Abstract
Transfer RNAs are the largest, most complex non-coding RNA family, universal to all living organisms. tRNAscan-SE has been the de facto tool for predicting tRNA genes in whole genomes. The newly developed version 2.0 has incorporated advanced methodologies with improved probabilistic search software and a suite of new gene models, enabling better functional classification of predicted genes. This chapter describes the use of the UNIX command-driven and online web versions, illustrating different search modes and options.
Keywords: Transfer RNA, Non-coding RNA, Gene prediction, Covariance model, RNA secondary structure
1. Introduction
tRNAscan-SE (1) has been the most widely adopted tool for predicting transfer RNA (tRNA) genes in genomic sequences over the last two decades. Its users include RNA biologists, sequencing centers, database annotators, and other basic researchers. tRNAscan-SE gene predictions can be found for over four thousand genomes in the Genomic tRNA Database (2). The tRNAscan-SE software employs covariance models (3) that capture the primary sequence and secondary structure information of tRNA training data to search for complete tRNA genes in query sequences. The results provide researchers with the genomic coordinates, predicted function (isotype and anticodon), and secondary structure of the predicted tRNA genes. To improve performance and prediction accuracy, the latest version of tRNAscan-SE integrates Infernal v1.1, the state-of-the-art covariance model search software (4), with updated models based on a much broader diversity of tRNA genes. The program achieves better functional classification by utilizing isotype-specific covariance models, and enables mitochondrial tRNA gene prediction in mammals and other vertebrates. While the full functionality of tRNAscan-SE is available as downloadable, standalone UNIX-based software, we have also developed a user-friendly web-based version (5) to increase accessibility to scientists who wish to search relatively short sequences (up to bacterial chromosome sizes) and may not have expertise with installing and running UNIX command-line software. In this chapter, we illustrate the use of both the online and command-driven versions for finding tRNAs encoded in DNA or RNA from any species.
2. Predicting tRNA genes using tRNAscan-SE
2.1. Using Online Version
The tRNAscan-SE web server (http://trna.ucsc.edu/tRNAscan-SE/) is a convenient, ready-for-use means to identify tRNA genes in one or more query sequences. The graphical interface also provides easy navigation to the details of prediction results and a quick way to learn about the features of the software without requiring familiarity with UNIX-based commands or installation on one’s own computer. Web-based analysis limits query sequences to a maximum of five million base pairs. The standalone version can be used for larger genomic sequences.
2.1.1. Enter Search Options
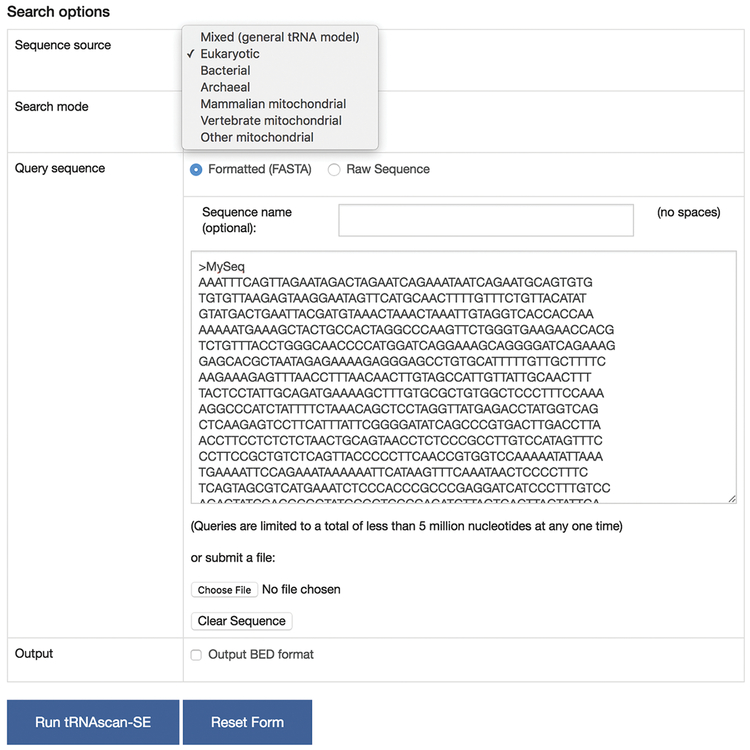
In addition to providing query sequence(s), the user is asked to select the source of the query sequence(s), if known (Fig. 1). One or more query sequences can be analyzed at a time, either typed or pasted into the text field, or uploaded as a FASTA-formatted file. The selected sequence source should correspond to the origin of the query sequences, namely sequences from eukaryotic, bacterial, archaeal or mitochondrial chromosomes (see Note 1). If the incorrect sequence source is given, the search still may identify tRNA genes, but the boundaries of the prediction may not be as accurate, and/or some low-scoring tRNAs could be missed. If the source of the query sequences is not known, for example a sequence from a metagenome, we recommend using the “Mixed (general tRNA model)” option. Alternatively, you could analyze query sequences with each of the possible sources one by one, and then only use the predictions that give the highest score for each identified gene.
Fig. 1.
Main search options for tRNAscan-SE online version. The minimum required options include the sequence source and the query sequence. BED file format can be optionally selected for output.
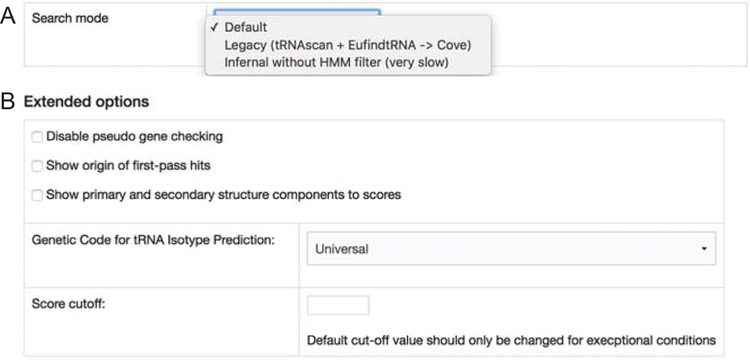
If you would like to obtain predictions and scores given by the original tRNAscan-SE v1.3 algorithm (for example, to match results found in older published predictions), select the Legacy search mode (Fig. 2A). This can be used in conjunction with the extended option of showing first-pass hit origin to check if the predictions are detected by tRNAscan and/or EufindtRNA – the fast first-pass screening algorithms that identify tRNA gene candidates in pre-2.0 tRNAscan-SE versions (Fig. 2B). If you require maximum search sensitivity and can accept much longer processing time, you may select the “Infernal without HMM filter” search mode (Fig. 2A). However, we do not recommend this mode except for non-standard mitochondrial tRNAs (e.g., those potentially missing tRNA stem-loops), as typical tRNAs are equally well identified by the much faster, efficient Default search mode.
Fig. 2.
(A) Search mode and (B) extended options for tRNAscan-SE online version.
tRNAscan-SE provides an overall bit score for each gene prediction. The higher the score, the more similar the prediction is to the consensus profile represented by the covariance model. The overall score can be split into the primary sequence score (i.e., conservation of the full linear sequence) and the secondary structure score (i.e., all base pairs expected in tRNAs). You can choose to show these detailed score components in the prediction results under the Extended Options (Fig. 2B). As part of the functional classification, tRNAscan-SE evaluates the gene predictions for possible pseudogenes based on characteristics commonly observed in non-functional tRNAs: a relatively weak overall score (<55 bits) and either a very low primary sequence score (<10 bits), or very low secondary structure score (<5 bits). This is important especially for many mammalian genomes known to have hundreds to thousands tRNA-derived short interspersed repeated elements (SINEs) (6–10). Under special circumstances where pseudogenes are not a concern, expert users may choose to disable the pseudogene checking (Fig. 2B). The default overall score cutoff is 20 bits, but can be overridden by entering a new score cutoff in the text field (Fig. 2B), with caution (see Note 2). For example, you may wish to lower the score cutoff to detect atypical mitochondrial tRNAs that lack the D-arm and/or T-arm.
Ciliate and mitochondrial genomes in some clades use alternate genetic codes for amino acids. If you use tRNAscan-SE to scan those genomic sequences, you can select the appropriate genetic code for tRNA isotype prediction under Extended Options (Fig. 2B). If the wrong genetic code is accidentally used, it will only affect the predicted “tRNA Type” (isotype), but no other aspects of the prediction.
To provide a method for visualizing the gene predictions in context using custom tracks in UCSC Genome Browser (11), you can obtain the results in the BED file format (see https://genome.ucsc.edu/FAQ/FAQformat.html), in addition to the default tRNAscan-SE output, by selecting “Output BED format” (Fig. 1).
2.1.2. Explore Search Results
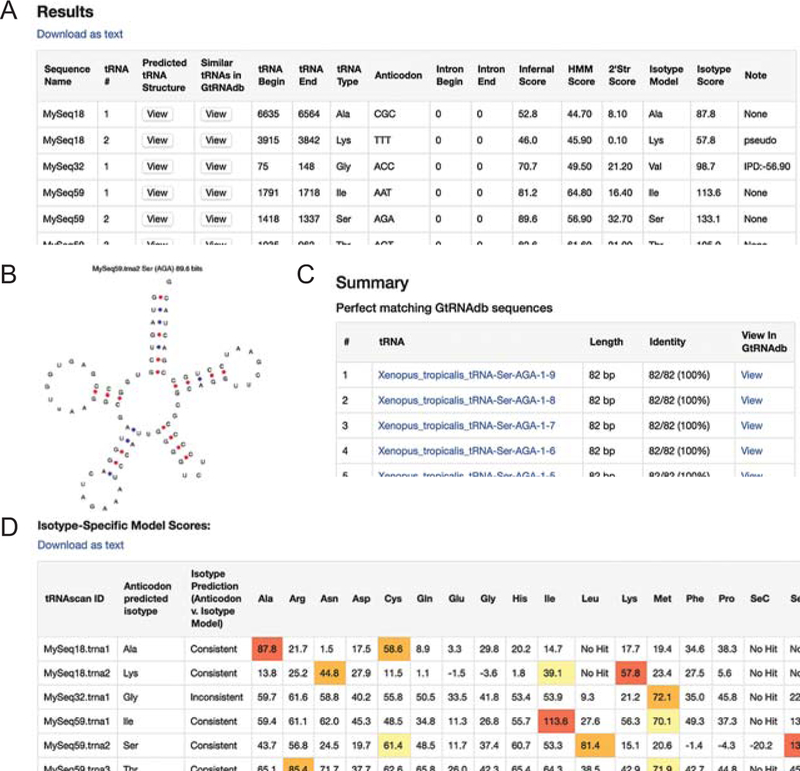
When the search is completed, the predicted tRNA genes will be listed in the first table of the results page (Fig. 3A). The table includes the coordinates, tRNA isotype, anticodon, location of intron if found, and the scores of the predicted genes. In addition, the sequences of the predictions are scanned with all 20+ isotype-specific models (e.g., tRNAs grouped by those decoding the same amino acid in translation) for additional classification information. The isotype-specific model that yielded the highest score against the prediction is shown in the Isotype Type column, followed by that model’s Isotype Score. If the highest scoring isotype model does not match with the anticodon-inferred isotype, this may indicate that the tRNA is a pseudogene, or a rare “hybrid” tRNA which theoretically could insert amino acids that do not correspond to the expected genetic code (this is very rare, but possible (12–14)). If there is disagreement, the score difference between the anticodon-expected isotype model, and the highest scoring isotype model will be listed in the Note column (see MySeq32.tRNA1 in Fig. 3A). The isotype scores of all the specific models are listed in the second table of the results page with the top three scores highlighted (Fig. 3D). You can click on the View button in the Predicted tRNA Structure column to view the secondary structure of the predicted tRNA (Fig. 3B). To check if the predicted genes are similar to those in GtRNAdb (2), click on the View button in the “Similar tRNAs in GtRNAdb” column and a list of results will be displayed on a separate page (Fig. 3C). You can also download the results in a text file by clicking on the Download as text link.
Fig. 3.
Search results of (A) predicted tRNA genes with scores, (B) secondary structure, (C) similar tRNA genes in GtRNAdb (2), and (D) scores of isotype-specific models for each predicted gene.
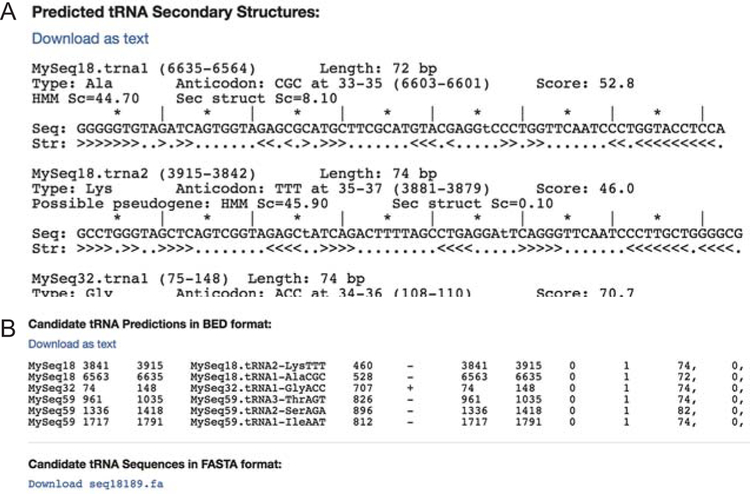
Run statistics that include the processing time, the number of predicted genes detected in the query sequences, and the number of predicted genes per isotype and anticodon are shown in the third section of the results page. The fourth section of the results page contains additional output formats. You can find the linear secondary structure of the predicted tRNA genes near the bottom of the page with a link for downloading in text format (Fig. 4A). If the output BED file format is selected as a search option, the BED output will follow (Fig. 4B). Finally, the sequences of predicted tRNA genes can be downloaded in a FASTA file (Fig. 4B).
Fig. 4.
Additional search outputs include (A) linear secondary structures of predicted tRNA genes, (B) output BED file format if selected, and sequences in FASTA file.
2.2. Using the UNIX Software
The UNIX command-line version of tRNAscan-SE offers the full functionality of the software. It is designed for researchers who have experience and access to UNIX-based computing environments, and would like to annotate tRNA genes in either large and/or many genomes. Institutions such as the National Center for Biotechnology Information (NCBI), the European Bioinformatics Institute (EBI), and others have integrated this software as part of a gene annotation pipeline for newly sequenced genomes.
2.2.1. Requirements and Installation
The source code of the standalone version can be downloaded at http://trna.ucsc.edu/tRNAscan-SE/. It can be used on a UNIX-based computer including Linux distributions such as CentOS and Ubuntu. However, it is not designed or tested for the Windows environment. The software should work on MacOS, provided that the computer has Perl 5, Apple Xcode, and Command Line Tools for Xcode pre-installed. You can follow the instructions in the INSTALL file of the source code package to build and install the tool. In addition, tRNAscan-SE requires the use of Infernal v1.1 (4) that must be installed in the same directory. The pre-built binaries and source code can be downloaded at http://eddylab.org/infernal/.
2.2.2. Apply Search Options
Similar to the online version, one or more query sequences (Fig. 1) are provided in a FASTA file by the user as the only required program argument to run the standalone version (Table 1) (see Note 3).
Table 1.
Main options for tRNAscan-SE command-line version
| Search Mode Options | |
|---|---|
-E |
search for eukaryotic tRNAs (default) |
-B |
search for bacterial tRNAs |
-A |
search for archaeal tRNAs |
-M <model> |
search for mitochondrial tRNAs (model: mammal or vert) |
-O |
search for other organellar tRNAs |
-G |
use general tRNA model (cytoslic tRNAs from all 3 domains included) |
--mt <model> |
use mito tRNA models for cytosolic/mito determination (if not specified, only cytosolic isotype-specific model scan will be performed) |
-I |
search using Infernal (default use with -E, -B, -A, or -G; optional for –O) |
--max |
maximum sensitivity mode - search using Infernal without hmm filter (very slow) |
-L |
search using the legacy method (tRNAscan, EufindtRNA, and COVE) (use with -E, -B, -A or –G) |
| -C or --cove | search using COVE analysis only (legacy, extremely slow) |
| -H or --breakdown | show breakdown of primary and secondary structure components to covariance model bit score |
| -D or --nopseudo | disable pseudogene checking |
| Output options | |
| -o or --output <file> | save final results in <file> |
| -f or --struct <file> | save tRNA secondary structures to <file> |
| -s or –isospecific <file> | save results of isotype-specific model scan in <file> |
| -m or --stats <file> | save statistics summary for run in <file> |
| -b or --bed <file> | save results in BED file format of <file> |
| -a or --fasta <file> | save predicted tRNA sequences in FASTA file format of <file> |
| -l or --log <file> | save log of program progress in <file> |
--detail |
display prediction outputs in detailed view |
--brief |
brief output format (no column headers) |
-? # |
‘#’ in place of <file> chooses default name for output files |
| -p or –prefix <label> | use <label> prefix for all default output file names |
| -d or --progress | display program progress messages |
| -q or --quiet | quiet mode (credits & run option selections suppressed) |
| -y or --hitsrc | show origin of hits (Ts=tRNAscan 1.4, Eu=EufindtRNA, Bo=Both Ts and Eu, Inf=Infernal) |
| Alternative cutoffs / data files | |
| -X or --score <score> | set cutoff score (in bits) for reporting tRNAs (default=20) |
| -g or –gencode <file> | use alternate genetic codes specified in <file> for determining tRNA type |
| Misc options | |
| -h or --help | print help message |
| -c or --conf <file> | tRNAscan-SE configuration file (default: tRNAscan-SE.conf) |
| -Q or --forceow | do not prompt user before overwriting pre-existing result files (for batch processing) |
| Advanced options | |
-U |
search for tRNAs with alternate models defined in configuration file |
--mid |
fast scan mode - search using Infernal with mid-level strictness of hmm filter |
| -F or –falsepos <file> | save first-pass candidate tRNAs in <file> that were then found to be false positives by second-pass analysis |
--thread <number> |
number of threads used for running infernal (default is to use available threads) |
$ tRNAscan-SE [-options] <FASTA file(s)>
To apply the correct tRNA covariance models for searching, the sequence source should be specified as an option, namely “-E“ for eukaryotes, “-B“ for bacteria, “-A” for archaea, “-M mammal” for mammalian mitochondria, “-M vert” for mitochondria in other vertebrates, “-G” for mixed (general), and “-O” the catch-all for all other organelles (by default, if not specified, the program will assume the source is eukaryotic; see Note 1).
To use the Legacy search mode as provided in the online version, you can specify the -L option with the -y option to show first-pass hit origin (Fig. 2A). The “Infernal without HMM filter” search mode in the online version is equivalent to the -- max option in the standalone version. Additionally, the -C option that selects COVE analysis is only available for backward compatibility. Similar to the --max option, -C provides the maximum sensitivity of the original COVE algorithm used in tRNAscan-SE v1.3 but is extremely slow in speed. We do not recommend it except for non-standard mitochondrial tRNAs when the --max (Infernal option) fails to provide expected results.
The -H option splits the overall score into the primary sequence and secondary scores that are provided in the output results (Fig. 2B). To disable the pseudogene checking as available in the online version, you can use the -D option. Similarly, the default overall score cutoff can be overwritten using the -X <score cutoff> option with caution (see Note 2). To specify alternative genetic codes for genomes such as ciliates and some mitochondria, you can use the -g option with a genetic code specification file included in the tRNAscan-SE package, or a custom specification file in the same format as the ones provided.
Many nuclear sequences of mitochondrial origin (NUMTs) have been discovered in large eukaryotic genomes, including the human genome (15). To determine if the predicted tRNA genes might be of mitochondrial origin, you can use the “--mt mammal” or “--mt vert” option for mammals or other vertebrates, respectively, to activate additional classification using the corresponding mitochondrial tRNA covariance model set. A Type column will be added in the result file describing if a predicted gene is “cytosolic” or “mito”.
The standalone version utilizes a configuration file for specifying the covariance models to be used in gene predictions, score cutoffs and other default values, alternative genetic code files, and locations of the temporary directory and installed directory for Infernal software (4). Advanced users who are familiar with the internals of the software can provide a custom configuration file in the same format. In addition, custom covariance models can be added to the configuration file in conjunction with the use of the -U option for alternate scan.
Be default, Infernal included in tRNAscan-SE will make use of all the available computing processing units to scan the query sequences. To limit the number of threads to be used, you can include the --thread option. This option is useful for users who include tRNAscan-SE as part of a gene prediction pipeline that shares computing resources with other applications (see Note 4).
2.2.3. Specify Result Output Options
Without specifying any output option, the standalone version of tRNAscan-SE will provide a list of predicted tRNA genes as standard output that is equivalent to the columns 1–2, 5–11, and 16 of the first result table in the online version (Fig. 3A). The HMM and 2’Str scores as columns 12 and 13 in the online version result will be included if the -H option is used. This primary output can be written into a text file by specifying the -O <file name> option. If you would like to obtain the isotype-specific model classification results equivalent to the Isotype Type and Isotype Score columns in the first online result table (Fig. 3A), the --detail option should be used. In addition, the isotype scores of all the specific models as provided in the second online result table (Fig. 3D) can be obtained by specifying the -s <file name> option.
To obtain additional analysis run statistics, use the -m <file name> option. Additional result output files can be generated with the following options: -f <file name> for linear secondary structure of predicted tRNA genes, -b <file name> for BED file format of predicted genes, and -a <file name> for the sequences of predicted genes in a FASTA file. A software execution progress log can also be obtained using the -l <file name> option.
If a specified output file already exists, a prompt will be provided asking for overwriting the file. To avoid having the prompt, you can include the “-Q” quiet option.
2.2.4. Obtain high confidence tRNA genes in large eukaryotes
As mentioned before, tRNA-derived SINEs are numerous in many large eukaryotic genomes (6–10). For example, the rat genome that has been previously reported with many tRNA pseudogenes that are caused by retrotransposon-driven repetitive elements (7), producing over 211,000 tRNA gene predictions. Although tRNAscan-SE classifies over 80% of the predictions from these mammals as pseudogenes, the remaining thousands still exceeds our expectation of true tRNA genes in any metazoan genome, given that there are only approximately 600 total human tRNA gene predictions. Therefore, the tRNAscan-SE package includes a post-filtering tool, namely EukHighConfidenceFilter, for determining the “high confidence” set of genes that we estimate are most likely to be involved in ribosomal translation (see Note 5). Currently, we recommend using this tool only on the predicted genes from large eukaryotic genomes such as mammalian or vertebrate genomes. To use the tool, you supply two output files from a completed tRNAscan-SE analysis run: the output result file and the secondary structure result file, with the -i and -s options respectively. Note that the “-- detail” option in tRNAscan-SE must be included to generate these result files. The outputs of the post-filtering tool will be written in the directory specified with the -o <dir path> option, with file names prefixed as specified with the -p <name> option.
In the default setting, the filtered tRNA hits will be tagged in the Note column of the result file using six categories (the first three categories include high-scoring predictions and the last three include potential repetitive elements): “high confidence set” (no issues), “isotype mismatch” for those that are high scoring but have inconsistent isotype/anticodon prediction (should be examined manually if possible), “unexpected anticodon” for those that are high scoring but with an anticodon typically not present in eukaryotic tRNA genes (should be examined manually if possible), “pseudo” for those that are classified as pseudogenes by tRNAscan-SE, “secondary filtered” for those that have low feature scores, and “tertiary filtered” for those that have an excessive number of gene copies for any single isotype which is common for tRNA-derived SINE families. To include in the results only the predicted genes in the first three high-scoring categories, you can specify the -r option.
3. Notes
tRNAscan-SE was not designed to identify tRNA genes in organelles except mitochondria in mammals and other vertebrates. Although the tool includes “general” and “organelle” search modes, it may not effectively detect all organellar genes especially those that are missing the D-arm and/or T-arm in the cloverleaf secondary structure. Other algorithms or additional covariance models may be needed to detect these genes.
Although tRNAscan-SE provides an option to overwrite the default score cutoff, a cutoff that is too low (permissive) may return many false positive predictions, while a cutoff that is too high (restrictive) may miss biologically active tRNA genes with slightly atypical features. The default score thresholds conservatively identify “typical” functional tRNAs (generally >55 bits), somewhat atypical tRNAs that may be active, but also include high-scoring pseudogenes (~40–55 bits), as well as likely tRNA-derived pseudogenes (20–40 bits). However, precise thresholds of functional versus non-functional tRNA genes are not known, as they almost certainly vary between species and tRNA isotype. The minimum threshold of 20 bits is meant to allow identification of all tRNA genes and all tRNA-derived sequences; cutoffs should be changed in only special situations, and with great caution by those able to inspect low-scoring predictions closely.
To list out all the options available for the standalone version of tRNAscan-SE and EukHighConfidenceFilter, you can use the --help or -h option. For getting help or reporting issues regarding the use of the tool, please send an email to our lab (trna@soe.ucsc.edu).
- Eukaryotes: -HQ -o# -f# -m# -s# -a# --detail -p <output prefix>
- Bacteria: -BHQ -o# -f# -m# -s# -a# --detail -p <output prefix>
- Archaea: -ADHQ -o# -f# -m# -s# -a# --detail -p <output prefix>
- Mammalian mitochondria: -M mammal -Q -o# -f# -m# -a# --detail –p <output prefix>
- Mitochondria in other vertebrates: -M vert -Q -o# -f# -m# -a# -- detail -p <output prefix>
Multiple options are available in EukHighConfidenceFilter for overwriting the default score cutoffs. We do not recommend users making changes to them in general, as we have done extensive testing on various large eukaryotic genomes. If you would like to apply the tool for filtering predicted genes in other genomes, the default score cutoffs may not work effectively and we suggest you to contact us for assistance.
References
- 1.Lowe TM and Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan PP and Lowe TM (2016) GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44, D184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy SR and Durbin R (1994) RNA sequence analysis using covariance models. Nucleic Acids Res 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nawrocki EP and Eddy SR (2013) Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe TM and Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44, W54–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels GR and Deininger PL (1985) Repeat sequence families derived from mammalian tRNA genes. Nature 317, 819–822. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera Holt R. A., Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Alba M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hubner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Payseur BA, Bourque G, Lopez-Otin C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S and Collins F (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521. [DOI] [PubMed] [Google Scholar]
- 8.Borodulina OR and Kramerov DA (1999) Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett 457, 409–413. [DOI] [PubMed] [Google Scholar]
- 9.Bao W, Kojima KK and Kohany O (2015) Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishihara H, Plazzi F, Passamonti M and Okada N (2016) MetaSINEs: Broad Distribution of a Novel SINE Superfamily in Animals. Genome Biol Evol 8, 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, Hinrichs AS, Haeussler M, Guruvadoo L, Navarro Gonzalez J, Gibson D, Fiddes IT, Eisenhart C, Diekhans M, Clawson H, Barber GP, Armstrong J, Haussler D, Kuhn RM and Kent WJ (2018) The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46, D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry J, Dai X and Zhao Y (2005) A mutation in the anticodon of a single tRNAala is sufficient to confer auxin resistance in Arabidopsis. Plant Physiol 139, 1284–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimata Y and Yanagida M (2004) Suppression of a mitotic mutant by tRNA-Ala anticodon mutations that produce a dominant defect in late mitosis. J Cell Sci 117, 2283–2293. [DOI] [PubMed] [Google Scholar]
- 14.Kollmar M and Muhlhausen S (2017) Nuclear codon reassignments in the genomics era and mechanisms behind their evolution. Bioessays 39. [DOI] [PubMed] [Google Scholar]
- 15.Simone D, Calabrese FM, Lang M, Gasparre G and Attimonelli M (2011) The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics 12, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]