Abstract
Background
Pancreatic β-cells adapt to high metabolic demand by expanding their β-cell mass and/or enhancing insulin secretion to maintain glucose homeostasis. Type 2 diabetes (T2D) is typically characterized by β-cell decompensation.
Scope of the review
The current review focuses on summarizing the “omics” and “epi-omics” approaches that particularly focus on addressing the β-cell adaptation to insulin resistance and T2D.
Major conclusions
The molecular mechanisms underlying successful versus compromised β-cell adaptation to insulin resistance are not entirely understood. The last decade has seen an exponential increase in the use of “omics” and “epi-omics” approaches to dissect pathophysiology of metabolic diseases. One recent example is the emergence of m6A mRNA methylation as a new layer of regulation of gene expression with the potential to impact diverse physiological processes in metabolic cells.
Keywords: β-cells, Insulin resistance, Genomics, Transcriptomics, Epigenomics, Epitranscriptomics
1. Introduction - β-cell failure in Type 2 diabetes
Type 2 diabetes (T2D) is currently estimated to affect over 415 million adults worldwide, and this number is expected to rise to 642 million by 2040 constituting a massive economic burden to healthcare systems worldwide [1]. Obesity, unhealthy dietary habits, lack of physical activity and genetic predisposition act concomitantly to induce systemic insulin resistance. Insulin resistance is one of the first physiological abnormalities that precede the development of T2D by many years (reviewed in [2]). Liver and skeletal muscle, two of the primary tissues that are involved in the control of glucose homeostasis are affected by insulin resistance manifested by increased liver gluconeogenesis and decreased glucose uptake in muscle [2]. Furthermore, insulin resistance has detrimental effects on non-canonical tissues including islet cells [2], [3], [4], [5]. A vast majority of patients with T2D exhibit a failure of β-cells to compensate for the ambient insulin resistance [6]. Nevertheless, T2D can occur without the presence of insulin resistance (reviewed in [7]). For example, insulin-secretory defects are a common characteristic of non-obese T2D that particularly impact Asian populations [7], [8]. The precise temporal control of the events and pathways that contribute to the lack of appropriate β-cell compensation to adapt for insulin resistance are not fully understood and are considered to vary greatly between individuals (reviewed in [9]). Despite the general acceptance among the scientific community that β-cell dysfunction is necessary for the development of overt T2D, the relative contributions of decreased β-cell function versus mass amid the uncertainties regarding the ability of human β-cells to expand has spawned a creative debate. Pancreatic β-cell mass increases during early life largely driven by β-cell proliferation and subsequently stabilizes after adolescence [9]. Furthermore, during periods of high metabolic demand such as in human pregnancy, β-cells have the capacity to increase their functional mass by increasing cell size and boosting proliferation while inhibiting pathways of cell loss such as apoptosis (reviewed in [10], [11]). Estimates of human β-cell mass have relied heavily on insulin immunostaining of post-mortem pancreas sections. The inability to measure β-cell mass in-vivo using non-invasive methods likely leads to imprecise estimation. This is further confounded by factors that could potentially influence the mass such as BMI, age and ethnicity [12]. Nonetheless, β-cell mass is believed to be reduced in T2D [13] despite great variability and overlap with non-diabetic individuals [9], [14], [15]. While it was initially thought that T2D is associated with increased cell death [13], recent work has demonstrated that during the progression of T2D, β-cells lose insulin expression and are accompanied by a decrease in expression of transcription factors and eventual loss of cell identity and maturation [16], [17], [18], [19], [20], [21]. Individuals who develop insulin resistance are at high risk of developing T2D, however, their progression from a prediabetic to diabetic state is not uniform; thus, while some individuals are able to mount a robust β-cell compensation to physiological metabolic demands and delay the development of overt disease [22] others fail to compensate and require clinical intervention. An understanding of the molecular differences that underlie a successful versus poor compensatory response to insulin resistance is important to gain insights into the disease process and to plan better therapeutics to counter its progression. The emergence of new “omics” approaches provides an opportunity to gain insights into this phenomenon and will be discussed in the following sections.
2. Genomics - genome-wide association studies (GWAS)
Type 2 diabetes has a strong genetic component and its complexity results from the interaction between genes and the environment. The reported heritability underlying T2D varies with the duration of the follow-up period and can reach up to 80% (reviewed in [23]). Notably, the concordance rate in monozygotic twins is 70% and in dizygotic twins between 20 and 30% [23]. In the early 2000s, the first linkage analyses and candidate approaches confirmed and identified several new genes associated with T2D, including PPARG [24] and TCFL2 [25]. Tcfl2 has a fundamental cell-autonomous role in regulating β-cell mass and function [26]. β-cell specific Tcfl2 KO exhibit a 30% decrease in β-cell mass and impaired glucose-stimulated insulin secretion (GSIS) [26]. With the development of more advanced next-generation sequencing and complex and extensive genome-wide association studies, additional loci were reported. The first set of GWAS studies identified a dozen new T2D associated loci and confirmed previously associated genes including SLC30A8, CDKAL1, and IGF2BP2 [27], [28], [29]. Zinc transporter 8 (ZnT8) is the product of SLC30A8 and is important in the regulation of insulin secretion by regulating zinc uptake into the insulin secretory granules (reviewed in: [30]). SLC30A8 haploinsufficiency confers protection for T2D in obese human individuals [31]. This first wave of GWAS studies was followed by meta-analyses combining data from multiple GWAS including thousands of patients [23]. Recently, genome-wide trans-ancestry meta-analysis has included non-European cohorts resulting in additional loci and adding robustness to some of the previous associations [32]. Despite this progress, the identification of several variants using GWAS can surprisingly explain only a small portion of the heritability of T2D. Thus the missing heritability of T2D is an important issue and it questionable whether rare variants play a fundamental role in the heritability. Fuchsberger and colleagues have performed the largest DNA sequencing study in T2D by sequencing over 12,000 individuals from 5 different ancestry groups and were able to detect the same associated variants that were previously identified by GWAS [33]. A recent approach to revealing casual effects of these genetic variants has been to perform integrative analysis of data from GWAS with expression quantitative locus (eQTL) [34], DNA methylation [35], or using multiple datasets including gene expression and DNA methylation data [36]. Although GWAS studies have identified more than 150 variants for T2D that correspond to over 120 different loci to date, the fact that this can explain less than 10% of the heritability of T2D has been termed a “geneticist's nightmare” [37].
3. Epigenomics
The short time frame of the T2D epidemic suggests that the environment is the main driver of the escalation in contrast to changes in the genetic pool. Epigenetics is the science that studies changes in gene expression without alterations in the nucleotide sequence. Epigenetic changes can be dynamic and environment can modulate genomes, reshape phenotypes and transmit epigenetic information via germline with the potential to impact the progeny. Epigenetic modifications encompass DNA methylation changes, histone modifications, and non-coding RNAs. DNA methylation can modulate gene expression and consist in the addition of a methyl group to the 5′ position of the cytosine pyrimidine ring of DNA. Histone modifications can alter chromatin accessibility and consequently modify affinity to DNA. They consist in the addition of covalent modifications to the N-terminal of histone tails such as lysine acetylation, arginine and lysine methylation, threonine and serine phosphorylation and lysine sumoylation and ubiquitination. Non-coding RNAs–ncRNAs contain a vast set of different RNAs including micro RNAs–miRNAs, small non-coding RNAs–sncRNAs, and long non-coding RNAs–lncRNAs among others. ncRNAs have different functional mechanisms but generally, they decrease mRNA by acting on poly(A) tails inducing mRNA decay (reviewed in: [38]). Several groups have explored the role of different epigenetic mechanisms in the regulation of islets and in particular the importance of epigenetic modifications in mediating the β-cell adaptation to T2D.
3.1. Epigenomics – DNA methylation
An initial set of candidate based studies were adequate to demonstrate the importance of DNA methylation in the pathophysiology of T2D. PPARGC1A regulates insulin secretion in human islets and its expression is downregulated in islets from T2D [39]. Furthermore, the promoter region of PPARGC1A is hypermethylated in T2D as compared to healthy controls and correlates negatively with its expression [39]. Insulin mRNA is downregulated in T2D human islets and a possible mechanism is the increased promoter methylation [40]. PDX1 is an essential transcription factor for β-cell identity and function and is downregulated in islets from T2D [41], [42], [43]. Furthermore, Pdx1 maintains β-cell identity by repressing alpha-cell genes such as MafB [44]. PDX1 enhancer and promoter regions have several hypermethylated CpGs in T2D islets that correlate with decreased gene expression in a process partially mediated by hyperglycemia [45]. The development of array-based methods to detect changes in DNA methylation identified many more differentially methylated genes in T2D islets. The first comprehensive DNA methylation profiling of human islets from controls and T2D identified more than 276 differentially methylated CpGs spanning the promoter regions of 254 genes. Many of these changes correlated with gene expression and were involved in pathways related to β-cell survival and function [46]. Volkov and colleagues went one step further by performing whole genome bisulfite sequencing (WGBS) in islets from healthy controls and T2D [47]. Their sequencing covered more than 80% of all genomic CpGs and identified 25,820 differentially methylated regions (DMRs); furthermore, DMRs spanned genes important for β-cell function such as PDX1 [47]. Finally, circulating blood-based DNA methylation biomarkers might be used in future to indirectly assess β-cell death and function in T2D [48] making DNA methylation an exciting field of study in islet biology.
3.2. Epigenomics – chromatin modifications and accessibility
Bhandare and colleagues performed the first genome-wide mapping of histone modifications in human islets by performing chromatin immunoprecipitation and sequencing (ChIP-sequencing). They focused on three histone marks associated with gene activation (H3K4me1; H3K4m2; and H3K4me3) and one with gene repression (H3K27me3) [49]. Human islet promoters have a high specificity of histone markers compared to other cell types. Furthermore, mature islets show bivalent histone markers and insulin and glucagon promoters show very low levels of H3K4me3 in contrast to genes like PDX1 and MAFB [49]. Recently, the development of an assay for transposase-accessible chromatin (ATAC-seq) high-throughput sequencing methods have allowed for a simpler and faster way of looking into regulatory genomic sites and tissue or disease-specific cis-regulatory networks [50]. Integrative analyses approach by combining ATAC-seq of human islet from controls and T2D with GWAS datasets [51] or DNA methylation and GWAS [52] datasets have helped gain insights into putative causal variants. These studies have allowed us to understand how the candidate genes alter chromatin accessibility resulting in gain- or loss-of-function of genes that are important for β-cell function and survival.
3.3. Epigenomics – ncRNAs
MicroRNAs provide an extra layer of gene regulation and are a major determinant of the regulation of gene expression in disease (Reviewed in [53]). The profile of miRNAs varies greatly among studies depending on the cell system used (cell lines or primary cells) and also vary depending on the islet cell type [53]. The Kaestner group identified one miRNAs cluster to be downregulated in T2D islets in a mechanism involving DNA methylation and TP53INP1. miRNAs that are highly expressed in islets and important for β-cell function include Let-7 and miR-375. Let-7 is among the first identified miRs that is important for cell development. Let-7 overexpression in β-cells induce impaired glucose tolerance and decreased insulin secretion [54]. miR-375 is highly expressed in rodent models of islet hyperplasia and mice lacking miR-375 in β-cells present impaired β-cell proliferation and decreased β-cell mass demonstrating the importance of microRNAs in controlling β-cell function and survival [55]. Once considered as “junk DNA” resulting from human evolution, lncRNAs control several biological functions and are candidates for disease targeting (reviewed in: [56]). The expression of the two lncRNAs βlinc2 and βlinc3 are among the 1500 lncRNAs expressed in islet cells from obese mice [57]. βlinc2 and βlinc3 are particularly highly enriched in β-cells and the expression of the human orthologue βlinc3 is altered in T2D and is associated with BMI [57]. Overexpression of βlinc3 in the mouse β-cell line Min6 cells induced an increase in apoptosis and validated the role of lncRNAs in controlling β-cell survival in obesity and T2D [57].
4. Transcriptomics
Oligonucleotide microarrays were among the first omics tools to study the transcriptomic changes in islets from patients with T2D. mRNA levels of HNF4α, IRS2, AKT2, INSR [58] and several other genes identified in GWAS including IGFBP2 [59] are downregulated in T2D as compared to controls. The accessibility to RNA-sequencing technology has considerably improved our knowledge regarding the overall importance of regulation of gene expression in T2D. For example, Fadista and colleagues conducted a tour de force genomic and transcriptomic analysis by performing exome and RNA sequencing in islets from 83 control and T2D [60] cases. This study presented enrichment in several GWAS SNPs associated with T2D and glycemic traits in eQTLs. Furthermore, several genetic variants presented allelic imbalance, some of which were associated with T2D and with changes in DNA methylation. This study was the most comprehensive performed in human islets and identified several eQTLs and sQTLs associated with β-cell function including variants in TMED, NT5E, PAK7, and TSPAN33 [60]. One limitation of these studies was the use of whole islets in their sequencing strategies. Considering islet cells are constituted by different cell types it is likely that the relative proportions of cell types are differentially altered in T2D compared to controls. This is particularly a problem with the identification of heterogeneous β-cells that exhibit different maturity and whose capacity for proliferation is variable [61]. An initial approach to circumventing this issue was to undertake fluorescence-activated cell sorting (FACS) strategies to sort and collect different types of β-cells [62]. An important breakthrough in the field was the use of single-cell RNA sequencing (scRNA-seq) to detect cell-type transcriptomic signatures in health and disease. scRNA-seq in human islets revealed the genetic programs of each individual cell types in the islets, and when applied to T2D, revealed genes impacted by T2D in a cell-type specific manner [63], [64], [65]. This approach revealed possible β-cell compensation transcriptomic signatures such as downregulation of FXYD2 and upregulation of GPD2 [64]. While upregulation of GPD2 might be potentiating insulin secretion by increasing mitochondrial respiration [64], β-cell-specific ablation of Fxyd2 in mice results in increased β-cell mass and hyperinsulinemia [66]. Gene “dropouts” are among the limitations of scRNA-seq analyses, which particularly impact genes that are expressed at low levels, and are captured randomly in some cell subpopulations and consequently artificially inflate cell heterogeneity [67]. Furthermore, due to relatively small sample sizes, differences in technical methods and data analyses, the comparisons between these datasets continues to reveal considerable heterogeneity that needs to be revised in future studies that include large cohorts of samples and improved bioinformatics analyses (reviewed in [67]).
5. Perspectives for a new era: epitranscriptomics
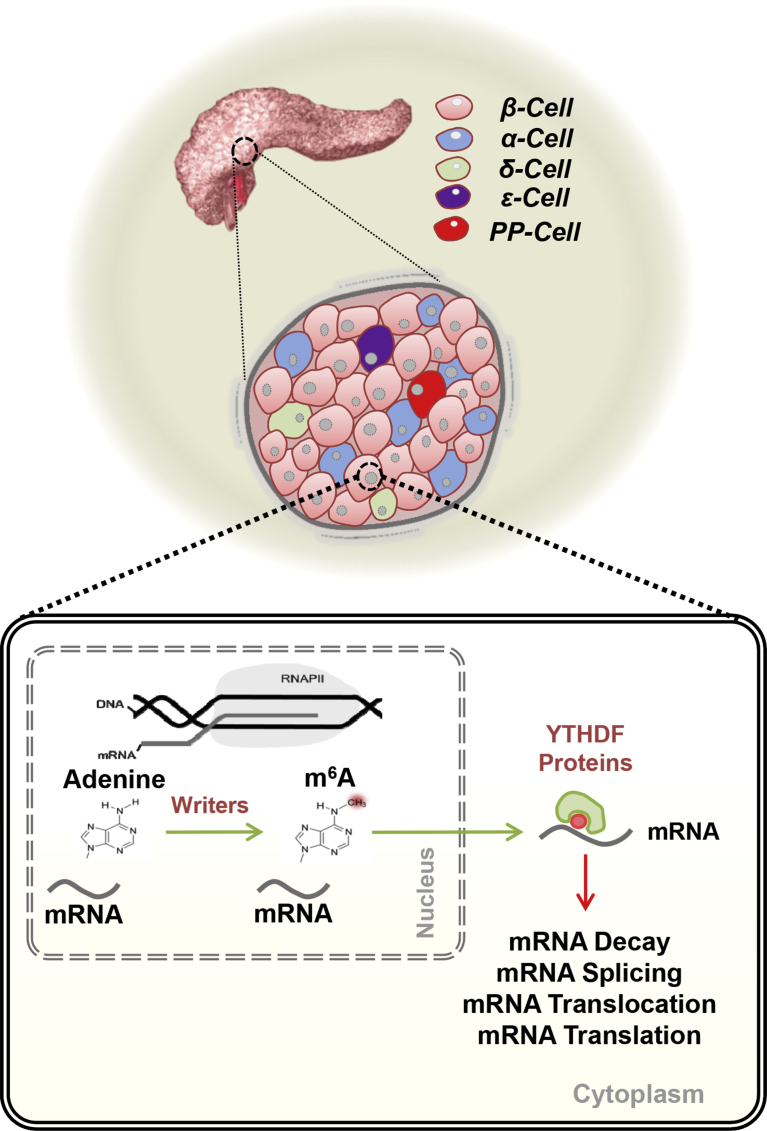
RNA transcription from DNA requires extensive regulation to guarantee proper gene expression. mRNA is transcribed from a genomic locus of DNA, and this process can be regulated by genomic and epigenetic events and impacted by diseases such as T2D. However, completion of the mRNA lifecycle involves capping, splicing, and polyadenylation before exportation to the cytoplasm for translation (reviewed in: [68]). Proper translation efficiency requires extensive chemical modifications of transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) [68]. Indeed, mRNA can be chemically modified [69], [70] and N6-methyladenosine (m6A) is the most abundant modification with a frequency of 1–2 per 1000 nucleotides [68]. The notion that mRNA methylation was not a stationary event but rather a dynamic phenomenon became clear with the identification of two mRNA m6A demethylases, namely fat mass- and obesity-associated protein (FTO) [71] and α-ketoglutarate-dependent dioxygenase AlkB family of proteins (ALKBH5) [72]. Interesting, polymorphisms in FTO are among the genes that are strongly associated with T2D and obesity. Today we know that m6A is dynamically regulated by ‘erasers’ and ‘writers’, complexes that include methyltransferase like 3 (METTL3) and methyltransferase like 14 (METTL14) (Figure 1) [73]. The development of an antibody-based m6A enrichment coupled with high-throughput sequencing was a breakthrough in the field and revealed that m6A is enriched at the 3′ UTR near stop codons and is conserved between human and mouse [74], [75]. m6A exerts its functions by two main mechanisms: 1) regulation of mRNA stability and 2) translation efficiency of a set of mRNA pools in response to dynamic environmental stimuli [68]. m6A impacts virtually all biological processes such as neurogenesis [76], [77], stem cell differentiation and maintenance [78], [79], oncogenesis [80], [81], [82], DNA damage response [83], development [84] and several signaling pathways including TGF-β [85] and AKT [86]. The last decade has seen rapid progress in sequencing technologies that have shed light on new layers of regulation that are conserved between different species and involved in the dynamic regulation of gene expression which includes the findings on m6A. Future studies on islet biology will be required to be integrative and include several different “omics” layers and necessarily include larger sample numbers. While scRNA-seq is extremely promising the field requires standard data analyses and improved cell-capture and sequencing technologies. It is desirable that findings arising from high-throughput analyses of islets from T2D patients are complemented with rodent models of insulin resistance in which one can determine ‘successful’ compensation that is reflected by the organism maintaining insulin secretion capacity in the face of insulin resistance. One such example is the liver-specific insulin receptor KO mouse (LIRKO) [87], [88], [89], [90], [91]. On the contrary, among models that exhibit insulin resistance but ‘fail’ to compensate include the db/db mouse [92], [93]. The availability of these genetic models will allow more careful time-course analyses and candidate validation studies that are otherwise not possible in human experiments. It is likely that the next decade will witness the importance of “omics” and “epi-omics” approaches to carefully address fundamental questions regarding the pathophysiology of pancreatic islet cells in T2D.
Figure 1.
N6-methyladenosine (m6A) mRNA methylation – a mechanism that can fine-tune the regulation of gene expression. m6A is the most abundant modification in mRNA. m6A controls mRNA decay, mRNA splicing, mRNA translocation and mRNA translation, thereby modulating diverse cellular functions and has the potential to contribute to the adaptation of β-cells to insulin resistance.
Funding
This article is part of a supplement entitled ‘Biomarkers of Beta-Cell Health and Dysfunction: Towards Personalised Diabetes Care. Proceedings of the 20th Servier-IGIS Symposium’. The Symposium and the publication of the Supplement have been made possible by an unrestricted educational grant from Institut la Conférence Hippocrate – Servier Group.
Acknowledgments
We apologize to those individuals whose work could not be cited due to space limitations. We acknowledge funding support from NIH Grants R01 DK067536, R01 DK103215, and UC4 DK116278.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nature Reviews Endocrinology. 2017;14:88. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.DeFronzo R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H. Type 2 diabetes mellitus. Nature Reviews Disease Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 3.Accili D. vol. 53. 2004. Lilly lecture 2003. The struggle for mastery in insulin action: from triumvirate to republic; pp. 1633–1642. [DOI] [PubMed] [Google Scholar]
- 4.Kulkarni R.N., Brüning J.C., Winnay J.N., Postic C., Magnuson M.A. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 5.Kawamori D., Kurpad A.J., Hu J., Liew C.W., Shih J.L. Insulin signaling in alpha cells modulates glucagon secretion in vivo. Cell Metabolism. 2009;9:350–361. doi: 10.1016/j.cmet.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shirakawa J., De Jesus D.F., Kulkarni R.N. Exploring inter-organ crosstalk to uncover mechanisms that regulate β-cell function and mass. European Journal of Clinical Nutrition. 2017;71:896. doi: 10.1038/ejcn.2017.13. [DOI] [PubMed] [Google Scholar]
- 7.Gerich J.E. Contributions of insulin-resistance and insulin-secretory defects to the pathogenesis of type 2 diabetes mellitus. Mayo Clinic Proceedings. 2003;78:447–456. doi: 10.4065/78.4.447. [DOI] [PubMed] [Google Scholar]
- 8.Rhee E.J. Diabetes in Asians. Endocrinology and metabolism (Seoul, Korea) 2015;30:263–269. doi: 10.3803/EnM.2015.30.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halban P.A., Weir G.C., Polonsky K.S., Bowden D.W., Hawkins M.A. β-Cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Journal of Clinical Endocrinology & Metabolism. 2014;99:1983–1992. doi: 10.1210/jc.2014-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rieck S., Kaestner K.H. Expansion of β-cell mass in response to pregnancy. Trends in Endocrinology and Metabolism. 2010;21:151–158. doi: 10.1016/j.tem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baeyens L., Hindi S., Sorenson R.L., German M.S. β-Cell adaptation in pregnancy. Diabetes, Obesity and Metabolism. 2016;18:63–70. doi: 10.1111/dom.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu K.C., Cohan P., Lee N.P., Chuang L.M. Insulin sensitivity differs among ethnic groups with a compensatory response in beta-cell function. Diabetes Care. 2000;23:1353–1358. doi: 10.2337/diacare.23.9.1353. [DOI] [PubMed] [Google Scholar]
- 13.Matveyenko A.V., Butler P.C. Relationship between β-cell mass and diabetes onset. Diabetes, Obesity and Metabolism. 2008;10:23–31. doi: 10.1111/j.1463-1326.2008.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donath M.Y., Halban P.A. Decreased beta-cell mass in diabetes: significance, mechanisms and therapeutic implications. Diabetologia. 2004;47:581–589. doi: 10.1007/s00125-004-1336-4. [DOI] [PubMed] [Google Scholar]
- 15.Rahier J., Guiot Y., Goebbels R.M., Sempoux C., Henquin J.C. Pancreatic β-cell mass in European subjects with type 2 diabetes. Diabetes, Obesity and Metabolism. 2008;10:32–42. doi: 10.1111/j.1463-1326.2008.00969.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim-Muller J.Y., Bouchi R., Accili D., Cinti F., Ratner L.E. Evidence of β-cell dedifferentiation in human type 2 diabetes. Journal of Clinical Endocrinology & Metabolism. 2016;101:1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter C.S., Stein R.W. Evidence for loss in identity, de-differentiation, and trans-differentiation of islet β-cells in type 2 diabetes. Frontiers in Genetics. 2017;8 doi: 10.3389/fgene.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swisa A., Avrahami D., Eden N., Zhang J., Feleke E. PAX6 maintains β cell identity by repressing genes of alternative islet cell types. Journal of Clinical Investigation. 2017;127:230–243. doi: 10.1172/JCI88015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez G.D., Bender A.S., Cirulli V., Mastracci T.L., Kelly S.M. Pancreatic β cell identity requires continual repression of non–β cell programs. Journal of Clinical Investigation. 2017;127:244–259. doi: 10.1172/JCI88017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spijker H.S., Song H., Ellenbroek J.H., Roefs M.M., Engelse M.A. Loss of β-cell identity occurs in type 2 diabetes and is associated with islet amyloid deposits. Diabetes. 2015;64:2928–2938. doi: 10.2337/db14-1752. [DOI] [PubMed] [Google Scholar]
- 21.Talchai C., Xuan S., Lin Hua V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mezza T., Muscogiuri G., Sorice G.P., Clemente G., Hu J. Insulin resistance alters islet morphology in nondiabetic humans. Diabetes. 2014;63:994–1007. doi: 10.2337/db13-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad R.B., Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes. 2015;6:87. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.-C. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature Genetics. 2000;26:76. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 25.Grant S.F.A., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38:320. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 26.Mondragon A., Hodson D.J., da Silva Xavier G., Rutter G.A., Mitchell R.K. Selective disruption of Tcf7l2 in the pancreatic β cell impairs secretory function and lowers β cell mass. Human Molecular Genetics. 2014;24:1390–1399. doi: 10.1093/hmg/ddu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I.W. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 29.The Wellcome Trust Case Control C, Burton P.R., Clayton D.G., Cardon L.R., Craddock N. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson H.W., Wenzlau J.M., O'Brien R.M. Zinc transporter 8 (ZnT8) and β cell function. Trends in Endocrinology and Metabolism. 2014;25:415–424. doi: 10.1016/j.tem.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syring K.E., Boortz K.A., Oeser J.K., Ustione A., Platt K.A. Combined deletion of Slc30a7 and Slc30a8 unmasks a critical role for ZnT8 in glucose-stimulated insulin secretion. Endocrinology. 2016;157:4534–4541. doi: 10.1210/en.2016-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, South Asian Type 2 Diabetes C, Mexican American Type 2 Diabetes C Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature Genetics. 2014;46:234. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V. The genetic architecture of type 2 diabetes. Nature. 2016;536:41. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics. 2016;48:481. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y., Zeng J., Zhang F., Zhu Z., Qi T. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nature Communications. 2018;9:918. doi: 10.1038/s41467-018-03371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue A., Wu Y., Zhu Z., Zhang F., Kemper K.E. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nature Communications. 2018;9:2941. doi: 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rich S.S. Still a geneticist's nightmare. Nature. 2016;536:37. doi: 10.1038/nature18906. [DOI] [PubMed] [Google Scholar]
- 38.De Jesus D.F., Kulkarni R.N. Epigenetic modifiers of islet function and mass. Trends in Endocrinology and Metabolism. 2014;25:628–636. doi: 10.1016/j.tem.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Ling C., Del Guerra S., Lupi R., Rönn T., Granhall C. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang B.T., Dayeh T.A., Kirkpatrick C.L., Taneera J., Kumar R. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA1c levels in human pancreatic islets. Diabetologia. 2011;54:360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachdeva M.M., Claiborn K.C., Khoo C., Yang J., Groff D.N. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nature Genetics. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 43.Brissova M., Shiota M., Nicholson W.E., Gannon M., Knobel S.M. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. Journal of Biological Chemistry. 2002;277:11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 44.Gao T., McKenna B., Li C., Reichert M., Nguyen J. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metabolism. 2014;19:259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B.T., Renström E., Nitert M.D., Volkov P.A., Malmgren S. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Molecular Endocrinology. 2012;26:1203–1212. doi: 10.1210/me.2012-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. The European Molecular Biology Organization Journal. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkov P., Bacos K., Ofori J.K., Esguerra J.L.S., Eliasson L. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66:1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 48.Willmer T., Johnson R., Louw J., Pheiffer C. Blood-based DNA methylation biomarkers for type 2 diabetes: potential for clinical applications. Frontiers in Endocrinology. 2018;9:744. doi: 10.3389/fendo.2018.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhandare R., Schug J., Le Lay J., Fox A., Smirnova O. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Research. 2010;20:428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raurell-Vila H., Ramos-Rodríguez M., Pasquali L. Assay for transposase accessible chromatin (ATAC-Seq) to chart the open chromatin landscape of human pancreatic islets. Methods in molecular biology. 2018;1766:197–208. doi: 10.1007/978-1-4939-7768-0_11. [DOI] [PubMed] [Google Scholar]
- 51.Khetan S., Kursawe R., Youn A., Lawlor N., Jillette A. Type 2 diabetes–associated genetic variants regulate chromatin accessibility in human islets. Diabetes. 2018;67:2466–2477. doi: 10.2337/db18-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thurner M., van de Bunt M., Torres J.M., Mahajan A., Nylander V. Integration of human pancreatic islet genomic data refines regulatory mechanisms at type 2 diabetes susceptibility loci. eLife. 2018;7 doi: 10.7554/eLife.31977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaPierre M.P., Stoffel M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Molecular Metabolism. 2017;6:1010–1023. doi: 10.1016/j.molmet.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost R.J.A., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singer R.A., Sussel L. Islet long noncoding RNAs: a playbook for discovery and characterization. Diabetes. 2018;67:1461–1470. doi: 10.2337/dbi18-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Motterle A., Gattesco S., Peyot M.-L., Esguerra J.L.S., Gomez-Ruiz A. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Molecular Metabolism. 2017;6:1407–1418. doi: 10.1016/j.molmet.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunton J.E., Kulkarni R.N., Yim S., Okada T., Hawthorne W.J. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 59.Marselli L., Thorne J., Dahiya S., Sgroi D.C., Sharma A. Gene expression profiles of beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. Public Library of Science one. 2010;5:e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bader E., Migliorini A., Gegg M., Moruzzi N., Gerdes J. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature. 2016;535:430. doi: 10.1038/nature18624. [DOI] [PubMed] [Google Scholar]
- 62.Dorrell C., Schug J., Canaday P.S., Russ H.A., Tarlow B.D. Human islets contain four distinct subtypes of β cells. Nature Communications. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xin Y., Kim J., Okamoto H., Ni M., Wei Y. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabolism. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 64.Segerstolpe Å., Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lawlor N., George J., Bolisetty M., Kursawe R., Sun L. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type–specific expression changes in type 2 diabetes. Genome Research. 2017;27:208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arystarkhova E., Liu Y.B., Salazar C., Stanojevic V., Clifford R.J. Hyperplasia of pancreatic beta cells and improved glucose tolerance in mice deficient in the FXYD2 subunit of Na, K-ATPase. Journal of Biological Chemistry. 2013;288:7077–7085. doi: 10.1074/jbc.M112.401190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y.J., Kaestner K.H. Single-cell RNA-seq of the pancreatic islets––a promise not yet fulfilled? Cell Metabolism. 2019;29:539–544. doi: 10.1016/j.cmet.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nachtergaele S., He C. Chemical modifications in the life of an mRNA transcript. Annual Review of Genetics. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lavi S., Shatkin A.J. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:2012–2016. doi: 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia G., Fu Y., Zhao X., Dai Q., Zheng G. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology. 2011;7:885. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roignant J.-Y., Soller M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends in Genetics. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 74.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 75.Meyer Kate D., Saletore Y., Zumbo P., Elemento O., Mason Christopher E. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoon K.-J., Ringeling F.R., Vissers C., Jacob F., Pokrass M. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171 doi: 10.1016/j.cell.2017.09.003. 877-889.e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y., Li Y., Yue M., Wang J., Kumar S. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nature Neuroscience. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nature Reviews Molecular Cell Biology. 2016;17:183. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 79.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 80.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millán-Zambrano G. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552:126. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z., Weng H., Su R., Weng X., Zuo Z. FTO plays an oncogenic role in acute myeloid leukemia as a N6-Methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31 doi: 10.1016/j.ccell.2017.02.013. 591-606.e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang Y., Laurent B., Hsu C.-H., Nachtergaele S., Lu Z. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature. 2018;555:256. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J., Eckert M.A., Harada B.T., Liu S.-M., Lu Z. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nature Cell Biology. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Okada T., Liew C.W., Hu J., Hinault C., Michael M.D. Insulin receptors in β-cells are critical for islet compensatory growth response to insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El Ouaamari A., Kawamori D., Dirice E., Liew Chong W., Shadrach Jennifer L. Liver-derived systemic factors drive β cell hyperplasia in insulin-resistant states. Cell Reports. 2013;3:401–410. doi: 10.1016/j.celrep.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Michael M.D., Kulkarni R.N., Postic C., Previs S.F., Shulman G.I. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Molecular Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 90.El Ouaamari A., Dirice E., Gedeon N., Hu J., Zhou J.-Y. SerpinB1 promotes pancreatic β cell proliferation. Cell Metabolism. 2016;23:194–205. doi: 10.1016/j.cmet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Georgia S., Hinault C., Kawamori D., Hu J., Meyer J. Cyclin D2 is essential for the compensatory beta-cell hyperplastic response to insulin resistance in rodents. Diabetes. 2010;59:987–996. doi: 10.2337/db09-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dalbøge L.S., Almholt D.L.C., Neerup T.S.R., Vassiliadis E., Vrang N. Characterisation of age-dependent beta cell dynamics in the male db/db mice. Public Library of Science one. 2013;8 doi: 10.1371/journal.pone.0082813. e82813-e82813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alarcon C., Boland B.B., Uchizono Y., Moore P.C., Peterson B. Pancreatic β-cell adaptive plasticity in obesity increases insulin production but adversely affects secretory function. Diabetes. 2016;65:438–450. doi: 10.2337/db15-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]