I-V and positive control NDUCF2 derived from WT and Ido1−/− macrophage mitochondria. Right: quantification of BN-PAGE complex I-V activities; n = 4 biologically independent samples per group, represented as the mean ± s.e.m.; **P = 0.0019, Student’s t-test.
See also Supplementary Fig. 2i.
We next examined mitochondrial morphology in mouse WT and Ido1−/− peritoneal macrophages using transmission electron microscopy (TEM; Fig. 2e). Loss of IDO1 disrupted the morphology and reduced numbers of mitochondria (Fig. 2f). Quantitative immunoblot analysis of the canonical proteins involved in mitochondrial dynamics and mitophagy were markedly altered in Ido1−/− and 1MT-treated macrophages. Significant reductions of the outer membrane fusion protein mitofusin-2 (MFN2) and the inner membrane fusion protein mitochondrial dynamin-like GTPase (OPA1) were observed along with increased oligomerization of the fission protein dynamin-related protein 1 (DRP1) (Fig. 2g,i). Additionally, the mitochondrial mass proteins voltage-dependent anion-selective channel 1 (VDAC1), protein transporter TIM17 (TIM17), and translocase of outer membrane 20 kDa subunit (TOM20) were markedly decreased and the 14-kDa form of LC3B (LC3BII) was increased, consistent with increased mitophagy (Fig. 2h).
We then investigated the function of the ETC using blue native polyacrylamide gel electrophoresis (BN-PAGE) (Fig. 2j). BN-PAGE allows the isolation of intact mitochondrial ETC protein complexes without dissociating them into individual protein subunits, permitting the analysis of mitochondrial complex enzyme activities. Ido1−/− mitochondria showed a selective deficit only in complex I activity, which requires NADH for electron transfer (Supplementary Fig. 2i).
Given the alterations in mitochondrial respiratory chain activity and morphology, we examined the overall changes to metabolite concentrations using untargeted LC-MS-based metabolomics (Supplementary Fig. 3). Between WT and Ido1−/− macrophages, 124 metabolites were significantly altered (false discovery rate (FDR) < 0.05), with decreases in protein dipeptides, suggesting lower protein catabolism, and increased fatty acids and acylcarnitines, suggesting impaired fatty acid oxidation. Taken together, these findings indicate that loss of IDO1 and KP activity decreases de novo NAD+ generation, complex I activity, and mitochondrial respiration, and alters cellular metabolism.
Loss of QPRT is a phenocopy of loss of IDO1 in macrophages.
We then tested whether loss of QPRT activity, which is downstream of IDO1 in the KP, would similarly disrupt mitochondrial function (Fig. 3 and Supplementary Fig. 4a–f). QPRT activity was inhibited genetically using Qprt−/− peritoneal macrophages, and pharmacologically in human MDMs using the inhibitor phthalic acid (PA)21. Loss of QPRT activity decreased NAD+ abundance, suppressed oxidative phosphorylation, and increased ECAR, changes that could be reversed with the administration of the NAD+ precursor β-nicotin-amide mononucleotide (NMN) (Fig. 3a–d and Supplementary Fig. 4a–d). Loss of QPRT activity increased mitochondrial reactive oxygen species (ROS; labeled with MitoSOX red dye) generation and disrupted membrane potential (Fig. 3e,f), mitochondrial morphology (Fig. 3g), and mitochondrial dynamics (Fig. 3h). BN-PAGE on mitochondria isolated from human MDMs treated with PA showed a selective deficit in complex I activity (Fig. 3i), similar to what was observed in Ido1−/− macrophages. Targeted metabolomics for Qprt−/− macrophages with or without NMN (Fig. 3j) and PA-treated human MDMs (Supplementary Fig. 4e,f) demonstrated that loss of QPRT activity elevates lactate, pentose phosphate pathway intermediates, and proinflammatory TCA intermediates that favor production of interleukin-1β (IL-1β) and IL-6 over anti-inflammatory cytokines such as tumor growth factor-β (TGF-β) and IL-42,22–24.
Fig. 3|. Loss of QPRT reduces cellular NAD+ and disrupts macrophage oxidative phosphorylation, dynamics, and metabolism.
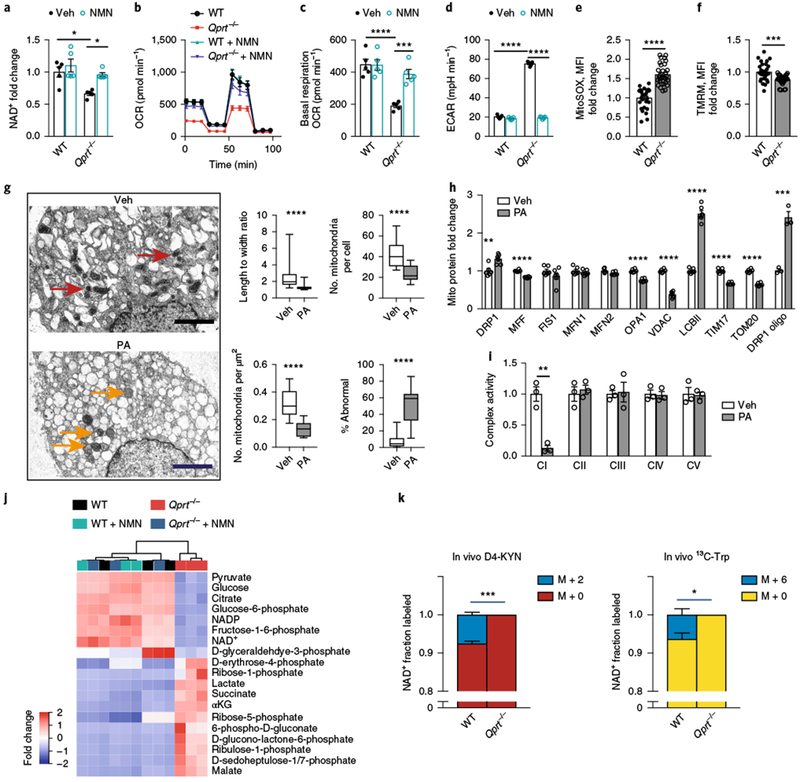
a-d WT and Qprt−/− macrophages were treated with vehicle or NMN 10 μM for 20 h. a, NAD+ levels were measured with LC-MS; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; *P = 0.0121 for WT vehicle versus Qprt−/− vehicle, *P = 0.0288 for Qprt−/− vehicle versus Qprt−/− + NMN, two-way ANOVA with Tukey’s post hoc test. b, Representative OCR trace from two independent experiments. c, Quantification of basal respiration; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001 and ***P < 0.001, two-way ANOVA with Tukey’s post hoc test. d, Quantification of ECAR; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-way ANOVA with Tukey’s post-hoc test. See Supplementary Fig. 4a–d. e, Quantification of mitochondrial ROS using MitoSOX in Qprt−/− and WT macrophages; n = 30 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. f, Quantification of mitochondrial membrane potential using TMRM; n = 30 biologically independent samples per group, represented as the mean ± s.e.m.; ***P = 0.001, two-tailed Student’s t-test. g, Human MDMs were treated with PA (500 μM, 20 h). Left: transmission electron micrographs of vehicle- and PA-treated human MDMs shows altered morphology of mitochondria (vehicle, red arrows; PA, orange arrows; scale bar, 2 μM). Right: quantification of mitochondrial length-to-width ratio, number of mitochondria per cell, density, and percentage abnormal mitochondria (n = 227 biologically independent samples per group, represented as a box and whisker plot (5–95 percentile); ****P < 0.0001, two-tailed Student’s t-test). h, Quantification of mitochondrial fission, fusion, and mass proteins in human MDMs treated with PA (500 μM, 20 h; n = 6 biologically independent samples per group; **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed Student’s t-test). i, BN-PAGE of complex I activity in PA-treated macrophages; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; **P = 0.0098, two-tailed Student’s t-test. j, Hierarchical clustering of targeted metabolomics for glycolysis, pentose phosphate shunt, and citric acid cycle metabolites in WT and Qprt−/− macrophages treated with vehicle or NMN (n = 3 biologically independent samples per group). See Supplementary Fig. 4e,f. k, WT and Qprt−/− mice were orally administered isotope-labeled KYN (D4-KYN) or Trp (13C-Trp) and de novo NAD+ synthesis was measured with LC-MS 4h later in peritoneal macrophages; n = 3 mice per group, represented as the mean ± s.e.m. *P = 0.0171, ***P = 0.0003, two-tailed Student’s t-test.
In vivo modeling of de novo NAD+ synthesis revealed a substantial incorporation of isotope-labeled D4-KYN or Trp (13C-Trp) into NAD+ in macrophages from WT but not Qprt−/− mice (Fig. 3k and Supplementary Fig. 4g). Mice were orally administered 8 mg kg−1 of D4-KYN or 13C-Trp; 4 h later, peritoneal macrophages were collected and subjected to LC-MS analysis. Basal in vivo labeling of de novo NAD+ was 7.56% for labeled KYN and 6.36% for Trp; however, there was no synthesis of NAD+ in Qprt−/− macrophages. These data indicate that the de novo pathway is active basally in vivo in macrophages and that it is dependent on QPRT activity.
De novo NAD+ synthesis regulates macrophage polarization.
The metabolic profile observed with the inhibition of QPRT suggested that de novo NAD+ synthesis may promote an anti-inflammatory polarization state. Recent data support the concept that mitochondrial bioenergetics regulate immune responses23–25. Accordingly, we tested whether inhibition of de novo NAD+ biosynthesis would alter macrophage function under basal conditions and in response to lipopolysaccharide (LPS). Here we assayed (1) surface markers using flow cytometry, (2) cytokine and chemokine generation, and (3) phagocytosis of Escherichia coli. QPRT inhibition by PA increased basal surface expression of the proinflammatory markers CD86 and CD64, and decreased the expression of the anti-inflammatory markers CD206 and CD23 as compared to vehicle controls, a change that could be prevented with supplementation of the NAD+ precursor NMN (Fig. 4a,b). The complex I inhibitors rotenone and piericidin A similarly shifted human MDM polarization toward a proinflammatory state (Supplementary Fig. 5b,c).
Fig. 4|. De novo NAD+ synthesis regulates basal and LPS-activated macrophage polarization, immune factor generation, and phagocytosis.
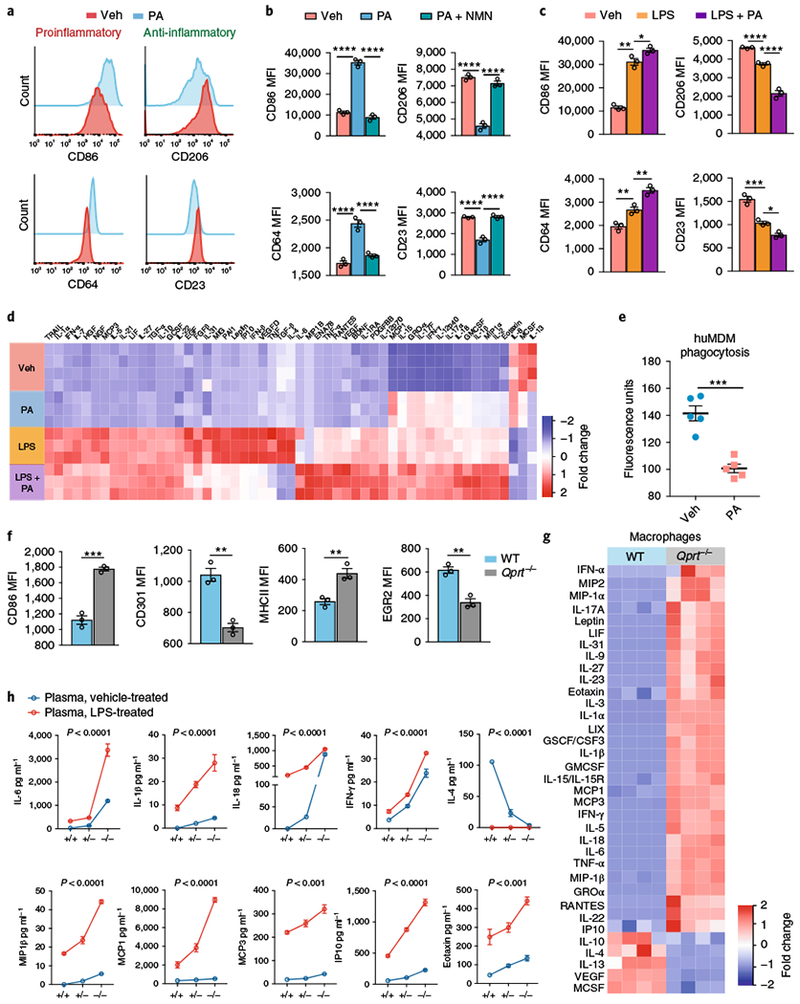
a–e Human MDMs were treated with the QPRT inhibitor PA (500 μM, 20 h) ± NMN (10 μM, 20 h) or ± LPS and assayed by flow cytometry for inflammatory surface markers and immune factors using Luminex multi-analyte measurements. a, Representative histograms of three independent experiments for the proinflammatory markers CD86 and CD64 and anti-inflammatory markers CD206 and CD23 in resting macrophages stimulated with PA. See Supplementary Fig. 5a,b. b, MFIs were quantified in human MDMs treated with PA and NMN; n = 3 biologically independent samples per group, 7,300–11,200 cells per group, represented as the mean ± s.e.m.; ****P < 0.0001, one-way ANOVA with Tukey’s post hoc test. c, MFIs for proinflammatory and anti-inflammatory markers in LPS-stimulated macrophages treated with PA; n = 3 biologically independent samples per group, 7,200–13,358 cells per group, represented as the mean ± s.e.m.; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Tukey’s post hoc test. See Supplementary Fig. 5c. d, Hierarchical clustering of immune factors in culture medium of human MDMs ± PA and ± LPS (n = 4 per group; P < 0.05 by ANOVA). See also Supplementary Figure 5d. e, Phagocytosis of fluorescein-labeled E. coli in human MDMs treated with PA or vehicle; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; ***P = 0.0002, two-tailed Student’s t-test. f, WT and Qprt−/− peritoneal macrophages were assayed by flow cytometry for proinflammatory CD86 and major histocompatibility complex class II (MHCII) and anti-inflammatory CD301 and early growth response protein 2 (EGR2); n = 3 biologically independent samples per group, 2,145–3,224 cells per group, represented as the mean ± s.e.m.; **P < 0.01, ***P <0.001, two-tailed Student’s t-test. g, Hierarchical clustering of immune factors produced by peritoneal macrophages from WT and Qprt−/− mice 20 h after isolation. h, WT and Qprt−/− mice were systemically stimulated with either vehicle or LPS (5 mg kg−1, intraperitoneal injection) and plasma was assayed at 20 h for immune factors. n = 3 biologically independent samples for the WT group, n = 4 biologically independent samples for the Qprt−/− group, represented as the mean ± s.e.m. The two-way ANOVA effect of LPS is P < 0.0001 for all immune factors; the effect of genotype is P < 0.0001 for all immune factors except for monocyte chemoattractant protein 1 (MCP1) and eotaxin, where P < 0.001.
The effect of LPS on proinflammatory and anti-inflammatory surface markers was potentiated with PA administration (Fig. 4c and Supplementary Fig. 5a). By multi-analyte assay, PA increased macrophage production of canonical proinflammatory factors, including growth-regulated oncogene-α (GRO-α), IL-17A, IL-12p40, interferon-γ (IFN-γ), IL-Iβ, IL-18, IL-2, and eotaxin (Fig. 4d and Supplementary Fig. 5d). LPS further amplified this proinflammatory response. Inhibition of QPRT also curtailed macrophage phagocytosis (Fig. 4e). In line with the enhanced proinflammatory state of PA-treated human MDMs, peritoneal macrophages derived from Qprt−/− mice also demonstrated elevated proinflammatory and reduced anti-inflammatory surface markers (Fig. 4f) and parallel changes in proinflammatory and anti-inflammatory immune factors (Fig. 4g). In vivo, in mice systemically stimulated with LPS, loss of one or both Qprt alleles increased plasma proinflammatory cytokines both basally and after LPS administration in a gene dose-dependent manner (Fig. 4h). We conclude that de novo NAD+ synthesis is essential in vitro and in vivo to maintain an anti-inflammatory and homeostatic macrophage phenotype basally and following immune challenge.
De novo NAD+ synthesis regulates SIRT3 activity.
Sirtuin 3 (SIRT3) is an NAD+-dependent protein deacetylase that regulates the activity of many mitochondrial proteins involved in the TCA cycle and oxidative phosphorylation. Mitochondrial SIRT3 activity was significantly reduced compared to controls in Ido1−/− mouse peritoneal macrophages and in human MDMs treated with PA (Fig. 5a,b). SIRT3 activity for both Ido1−/− macrophages and PA-treated human MDMs was restored to that of controls with NAD+ supplementation (Supplementary Fig. 5a,b), indicating that low SIRT3 activity was due to limiting amounts of mitochondrial NAD+. Inhibition with PA had no effect on SIRT3 protein abundance (Supplementary Fig. 5c).
Fig. 5|. De novo NAD+ synthesis regulates SIRT3 deacetylation of complex I subunits and SOD2.
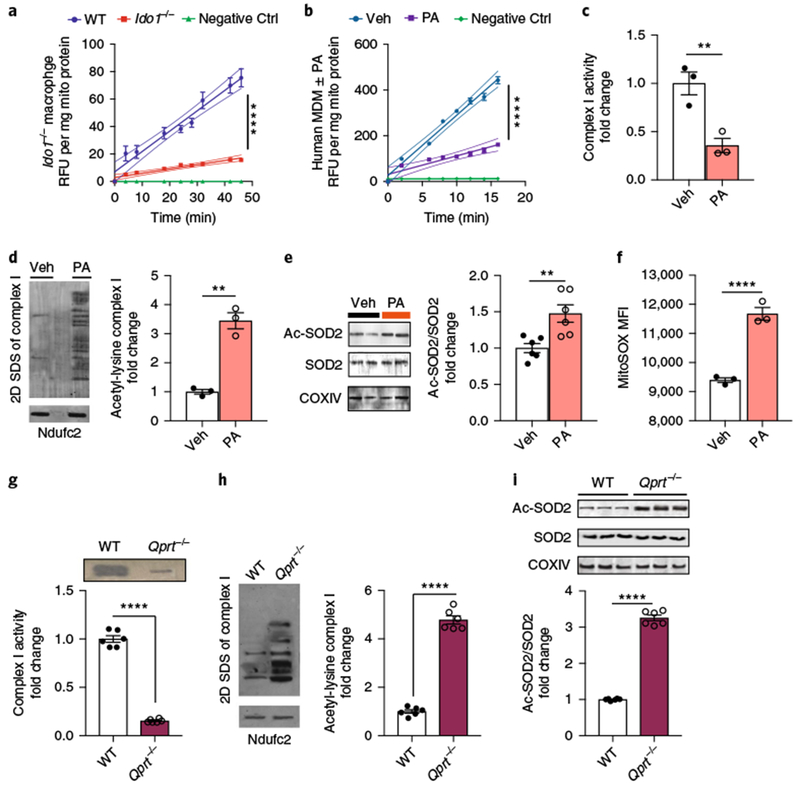
a, WT and Ido1−/− mouse peritoneal macrophages were assayed for SIRT3 steady-state kinetics; n = 3 biologically independent samples per group, represented as the mean ± s.e.m. with curved thin lines denoting 95% confidence intervals (CIs); ****P < 0.0001 via linear regression analysis. See also Supplementary Fig. 6a. b-f, Human MDMs were treated with PA (500 μM for 20 h) and assayed for SIRT3 activity, acetylation of complex I subunits and SOD2, and mitochondrial ROS. b, SIRT3 steady-state kinetics; n = 3 biologically independent samples per group, represented as the mean ± s.e.m. with curved thin lines denoting 95% CIs; ****P < 0.0001 via linear regression analysis. See also Supplementary Fig. 6b. c, BN-PAGE quantification of complex I activity; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; **P = 0.0098, two-tailed Student’s t-test. d, Representative 2D SDS gel electrophoresis of three independent experiments of complex I with quantification of acetyl-lysine in vehicle versus PA-treated human MDMs, n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; **P = 0.0010, two-tailed Student’s t-test. e, Representative immunoblot of two independent experiments of acetylated-SOD2 (ac-SOD2) and SOD2 and quantification of the ac-SOD2/total SOD2 ratio, n = 6 biologically independent samples per group, represented as the mean ± s.e.m; **P = 0.0056, two-tailed Student’s t-test. f, Mean fluorescence index of mitochondrial ROS; n = 3 biologically independent samples per group, 14,700-15,200 cells per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. g-i, WT and Qprt−/− peritoneal macrophages were assayed for complex I activity and SIRT3 deacetylation of complex I subunits and SOD2. g, Complex I activity is reduced in Qprt−/− macrophages; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. h, Quantification of acetyl-lysine residues in complex I subunits; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. i, Ratio of ac-SOD2 to total SOD2; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test.
SIRT3 regulates mitochondrial function in part through its deacetylation of complex I subunits26. Accordingly, the acetylation state of complex I subunits in human MDMs treated with PA and Qprt−/− macrophages was investigated using two-dimensional (2D) BN-SDS-PAGE, wherein complex I was run in a second dimension and individual subunits were assayed for lysine acetylation27 (Fig. 5c,d, g,h). While complex I protein abundance was not altered with the inhibition of QPRT activity, subunit acetylation content was elevated in PA-treated human MDMs and in Qprt−/− mouse macrophages compared to control cells, consistent with a role for de novo NAD+ synthesis in regulating complex I activity by providing NAD+ for SIRT-dependent deacetylation.
SIRT3 also plays a critical role in countering increases in mitochondrial ROS by deacetylating and activating superoxide dismutase 2 (SOD2) in mitochondria28,29. We tested whether loss of de novo NAD+ synthesis, either in PA-treated human MDMs or in Qprt−/− macrophages, would disrupt SOD2 deacetylation and function. Quantitative immunoblot analysis for K68 acetyl-lysine showed increased acetylation of SOD2 in PA-treated human MDMs and Qprt−/− macrophages (Fig. 5e,i). Flow cytometry analysis revealed an elevation in MitoSOX signal (Fig. 5f). The combined impairments in deacetylation of complex I subunits and SOD2 highlight the importance of de novo NAD+ in macrophage mitochondrial function and redox homeostasis.
LPS challenge suppresses de novo NAD+ synthesis.
Previous studies have demonstrated that IDO1 expression is highly induced following innate immune challenge10,11. We next tested whether LPS led to a systemic induction of the KP in vivo. Indeed, LPS triggered increases in plasma concentrations of KYN and the downstream KP metabolites 3-hydroxykynurenine (3-HK), 3-hydroxyanthranilic acid (3-HANA), and QA 20 h after challenge with LPS (Fig. 6a). This induction was entirely dependent on IDO1 and was muted in a gene dose-dependent manner in Ido1+/− and Ido1−/− mice.
Fig. 6|. LPS suppresses QPRT expression and de novo NAD+ synthesis in human MDMs.
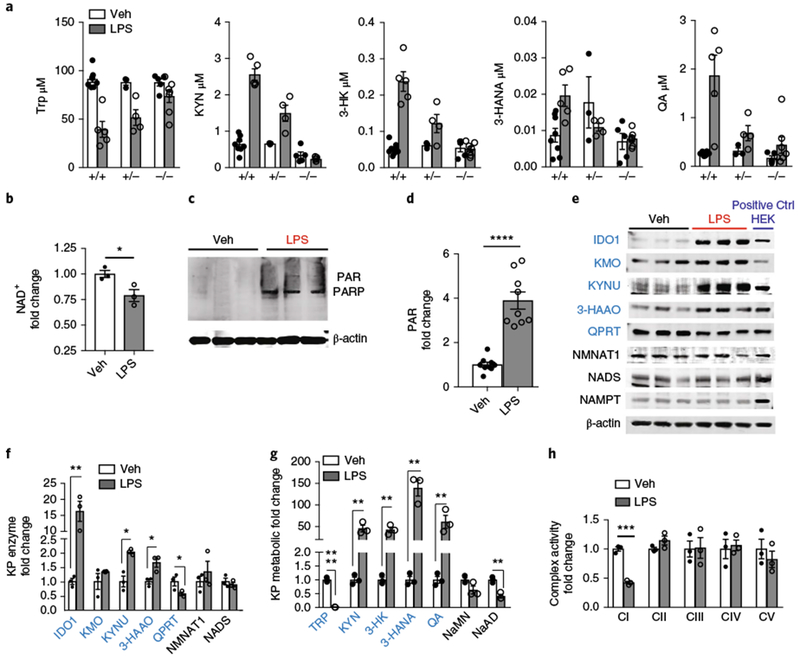
a, WT, Ido1+/−, and Ido1−/− mice were stimulated with LPS (5 mg kg−1 intraperitoneally) and KP metabolites were measured 20 h later in plasma using LC-MS, n = 3–8 per group, represented as the mean ± s.e.m.; two-way ANOVA, effects of genotype are: P < 0.05, 0.0001, 0.0001, 0.05, and 0.001 for Trp, KYN, 3-HK, 3-HANA, and QA, respectively. b-h, Human MDMs were treated with LPS (100 ng ml−1) or vehicle for 20 h. b, Representative LC-MS of two independent experiments of cellular NAD+ levels, n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; *P = 0.0231, two-tailed Student’s t-test. c, Representative immunoblot of three independent experiments of human MDM cell lysates showing PARylated PARP (molecular weight = 116 kDa). d, Quantification of polyADP-ribosylation (PAR) in LPS versus vehicle-treated cells, normalized to vehicle; n = 9 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. e, Representative immunoblot of three independent experiments for KP enzymes in human MDMs stimulated with vehicle or LPS for 20 h. Far right lane: positive Ctrl (+ Ctrl HEK) consists of 20 μg lysates derived from HEK cells transiently transfected with the respective KP enzyme complementary DNA. f, Quantification of KP enzyme levels in LPS and vehicle-stimulated human MDMs at 20 h, normalized to loading control β-actin and then normalized to vehicle-treated; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; *P < 0.05, **P < 0.01, two-tailed Student’s t-test. See also Supplementary Fig. 6f,g. g, LC-MS of human MDM cell lysates for KP and de novo NAD+ metabolites; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; **P < 0.01, two-tailed Student’s t-test. See also Supplementary Fig. 10g. h, BN-PAGE of complex I activity in LPS-treated human MDMs; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; ***P = 0.0002, two-tailed Student’s t-test.
We then tested whether induction of the KP following LPS translated into activation of de novo NAD+ synthesis. However, in human MDMs stimulated with LPS, NAD+ abundance fell significantly compared to vehicle-treated human MDMs (Fig. 6b). This reduction was associated with substantial accumulation of poly(ADP-ribose), consistent with an increase in NAD+ consumption by PARPs (Fig. 6c,d). Since NAD+ abundance reflects the balance between NAD+ generation and consumption, we measured expression of KP and salvage enzymes (Fig. 6e,f). Although amounts of the salvage pathway enzyme NAMPT did not change, KP enzyme abundance was dynamically upregulated following LPS, with the notable exception of QPRT, which began to decline by 4 h after LPS stimulation (Supplementary Fig. 6g). By 20 h after LPS stimulation, IDO1 was increased 16-fold and downstream enzymes, including KYNU and 3-hydroxyanthranilic acid oxygenase, were also increased; however, QPRT expression decreased by ~40%. LC-MS quantification of KP metabolites revealed nearly total consumption of Trp following LPS and large increases in KP metabolites upstream of QPRT, including KYN (up 45-fold), 3-HK (up 43-fold), 3-HANA (up 140-fold), and QA (up 61-fold) (Fig. 6g and Supplementary Fig. 6h), increases that were consistent with changes in their respective KP enzyme abundance. Reduced QPRT expression was associated with decreases in the immediate NAD+ precursors NaMN and NaAD (Fig. 6g) and NAD+ (Fig. 6b). In parallel, LPS suppressed oxidative phosphorylation, increased ECAR (Supplementary Fig. 6d–f), and caused selective loss of complex I activity (Fig. 6h and Supplementary Fig. 6i), in line with the reductions in complex I activity observed in Ido1−/− and PA-treated macrophages.
Taken together, LPS stimulation of macrophages increased NAD+ consumption by PARPs, reduced NAD+ abundance, and suppressed complex I activity and oxidative phosphorylation. Although LPS triggered a substantial increase in KP enzyme abundance and metabolites, QPRT expression instead declined, limiting de novo NAD+ synthesis. Since, de novo NAD+ regulates macrophage polarization, inflammatory response, and phagocytosis basally and in the context of immune challenge, we hypothesized that abundance of QPRT may directly regulate macrophage immune function.
QPRT regulates mitochondrial and immune functions.
We tested whether engineered modulation of QPRT could alter the macrophage response to LPS. We ectopically expressed QPRT in human MDMs (Fig. 7a) and assayed mitochondrial respiration and immune responses (Fig. 7b–m). Compared to control (green fluorescent protein (GFP)-expressing) macrophages, QPRT-overexpressing human MDMs generated higher NAD+ amounts under both basal and LPS conditions (Fig. 7b). Isotope labeling using 13C-Trp demonstrated a significant incorporation of M+6 NAD+ basally and on ectopic expression of QPRT (Fig. 7c). Under resting conditions, de novo NAD+ accounted for 8% of the NAD+ pool, doubling to 16% with QPRT overexpression. LPS stimulation of control macrophages decreased NAD+ labeling, in line with the observed reduction in QPRT expression following LPS. Remarkably, the labeled fraction of NAD+ increased to 34% in QPRT-expressing human MDMs stimulated with LPS. The increase in de novo generation of NAD+ restored mitochondrial respiration, ECAR, and complex I activity (Fig. 7d,e and Supplementary Figs. 6j,k and 8a,b). Ectopic expression of QPRT restored SIRT3 activity in LPS-treated human MDMs to basal levels (Fig. 7f), rescued deacetylation of complex I subunits and SOD2 (Fig. 7g,h and Supplementary Fig. 8c), and normalized MitoSOX and membrane potential (Fig. 7i,j). In line with these changes, glycolytic and TCA cycle intermediates were restored to homeostatic abundance in QPRT-overexpressing macrophages stimulated with LPS (Fig. 7k and Supplementary Fig. 7). These data indicate that in immune-activated macrophages, QPRT activity and de novo NAD+ synthesis are limiting for cellular respiration, SIRT3 activity, and mitochondrial function.
Fig. 7|. Increasing QPRT expression in LPS-treated human MDMs restores mitochondrial metabolism and immune responses.
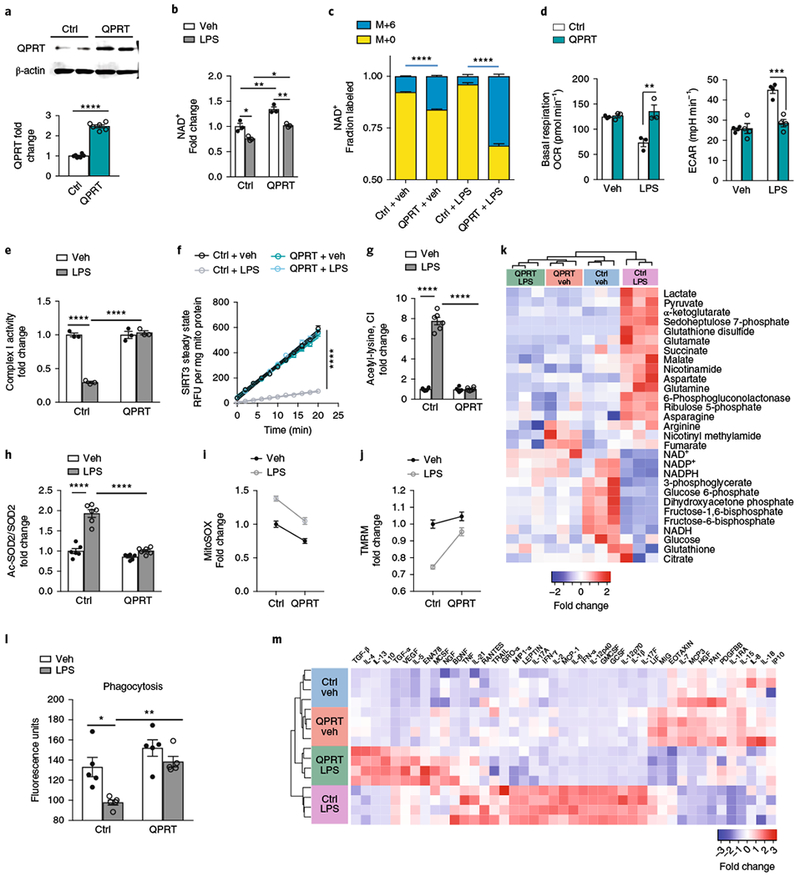
Human MDMs transfected with QPRT or control (GFP) vector were stimulated with either vehicle or LPS (100 ng ml−1) and assayed at 20 h. a, Representative quantitative immunoblot analysis of three independent experiments of control (Ctrl) and QPRT-overexpressing human MDMs; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. b, LC-MS of NAD+ in control and QPRT-overexpressing human MDMs ± LPS; n GFP = 3 biologically independent samples per group, represented as the mean ± s.e.m. Two-way ANOVA, effect of QPRT P < 0.001 and effect of LPS P < 0.01; Tukey’s post hoc test, *P < 0.05, **P < 0.01. c, Isotope-labeled 13C-Trp was added to Ctrl and QPRT human MDMs ± LPS; the fraction of labeled M+6 NAD+ was quantified with LC-MS; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. d, Quantification of basal respiration and ECAR in control and QPRT-overexpressing human MDMs ± LPS; n = 3 biologically independent samples per group for basal respiration: two-way ANOVA, effect of QPRT P < 0.01 and effect of LPS P < 0.05; Tukey’s post hoc test **P = 0.0021; n = 4 per group for ECAR: two-way ANOVA effect of QPRT P < 0.001 and effect of LPS P < 0.0001, Tukey’s post hoc test ***P = 0.00031; data are represented as the mean ± s.e.m. See also Supplementary Fig. 6j,k. e, Complex I activity in LPS-stimulated macrophages overexpressing QPRT, as assayed by BN-PAGE; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA, effects of QPRT and LPS P < 0.0001; Tukey’s post hoc test ****P < 0.0001. See Supplementary Fig. 8a,b. f, SIRT3 steady-state kinetics in LPS-stimulated human MDMs ± QPRT overexpression; n = 6 biologically independent samples per group, represented as the mean ± s.e.m. with curved lines denoting 95% CIs; ****P < 0.0001 by linear regression analysis for Ctrl + LPS versus the other groups. g, Deacetylation of complex I subunits in LPS-stimulated macrophages ± QPRT overexpression; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA effects of QPRT and LPS, P < 0.0001; Tukey’s post hoc test, ****P < 0.0001. h, Ratios of ac-SOD2 to total SOD2 in LPS-stimulated human MDMs ± QPRT overexpression; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA effects of QPRT and LPS, P < 0.0001; Tukey’s post hoc test ****P < 0.0001. See Supplementary Fig. 8c. i, Mitochondrial ROS in LPS-stimulated human MDMs ± QPRT overexpression; n = 30 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA effects of QPRT and LPS, P < 0.0001. j, TMRM in LPS-stimulated human MDMs ± QPRT overexpression; n = 30 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA effects of QPRT and LPS, P < 0.0001. k, Hierarchical clustering of targeted metabolomics of control and QPRT-overexpressing human MDMs ± LPS. See also Supplementary Fig. 7 for untargeted metabolomics analyses. l, Phagocytosis of fluorescein-labeled E. coli is rescued in LPS-treated human MDMs with QPRT overexpression; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA, effects of QPRT and LPS, ***P = 0.0006 and **P < 0.003, respectively; Tukey’s post hoc test, *P < 0.05 and **P < 0.01. m, Hierarchical clustering of immune factors in control and QPRT human MDMs ± LPS, n = 4 per group; P < 0.05 by ANOVA with multiple comparisons. See also Supplementary Fig. 8d,e.
To determine the effect of restoring de novo NAD+ synthesis on functional responses to LPS, we measured changes in phagocytosis and cytokine generation in control and QPRT-overexpressing human MDMs stimulated with vehicle or LPS (Fig. 7l,m and Supplementary Fig. 8d,e). At 20 h after LPS challenge, phagocytic function was significantly suppressed in control macrophages; however, this suppression was fully reversed with QPRT overexpression (Fig. 7l). After LPS treatment, 22 cytokines remained elevated in control human MDMs. QPRT overexpression normalized expression of most of these factors, including canonical proinflammatory cytokines IL-1β, TNF, IL-12, macrophage inflammatory protein 1α, and IL-6. Conversely, anti-inflammatory cytokines that were suppressed with LPS, including IL-4 and TGF-β, increased in LPS-treated QPRT-overexpressing macrophages. Thus, by replenishing QPRT, de novo NAD+ reprograms the polarization state of macrophages away from a proinflammatory state with reduced phagocytic potential toward a phagocytosis-competent and anti-inflammatory state.
De novo NAD+ synthesis in aging macrophages.
NAD+ abundance declines significantly in aging tissues3,7,8, and aging is characterized by a persistent and low-grade inflammatory response. We hypothesized that de novo NAD+ may similarly control mitochondrial metabolism and inflammatory activation state in aging macrophages. Quantification of QPRT expression in human MDMs derived from individuals ≤35 years old (young) and ≥65 years old (aged) demonstrated a significant decline in QPRT in aged macrophages (Fig. 8a and Supplementary Fig. 9a,b). This decline in QPRT expression was associated with an induction of upstream KP metabolites culminating in an accumulation of QA but decreased abundance of NaMN, NaAD, and NAD+ (Fig. 8b), a profile similar to that observed in LPS-stimulated macrophages.
Fig. 8|. De novo NAD+ synthesis is reduced in aging human MDMs and in vivo in aging mouse macrophages.
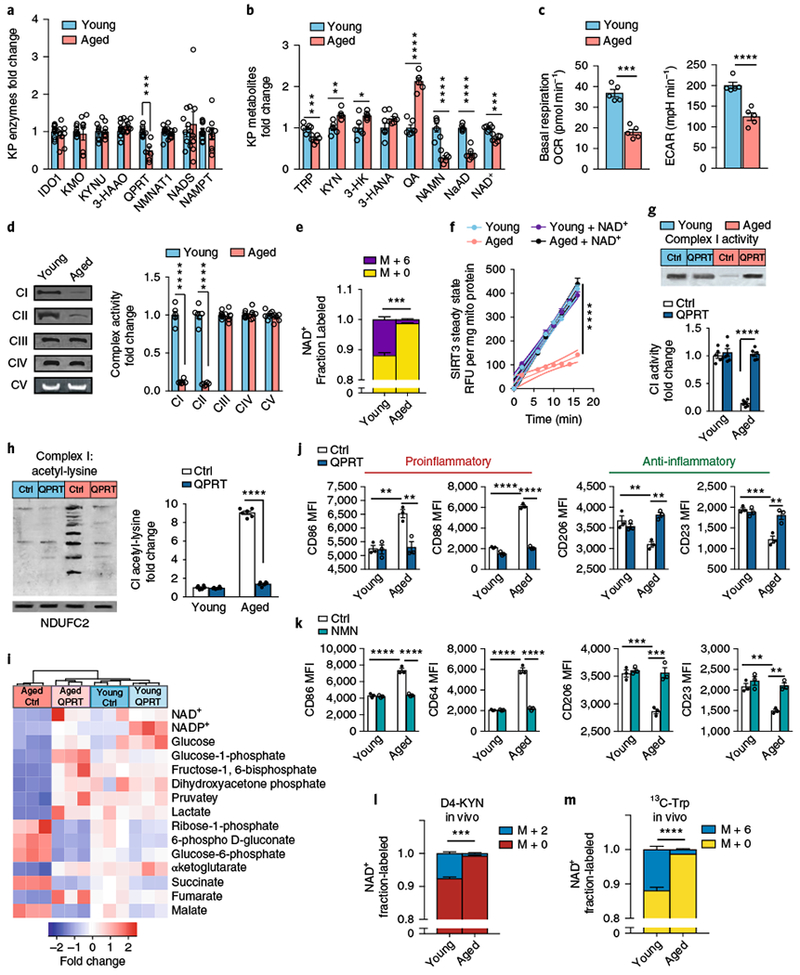
a–f, Young and aged human MDMs were derived from individuals ≤35 and ≥65 years old, respectively, and analyzed for QPRT levels, KP and NAD+ metabolites, mitochondrial respiration, and immune polarization state. See Supplementary Figs. 9,10. a, Quantitative immunoblot analysis of human MDMs of QPRT expression in aging versus young macrophages; n = 9 biologically independent samples per group for IDO1 and NADS, n = 10 biologically independent samples per group for 3-HAAO, n = 8 biologically independent samples per group for all other enzymes, represented as the mean ± s.e.m.; ***P = 0.0006, two-tailed Student’s t-test. b, LC-MS quantification of KP metabolites, NAD+ precursors, and NAD+ in young versus aged macrophages; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed Student’s t-test. c, Basal respiration and ECAR in young compared to aged macrophages; n = 5 biologically independent samples per group, represented as the mean ± s.e.m.; ***P = 0.0002, ****P < 0.0001, two-tailed Student’s t-test. d, Left: representative BN-PAGE blot of complex I and II activities in young versus aged human MDMs. Right: quantification of complex activities; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; ****P < 0.0001, two-tailed Student’s t-test. e, 13C-Trp was added to young and aged human MDMs and the fraction of labeled M+6 NAD+ was quantified with LC-MS; n = 3 biologically independent samples per group, represented as the mean ± s.e.m.; ***P = 0.0004, two-tailed Student’s t-test. f, Young and aged human MDMs were assayed for SIRT3 steady-state activity; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; the curved thin lines denote 95% CIs; ****P < 0.0001 via linear regression analysis. g-j, Young and aged human MDMs were transfected with either control vector (Ctrl) or QPRT vector (QPRT) and assayed for complex activities, targeted metabolomics, and immune polarization. g, Top: representative BN-PAGE blot of complex I activity in young and aged human MDMs transfected with either control or QPRT vectors. Bottom: quantification of complex I activity; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA, effect of age and QPRT, ****P < 0.0001; Tukey’s post hoc test, ****P < 0.0001. h, Left: representative 2D SDS blot of complex I from young and aged human MDMs ± QPRT overexpression assayed for acetyl-lysines. Right: quantification of acetyl-lysines in complex I; n = 6 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA, effect of age and QPRT, ****P < 0.0001; Tukey’s post hoc test, ****P < 0.0001). i, Hierarchical clustering of targeted metabolomics in young versus aged human MDMs transfected with control or QPRT vector (n = 3 biologically independent samples per group). j, MFIs of the proinflammatory markers CD86 and CD64 and anti-inflammatory markers CD206 and CD23 in aged and young human MDMs transfected with control or QPRT vector. N = 3 biologically independent samples per group, 8,374–12,576 cells per group, represented as the mean ± s.e.m.; two-way ANOVA effect sizes: CD86, effect of age and QPRT, P < 0.01; CD64, effect of age and QPRT, P < 0.0001; CD206, effect of QPRT, P < 0.01; CD23, effect of age, P < 0.001, and effect of QPRT, P<0.01; Bonferroni post hoc test: **P < 0.01, ***P < 0.001, ****P < 0.0001. k, Effect of NMN supplementation (10 μM, 20 h). N = 3 biologically independent samples per group, represented as the mean ± s.e.m.; two-way ANOVA effect sizes: CD86, effect of age and NMN, P < 0.0001; CD64, effect of age and NMN, P < 0.0001; CD206, effect of age and NMN, P < 0.001; CD23, effect of age and NMN, P < 0.01; Bonferroni post hoc test: **P < 0.01, ***P < 0.001, ****P < 0.0001. l-m, De novo NAD+ synthesis is reduced in aging mouse macrophages in vivo; n = 3 biologically independent samples per group; ***P = 0.0002 and ****P < 0.0001 for young versus aged, two-tailed Student’s t-test. Young (3 months) and aged (16–18 months) C57B/6J mice were administered 8 mg of D4-KYN (l) or 13C-Trp (m) by oral gavage, and de novo NAD+ levels were measured in peritoneal macrophages 4 h later with LC-MS. Data are represented as the mean ± s.e.m.
Oxygen consumption in aged versus young macrophages was notable for a decrease in both basal respiration and ECAR (Fig. 8c) and a marked reduction in complex I and II activities (Fig. 8d). Metabolic tracing studies using 13C-Trp revealed a marked decline in de novo NAD+ synthesis in aged compared to young human MDMs (Fig. 8e) that was associated with a decline in mitochondrial SIRT3 activity (Fig. 8f). Similar age-related changes in mitochondrial respiration, complex I and II activities, and mitochondrial SIRT3 activity were observed in primary mouse peritoneal macrophages (Supplementary Fig. 10a–d). Reduced SIRT3 steady-state activity in aged human MDMs and mouse macrophages was reversed with NAD+ supplementation (Fig. 8f and Supplementary Fig. 10d), indicating that NAD+ is limiting. In human MDMs, we observed an associated increase in proinflammatory polarization state in aged compared to young macrophages basally and following LPS (Supplementary Fig. 9c).
To test whether increasing de novo NAD+ synthesis in aged macrophages could revert their metabolic and immune phenotypes to those of young macrophages, we examined the effect of increasing expression of QPRT (Supplementary Fig. 9d). QPRT overexpression in aged macrophages prevented the accumulation of QA and restored NAD+ abundance similar to that observed in young macrophages (Supplementary Fig. 9e), and increased oxidative phosphorylation, ECAR, and complex I and II activities (Fig. 8g and Supplementary Fig. 9f–h). Deacetylation of complex I and II subunits was also restored (Fig. 8h and Supplementary Fig. 9i), consistent with a rescue of NAD-dependent mitochondrial SIRT3 activity. Increased QPRT in aged human MDMs normalized glycolytic and TCA metabolites, including the proinflammatory succinate and malate and the anti-inflammatory α-ketoglutarate30 (Fig. 8i), and shifted macrophage activation state toward a less proinflammatory and more homeostatic polarization state (Fig. 8j). Similar effects were observed following NMN supplementation of aged human MDMs (Fig. 8k and Supplementary Fig. 9l). Interrogation of complex II activity using the inhibitor malonate also shifted macrophage polarization toward a more proinflammatory state (Supplementary Fig. 9j,k). Thus, restoring de novo NAD+ synthetic capacity in aged macrophages rejuvenated their metabolic and immune phenotypes and resulted in increased glycolysis and oxidative phosphorylation, and resumption of an anti-inflammatory polarization state.
In vivo modeling in young and aged mice revealed a substantial incorporation of isotope-labeled D4-KYN or 13C-Trp into NAD+ in young mice but not in aged mice (Fig. 8l,m). Although peritoneal macrophages derived from young mice demonstrated robust 7.6% and 11.91% incorporation of labeled KYN and Trp, respectively, aged macrophages showed a tenfold decrease in NAD+ synthetic capacity (0.74% and 1.34% for labeled KYN and Trp, respectively). These findings indicate that the de novo NAD+ pathway contributes to the maintenance of macrophage NAD+ abundance in vivo. This capability is significantly lost with aging, resulting in a proinflammatory activation state.
Discussion
To date, studies of the KP have focused on the effects of intermediate immunomodulatory and neuroactive metabolites in cancer12,20, immune tolerance11,13, and neurodegeneration31–37. Our findings uncover a fundamental role for the KP in regulating macrophage immune function through cell-autonomous generation of NAD+ under basal conditions, with immune challenge, and in aging. In macrophages at rest, we demonstrate that KP activity and de novo generation of NAD+ are necessary to maintain NAD+ levels and mitochondrial respiration; this metabolic state in turn directs an anti-inflammatory homeostatic state with robust phagocytic capability. However, in LPS-activated macrophages, a more complex interaction between metabolic state and immune function emerges, wherein the highly induced KP unexpectedly fails to replenish NAD+ stores because the level of QPRT, which metabolizes QA to the NAD+ precursor NaMN, becomes limiting. In this situation, NAD+ depletion from increased activity of NAD-consuming enzymes is not countered by increased de novo NAD+ synthesis; mitochondrial respiration is suppressed in part through deficient SIRT3 activity and defective complex I function, leading to impaired phagocytic capacity and a proinflammatory phenotype. Replenishment of QPRT restores de novo NAD+ synthesis and reverts LPS-stimulated macrophages to a respiration- and phagocytosis-competent state with a lower inflammatory cytokine profile. Remarkably, aged macrophages demonstrate similar upstream KP activation in tandem with suppressed QPRT and de novo NAD+ synthesis; this in turn decreases mitochondrial respiration and induces a proinflammatory shift in activation state. De novo macrophage NAD+ synthetic capacity decreases significantly in aging mice, highlighting a role for KP-derived NAD+ in regulating macrophage metabolism and function not only in immune contexts but also in dysregulated immune responses characteristic of aging.
Recent studies indicate that cellular metabolism can direct macrophage polarization state via the accumulation of TCA cycle intermediates that regulate proinflammatory responses1,2,22,38–40. In line with these findings, blockade of de novo NAD+ synthesis led to accumulation of the proinflammatory TCA intermediates succinate and malate. Succinate accumulation increases proinflammatory cytokines IL-1β and IL-6 and suppresses anti-inflammatory IL-4 and IL-10 through the inhibition of prolyl-hydroxylase domains responsible for regulating hypoxia-inducible factor-1α, a transcriptional activator of canonical proinflammatory cytokines. The proinflammatory state resulting from blockade of de novo NAD+ synthesis may be relevant to current use of IDO1 inhibitors in cancer to reduce KYN-mediated immune suppression14. Since anti-inflammatory macrophage activity may be an important mechanism by which tumors escape immune surveillance, IDO1 inhibitors may promote immunotherapy by impairing macrophage de novo NAD+ synthesis and enhancing their proinflammatory responses. On the other hand, we also found that blockade of de novo NAD+ synthesis suppressed phagocytic capacity, an immune function important in macrophage clearance of tumor cells.
A unique property of macrophages is the ability to rapidly launch and sustain an effective and self-limited immune response. A variety of immune danger signals, including LPS and IFN-γ, strongly increase the expression of IDO1, so activation of the KP to generate de novo NAD+ represents a reasonable strategy to manage an increase in bioenergetic and metabolic demand. However, although immune challenge with LPS triggered upstream KP activation, it limited de novo NAD+ synthesis by reducing QPRT expression and impeding conversion of QA to NAD+, prolonging the inflammatory state and suppressing phagocytosis. This could represent a normal response designed to sustain a high proinflammatory state or, alternatively, an inability of the macrophage to autonomously resolve the inflammatory response and return to homeostasis.
To date, there is limited understanding of QPRT regulation. Several studies indicate that QPRT is subject to post-translational ubiquitination that leads to degradation41–43. Further investigation may reveal whether increased QPRT activity is a physiological trigger for the return of macrophages to the resting, noninflammatory state, in which case QPRT activity may function as a novel metabolic switch that regulates macrophage effector responses. The nearly identical change in levels of QPRT and KP metabolites in aged macrophages, compared to LPS-challenged macrophages, suggests that de novo NAD+ synthesis may be a conserved mechanism to program macrophage immune responses across acute and chronic inflammatory conditions. In vivo, in aged mice, a clear suppression of endogenous NAD+ synthetic capacity was observed, indicating that KP-derived NAD+ is critical in sustaining cellular NAD+concentrations in aging. Age-associated loss of de novo NAD+ generation probably contributes to the proinflammatory shift in immune polarization state and to changes in essential immune functions, including phagocytosis and the ability to return to immune homeostasis.
The observed elevation of QA and breakdown of de novo NAD+ synthesis following immune challenge may also bear on the pathophysiology of infectious and inflammatory brain diseases35, where brain expression of QA from brain microglia44 or infiltrating macrophages are highly elevated. QA is an agonist of the N-methyl-d-aspartate receptor, and elevated abundance of QA may elicit neurotoxic effects in vivo through over-activation of these receptors45. Based on our findings, the elevation in QA may be a marker more generally of immune cell dysfunction and reflect a state of myeloid NAD+ depletion that drives proinflammatory and neurotoxic responses. Strategies to enhance the metabolism of QA to NAD+, for example, by activating existing QPRT or increasing QPRT expression levels, could return myeloid cells to a more homeostatic and neurotrophic state.
Recent studies have identified a critical role for cellular bioenergetics in determining macrophage activation and functional states1,2,25,46,47. Additional advances have highlighted the importance of NAD+ metabolism in regeneration, aging, and disease4,8,48–50, where immune responses play a prominent role. In this context, we propose a novel function for the KP where its contribution of de novo NAD+ synthesis, as determined by the activity of QPRT, regulates homeostatic, immune-challenged, and aging macrophage immune responses through effects on NAD-dependent signaling and cellular metabolism.
Methods
Animals.
This study was conducted in accordance with National Institutes of Health (NIH) guidelines; protocols were approved by the Institutional Animal Care and Use Committee at Stanford University. All mice were housed in an environmentally controlled, pathogen-free barrier facility on a 12 h light-dark cycle, temperature, and humidity, with food and water available ad libitum. Ido1−/− mice backcrossed onto a C57BL/6J background were obtained from the Jackson Laboratory (Ido1tmLAlm; donating investigator: A. Mellor, Medical College Georgia) and heterozygous Ido1+/− mice were intercrossed to generate the Ido1+/+ and Ido1−/− genotypes. Qprt−/− mice on a C57BL/6J-129SvEv background were provided by R. Schwarcz at the University of Maryland (generated by Lexicon Pharmaceuticals; Qprt−/− mice available at Taconic Biosciences).
Materials.
FK866 (Cayman Chemical) and L-kynurenine (Sigma-Aldrich) were used in cell culture at 10 and 25 μM, respectively, for 20 h. The IDO1-inhibitor 1MT (Sigma-Aldrich) and QPRT inhibitor PA (Sigma-Aldrich) were used in cell culture at 200 μM 1MT or 500 μM PA for 20 h. Endotoxin-free LPS (055:B5 E. coli; Merck) was used in vitro at 100 ng ml−1 and in vivo at 5 mg kg−1 by intraperitoneal injection. Flow cytometry and immunoblot antibodies are described in Supplementary Tables 1 and 2.
Mouse peritoneal macrophage culture.
Peritoneal macrophages were collected from 6- to 7-month-old Ido1−/−, Qprt−/−, and WT mice. Mice were injected intraperitoneally with 1.5 ml 3% (w/v) thioglycolate medium (BD Biosciences), and primary macrophages were isolated 3–4 days later by flushing with ice-cold 1 × PBS buffer (Corning). Cells were seeded at a density of 3 × 106 cells per well in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), 100 U ml−1 penicillin and streptomycin, and maintained at 5% CO2 at 37 °C. After overnight culture, cells were washed twice with medium to remove nonadherent cells.
Human macrophage culture.
Peripheral blood mononuclear cells from de-identified healthy donors (35–65 years old) were obtained from the Stanford Blood Center and transferred to 50 ml conical tubes. Samples were diluted with 20 ml PBS and layered onto 10 ml of Ficoll-Paque (GE Healthcare) using a Pasteur pipette. Tubes were centrifuged at 1,500 r.p.m. for 25 min without brake at 20 °C. The mononuclear cell layer was transferred to a new 50 ml conical tube, resuspended in 50 ml 1 × PBS and centrifuged at 1500 r.p.m. for 10 min, repeated twice. After centrifugation, monocytes were isolated using a Monocyte Isolation Kit, human (MACS; Miltenyi Biotech). Cells were then plated 10 × 106 per 10-cm petri dish and differentiated for 7 days in Roswell Park Memorial Institute (RPMI) media supplemented with 10% FBS, 1% penicillin-streptomycin, and 50 ng ml−1 M-CSF (Peprotech).
Quantitative immunoblotting.
Quantitative immunoblotting was carried out as previously described51,52. Mouse anti-β-actin (1:10,000; Sigma-Aldrich) was used as an internal loading control. Densitometry quantification was carried out using ImageJ (NIH). Antibodies and their concentrations are listed in Supplementary Table 1. Specificity of antibodies to KP enzymes was validated using human embryonic kidney (HEK)-transfected cells for respective enzyme complementary DNA (for a list of vectors, see Supplementary Table 5).
LC-MS measurement of KP, NAD+ precursors, and NAD+.
Isotope labeling was performed as previously described53. Labeled compounds L-kynurenine D4 (Buchem BV), L-tryptophan (13C11; Cambridge Isotope Laboratories), or unlabeled compounds L-kynurenine (Sigma-Aldrich), L-tryptophan (Sigma-Aldrich) were added to customized RPMI media lacking L-tryptophan (customized RPMI 1640 Medium + Gibco GlutaMAX supplement + HEPES; Thermo Fisher Scientific).
Human MDMs were grown on 10-cm plates (Corning). For steady-state labeling of metabolites U-13C-tryptophan (0.025 mM) or l-kynurenine D4 (0.025 mM), labeled medium was replaced every day and then 2 h before metabolic analysis at which point cellular metabolism was quenched by rapidly cooling cells on dry ice. Cells were washed with 1 × PBS twice by aspirating media and immediately adding 1 ml −80 °C 80:20 methanol/water. After 20 min of incubation on dry ice, the resulting mixture was scraped, collected into a centrifuge tube, and centrifuged at 10,000g for 5 min at 4 °C. Pellets were then extracted again with 500 μl −80 °C 80:20 methanol/water and incubated for 5 min, centrifuged at 10,000g for 5 min at 4 °C. Both extractions were combined into a 1.5 ml microcentrifuge tube.
LC-MS was performed as previously described54,55. In brief, the LC-MS method involved hydrophilic interaction chromatography (HILIC) coupled to a Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The LC separation was performed on a XBridge BEH Amide Column (150 mm × 2.1 mm, 2.5 μm particle size; Waters). Solvent A is 95:5 water:acetonitrile with 20 mM ammonium bicarbonate; solvent B is acetonitrile. The gradient was: 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; and 30 min, 85% B. Other LC parameters are: flow rate 150 μl min−1, column temperature 25 °C, injection volume 5 μl The mass spectrometer was operated in positive ion mode for the detection of NAD+ and negative ion mode for KP metabolites (Trp, KYN, 3-HANA, QA). Other MS parameters were: resolution of 140,000 at m/z 200, automatic gain control target at 3e6, maximum injection time of 30 ms and scan range of m/z 75–1,000. All isotope labeling patterns were corrected for natural abundance.
Real-time oxygen consumption rate (OCR) and ECAR.
Cells were plated at 2.7 × 105 cells per well in a Seahorse XF24 Cell Culture Microplate (Agilent). Cells were then treated with LPS (100 ng ml−1), 1MT (200 μM), PA (500 μM), or combination for 20 h. Cells were washed twice with Agilent Seahorse XF Media (Agilent) supplemented with 1 mM pyruvate, 2 mM L-glutamine, and 2 mM D-glucose; a final volume of 525 μl was placed in each well. Cells were then incubated in a 0% CO2 chamber at 37 °C for 1 h before being placed into a Seahorse XFe24 Analyzer (Agilent). For OCR and ECAR experiments, cells were treated with 1 μM oligomycin, 2 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin. A total of three OCR and pH measurements were taken after each compound was administered. All Seahorse experiments were repeated at least three times.
In situ complex activity.
Primary mouse macrophages or differentiated human MDMs were plated at 2.7 × 105 cells per well in a Seahorse XF24 Culture Microplate. Cells were washed twice with mannitol sucrose-bovine serum albumin (MAS-BSA) solution and then a final volume of 525 μl was placed in each well. Cells were incubated in 0% CO2 for 10 min before being placed into a Seahorse XFe24 Analyzer. To measure ETC complexes in situ, cells were permeabilized using saponin (25 μg ml−1; Sigma-Aldrich) and stimulated with specific complex substrates: complex I, pyruvate (5 mM) + malate (2.5 mM) + ADP (1 mM); complex II, succinate (10 mM) + rotenone (1 μM) + ADP (1 mM); complex III, duroquinol (10 mM) + ADP (1 mM); complex IV, TMPD (0.5 mM) + ascorbate (2 mM). A total of three OCR measurements were taken after each compound was administered. All Seahorse experiments were repeated at least three times.
Tetramethylrhodamine methyl ester (TMRM) staining.
TMRM (Thermo Fisher Scientific) was loaded onto cells at 150 nM in assay buffer (80 mM NaCl, 75 mM KCl, 25 mM D-glucose, 25 mM HEPES, pH 7.4); cells were incubated at 37 °C for 10 min, washed four times in PBS and analyzed using a SpectraMax M2e microplate reader (Molecular Devices; excitation: 544 nm and emission: 590 nm; bottom reading with 50 flashes per well).
TEM.
Primary mouse macrophages (Ido1+/+ and Ido1−/−) or differentiated human MDMs ± PA (500 μM for 20 h) were plated in 6-well plates at 2 × 106 cells per well; 2 × 106 cells ml−1 were isolated for electron microscopy processing. Cells were fixed in 2% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate (pH 7.4), treated with 10% gelatin solution in sodium cacodylate buffer, and incubated with 2% osmium tetroxide. Cell pellets were stained with 1% uranyl acetate, dehydrated in ethanol, and embedded in resin (EPON epoxy resin). Sections were generated using a Leica EM UC7 ultramicrotome (Leica Microsystems) at 50 nm and placed within grids stained with a 1:1 mix of 3% uranyl acetate, 50% acetone for 30–60 s. Grids were imaged with a JEM 1400 transmission electron microscope (JEOL) at ×1,200 for low magnification and ×12,000 for high magnification.
BN-PAGE and 2D SDS-PAGE.
Mitochondria were isolated from human MDMs and mouse peritoneal macrophages, as previously described56. In brief, two 10-cm plates containing 20 × 106 cells were harvested with 0.25% trypsin-EDTA (Thermo Fisher Scientific), washed twice with cold PBS (Corning) and resuspended at a concentration of 1 × 106 cells in 100 μl PBS in a 1.5 ml Eppendorf tube and 100 μl digitonin solution (8 mg ml−1; Sigma-Aldrich). Cells were incubated on ice for 10 min and 1 ml of PBS was added to dilute samples. Cells were then spun for 5 min at 10,000g at 4 °C. After removing the supernatant, the pellet was washed in 1 ml cold PBS. Pellets were solubilized in 100 μl buffer containing 1.5 M 6-aminohexanoic acid, 50 mM Bis-Tris-HCl buffer (pH 7.0), and 0.5 mM EDTA; 20 μg of mitochondrial protein was loaded into a 4.5–13% gradient gel for electrophoresis. Samples were run at 30 V for 30 min, after which the voltage was increased to 80 V. Individual lanes were removed and incubated in the respective substrates (Supplementary Table 3) for each complex of the ETC overnight. Densitometry analysis was carried out for each complex using ImageJ; 2D SDS was performed as previously described56,57. In brief, individual complexes were isolated from BN gels and placed in reducing SDS buffer prepared as previously described for 30 min56. After, gels were placed on a second dimension gel (NuPAGE 4–12% Bis-Tris Protein Gel, 1.0 mm, 2D-well; Thermo Fisher Scientific) and run at 80 V for 2.5 h. Gels were then assayed for acetyl-lysine, acetyl-SOD2 (ac-SOD2), and SOD2 quantitative immunoblot analyses.
Telomere quantification.
Genomic DNA was extracted from de-identified young (<35 years old) and aged (>65 years old) differentiated human MDMs (5 × 106 cells per de-identified sample) using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific). Genomic DNA was assessed for purity using the NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific). The quantity of genomic DNA was assayed using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific) on the Qubit 3.0 Fluorometer (Thermo Fisher Scientific); 35 ng of genomic DNA from each sample with a purity A260/280 ratio = 1.80 was used in quantitative PCR (qPCR) analysis. The DNA oligomers for the telomeres are listed in Supplementary Table 4. The DNA abundance for each target gene was quantified by SYBR Green-based qPCR using the PowerUp SYBR Green PCR reagent (Thermo Fisher Scientific) on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). Melting curve analysis confirmed the specificity of each reaction. The reaction was performed using 35 ng of genomic DNA, 0.5 μl each of forward and reverse primer, and 5 μl PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) with a final volume of 10 μl Quantification was performed using the standard curve method.
Untargeted metabolomics by LC-MS.
Metabolites were extracted from macrophages and analyzed using a broad spectrum LC-MS platform as previously described58,59.
A solvent mixture of 80:20 methanol/water (500 μl) containing seven internal standards was used to resuspend the pellets that were composed of 5 × 106 cells. Cell suspensions were vortexed for 30 s, sonicated in a water bath (30 s sonication, 30 s on ice, repeated 3 times), vortexed for 30 s and incubated for 2 h at −20°C to allow protein precipitation. The supernatant was collected after centrifugation at 10,000 r.p.m. for 10 min at 4 °C and evaporated to dryness under nitrogen. The dry extracts were then reconstituted with 100 μl of 50:50 methanol/water before analysis.
Metabolic extracts were analyzed four times using HILIC and reverse phase LC (RPLC) separations in both positive and negative ionization modes. Data were acquired on a Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer for HILIC and a Q Exactive mass spectrometer (Thermo Fisher Scientific) for RPLC. Both instruments were equipped with a HESI-II probe and operated in full MS scan mode. MS/MS data were acquired on a pool sample. HILIC experiments were performed using a ZIC-HILIC column (2.1 × 100 mm, 3.5 μm, 200 Å; Merck Millipore) and mobile phase solvents consisting of 10 mM ammonium acetate in 50:50 acetonitrile:water (A) and 10 mM ammonium acetate in 95:5 acetonitrile:water (B)59. RPLC experiments were performed using a Zorbax SB-aq column (2.1 × 50 mm, 1.7 μm, 100 Å; Agilent) and mobile phase solvents consisting of 0.06% acetic acid in water (A) and 0.06% acetic acid in methanol (B).
Data quality was ensured by (1) sample randomization for metabolite extraction and data acquisition, (2) injection of 12 and 5 pool samples to equilibrate the LC-MS system before running the sequence for HILIC and RPLC, respectively, (3) injection of pool samples every 10 injections to control for signal deviation with time, and (4) checking mass accuracy, retention time, and peak shape of internal standards in every samples.
Metabolic data were processed using an in-house data analysis pipeline. Briefly, metabolic features (characterized by a unique mass/charge ratio and retention time) were extracted, aligned, and quantified with the ‘xcms’ package (version 1.39.4) in R (version 3.0.1). Grouping and annotation were performed with the CAMERA package (version 1.16.0). Features from blanks, that did not present sufficient linearity on serial dilution of a pool sample and that were not present in at least 66% of the samples were discarded to limit noise in the dataset. The signal drift with time was corrected by applying locally estimated scatterplot smoothing normalization using the pool samples injected repetitively along the sequence. A parametric two-tailed Welch’s t-test was used for statistical comparison. P values were corrected for multiple hypothesis testing using q value correction. An FDR of < 0.05 was considered significant. Significant metabolites were formally identified by matching fragmentation spectra to public spectral libraries or by matching retention time and fragmentation spectra to authentic standards when possible. Enrichment analysis was carried out using MBROLE 2.060.
SIRT3 enzymatic assay.
Peritoneal macrophages or human MDMs were plated at a confluency of 10 × 106 cells per 10-cm petri dish and treated with 1MT (200 μM) or PA (500 μM), 20 h. Mitochondria were isolated as described previously56. All reactions were performed in low-binding 96-well microtiter black plates with 10 μg of mitochondrial protein (Corning) in triplicate in each assay. Control wells without mitochondrial lysate were included in each plate as negative controls, while purified recombinant bovine SRT3 served as a positive control. SIRT3 enzymatic activity and steady state were measured using the SIRT3 Activity Assay kit (fluorometric) according to manufacturer’s instruction, with and without the inclusion of exogenous NAD+ supplied ((NAD+) = 2 mM) (Abcam). Difference in steady state between treatments was determined using a least squares regression line in Prism 7 (GraphPad) with plotted residuals.
Flow cytometry.
Human MDMs were plated in 10-cm plates at 10 × 106 cells per well and were collected using 0.25% trypsin-EDTA at 37 °C. Cells were then washed with flow cytometry buffer (PBS with 2% FCS, 2 mM EDTA, and 25 mM HEPES, pH7.4), followed by incubation with blocking buffer (5% mouse serum in flow cytometry buffer) for 15 min at 4 °C. Cells were then stained with the desired antibody combinations for 30 min at 4 °C. Dead cells were identified and excluded with 0.5 μg ml−1 propidium iodide. The anti-human antibodies used for flow cytometry of human MDMs (Supplementary Table 2) are as follows (all are manufactured by BD Biosciences): CD14 APC-Cy7 (clone MφP9); FITC mouse anti-human CD64 (clone 10.1); BV421 mouse anti-human CD206 (clone 19.2); PE-Cy 7 mouse anti-human CD68 (clone Y1/82A); BV421 mouse anti-human CD23 (clone EBVCS-5); BV711 mouse anti-human CD163 (clone GHI/61); APC mouse anti-human CD86 (clone FUN-1). For the determination of mitochondrial superoxide by flow cytometry, cells were incubated with MitoSOX (3 μM) (Thermo Fisher Scientific) at 37 °C for 15 min. The media were then removed and cells were washed with Hanks’ Balanced Salt Solution/Ca/Mg twice. The cells were then collected in Eppendorf tubes using Cellstripper (Corning) for 5 min at 37 °C. Cells were centrifuged at 1000 r.p.m. for 5 min and the cell pellet was resuspended in RPMI supplemented with 3% FBS. The following controls were used: unstained cells; single-stained cells; and dead cells. The cells were gated using forward and side scatter, as well as live/dead staining using 4,6-diamidino-2-phenylindole (Thermo Fisher Scientific). Cells were analyzed on a BD FACSAria II (BD Biosciences). Raw FCS files were analyzed with the FlowJo software.
Nucleofection of human MDMs.
Plasmid-containing human QPRT (pCMV6-QPRT-Myc-DDK-tagged clone; OriGene) or control GFP (2 μg) (pCMV6-A-GFP-tagged cloning vector; OriGene) were incubated with a mixture of nucleofection solution and P4 primary cell supplement (82 μM8 μl nucleofection solution:supplement) and placed in nucleofection cuvettes. A total of 1 × 106 human MDMs were added to each cuvette and subjected to program Y-010 for the Nucleofector 2b Device (Lonza). Immediately afterwards, 500 μl of DMEM (preincubated at 37 °C under 5% CO2 and supplemented with 20% FBS and 1% penicillin-streptomycin) was added. Cells were then plated in 10-cm plates and incubated 37 °C under 5% CO2 for 24 h before QPRT protein expression was analyzed.
Chemokine and cytokine assay.
Human MDM cell lysates or mouse plasma were stored at —80 °C and cytokine analysis was carried out at the Human Immune Monitoring Core (Stanford University) using Luminex human 62-plex or Luminex mouse 39-plex kits. Plates were read using a Luminex LabMap200 instrument with a lower bound of 100 beads per sample per measured cytokine. Each sample was tested in triplicate. Mean fluorescence intensity (MFI) was averaged over duplicate wells for each cytokine per sample on each plate.
Phagocytosis assay.
Human MDMs were grown in 10-cm plates at 10 × 106 cells per well and then trypsinized using 0.25% trypsin-EDTA at 37 °C. Cells were then plated in 96-well plates at 80,000 cells per well and the Vybrant Phagocytosis Assay Kit (Thermo Fisher Scientific) with E. coli particles was carried out following the manufacturer’s protocol.
Statistical analyses.
Data are expressed as the mean ± s.e.m. Statistical comparisons were made in the Prism software using a Student’s t-test (for two groups meeting the normal distribution criteria, according to the Shapiro-Wilk normality test), Mann-Whitney U-test (for two groups not meeting the normal distribution criteria), or analysis of variance (ANOVA) with Bonferroni correction or Tukey’s multiple comparison test (for groups across variables, with multiple comparisons between groups). Data were subjected to Grubbs’ test to identify the presence or absence of outlier data points. For all tests, P < 0.05 was considered significant, except for untargeted and targeted metabolomics in which Q < 0.05.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by grant no. RO1AG048232 (K.I.A.), grant no. RF1AG058047 (K.I.A.), grant no. 1P50 AG047366 (K.I.A.), Bright Focus (K.I.A.), The Paul and Daisy Soros Fellowship for New Americans (P.S.M.), the Gerald J. Lieberman Fellowship (P.S.M.), grant no. DP1DK113643 (L.L. and J.D.R.), grant no. 5U54DK10255603 (K.C., B.A.L., and M.P.S.), grant no. 5R01CA188055 (C.D. and R.M.), R37 AA11147 MERIT (A.J. and D.M.R.), the Takeda Pharmaceuticals’ Science Frontier Fund (D.M.R.), and grant no. 5T32HL094274 (M.C. and D.B.). The authors would like to thank L. Alexandrova at the Stanford University Mass Spectrometry Core, J. Perrino at the Stanford Microscopy Facility (supported by NIH grant no. 1S10RR02678001), and the Stanford Human Immune Monitoring Center.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41590-018-0255-3.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41590-018-0255-3.
References
- 1.Pearce EL & Pearce EJ Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills EL, Kelly B & O’Neill LAJ Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Verdin E NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Katsyuba E & Auwerx J Modulating NAD+ metabolism, from bench to bedside. EMBO J. 36, 2670–2683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y & Sauve AA NAD+ metabolism: bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 1864, 1787–1800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belenky P, Bogan KL & Brenner C NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Camacho-Pereira J et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 23, 1127–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai S & Guarente L NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grahnert A et al. Review: NAD+: a modulator of immune functions. Innate Immun. 17, 212–233 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Yeung AW, Terentis AC, King NJ & Thomas SR Role of indoleamine 2,3-dioxygenase in health and disease. Clin. Sci. 129, 601–672 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Munn DH & Mellor AL Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34, 137–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opitz CA et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Mellor AL et al. Prevention of T cell-driven complement activation and inflammation by tryptophan catabolism during pregnancy. Nat. Immunol. 2, 64–68 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Prendergast GC, Malachowski WP, DuHadaway JB & Muller AJ Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 77, 6795–6811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bender DA & Olufunwa R Utilization of tryptophan, nicotinamide and nicotinic acid as precursors for nicotinamide nucleotide synthesis in isolated rat liver cells. Br. J. Nutr. 59, 279–287 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Nishizuka Y & Hayaishi O Studies on the biosynthesis of nicotinamide adenine dinucleotide. I. Enzymic synthesis of niacin ribonucleotides from 3-hydroxyanthranilic acid in mammalian tissues. J. Biol. Chem. 238, 3369–3377 (1963). [PubMed] [Google Scholar]
- 17.Liu L et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellicciari R et al. α-Amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) inhibitors as novel modulators of de novo nicotinamide adenine dinucleotide (NAD+) biosynthesis. J. Med. Chem. 61, 745–759 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Baban B et al. Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. J. Reprod. Immunol. 61, 67–77 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Hou DY et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 67, 792–801 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Malik SS, Patterson DN, Ncube Z & Toth EA The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid. Proteins 82, 405–414 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Mills EL et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470. e413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan DG & O’Neill LAJ Krebs cycle rewired for macrophage and dendritic cell effector functions. FEBS Lett. 591, 2992–3006 (2017). [DOI] [PubMed] [Google Scholar]
- 24.O’Neill LAJ, Kishton RJ & Rathmell J A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly B & O’Neill LA Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn BH et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl Acad. Sci. USA 105, 14447–14452 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrero-Castillo S et al. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 25, 128–139 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Qiu X, Brown K, Hirschey MD, Verdin E & Chen D Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12, 662–667 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Bell EL, Emerling BM, Ricoult SJH & Guarente L SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30, 2986–2996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu PS et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Zwilling D et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell 145, 863–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillemin GJ Quinolinic acid, the inescapable neurotoxin. FEBS J 279, 1356–1365 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Kim H et al. Brain indoleamine 2,3-dioxygenase contributes to the comorbidity of pain and depression. J. Clin. Invest. 122, 2940–2954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgini F et al. Targeted deletion of kynurenine 3-monooxygenase in mice: a new tool for studying kynurenine pathway metabolism in periphery and brain. J. Biol. Chem. 288, 36554–36566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyes MP et al. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain 115, 1249–1273 (1992). [DOI] [PubMed] [Google Scholar]
- 36.O’Connor JC et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol. Psychiatry 14, 511–522 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarcz R & Stone TW The kynurenine pathway and the brain: challenges, controversies and promises. Neuropharmacology 112, 237–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jha AK et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Lampropoulou V et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tannahill GM et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z et al. Quinolinate phosphoribosyltransferase is an antiviral host factor against hepatitis C virus infection. Sci. Rep. 7, 5876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn HS et al. Structural insights into the quaternary catalytic mechanism of hexameric human quinolinate phosphoribosyltransferase, a key enzyme in de novo NAD biosynthesis. Sci. Rep. 6, 19681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H et al. Structural and kinetic characterization of quinolinate phosphoribosyltransferase (hQPRTase) from Homo sapiens. J. Mol. Biol. 373, 755–763 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyes MP et al. Human microglia convert l-tryptophan into the neurotoxin quinolinic acid. Biochem. J. 320, 595–597 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyes MP et al. Elevated cerebrospinal fluid quinolinic acid levels are associated with region-specific cerebral volume loss in HIV infection. Brain 124, 1033–1042 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Biswas SK & Mantovani A Orchestration of metabolism by macrophages. Cell Metab. 15, 432–437 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Ganeshan K & Chawla A Metabolic regulation of immune responses. Annu. Rev. Immunol. 32, 609–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown K et al. SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonkowski MS & Sinclair DA Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17, 679–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiarugi A, Dölle C, Felici R & Ziegler M The NAD metabolome: a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Johansson JU et al. Prostaglandin signaling suppresses beneficial microglial function in Alzheimer’s disease models. J. Clin. Invest. 125, 350–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodling NS et al. Cyclooxygenase inhibition targets neurons to prevent early behavioural decline in Alzheimer’s disease model mice. Brain 139, 2063–2081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su X, Lu W & Rabinowitz JD Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Chen L, Liu L, Su X & Rabinowitz JD Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 139, 14368–14371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hui S et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nijtmans LG, Henderson NS & Holt IJ Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26, 327–334 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Schägger H & von Jagow G Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 (1991). [DOI] [PubMed] [Google Scholar]
- 58.Piening BD et al. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 6, 157–170. e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Contrepois K, Jiang L & Snyder M Optimized analytical procedures for the untargeted metabolomic profiling of human urine and plasma by combining hydrophilic interaction (HILIC) and reverse-phase liquid chromatography (RPLC)-mass spectrometry. Mol. Cell. Proteomics 14, 1684–1695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López-Ibáñez J, Pazos F & Chagoyen M MBROLE 2.0: functional enrichment of chemical compounds. Nucleic Acids Res. 44, W201–W204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


