Abstract
Rationale:
Activated fibroblasts are the major cell type that secrete excessive extracellular matrix in response to injury, contributing to pathological fibrosis and leading to organ failure. Effective anti-fibrotic therapeutic solutions, however, are not available due to the poorly defined characteristics and unavailability of tissue-specific fibroblasts. Recent advances in single-cell RNA-sequencing (scRNA-seq) fill such gaps of knowledge by enabling delineation of the developmental trajectories and identification of regulatory pathways of tissue-specific fibroblasts among different organs.
Objective:
This study aims to define the transcriptome profiles of tissue-specific fibroblasts using recently reported mouse scRNA-seq atlas, and to develop a robust chemically defined protocol to derive cardiac fibroblasts (CFs) from human induced pluripotent stem cells (iPSCs) for in vitro modeling of cardiac fibrosis and drug screening.
Methods and Results:
By analyzing the single-cell transcriptome profiles of fibroblasts from 10 selected mouse tissues, we identified distinct tissue-specific signature genes, including transcription factors that define the identities of fibroblasts in the heart, lungs, trachea, and bladder. We also determined that CFs in large are of the epicardial lineage. We thus developed a robust chemically-defined protocol that generates CFs from human iPSCs. Functional studies confirmed that iPSC-derived CFs preserved a quiescent phenotype and highly resembled primary CFs at the transcriptional, cellular, and functional levels. We demonstrated that this cell-based platform is sensitive to both pro- and anti-fibrosis drugs. Finally, we showed that crosstalk between cardiomyocytes and CFs via the atrial/brain natriuretic peptide-natriuretic peptide receptor 1 pathway is implicated in suppressing fibrogenesis.
Conclusions:
This study uncovers unique gene signatures that define tissue-specific identities of fibroblasts. The bona fide quiescent CFs derived from human iPSCs can serve as a faithful in vitro platform to better understand the underlying mechanisms of cardiac fibrosis and to screen anti-fibrotic drugs.
Keywords: Fibroblast, single-cell RNA sequencing, induced pluripotent stem cells, fibrosis, development, crosstalk
Subject Terms: Cellular Reprograming, Development Biology, Fibrosis, Myocardial Biology, Stem Cells
Graphical Abstract
INTRODUCTION
Fibroblasts are highly plastic and can transdifferentiate into myofibroblasts after tissue injury. Myofibroblasts are distinct from quiescent fibroblasts by demonstrating a contractile phenotype with a greater synthetic ability to produce extracellular matrix (ECM) proteins.1 Excessive deposition of ECM, a hallmark of tissue fibrosis, can take place in various organs, and may ultimately lead to organ failure.
Because the mechanisms underlying tissue fibrosis remain poorly understood, effective anti-fibrosis therapeutic strategies are not yet available. However, accumulating evidence suggests that the regulatory networks governing fibrogenesis are tissue-specific.2 Therefore, it would be more effective to develop tissue-specific anti-fibrotic therapies rather than “one-size-fits-all” drugs. However, gene markers that identify fibroblasts from various organs are largely undetermined, making it challenging to delineate the mechanisms of fibrosis in a tissue-specific manner.
As an emerging and powerful tool for comprehensive transcriptome profile analysis, single-cell RNA-sequencing (scRNA-seq) technology has provided profound insights into the diversity of cell populations, the heterogeneity of gene expression patterns within the same cell type, and the complex developmental trajectories of different cell types.3 This powerful technology may help uncover tissue-specific transcriptome profiles of fibroblasts.
Due to the limited availability of primary human cells and challenges to propagate them long-term in vitro, as well as the limited translational value of animal models in recapitulating phenotypic hallmarks of diseases in humans, human induced pluripotent stem cell (iPSC) technology has been increasingly used for disease modeling and drug screening.4 However, until now a reliable protocol to generate human iPSC-derived cardiac fibroblasts (iPSC-CFs) for cardiac fibrosis research has not been available.
By analyzing the recently reported Tabula Muris,5 a mouse atlas of single-cell transcriptome from >100,000 cells across 20 tissues, we report here that fibroblasts carry tissue-specific transcriptomic signatures that are conserved in humans. We found that CFs in large are of the epicardial lineage based on specifically expressed epicardial transcription factors (TFs) in these cells. We used these insights to develop a robust protocol for generating human iPSC-CFs that closely resemble quiescent human primary CFs at the transcriptional, cellular, and functional levels. We further showed that human iPSC-CFs can serve as a powerful in vitro platform to better understand the mechanisms of cardiac fibrosis and to screen anti- and pro-fibrotic drugs. Collectively, our study used comprehensive scRNA-seq data to define tissue-specific genes and pathways in fibroblasts. Importantly, the human iPSC-based induction protocol we developed can be adapted to generate patient-specific CFs, which may pave the way to reveal the complexity of cardiac fibrosis and thereby offer a new direction for precision medicine.
METHODS
A detailed Methods section is available in the Online Data Supplement. All data and materials supporting the findings of this study are available from the corresponding author upon request.
RESULTS
Mouse Single-Cell Transcriptome Reveals Tissue-Specific Gene Markers for Fibroblasts are Conserved in Humans.
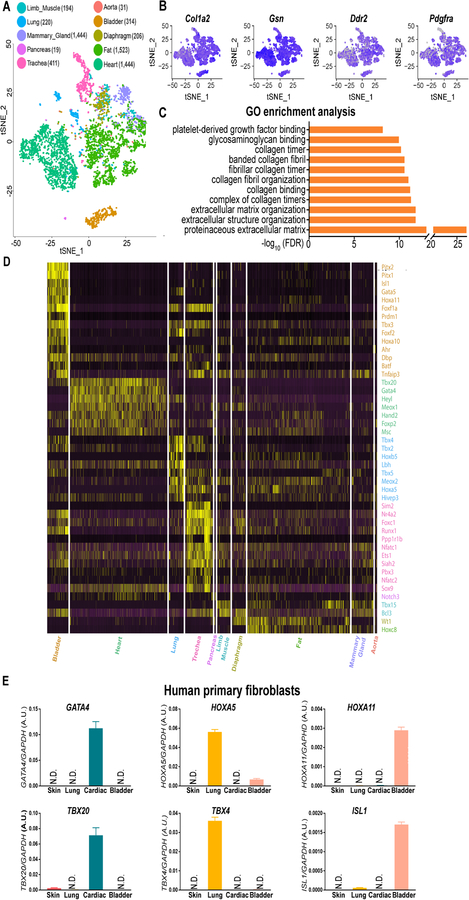
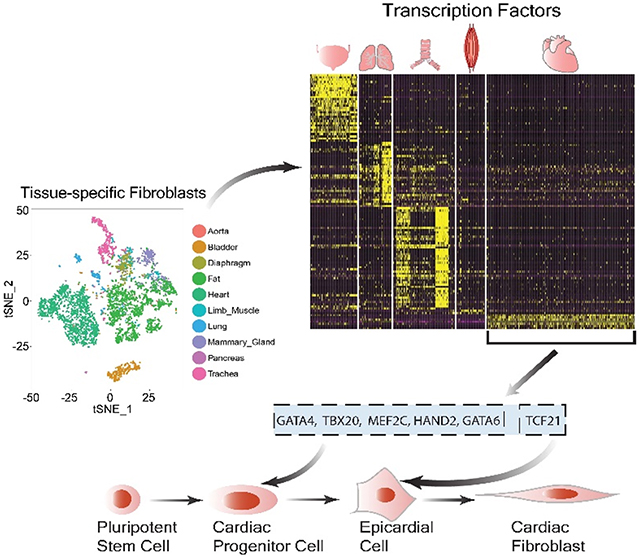
Because specific genes that can distinguish fibroblasts from other cell types are not yet available, we first subtracted the transcriptome profiles of fibroblast-containing cell clusters in 10 selected tissues from the Tabula Muris database (Online Figure IA).5 We then mapped these fibroblast-containing cell clusters on a t-distributed stochastic neighbor embedding (t-SNE) plot (Online Figure IB), and refined the fibroblast population in each tissue by selecting cells that are positive for genes (Col1a2, Ddr2, Dcn, Timp2, and Pdgfra) highly expressed in fibroblasts (Online Figure IC), and by excluding those expressing any of the gene markers for endothelial cells (Pecam1 and Cdh5), smooth muscle cells (Tagln and Cnn1), or immune cells (Ptprc) (Online Figure ID). Ultimately, we identified a total of 4,685 out of 44,949 cells as fibroblasts in the Tabula Muris dataset, and mapped them on a new t-SNE plot (Figure 1A). As expected, these cells express common fibroblast marker genes (Figures 1B and Online Figure II). Moreover, gene ontology (GO) enrichment analysis revealed that these genes are enriched in fibroblast-related signaling pathways, such as proteinaceous extracellular matrix, extracellular structure organization, and platelet-derived growth factor binding (Figure 1C).
Figure 1. Mouse single-cell transcriptome reveals tissue-specific gene markers for fibroblasts are conserved in humans.
A, A t-SNE (t-distributed stochastic neighbor embedding) plot showing the distribution patterns of 4, 685 fibroblasts derived from 10 tissues of healthy adult mice. The numbers of tissue-specific fibroblasts used for transcriptome analysis are listed in the brackets next to the individual tissue types. B, Representative t-SNE plots showing tissue-specific fibroblast subpopulations express genes reported to be detected in fibroblasts with high abundance. C, Gene ontology (GO) enrichment analysis reveals that all the cell clusters in (A) possess fibroblast-specific biological functions. D, A heatmap comparing the most specifically expressed (25% fibroblasts expressed, logFC≥1.5, and FDR adjusted P-value <1%; Wilcoxon rank sum test) transcription factors in 10 tissue-specific mouse fibroblast subpopulations. E, Cross-species validation of cardiac (GATA4 and TBX20), lung (HOXA5 and TBX4), and bladder (HOXA11 and ISL1) specific genes in human primary cardiac, lung, and bladder fibroblasts. Primary skin fibroblasts are used as a negative control. N.D., not detected. Data are presented as mean ± SEM.
Next, we examined which factors are responsible for separating fibroblasts into different clusters (Figure 1A). After pooling fibroblasts from each mouse and mapping them on a new t-SNE plot, we did not observe any donor effects that contributed to the specificity of fibroblast clustering in different tissues (Online Figure IIIA). However, fibroblasts in the heart and fat tissues were separated by sex (Online Figure IIIB). Further analysis revealed that the sex-polarized gene expression patterns (Online Figure IVA and IVB) might be associated with higher expression levels of sex hormonal receptors in the heart and fat tissues compared to other organs (Online Figure IVC).
We then compared the transcriptome profiles between tissue-specific fibroblasts, and observed that fibroblasts derived from the heart, lungs, bladder, and trachea demonstrated distinct gene signatures (Online Figure V and Online Table I). Moreover, we observed that fibroblasts in these four organs also exhibit a tissue-specific distribution pattern of TFs (Figure 1D and Online Table II). TFs have been suggested to play a critical role in defining cell identities.6, 7 Tbx20, Gata4, and Hand2 are specifically enriched in CFs, whereas Tbx2, Tbx4, and Hoxa5 are highly expressed in lung fibroblasts. Fibroblasts from the aorta, diaphragm, fat, mammary gland, and limb muscle did not show an apparent tissue-specific pattern for TFs (Figure 1D). Intriguingly, human primary fibroblasts isolated from the heart, lungs, and bladder also specifically expressed GATA4/TBX20, HOXA5/TBX4, and HOXA11/ISL1, respectively (Figure 1E). Together, the single-cell transcriptome suggests that the identities of fibroblasts in different mouse tissues are distinguished and probably determined by a handful of conserved tissue-specific signature genes.
Generation Quiescent Human iPSC-CFs.
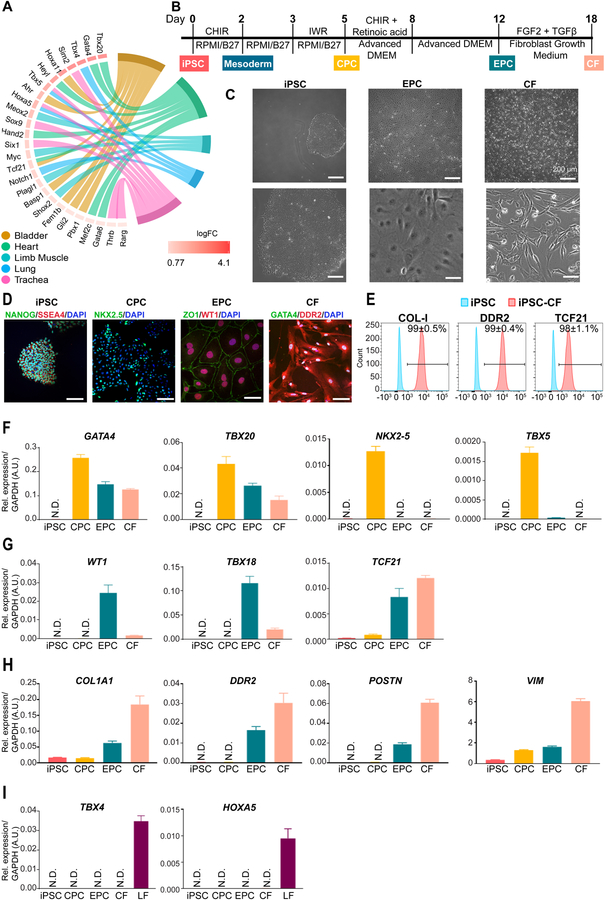
By performing the GO pathway enrichment analysis on TFs, we observed that some of these genes are enriched in a tissue-specific development pathway (Figure 2A). For example, we identified that Gata4, Gata6, Tbx20, Heyl, Hand2, Mef2c, and Tcf21 are enriched in the cardiac development pathway (Figure 2A). Because GATA4 and TBX20 are highly expressed in cardiac mesoderm,8, 9 and TCF21 is a marker for epicardium,10 we hypothesized that CFs could be generated from human iPSCs based on this developmental program. Accordingly, we modified an established protocol11 designed to generate human iPSC-derived epicardial cells (EPCs) by continuing to differentiate the intermediate cells in a commercial fibroblast growth medium in the presence of fibroblast growth factor (FGF2) and transforming growth factor beta (TGF-β) inhibitor SB431542 (SB) (Figure 2B). After six days of extended culture (i.e., day 18 of differentiation), the cells showed a typical fibroblast morphology (Figure 2C) and expressed markers (GATA4 and DDR2) that are highly expressed in primary CFs (Figure 2D). Notably, flow cytometry analysis revealed that >90% of the cells were positive for COL-I, DDR2, and TCF21 compared with undifferentiated iPSCs (Figure 2E).
Figure 2. Generation human induced pluripotent stem cell-derived cardiac fibroblasts.
A, Developmental pathway enrichment analysis (FDR adjusted P-value <1%; Hypergeometric test) showing tissue-specific transcription factors (≥25% fibroblasts, logFC≥log2 and FDR adjusted P-value <1%; Wilcoxon rank sum test) in fibroblasts of different tissue origins. B, A schematic diagram showing the protocol for small molecule-directed differentiation from human iPSCs to iPSC-CFs. iPSC, induced pluripotent stem cell; CPC, cardiac progenitor cell; EPC, epicardial cell; CF, cardiac fibroblast; FGF2, fibroblast growth factor-2; TGF-β, transforming growth factor-β. C, Representative bright-field images showing stage-specific cell morphology changes during the differentiation of human iPSC-CFs. Scale bars, 200 μm. D, Representative immunofluorescent images showing stage-specific cells express pluripotency genes (NANOG and SSEA4), cardiac mesodermal gene (NKX2–5), epicardial genes (ZO1 and WT1), and CF genes (GATA4 and DDR2) during differentiation. Scale bars, 100 μm. E, Representative flow cytometric histograms showing protein levels of COL-I, DDR2, and TCF21 in human iPSC-CFs on day 18 of differentiation (red) versus undifferentiated iPSCs (blue). F, The expression levels of cardiac-specific, early development-related genes (GATA4, TBX20, NKX2–5, and TBX5) at individual differentiation stages. N.D., not detected. G, The expression levels of epicardial markers (WT1, TBX18, and TCF21) at individual differentiation stages of iPSC-CFs. H, The expression levels of genes that are highly expressed in fibroblasts (COL1A1, DDR2, POSTN, and VIM) in stage-specific cells during iPSC-CF differentiation. I, The expression levels of lung-specific genes (TBX4 and HOXA5) in stage-specific cells during iPSC-CF differentiation. Primary LFs (lung fibroblasts) are used as a positive control. Data in (E-I) were generated based on three independent differentiations. Data are presented as mean ± SEM.
We then examined the dynamic changes of the transcriptional profiles of cardiac and fibroblast-specific genes at various stages of differentiation. We observed that GATA4 and TBX20, two early cardiac-specific developmental marker genes, showed relative higher expression levels in iPSC-derived cardiac progenitor cells (CPCs) than in iPSC-EPCs and iPSC-CFs (Figure 2F). By contrast, NKX2–5 and TBX5 were only transiently expressed in iPSC-CPCs. Epicardial markers WT1 and TBX18 became highly expressed in iPSC-EPCs but decreased in iPSC-CFs (Figure 2G). It has been reported that TCF21-expressing EPCs are initially multipotent, and eventually become committed to the fibroblast lineage with the upregulation of TCF21.12 Consistently, TCF21 was found to be substantially increased in iPSC-EPCs and became more highly expressed in iPSC-CFs (Figure 2G). As expected, the expression levels of typical fibroblast markers (COL1A1, DDR2, POSTN, and VIM) were gradually increased during differentiation (Figure 2H), whereas lung-specific development-related genes such as TBX4 and HOXA5 were not detectable at any stage of differentiation (Figure 2I). Taken together, we confirm that iPSC-CFs obtained through the differentiation protocol used in this study express both cardiac and fibroblast-specific markers.
Human iPSC-CFs Preserve Their Cell Identity Profile During Passaging.
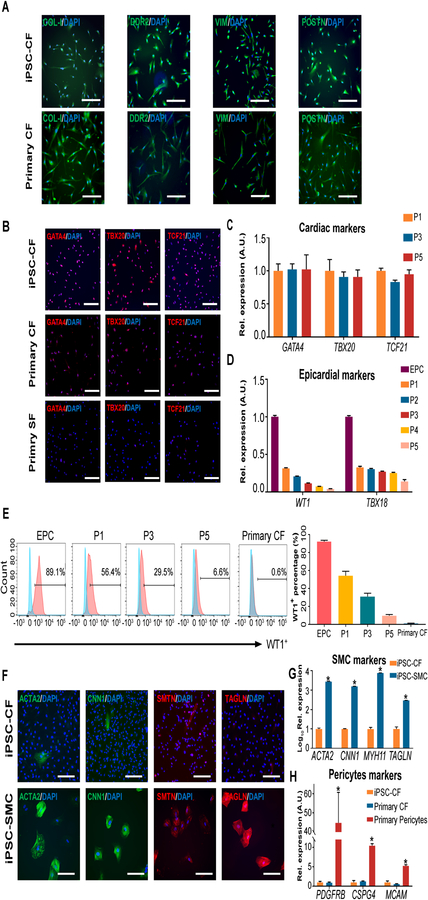
In order to verify whether iPSC-CFs are bona fide cardiac fibroblasts, we next compared their transcriptomes and proteomes with primary cultures. Intriguingly, human iPSC-CFs expressed similar protein levels of typical fibroblast markers (COL-I, DDR2, VIM, and POSTN) and cardiac markers (GATA4, TBX20, and TCF21) as primary fibroblasts (Figure 3A and 3B). We also found that the human iPSC-CFs could be expanded for at least five passages without losing their cardiac fibroblast identity (Figure 3C). As expected, the expression levels of TBX18 were persistently much lower in iPSC-CFs during serial passaging than in iPSC-EPCs (Figure 2G and 3D). While WT1 is hardly detectable in adult primary CFs, it decreased dramatically to less than 10% in human iPSC-CFs at passage 5 (Figure 3D, 3E, and Online Figure VI). Taken together, these data indicate that iPSC-CFs show a high similarity to primary CFs in terms of cell identity profiles.
Figure 3. Human iPSC-derived cardiac fibroblasts are highly pure and preserve their cardiac identity during passaging.
A, Representative immunofluorescent images showing human iPSC-CFs are positive for gene makers (COL-I, DDR2, VIM, and POSTN) that are highly expressed in primary CFs. Scale bars, 100 μm. iPSC, induced pluripotent stem cell; CF, cardiac fibroblast. B, Representative immunofluorescent images showing the expression levels of cardiac-specific genes (GATA4, TBX20, and TCF21) in human iPSC-CFs, primary cardiac and skin fibroblasts. SF, skin fibroblast. Scale bars, 100 μm. C, The expression levels of cardiac-specific genes (GATA4, TBX20, and TCF21) in human iPSC-CFs at different passages. D, The expression levels of epicardial marker genes (WT1 and TBX18) in human iPSC-EPCs and iPSC-CFs at different passages. EPC, epicardial cell. E, Representative flow cytometric histograms showing the percentage of WT1+ population in human iPSC-EPCs and iPSC-CFs at different passages. Primary CFs are used as a control. The quantitative data are shown on the right. F, Representative immunofluorescent images showing negligible expression of SMC (smooth muscle cell)-specific genes (ACTA2, CNN1, SMTN, and TAGLN) in iPSC-CFs. The same iPSC lines were used to derive SMCs of the epicardial lineage, which are used as a positive control. Scale bars, 100 μm. G, The expression levels of SMC-specific genes (ACTA2, CNN1, MYH11, and TAGLN) in human iPSC-CFs versus iPSC-SMCs of the epicardial lineage. H, The expression levels of pericyte-specific genes (PDGFRB, CSPG4, and MCAM) in human iPSC-CFs, primary cardiac fibroblasts, and brain pericytes. Data in (C, D, E, G, and H) are generated from three independent iPSC lines, and are presented as mean ± SEM. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons.
Because cardiac smooth muscle cells and pericytes are also derived from the epicardial lineage,13, 14 we blocked these potential differentiation trajectories by adding SB431542 (SB), a TGF-β inhibitor, during CF differentiation (Figure 2B). As a result, human iPSC-CFs expressed negligible levels of smooth muscle marker genes (ACTA2, CNN1, TAGLN, SMTN, and MYH11) at both transcriptional and protein levels (Figure 3F and 3G). We also observed that human iPSC-CFs expressed higher levels of myogenic repressor genes ELK1 and KLF415, 16 than derived smooth muscle cells of the epicardial lineage (Online Figure VII). Moreover, human iPSC-CFs expressed significantly lower levels of pericyte markers (PDGFRB, CSPG4, and MCAM) compared to primary pericytes (Figure 3H). Therefore, the human iPSC-CFs generated in this study are highly pure cultures and can preserve cardiac fibroblast identity during serial passaging.
Human iPSC-CFs Closely Resemble Primary CFs.
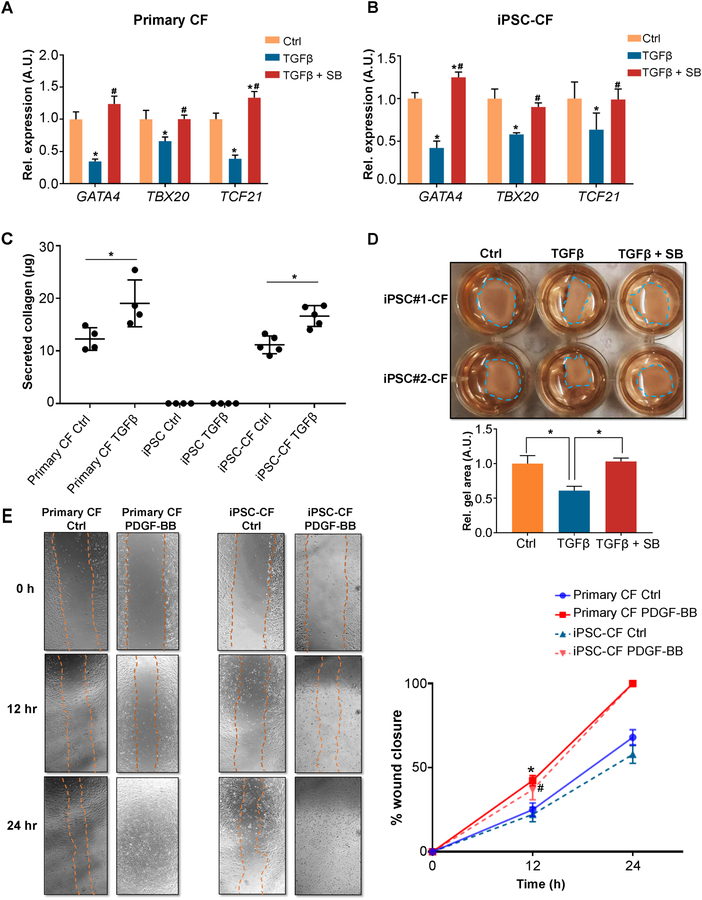
Next, we conducted a series of functional studies comparing human primary and iPSC-CFs. Under pathological conditions, upregulated TGF-β is one of the potent effectors that can stimulate fibroblast-to-myofibroblast transdifferentiation and thereby contribute to tissue fibrosis.17 Interestingly, we observed that the expression levels of cardiac-specific genes (GATA4, TBX20, and TCF21) were significantly downregulated in TGF-β-treated primary CFs as compared with vehicle-treated cells (Figure 4A). A similar phenotype was also observed in human iPSC-CFs after TGF-β treatment (Figure 4B). Because a previous study reported that fibroblast-restricted depletion of Tbx20 is associated with thicker scar tissues in infarcted mouse hearts,18 the downregulation of these cardiac-specific TFs may be involved in promoting CF transdifferentiation and cardiac fibrosis. However, the precise role of cardiac-specific TFs in cardiac fibrosis warrants further investigation.
Figure 4. Human iPSC-derived cardiac fibroblasts demonstrate similar biological characteristics to primary cardiac fibroblasts.
A, Expression levels of cardiac-specific genes (GATA4, TBX20, and TCF21) in human primary CFs in the absence or presence of TGF-β ± a TGF-β inhibitor SB431542 (SB). CF, cardiac fibroblast. *P<0.05 vs. vehicle control, #P<0.05 vs. TGF-β group by one-way ANOVA followed by Bonferroni multiple comparisons. B, Expression levels of cardiac-specific genes (GATA4, TBX20, and TCF21) in human iPSC-CFs in the absence or presence of TGF-β ± SB. iPSC, induced pluripotent stem cell. *P<0.05 vs. vehicle control, #P<0.05 vs. TGF-β group by one-way ANOVA followed by Bonferroni multiple comparisons. C, Secreted collagen contents (measured by Sirius red) are increased in both human iPSC-CFs and primary CFs after TGF-β treatment. Human iPSCs were used as a negative control. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons. D, Representative collagen gel contraction responses in two independent human iPSC-CFs in the absence or presence of TGF-β ± SB. The residual surface areas of collagen gel in each group were used for quantification (shown below the representative image), and normalized to that of the vehicle control. The size of the collagen gel area is inversely proportional to the contractility. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons. E, Representative images showing human iPSC-derived and primary CF migration in the absence or presence of PDGF-BB (25 ng/ml) at different time points over the 24-hour observation frame using a wound-healing assay. Quantitative data showing similar migration rate (wound closure area) between human iPSC-CFs and primary CFs under the same conditions. *P<0.05 indicates PDGF-BB treatment vs. vehicle control in primary CF, #P<0.05 indicates PDGF-BB treatment vs. vehicle control in iPSC-CF, by one-way ANOVA followed by Bonferroni multiple comparisons. All data were generated from three or four independent iPSC lines, and are presented as mean ± SEM.
Aberrant collagen synthesis contributes to tissue fibrosis. Using a Sirius red-based colorimetric assay kit, we observed that human iPSC-CFs and primary CFs secreted comparable basal levels of collagen into culture media, which were not detectable in human iPSCs (Figure 4C). A similar rise in collagen content was detected in both groups of CFs upon TGF-β treatment (Figure 4C). Increased contractile activity is also a specific measure of fibroblast transdifferentiation. We seeded human iPSC-CFs on collagen membranes, and observed a stronger collagen gel contraction effect in the TGF-β treated group than the vehicle group, whereas this effect was completely abrogated by SB (Figure 4D). Human iPSC-CFs and primary CFs responded similarly to platelet-derived growth factor-BB-induced cell migration (Figure 4E). It has been shown that cardiac myofibroblasts could lead to myosin heavy chain 7 re-expression during cardiac fibrosis in vivo.19 Interestingly, this phenotype was successfully recapitulated by co-culturing human iPSC-derived cardiomyocytes (iPSC-CMs) with iPSC-CFs in the presence of TGF-β (Online Figure VIII). Taken together, these functional studies confirm that the human iPSC-CFs generated in this study are bona fide cardiac fibroblasts, which resemble the biological and physiological properties of their primary counterparts.
Quiescent Human iPSC-CFs Provide an in vitro Platform to Model Fibrosis.
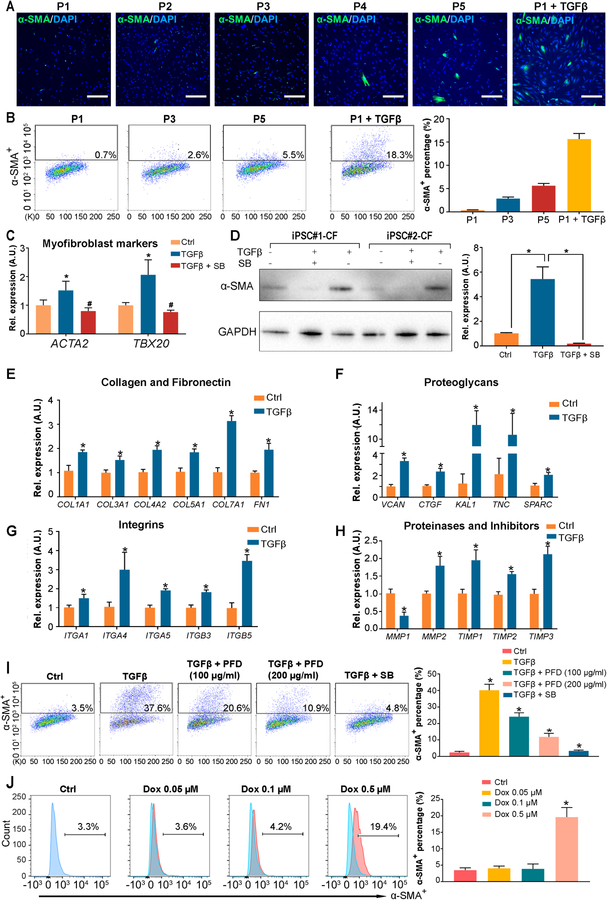
Previous studies have shown that primary fibroblasts can undergo spontaneous transdifferentiation into myofibroblasts during in vitro culturing.20 We hypothesized that inhibiting the TGF-β pathway during iPSC-CF differentiation may preserve their quiescent phenotype and suppress transdifferentiation. Using the α-smooth muscle actin (α-SMA) level as an indicator for the extent of fibroblast activation, we observed that the generated human iPSC-CFs were quiescent cells when the TGF-β pathway was inhibited (Figure 5A and 5B). Notably, most of these cells remained quiescent after several passages, and only about 5% of iPSC-CFs at passage 5 were positive for α-SMA (Figure 5A and 5B). Therefore, we used iPSC-CFs between passages 1–4 as a reliable cell model for the subsequent fibrosis studies in vitro.
Figure 5. Human iPSC-derived cardiac fibroblasts are quiescent fibroblasts and therefore provide an ideal in vitro platform to model fibrosis.
A-B, Representative immunofluorescent (A) and flow cytometry (B) images showing the degree of spontaneous transdifferentiation of iPSC-CFs to myofibroblasts (α-SMA+, smooth muscle α-actin) during passages 1–5. TGF-β treated CFs were used as a positive control. The percentage of SMA+ myofibroblasts in each passage of iPSC-CFs are calculated based on the flow cytometry (B, right). iPSC, induced pluripotent stem cell; CF, cardiac fibroblast. Scale bars, 100 μm. C, The expression levels of myofibroblast markers (ACTA2 and POSTN) in human iPSC-CFs cultured in the absence or presence of TGF-β ± a TGF-β inhibitor SB431542 (SB). *P<0.05 vs. vehicle control, #P<0.05 vs. TGF-β group by one-way ANOVA followed by Bonferroni multiple comparisons. D, A representative immunoblot showing the α-SMA levels in CFs derived from two independent iPSC lines in the absence or presence of TGF-β ± SB. The corresponding densitometric quantification data are shown next to the immunoblot. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons. E-H, The expression levels of genes coding extracellular matrix proteins (collagen, fibronectin, proteoglycans, integrins, and matrix proteinases and inhibitors) in human iPSC-CFs cultured in the absence or presence of TGF-β. *P<0.05 vs. vehicle control by one-way ANOVA followed by Bonferroni multiple comparisons. I, Representative flow cytometry images and quantitative data showing pirfenidone (a commercial anti-fibrosis drug) or SB significantly suppresses TGF-β induced propagation of the α-SMA+ population in iPSC-CFs. PFD, pirfenidone. *P<0.05 vs. TGF-β group by one-way ANOVA followed by Bonferroni multiple comparisons. J, Representative flow cytometric histograms and quantitative data showing a dose-dependent induction of the α-SMA+ population after human iPSC-CFs were treated with doxorubicin. *P<0.05 vs. ctrl group by one-way ANOVA followed by Bonferroni multiple comparisons. Data in (B-J) were generated from three independent iPSC lines, and are presented as mean ± SEM.
We first confirmed that the expression levels of ACTA2 and POSTN were significantly increased in human iPSC-CFs upon TGF-β treatment as compared with the vehicle treatment (Figure 5C). As expected, the addition of SB effectively inhibited the upregulation of these two myofibroblast markers (Figure 5C). The α-SMA protein levels were also significantly increased in CFs derived from different human iPSC lines after TGF-β treatment, which were abolished by SB (Figure 5D). Because the transdifferentiation from fibroblasts to myofibroblasts is strongly associated with the dynamic changes of ECM genes, we next performed a comprehensive ECM transcriptomic analysis on human iPSC-CFs after treatment with vehicle or TGF-β. We observed that the expression levels of ECM genes such as COL1A1, COL3A1, COL4A2, and FN1 were significantly increased by TGF-β (Figure 5E). Similarly, several proteoglycans (VCAN, CTGF, KAL1, TNC, and SPARC) and cell-matrix focal adhesion molecules (ITGA4, ITGA5, and ITGB5) also showed upregulated expression in the presence of TGF-β (Figure 5F and 5G). Matrix metalloproteinases (MMPs) and their endogenous inhibitors (TIMPs) regulate the dynamic turnover of ECM.21 We observed an upregulation of TIMP1, TIMP2, and TIMP3 concurrently with downregulation of MMP1 by TGF-β (Figure 5H). Because MMP2 has been shown to be involved in activating TIMP2,22 it is not surprising that we observed an upregulation of MMP2 in iPSC-CFs after TGF-β treatment (Figure 5H).
Next, we tested if the FDA-approved anti-idiopathic lung fibrosis drug pirfenidone23 could suppress TGF-β-induced transdifferentiation of human iPSC-CFs. Pirfenidone showed a dose-dependent inhibitory effect on TGF-β-induced upregulation of α-SMA (Figure 5I). Doxorubicin, a widely used chemotherapy drug, has been shown to cause several cardiac side effects, including cardiac fibrosis.24 Using human iPSC-CFs, we recapitulated doxorubicin-induced pro-fibrosis phenotype in vitro, which also showed a dose-dependent pattern (Figure 5J). Together, these results suggest that the human iPSC-CFs generated in this study preserve a quiescent phenotype after differentiation and during early passages, making them sensitive to pro-fibrogenic stimuli. Therefore, these cells can serve as a faithful in vitro platform to screen drugs that can promote or suppress fibrosis.
Crosstalk Between Human iPSC-CMs and iPSC-CFs Through the ANP/BNP-NPR1 Signaling Pathway Represents a Novel Strategy for Anti-Fibrotic Therapy.
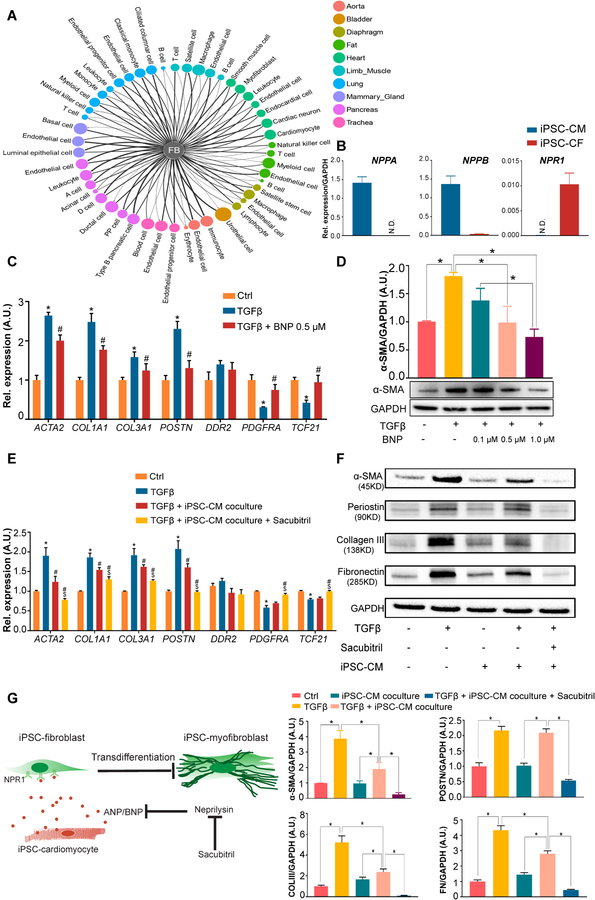
Cardiac fibroblasts are a key component of the heart that play a critical role in cardiac remodeling through dynamic cell-cell interactions.25 By carrying out complementary receptor-ligand interaction analysis (Figure 6A and Online Table III), we observed that CFs are capable of interacting with a broad spectrum of cell types such as CMs, endothelial cells, smooth muscle cells, and immune cells through different signaling pathways. For example, we identified a novel interaction between ANP/BNP, the commonly used hypertrophic markers in CMs,26 and their cognate receptor NPR1 in CFs (Online Table III). We further verified that natriuretic peptide type A/B (NPPA/NPPB) and NPR1 are expressed in human iPSC-CMs and iPSC-CFs, respectively (Figure 6B).
Figure 6. Human iPSC-derived cardiomyocytes attenuate TGF-β-induced fibrotic responses in iPSC-derived cardiac fibroblasts through a paracrine mechanism.
A, Computational analysis showing broad intercellular communications between fibroblasts and a diverse array of cell types through different complementary ligand-receptor signaling pathways. The threshold for defining a ligand/receptor expressed in a cluster is ≥25% with gene expression > 0. FB, fibroblast. B, The basal expression levels of ligands NPPA (natriuretic peptide type A) and NPPB and their cognate receptor NPR1 (natriuretic peptide receptor 1) in human iPSC-CMs and iPSC-CFs. iPSC, induced pluripotent stem cell; CM, cardiomyocyte; CF, cardiac fibroblast. C, Expression levels of CF and myofibroblast markers in human iPSC-CFs in the absence or presence of TGF-β ± BNP. *P<0.05 vs. vehicle control, #P<0.05 vs. TGF-β group by one-way ANOVA followed by Bonferroni multiple comparisons. D, A representative immunoblot showing α-SMA (smooth muscle α-actin) levels in human iPSC-CFs treated with TGF-β and escalating concentrations of BNP. Densitometric quantifications are shown above the immunoblot. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons. E, Expression levels of CF and myofibroblast markers in human iPSC-CFs after they were indirectly co-cultured with or without iPSC-CMs in the absence or presence of TGF-β ± sacubitril. Sacubitril is an inhibitor for ANP (atrial natriuretic peptide) and BNP-degrading enzyme. *P<0.05 vs. vehicle control, #P<0.05 vs. TGF-β group, $P<0.05 vs. iPSC-CM co-culture group by one-way ANOVA followed by Bonferroni multiple comparisons. F, Representative immunoblots showing the levels of α-SMA, periostin (POSTN), collagen III (COLIII), and fibronectin (FN) in human iPSC-CFs after they were indirectly co-cultured with or without iPSC-CMs in the absence or presence of TGF-β ± sacubitril. Densitometric quantifications are shown below the immunoblots. *P<0.05 by one-way ANOVA followed by Bonferroni multiple comparisons. G, Schematic of the proposed mechanisms for the role of cardiomyocytes in suppressing TGF-β-induced CF transdifferentiation through the ANP/BNP-NPR1 signaling pathway. Sacubitril, a pharmacological drug, can further enhance the anti-fibrotic effect by suppressing the activity of ANP- and BNP-degrading enzyme, neprilysin. Data in (B-F) were generated based on three independent differentiations, and are presented as mean ± SEM.
To understand whether the interaction through the ANP/BNP-NPR1 plays a role in cardiac fibrosis, we treated human iPSC-CFs with BNP in the presence of TGF-β. Interestingly, CFs expressed significantly lower mRNA levels of myofibroblast markers and ECM genes (ACTA2, POSTN, COL1A1, and COL3A1), and higher levels of CF markers (PDGFRA and TCF21) with BNP treatment (Figure 6C). α-SMA protein levels were also significantly suppressed with escalating concentrations of BNP in a dose-dependent manner (Figure 6D). We then co-cultured human iPSC-CFs with iPSC-CMs in the presence of TGF-β, and observed a similar expression pattern of these genes in co-culture as was seen with BNP treatment (Figure 6E). The protein levels of myofibroblast markers and ECM proteins (α-SMA, collagen III, and fibronectin) in human iPSC-CFs were also significantly lower in the co-cultured than non-co-cultured condition (Figure 6F).
ANP and BNP are known to be upregulated in cardiac hypertrophy and fibrosis.26 However, evidence shows that ANP and BNP have relatively short half-lives (3 minutes and 20 minutes, respectively) due to the ubiquitous presence of their degrading enzyme, neprilysin, in the circulatory system.27 As such, ANP/BNP may not be able to exert a long-lasting anti-fibrotic effect through the interaction with their receptor NPR1 expressed on CFs. Sacubitril, a neprilysin inhibitor, has been shown to ameliorate heart failure when used together with valsartan.28 Therefore, we further examined whether the addition of sacubitril to the co-culture system would further suppress CF transdifferentiation. Indeed, we observed that sacubitril further decreased the mRNA and protein levels of myofibroblast markers and ECM genes in the co-cultured condition (Figure 6E and 6F). In particular, the myofibroblast marker periostin was significantly decreased only when sacubitril was added to the co-culture system. Together, these data suggest that targeting the ANP/BNP-NPR1 signaling pathway may be a new strategy for anti-cardiac fibrosis therapy (Figure 6G).
DISCUSSION
It is increasingly recognized that fibroblasts in different tissues are heterogeneous as evident by their distinct gene expression patterns.18, 29 However, the inability to characterize the full transcriptomic picture of fibroblasts in a tissue-specific manner has impeded the development of mechanism-oriented, organ-specific anti-fibrotic therapies.30 In this study, by performing an in-depth analysis of mouse single-cell transcriptomics data in combination with several assays, we identified a group of tissue-specific signature genes that are conserved in mouse and human primary fibroblasts. After identifying that CFs in large are of the epicardial lineage, we successfully developed a robust protocol to generate quiescent bona fide CFs from human iPSCs that could recapitulate most of the fundamental biological features of primary CFs (Figure 7). We further demonstrated that human iPSC-CFs could be used as a faithful in vitro cell model to test drugs with pro- or anti-fibrosis potentials. Moreover, we identified that crosstalk between iPSC-CMs and iPSC-CFs through the ANP/BNP-NPR1 pathway might represent a novel route for anti-cardiac fibrosis therapy. Our study also highlights the value of scRNA-seq in developing new candidate therapeutic strategies for treating tissue-specific fibrosis.
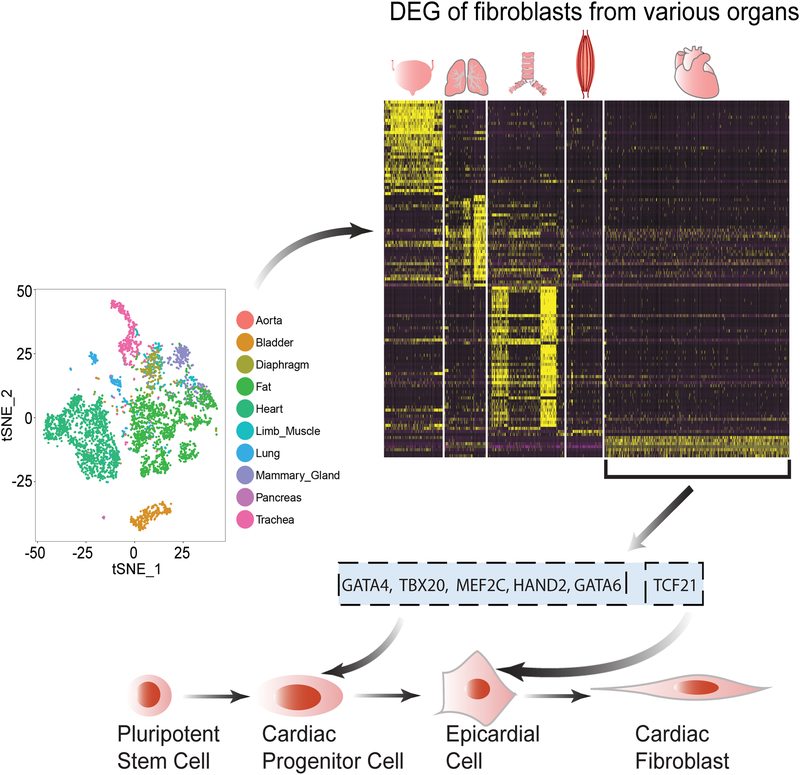
Figure 7. A logical flow diagram demonstrates the process of developing the human iPSC-derived cardiac fibroblast differentiation protocol.
The differentiation protocol for human iPSC -CFs developed in this study is based on the recognition of tissue-specific marker genes expressing in fibroblast subpopulations, as revealed by the mouse single-cell transcriptomic data. Further DEG (differentially expressed genes) analysis highlights the critical roles of tissue-specific transcription factors in regulating the development trajectories of fibroblast subpopulations, and also suggests that CFs may be derived from the epicardial lineage. As such, human iPSC-CFs were successfully generated after intermediate cell types of cardiac progenitor cells and epicardial cells being sequentially differentiated. iPSC, induced pluripotent stem cell; CF, cardiac fibroblast.
Although questions on how tissue-specific fibroblast identities are defined during development and organogenesis remain unanswered, accumulating evidence from animal studies suggests that cell lineage specification is governed by a handful of tissue-specific TFs.6, 7, 31 In this study, we observed that fibroblasts express high levels of tissue-specific, development-related TFs, raising the possibility that these genes may help define fibroblast tissue identity. Animal studies support this idea. Deficiency of Gata4 leads to failure to initiate proepicardium formation during cardiogenesis,32 and ablation of Tcf21 blocks the differentiation of CFs from proepicardial cells by suppressing epithelial-mesenchymal transition.12 In line with these in vivo findings, we observed a significant rise in GATA4 and TCF21 expression in human iPSC-CPCs and iPSC-EPCs, respectively. Collectively, these data suggest that a sequential expression of these lineage-predominant TFs may play a critical role in defining the fate of CFs.
Driven by this hypothesis, we successfully differentiated human iPSCs into CFs by modifying a protocol initially developed to generate iPSC-EPCs.11 Importantly, we demonstrated that our human iPSC-CFs can serve as a faithful in vitro cell model to explore the underlying mechanisms of cardiac fibrosis and to screen anti-fibrotic drugs due to the following advantages. First, human iPSC-CFs highly resemble primary cells at the genetic and functional levels, and preserve a quiescent phenotype until passage 4. Therefore, the derived cells are sensitive to pro-fibrotic stimuli. Indeed, using this powerful in vitro platform, we demonstrated that human iPSC-CFs are highly responsive to both anti- and pro-fibrotic drugs. Second, human iPSCs can be used as an unlimited cell source to generate a large quantity of CFs for mechanistic studies and high throughput anti-fibrotic drug discovery. Third, the induction protocol for human iPSC-CF developed in this study will allow for testing of sex- and patient-specific cellular responses to candidate anti-fibrotic drugs in vitro.
A substantial body of evidence shows that crosstalk between fibroblasts and inflammatory cells regulates fibrosis in different injured tissues, such as the heart,33 lungs,34 and liver.35 Interestingly, using the complementary receptor-ligand analysis tool, intercellular communication networks generated using mouse scRNA-seq data suggest that fibroblasts may reciprocally interact with a wide range of cell types other than inflammatory cells to regulate their cellular and biological functions. By coculturing human iPSC-CMs and iPSC-CFs, we identified that crosstalk between these two cell types through the ANP/BNP-NPR1 pathway may play a fundamental role in suppressing cardiac fibrosis. Importantly, we further showed that the anti-fibrotic effect seen in co-culture could be significantly enhanced by treating these cells with sacubitril, a drug that inhibits the activity of ANP/BNP-degrading enzyme neprilysin. Sacubitril has been used in combination with an angiotensin II type 1 receptor blocker called valsartan (a drug named LCZ696) to reduce the blood pressure and improve mortality in patients with heart failure.28 However, evidence shows that LCZ696 improved isoproterenol-induced cardiac fibrosis more significantly than valsartan alone in rats,36 suggesting the anti-fibrotic effect of neprilysin inhibitor beyond its diuretic function. In agreement with the animal study, a recent large-scale prospective cohort study showed that compared with baseline, the administration of LCZ696 for eight months drastically attenuated profibrotic signaling in heart failure patients with reduced ejection fraction.37 Collectively, these results suggest that modulating fibrosis-related intercellular signaling cascades such as the ANP/BNP-NPR1 signaling pathway may represent another promising route for anti-fibrotic therapies in addition to suppressing fibroblast activation directly.
In conclusion, the heterogeneity of fibroblasts in different tissues is associated with the expression of tissue-specific signature genes during development. The induction protocol for bona fide quiescent human iPSC-CFs developed in this study allowed us to identify and validate signaling pathways that may be involved in cardiac fibrosis. This study also sheds light on the significance of using human iPSC-CFs as a robust cell platform to test the efficacy of candidate anti-fibrotic drugs in a patient-specific manner.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Fibroblasts from different organs express tissue-specific transcriptome profiles.
Activated cardiac fibroblasts (CFs) are the predominant cell source that contributes to cardiac fibrosis and dysfunction.
What New Information Does This Article Contribute?
Murine single-cell transcriptome data reveal that tissue-specific fibroblasts can be distinguished by a handful of transcription factors.
Human iPSC-derived CFs closely resemble primary CFs at the transcriptional, cellular, and functional levels.
Crosstalk between cardiomyocytes and CFs through natriuretic peptides and their cognate receptor suppresses CF activation, thereby representing a new direction of anti-fibrotic therapy.
CFs, once activated in injured hearts, can contribute to cardiac fibrosis and heart failure. Due to the limited availability of primary human CFs and limited translational value of animal models, the precise mechanisms for cardiac fibrosis is largely unknown and therefore, effective anti-fibrotic therapies are not yet available. Using murine single-cell transcriptome data, we uncovered key transcription factors that are involved in the development trajectory of CFs. Based on this knowledge, we developed an induction protocol that can derive bona fide cardiac fibroblasts from human iPSCs. This robust protocol allowed us to generate unlimited amount of human iPSC-derived CFs, which were used to explore the mechanisms of cardiac fibrosis. We discovered that the interaction between natriuretic peptide type A and B expressed in cardiomyocytes and natriuretic receptor-1 expressed in CFs as a novel, potential anti-fibrosis pathway. Notably, this in vitro iPSC-based platform will help us gain a better understanding of the complexity of cardiac fibrosis by using patient-specific iPSC-derived CFs, which may advance the development of more effective anti-fibrotic therapies in the context of precision medicine.
ACKNOWLEDGMENTS
We thank Ke Yuan from Stanford Pulmonary Critical Care Medicine Division for gifting us primary brain pericytes.
SOURCES OF FUNDING
This work was supported by research grants from National Institutes of Health (NIH) K99 HL135258 (M.G.), K99 HL133473 (H.W.), T32 EB009035 (D.T.P.), R01 HL141371, and UG3 TR002588 (J.C.W.).
Nonstandard Abbreviations and Acronyms:
- ANP
atrial natriuretic peptide
- BNP
brain natriuretic peptide
- CF
cardiac fibroblast
- CM
cardiomyocyte
- CPC
cardiac progenitor cell
- ECM
extracellular matrix
- EPC
epicardial cell
- FGF2
fibroblast growth factor 2
- GO
gene ontology
- iPSC
induced pluripotent stem cell
- NPPA/NPPB
natriuretic peptide type A/B
- NPR1
natriuretic peptide receptor 1
- α-SMA
α-smooth muscle actin
- SB
SB431542
- scRNA-seq
single-cell RNA-sequencing
- TF
transcription factors
- TGF-β
transforming growth factor-β
- t-SNE
t-distributed stochastic neighbor embedding
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C and Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–63. [DOI] [PubMed] [Google Scholar]
- 2.Zeisberg M and Kalluri R. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am J Physiol Cell Physiol. 2013;304:C216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee JW, Kim Y, Wirka RC, Buikema JW, Wu SM, Red-Horse K, Quertermous T and Wu JC. Large-scale single-cell RNA-seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ Res. 2018;123:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Inoue H, Wu JC and Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf T and Enver T. Forcing cells to change lineages. Nature. 2009;462:587–94. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Du Y and Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–34. [DOI] [PubMed] [Google Scholar]
- 8.Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J and Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development (Cambridge, England). 2005;132:2697–707. [DOI] [PubMed] [Google Scholar]
- 9.Molkentin JD, Lin Q, Duncan SA and Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes & Development. 1997;11:1061–72. [DOI] [PubMed] [Google Scholar]
- 10.Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM and Conlon FL. Tcf21 regulates the specification and maturation of proepicardial cells. Development. 2013;140:2409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Sinha S and Talasila A. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development. 2016;143:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, Olson EN and Tallquist MD. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Talasila A and Sinha S. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development. 2015;142:1528–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armulik A, Genove G and Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Developmental Cell. 2011;21:193–215. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A and Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–9. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH and Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. The Journal of Biological Chemistry. 2005;280:9719–27. [DOI] [PubMed] [Google Scholar]
- 17.Schafer S, Viswanathan S, Widjaja AA, Lim WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E, Tan J, Chothani SP, Ye L, Rackham OJL, Ko NSJ, Sahib NE, Pua CJ, Zhen NTG, Xie C, Wang M, Maatz H, Lim S, Saar K, Blachut S, Petretto E, Schmidt S, Putoczki T, Guimaraes-Camboa N, Wakimoto H, van Heesch S, Sigmundsson K, Lim SL, Soon JL, Chao VTT, Chua YL, Tan TE, Evans SM, Loh YJ, Jamal MH, Ong KK, Chua KC, Ong BH, Chakaramakkil MJ, Seidman JG, Seidman CE, Hubner N, Sin KYK and Cook SA. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, Park A, Snider P, Chandran A, Harvey RP, Boyd R, Conway SJ, Pearson J, Kaye DM and Rosenthal NA. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114:1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandya K, Kim HS and Smithies O. Fibrosis, not cell size, delineates beta-myosin heavy chain reexpression during cardiac hypertrophy and normal aging in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaney JS, Roth DM, Olson ER, Naugle JE, Meszaros JG and Insel PA. Inhibition of cardiac myofibroblast formation and collagen synthesis by activation and overexpression of adenylyl cyclase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan D, Takawale A, Lee J and Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis & Tissue Repair. 2012;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Juttermann R and Soloway PD. TIMP-2 is required for efficient activation of proMMP-2 in vivo. The Journal of Biological Chemistry. 2000;275:26411–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taniguchi H, Ebina M, Kondoh Y, Ogura T, Azuma A, Suga M, Taguchi Y, Takahashi H, Nakata K, Sato A, Takeuchi M, Raghu G, Kudoh S and Nukiwa T. Pirfenidone in idiopathic pulmonary fibrosis. The European Respiratory Journal. 2010;35:821–9. [DOI] [PubMed] [Google Scholar]
- 24.Levick SP, Soto-Pantoja DR, Bi J, Hundley WG, Widiapradja A, Manteufel EJ, Bradshaw TW and Melendez GC. Doxorubicin-induced myocardial fibrosis involves the Neurokinin-1 receptor and direct effects on cardiac fibroblasts. Heart, lung & circulation. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard CM and Baudino TA. Dynamic cell-cell and cell-ECM interactions in the heart. J Mol Cell Cardiol. 2014;70:19–26. [DOI] [PubMed] [Google Scholar]
- 26.Kerkela R, Ulvila J and Magga J. Natriuretic peptides in the regulation of cardiovascular physiology and metabolic events. J Am Heart Assoc. 2015;4:e002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potter LR. Natriuretic peptide metabolism, clearance and degradation. The FEBS journal. 2011;278:1808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh JSS, Burrell LM, Cherif M, Squire IB, Clark AL and Lang CC. Sacubitril/valsartan: beyond natriuretic peptides. Heart. 2017;103:1569–1577. [DOI] [PubMed] [Google Scholar]
- 29.Horie M, Miyashita N, Mikami Y, Noguchi S, Yamauchi Y, Suzukawa M, Fukami T, Ohta K, Asano Y, Sato S, Yamaguchi Y, Ohshima M, Suzuki HI, Saito A and Nagase T. TBX4 is involved in the super-enhancer-driven transcriptional programs underlying features specific to lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314:L177–L191. [DOI] [PubMed] [Google Scholar]
- 30.Weiskirchen R, Weiskirchen S and Tacke F. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol Aspects Med. 2018. [DOI] [PubMed] [Google Scholar]
- 31.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI and Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watt AJ, Battle MA, Li J and Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proceedings of the National Academy of Sciences. 2004;101:12573–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan X, Zhang H, Fan Q, Hu J, Tao R, Chen Q, Iwakura Y, Shen W, Lu L, Zhang Q and Zhang R. Dectin-2 deficiency modulates Th1 differentiation and improves wound healing after myocardial infarction. Circ Res. 2017;120:1116–1129. [DOI] [PubMed] [Google Scholar]
- 34.Hartupee J and Mann DL. Role of inflammatory cells in fibroblast activation. J Mol Cell Cardiol. 2016;93:143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlmark KR, Weiskirchen R, Zimmermann HW, Gassler N, Ginhoux F, Weber C, Merad M, Luedde T, Trautwein C and Tacke F. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50:261–74. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi T, Nakamura K, Miura D, Yoshida M, Saito Y, Akagi S, Ohno Y, Kondo M and Ito H. Effect of LCZ696, a dual angiotensin receptor neprilysin inhibitor, on isoproterenol-induced cardiac hypertrophy, fibrosis, and hemodynamic change in rats. Cardiology Journal. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zile MR, O’Meara E, Claggett B, Prescott MF, Solomon SD, Swedberg K, Packer M, McMurray JJV, Shi V, Lefkowitz M and Rouleau J. Effects of Sacubitril/Valsartan on biomarkers of extracellular matrix regulation in patients With HFrEF. J Am Coll Cardiol. 2019;73:795–806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.