Abstract
Background
Obesity and type 2 diabetes (T2D) are major public health issues worldwide, and put a significant burden on the healthcare system. Genetic variants, along with traditional risk factors such as diet and physical activity, could account for up to approximately a quarter of disease risk. Epigenetic factors have demonstrated potential in accounting for additional phenotypic variation, along with providing insights into the causal relationship linking genetic variants to phenotypes.
Scope of review
In this review article, we discuss the epidemiological and functional insights into epigenetic disturbances in obesity and diabetes, along with future research directions and approaches, with a focus on DNA methylation.
Major conclusions
Epigenetic mechanisms have been shown to contribute to obesity and T2D disease development, as well as potential differences in disease risks between ethnic populations. Technology to investigate epigenetic profiles in diseased individuals and tissues has advanced significantly in the last years, and suggests potential in application of epigenetic factors in clinical monitoring and as therapeutic options.
Keywords: Epigenetics, DNA methylation, Obesity, Type 2 diabetes
Highlights
-
•
Aberrance in epigenetics associated with metabolic disturbances.
-
•
Whole-genome bisulfite sequencing provides improved coverage at lower precision relative to arrays.
-
•
Demethylating therapies could play a role in future obesity and diabetes treatment.
1. Introduction
Obesity is a major public health problem in all regions of the world, with approximately 1.5 billion people worldwide overweight or affected by obesity [1], and at risk for a wide range of diseases including type 2 diabetes (T2D) and cardiovascular disease (CVD), as well as related metabolic and inflammatory disturbances [2]. As a direct result of the increase in obesity, the number of people affected by T2D worldwide is projected to increase from 366 million in 2011 to 552 million by 2030 [3]. This increase in risk of T2D, as well as CVD with obesity, has been shown by prospective population studies to occur across all age groups and ethnicities [4], [5], [6].
Although genetic variation that affects obesity, measured by body mass index (BMI), has been identified at more than 200 different loci, these known variants together account for only 3–4% of phenotypic variation [7], [8], [9], [10]. Even with the possibility of more genes being added as research advances, according to modelling by Locke et al., no more than 30% of the variation could be attributable to common variants [7], [11]. Diet and physical activity are also important determinants of adiposity, but do not explain variability in susceptibility to obesity. Similarly, in T2D, more than 200 genetic loci has been identified to date via genome-wide association studies (GWAS), with these genetic variants explaining less than 20% of observed T2D risk [12]. Further, genetic variation, even along with traditional risk factors such as diet and physical activity, does not account for the differences in risk observed between ethnic populations [13]. For example, Indian Asians are at four-fold higher risk of T2D compared with Europeans [14], along with higher likelihood of early development of T2D [15].
Recent studies suggested that epigenetic factors, in particular DNA methylation, may play a central role in adiposity and T2D. In this review article, we discuss the epidemiological and functional insights into epigenetic disturbances in obesity and diabetes, along with future research directions and approaches, with a focus on DNA methylation.
2. Epigenetics
Epigenetics refers to the study of mechanisms that control gene expression in a heritable fashion without affecting the underlying genomic sequences. In contrast to the genome, which is largely static, the epigenome is much more dynamic and displays variation across cell types [16]. Epigenomic variation, including DNA methylation, histone modifications, and chromatin binding, contributes to cellular phenotypes as well as responsiveness to external signals [17]. Epigenetic changes are preserved during somatic cell division, and may also be transmitted from the parental germline to the offspring. This transgenerational epigenetic inheritance is documented in a wide range of organisms, including prokaryotes, plants, and animals [18].
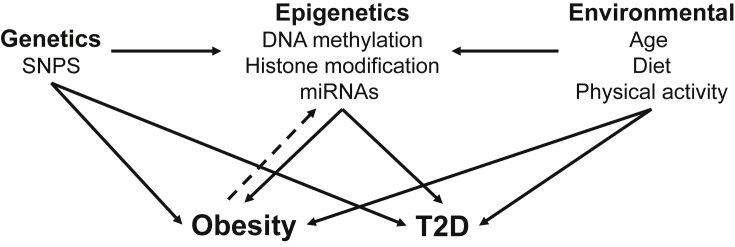
DNA methylation, a key regulator of gene expression and molecular phenotype, is one of the most well-characterized epigenetic modifications, involving the conversion of cytosine to 5-methylcytosine via the covalent transfer of a methyl group to the fifth carbon position of cytosine [19]. This epigenetic mark remains stable during cell division and acts as a form of cellular memory, regulating a wide range of cellular activities including transcription and chromosomal stability, and plays a crucial role in embryonic development, genomic imprinting and X-chromosome inactivation. Methylation patterns are established and modified by specific DNA methyltransferases, such as DNMT1 which transfers patterns of methylation to a newly synthesized strand after DNA replication [20]. DNA methylation is also functionally linked in transcription to histone modification, another major epigenetic regulator in mammalian cells [17]. Together, these two epigenetic processes may provide the underlying mechanism for the stable propagation of gene activity from one generation of cells to the next (Figure 1) [21]. In addition, microRNAs (miRNA) and other small RNAs also provide post-transcriptional regulation for gene and protein expression [22].
Figure 1.
Role for epigenetic mechanisms in obesity and type 2 diabetes.
Evidence has consistently pointed to the role of epigenetic modification in response to environmental exposure. For example, it is well established that dietary manipulation in Avy mice such as a methyl-poor diet impacts upon its coat colour, weight and propensity to develop diabetes and cancer [23]. In rats, maternal protein restriction has been shown to lead to impaired glucose tolerance and insulin resistance in the adult offspring, accompanied by changes in expression for genes involved in insulin-signalling [24]. Similarly, intrauterine growth restriction (IUGR) in rats has resulted in histone code modifications repressing glucose transporter 4 (glut4) expression in the offspring and widespread alterations of DNA methylation in pancreatic islets, and a resultant, increased risk of T2D [25]. Similarly, in humans, maternal under-nutrition and low birth-weight have been associated, following IUGR, with increased risk of T2D in the offspring, with changes in DNA methylation at the HNF4A gene locus, a known T2D susceptibility locus [26], [27], [28]. Data from the Dutch Hunger Winter Families study has also demonstrated that individuals exposed to famine in utero during World War II have less methylation of the IGF2 gene in blood DNA and tended to develop obesity later in life at higher rates compared to unexposed same-sex siblings [29].
3. DNA methylation in obesity and type 2 diabetes
Aberrant DNA methylation has been found to be associated with various complex human diseases, in particular with metabolic disturbances such as obesity and T2D [30], [31], [32], [33], [34], [35], [36], [37]. Using the comprehensive array-based relative methylation (CHARM) analysis in 74 individuals, Feinberg and colleagues reported a total of 227 variably methylated regions across the genome enriched for developmental genes. Among these regions, half were stable within individuals for more than 10 years on average, of which four showed consistent covariation with BMI, and were located in or near genes previously implicated in regulating body weight or T2D [30]. Taking an epigenome-wide approach with the Illumina Infinium HumanMethylation450 (450K) array, using peripheral blood samples with a modest sample size of 48 obese and 48 lean African-American youths, Xu et al. found >20,000 differentially methylated CpG sites at a false discovery rate (FDR) of less than 0.05, accompanied by significant enrichment of genes identified by previous GWAS to be associated with obesity as well as T2D and other related diseases such as hypertension and dyslipidemia [31].
In European individuals, Dick et al. reported an increase in methylation at the HIF3A locus to be associated with increase in BMI of adults, both in blood and in adipose tissue. The authors also observed a significant inverse correlation between methylation and gene expression of HIF3A in adipose tissue, providing further support for the role of hypoxia inducible transcription factor pathways in obesity [32]. This association of methylation changes in HIF3A was also corroborated by another study conducted in ∼2,000 African American adults from the Atherosclerosis Risk in Communities (ARIC) study, suggesting that disturbances in DNA methylation associated with adiposity traits are likely stable across tissue type and ethnicity [33]. He and colleagues reported that DNA methylation changes that impact upon adolescent body weight in healthy individuals coincided with methylation patterns at genes previously implicated in obesity, suggesting that the observed changes in methylation profiles occurring early in life at adolescence may likely impact upon increased risk for cardiometabolic diseases later in adulthood [37].
Wahl et al. demonstrated that BMI is associated with widespread changes in DNA methylation in blood across Europeans and Indian Asians (n∼10,000), with these changes largely a consequence rather than cause of adiposity [35]. The identified methylation loci were not only enriched for functional genomic features in multiple tissues and identified genes involved in important pathways such as lipid and lipoprotein metabolism, but also identified gene expression signatures across multiple loci. In addition, a methylation risk score calculated across the loci associated with BMI was strongly predictive of future T2D. Interestingly, it has also been separately shown that approximately half of all T2D associated SNPs affect DNA methylation in blood, further suggesting that aberrant methylation is part of a causal pathway towards development of T2D [38].
To investigate the contribution of DNA methylation to T2D risk in Indian Asians and its contribution to the increased risk relative to Europeans, Chambers et al. conducted a large prospective nested case–control study of Indian Asians and Europeans with incident T2D [34]. Via epigenome-wide association analyses, the authors identified and replicated an independent association between DNA methylation and future T2D incidence across 5 loci. Taking a methylation risk score approach, a 3.5 times higher risk for future T2D between upper and lower quartiles of methylation was revealed, and it was further found that methylation patterns among Indian Asians compared with Europeans are indeed associated with the increased risk of developing T2D.
Barajas-Olmos and colleagues interrogated the methylation profiles in liver tissue (n = 16), visceral tissue, and subcutaneous adipose tissues (n = 30), as well as peripheral blood (n = 38) from obese individuals with and without T2D to investigate the role of alterations of DNA methylation influencing T2D pathogenesis among obese individuals [39]. The authors replicated the findings of previous studies, and also identified novel differential methylation in genes such as LCAT, FOXA2, PON1 and FGF21 that have been previously associated with metabolic traits in genetic studies [40].
DNA methylation has also demonstrated potential in contributing to T2D risk and differences in risk between ethnic populations [34], [41], [42], [43], [44]. Most of these studies investigated DNA methylation and T2D in metabolically relevant tissues such as adipose, muscle, and pancreas from small sample series [42], [43], [44]. Specifically, applying the Illumina Infinium HumanMethylation27 array on pancreatic islets from T2D and non-diabetic donors, Volkmar and colleagues discovered a total of 276 differentially methylated CpG loci across 254 genes, with these epigenetic changes appearing to be specific to the pancreatic islets and not present in blood [44]. Functional annotation of these genes suggested involvement in β-cell survival and function, as well as cellular dysfunction and response to stress.
Leveraging on the unique property of monozygotic twins discordant for T2D to decipher the contribution of environment versus that of genetics on T2D traits, Ribel-Madsen and colleagues interrogated the methylation profile of skeletal muscle (n = 11 pairs) and subcutaneous adipose tissue (n = 5 pairs) biopsies between these twin pairs [43]. The authors found methylation changes related to known T2D-related genes, including PPARGC1A in muscle and HNF4A in subcutaneous adipose tissue in a targeted analysis. Taking an unbiased epigenome-wide analysis approach, one CpG site in muscle (IL8) and 7 sites in adipose tissue (ZNF668, HSPA2, C8orf31, CD320, SFT2D3, TWIST1, MYO5A) were statistically significant after permutation correction. Following a similar design centered upon monozygotic twins, along with same-sex dizygotic twins and additional independent case–control cohorts, Nilsson et al. reported a total of 1,410 and 15,627 CpG sites to be differentially methylated between monozygotic twins and unrelated case–control individuals respectively [42].
Toperoff and colleagues undertook a stepwise study design to explore the contribution of DNA methylation to T2D, whereby they performed initial pool-based, epigenome-scale screening followed by targeted analyses at selected top-ranking regions. Hypomethylation at FTO, a well-known gene linked to obesity and T2D from genetic studies, was found to be associated with progression to impaired glucose metabolism and prevalent T2D, with the odds of belonging to the T2D group increasing by 6.1% for every 1% decrease in methylation (OR = 1.061, 95% CI: 1.032–1.090) [45]. An epigenome-wide association study (EWAS) of fasting measures of glucose, insulin, and HOMA-IR among 837 non-diabetic participants reported significant association for two CpG sites within ABCG1 gene to be associated with fasting insulin and HOMA-IR [46].
4. Alternative epigenetic mechanisms in obesity and type 2 diabetes
4.1. Histone modification
Despite its strong functional link to DNA methylation, there exists less conclusive evidence in the role that histone modification plays with respect to obesity and T2D. To date, there are only a handful of observational studies in T2D, conducted on a small number of subjects. Significantly higher levels of histone H3K9me2 were reported around the interleukin-1A promoter and PTEN coding regions in monocytes from T2D subjects relative to controls [47], along with elevated histone H3 acetylation at the tumor necrosis factor-alpha (TNF-α) and cyclooxygenase-2 (COX-2) gene promoter regions [48]. In addition, subjects with prevalent T2D displayed Set7-dependent monomethylation of H3K4me1 in the NF-kB promoter region, potentially contributing to underlying vascular dysfunction [49]. In an effort to further understand the dynamics of histone marks underlying obesity and T2D, Nie and colleagues undertook a mass spectrometry-based label-free and chemical stable isotope labeling quantitative proteomic approach to systematically profile liver histone post-translational modifications in a prediabetic high-fat diet-induced obese (DIO) mouse model, and reported fifteen histone marks differing in abundance in DIO mouse liver compared with liver from chow-fed mice in label-free quantification, and six histone marks in stable isotope labeling quantification. Interestingly, metformin was able to reverse DIO-stimulated histone H3K36me2, providing support that the histone modification is potentially associated with T2D development [50].
4.2. MicroRNA (miRNA)
MicroRNAs (miRNAs) are a class of small non-coding RNAs of 20–24 nt in length involved in the regulation of gene expression at the post-transcriptional level [51]. miRNAs act by degrading their target mRNAs and/or inhibiting their translation, and are involved in the maintenance of normal cellular physiology and regulation of numerous biological processes including cell proliferation and differentiation. In contrast to research on the contribution of histone modifications to obesity and T2D, there has been significantly more work done with respect to the role of miRNAs.
Heneghan et al. found unique miRNA expression profiles for omentum and subcutaneous adipose tissues, along with significantly different levels of two miRNAs (miR-17-5p and miR-132) in omental adipose tissue between obese and non-obese subjects. The authors also observed that the miRNA expression in both omental fat and blood form obese individuals correlated with a range of adiposity and glycaemic traits, including BMI, fasting blood glucose, and glycosylated hemoglobin [52].
In plasma, for both adults and children, circulating levels of miRNAs are associated with obesity and other anthropometric measurements, including percentage fat mass and waist/hip circumference [53]. There is a substantial overlap in the panel of miRNAs reported, namely in increased levels of miR-140-5p and -142-3p, and decreased levels of miR-532-5p, −125b, −221, −130b and -423-5p. These miRNAs were identified from the comparison of morbidly obese with lean individuals [53], and in the comparison of lean versus obese children [53]. In another recent study that investigated plasma miRNAs in insulin resistance phenotypes in females with and without obesity across 175 miRNAs, ∼60% of the miRNAs were found to be significantly different between controls and at least one obesity phenotype (adjusted P ≤ 0.05), of which two of the miRNAs (miR-378a and miR-122) were observed to be perturbed in metabolically relevant tissues such as visceral adipose tissue and pericardial fat in a murine model of obesity [54].
Comparison of miRNA profile of skeletal muscle tissue between prevalent T2D individuals versus controls reported widespread changes, of which 15% of the differentially expressed miRNAs already showed changes in individuals with impaired glucose tolerance, indicative of early involvement of these miRNAs in T2D development [55]. To explore the potential of circulating miRNA profiles in T2D patients as a convenient biomarker, Zampetaki and colleagues carried out a systematic strategy of comprehensive miRNA profiling via an array-based approach on pooled samples. The authors further identified and validated a plasma miRNA signature of five miRNAs (miR-15a, miR-28-3p, miR-29b, miR-126 and miR-223) that displayed a characteristic deregulation in T2D patients, with this change in miRNA profile already observable 5–10 years before disease onset [56]. Other studies that undertook either a microarray profiling approach or a more targeted qPCR strategy in serum, peripheral blood mononuclear cells and whole blood have discovered additional miRNA markers associated with T2D and other glycaemic traits [57], [58], [59], [60], [61], [62], [63], [64], [65]. In particular, de Candia et al. recently quantified circulating miRNAs in incident diabetic individuals, allowing them to differentiate, for the first time, between pre-diabetic subjects that progress to T2D or not based on miRNA profiles [66]. Interestingly, a number of these microRNAs also significantly correlated with measures of cholesterol metabolism.
5. Functional insights and potential mechanisms
DNA methylation at CpG sites regulates gene expression and mediates biological response to environmental exposures. Obesity has been suggested to be a form of systemic, low-grade inflammatory state in adipose tissue, characterized by proinflammatory macrophage infiltration and oxidative stress, which promotes insulin resistance [67]. In view of the critical role that hypoxia plays in the regulation of inflammation and reactive oxygen species production [67], [68], [69], [70], [71], [72], it is natural to hypothesize that hypoxia and cellular hypoxic responses may provide mechanistic insight into the causal mechanisms underlying obesity, inflammation, and insulin resistance. Indeed, in fat tissue, a direct role of hypoxia in triggering adipose tissue dysfunction underlying obesity, both in adipose and non-adipose cells, has been demonstrated across in vivo and in vitro studies, as well as in animal models [73], [74], [75], [76]. To date, there have been three HIF-α subunits identified, namely HIF-1α, -2α, and -3α. The role that HIF-1α and HIF-2α plays in obesity and glycemic traits such as insulin resistance has been extensively characterized. The two isoforms mediate adaptation and survival to hypoxia through activation of genes involved in angiogenesis, glucose uptake and glycolysis, with non-overlapping targets [67]. Interestingly, differential methylation in HIF3A, which encodes the less well interrogated subunit HIF-3α, has been identified in methylation studies for obesity in both adults and children, and even neonates [32], [33], [77], [78], [79].
Taking a Mendelian Randomization approach, Wahl et al. provided evidence supporting the view that the observed changes in DNA methylation across majority of the 187 identified loci are likely a consequence, and not the cause of adiposity [35]. The methylation loci were enriched for sites of open chromatin in multiple tissues, consistent with the presence of constitutive cis enhancers. In addition, the candidate genes at these loci consisted of genes previously reported to be involved in lipid metabolism, amino acid and small molecule transport, and inflammation, as well as metabolic, cardiovascular, respiratory and neoplastic disease, including ABCG1, which was also identified in earlier EWASs for T2D [34], [46]. The only CpG site found in the study that suggested a potential causal role of methylation on BMI was cg26663590. Methylation levels at the locus measured at baseline was predictive of weight gain in longitudinal population studies. This locus also contains the gene encoding SH2B1, which has been previously linked with obesity in GWAS, and reported to be involved in energy and glucose homeostasis [80], [81].
In the T2D EWAS conducted across Indian Asian and European incident T2D cases, in addition to ABCG1, significant association was identified for differential methylation and risk of future T2D at four other epigenetic loci, namely PHOSPHO1, SOCS3, SREBF1, and TXNIP [34]. Consistent with being an early marker for impaired glucose homoeostasis, TXNIP expression was found to be highly sensitive to glucose concentration, while methylation changes at ABCG1, PHOSPHO1, SOCS3 and SREBF1 were associated with measure of adiposity and T2D-related traits such as BMI, waist circumference, insulin concentrations, and HOMA-IR.
Although the exact mechanisms accounting for these early changes in methylation levels prior to T2D onset are unclear, it is worth noting that apart from PHOSPHO1, a bone specific phosphatase with a recognized role in bone mineralization [82], the remaining four genes lie within key pathways underlying T2D and associated metabolic disturbances. ABCG1 is involved in macrophage cholesterol and phospholipid transport, promotes cholesterol efflux to HDL, and regulates cellular lipid homeostasis in various cell types, including pancreatic β-cells. Abcg1−/− mice have impaired glucose tolerance and insulin secretion with normal insulin sensitivity [83], while ABCG1 expression has been shown to be downregulated in humans with diabetes and upregulated by the use of insulin sensitizing agents [84].
SOCS3, a cytokine-inducible negative regulator of cytokine signaling, is a major negative regulator of insulin signaling, and has been implicated in the pathogenesis of obesity and associated metabolic abnormalities. SOCS3 expression is increased in skeletal muscle in the setting of diet-induced and genetic obesity, inflammation, and hyperlipidemia, along with impaired systemic and muscle-specific glucose homeostasis and insulin action in the case of muscle-specific overexpression of SOCS3 despite unchanged body weight [85]. On the other hand, SOCS3−/− mice demonstrate protection against obesity-induced hyperinsulinemia and insulin resistance [86]. SREBPF1 is the master transcriptional regulator of hepatic lipogenesis. Insulin activates SREBPF1 by increasing SREBP1 transcription, and the processing of SREBPF1 from an inactive membrane-bound precursor to a soluble fragment capable of translocating to the nucleus to activate transcription. SREBPF1 is decreased in insulin-deficient states such as fasting, but increased in feeding, obesity and insulin resistance [87].
TXNIP, a key component of pancreatic β-cell biology, nutrient sensing, energy metabolism, and regulation of cellular redox, has its expression highly induced by glucose through activation of the carbohydrate response element-binding protein, which binds the TXNIP promoter [88]. TXNIP downregulates GLUT1, a major transmembrane glucose transporter, thereby acting as a negative feedback loop to regulate glucose entry and mitochondrial oxidative stress. In fact, TXNIP is one of the most glucose-responsive genes expressed in human islets, acting as a mediator of glucotoxic β-cell death in animal models. TXNIP downregulation has a protective effect against obesity-induced diabetes by preventing β-cell apoptosis and preserving β-cell mass [89].
Apart from the above candidate genes, functional annotation of aberrantly methylated genes identified from an EWAS on pancreatic islets from T2D and non-diabetic donors, along with additional RNAi experiments highlighted pathways implicated in β-cell survival and function, as well as cellular dysfunction and adaptation to stressors [44]. A separate study on rat islets and clonal β-cells also found Hdac7 expression to result in impaired mitochondrial function and insulin secretion [90], whereby HDAC7 was one of the differentially methylated loci identified from human islet EWAS [91].
In terms of the role of miRNAs, obesity-induced overexpression of miR-143 has been shown to inhibit insulin-stimulated AKT activation and glucose homeostasis in mice, while mice deficient for the miR-143-145 cluster are protected from the development of obesity-associated insulin resistance. Together, these provide support that the dysregulation of post-transcriptional gene silencing contributes to the development of obesity-induced insulin resistance [92].
MiR-126, first discovered by Zampetaki et al. in the comparison miRNA profiles between prevalent T2D cases and control within the population-based Bruneck study [56], plays a key role in endothelial cells and contributes to the maintenance and repair of vascular integrity and angiogenesis [93]. This finding was subsequently corroborated by Zhang et al., where it was found to be the only microRNA with a significantly reduced expression in patients with T2D compared to normoglycemic individuals, but also in individuals with impaired fasting glucose relative to normal subjects, suggesting a role in the development of T2D [94].
6. Existing challenges and future directions
Recent advancements in technology has now rendered it possible to investigate the link between DNA methylation and various human phenotypes in a high-throughput fashion. To date, most of the published EWASs were performed on methylation arrays. Although the development of methylation arrays has made DNA methylation analyses much more affordable, these arrays remain largely inefficient, covering less than 3% of the CpG sites in the human genome, even on the latest Illumina MethylationEPIC Beadchip [95]. In addition, as the contents of these arrays were determined by expert panels, the selected CpG sites present a biased representation of the genome.
In contrast, whole genome bisulfite sequencing (WGBS) is able to reveal methylation status at each cytosine across the whole genome, with approximately 95% of all CpG sites in the human genome assessable via WGBS [95]. In fact, despite the value of higher resolution and unbiased coverage, the widespread utilization of WGBS has been hindered primarily by its high cost and the large DNA input required, compounded by the extensive computational power and expertise necessary for its accurate interpretation. The value of WGBS in the study of diabetes has been previously demonstrated by Jeon et al. who found a 10 kb stretch in the MSI2 gene displaying methylation differences strongly related to hyperglycemia in islet preparations from two donors with T2D and 16 non-diabetic donors [96], and Volkov et al. who identified >25,000 differentially methylated regions in islets from individuals with T2D covering loci with known islet function and binding sites previously identified by ChIP-seq for islet-specific transcription factors as well as enhancer regions [97].
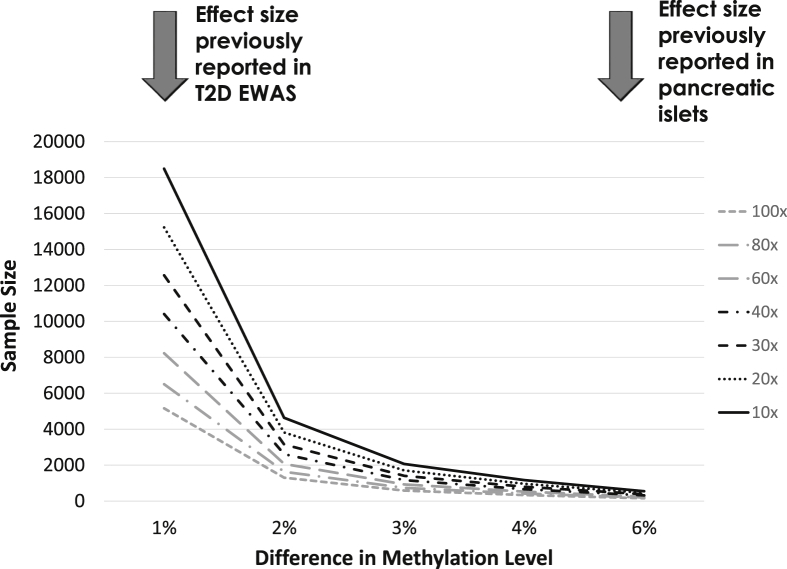
With maturation of the next generation sequencing (NGS) technology and improvements in library preparation methods [98], both the cost of WGBS and the DNA input amount necessary have greatly reduced, rendering this technology increasingly affordable for usage in EWAS and other studies. In our benchmarking study where we systematically compared the performance of three WGBS library preparation methods with low DNA input requirement (Swift Biosciences Accel-NGS, Illumina TruSeq and QIAGEN QIAseq) on two state-of-the-art sequencing platforms (Illumina NovaSeq and HiSeq X), we demonstrated that the Swift Accel-NGS library preparation method presented the best overall data quality, along with similar performance between NovaSeq and HiSeq X platforms [99]. When we assessed concordance between data generated by WGBS and methylation arrays, we discovered systematic biases between WGBS and methylation arrays, with lower precision observed for WGBS across all depths investigated (10–120x). To achieve a level of precision broadly comparable to the methylation array, a minimum coverage of 100x is recommended. Given the differential cost for sequencing at different depths, this has important implications for the design of future WGBS studies, in particular population-based studies on large cohorts. For example, to detect a difference of 1% in methylation levels between cases and controls, as was observed in previous T2D EWAS, a sample size of ∼4,500 will be needed on the methylation array, and ∼5,000 for WGBS at 100x coverage. For an average detectable difference of ∼6% as reported to be observed in pancreatic islets, a sample size of 158 will suffice at 100x, or 384 at a more modest 30x coverage (Figure 2).
Figure 2.
Sample size estimation for WGBS studies.
One major drawback of WGBS is the bisulfite conversion step itself, in view of the harsh chemical reaction that degrades up to 99% of the input DNA. This is particularly challenging for clinical application, in view of the limited amount of available input material. Liu et al. successfully developed a bisulfite-free method that directly interrogates 5-methylcytosine (5 mC) and 5-hydroxymethylcytosine (5hmC) at single-base resolution [100]. The method claims to achieve higher sequencing quality for cytosines and guanine base pairs, along with more even coverage of the genome and shorter computational time for analysis. Given the lower DNA input necessary, this method for single-base resolution whole genome sequencing of the epigenome could be the next step forward in epigenetic studies.
7. Potential therapeutics
Epigenetic-based therapy, or “epidrugs” as coined by Berdasco and Esteller in their recent review, is one potential focus area for translational epigenetics research [101]. Although all the epidrugs approved for clinical use today are in the field of oncology, it is clear from this that epigenetic-based therapies are currently being explored in preclinical studies as well as in clinical trials. It is also worth noting that although the currently approved epidrugs consist of only DNA methyltransferase inhibitor (DNMTi) or histone deacetylase inhibitor (HDACi), other alternative targets are being investigated, including methyltransferase inhibitors (HMTi) and histone demethylase inhibitors (HDMi).
One potential candidate for the prevention of T2D is HDAC7 which encodes a histone deacetylase (HDAC). In an EWAS on islets from 15 T2D and 34 non-diabetic donors, HDAC7 was found to be hypomethylated and overexpressed in islets from donors with T2D [91]. A follow-up study in in rat islets and clonal β-cells subsequently found increased Hdac7 expression to impact upon impaired mitochondrial function and insulin secretion, suggesting that changes in HDAC7 methylation and expression may perturb β-cell function [90]. Therefore, it was suggested that HDAC inhibitors could serve as a potential novel therapy by restoring the observed defects described above in Hdac7-overexpressing β-cells [102]. In addition, Set7, a histone methyltransferase, has also been suggested as a novel therapeutic approach to prevent atherosclerotic vascular disease in T2D patients, in view of the impact on vascular dysfunction by Set7-induced epigenetic changes in T2D [49].
MiRNAs could also serve as potential therapeutic agents for obesity and/or T2D. Although their current use is again restricted to cancer, there are on-going clinical trials that are evaluating miRNA mimetics in other pathologies. One example is a mimetic of miR-34a, which is inhibited in most cancers and has been demonstrated to inhibit fat browning in obesity in part by suppressing the browning activators fibroblast growth factor 21 (FGF21) and SIRT [103]. This miR-34a mimetic is currently in Phase I clinical trial [104].
Last but not least, demethylating agents might play a critical role in therapeutic options for the treatment and prevention of obesity, T2D and associated complications. The two demethylating agents currently in clinical use are 5-azacytidine and decitabine, both of which are approved only for the treatment of specific forms of myelodysplastic syndrome and acute myeloid leukemia [105]. However, given the broad demethylating properties of both agents and their corresponding high cytotoxity when incorporated into DNA, it is likely safe to say that these therapies will not be applicable in chronic non-life threatening diseases such as T2D and its associated complications. Less toxic and more-targeted demethylating therapies are needed before it will become possible to incorporate demethylating agents in routine weight management and diabetes treatment.
Funding
This study was supported by the NMRC STaR grant (L0465301.010.710079).
This article is part of a supplement entitled ‘Biomarkers of Beta-Cell Health and Dysfunction: Towards Personalised Diabetes Care. Proceedings of the 20th Servier-IGIS Symposium’. The Symposium and the publication of the Supplement have been made possible by an unrestricted educational grant from Institut la Conférence Hippocrate – Servier Group.
Conflict of interest
None declared.
References
- 1.Wang Y.C., McPherson K., Marsh T., Gortmaker S.L., Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378(9793):815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 2.Hruby A., Manson J.E., Qi L., Malik V.S., Rimm E.B., Sun Q. Determinants and consequences of obesity. American Journal of Public Health. 2016;106(9):1656–1662. doi: 10.2105/AJPH.2016.303326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whiting D.R., Guariguata L., Weil C., Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Research and Clinical Practice. 2011;94(3):311–321. doi: 10.1016/j.diabres.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Kuulasmaa K., Tunstall-Pedoe H., Dobson A., Fortmann S., Sans S., Tolonen H. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 2000;355(9205):675–687. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- 5.Wong N.D. Epidemiological studies of CHD and the evolution of preventive cardiology. Nature Reviews Cardiology. 2014;11(5):276–289. doi: 10.1038/nrcardio.2014.26. [DOI] [PubMed] [Google Scholar]
- 6.Meisinger C., Doring A., Thorand B., Heier M., Lowel H. Body fat distribution and risk of type 2 diabetes in the general population: are there differences between men and women? The MONICA/KORA Augsburg cohort study. American Journal of Clinical Nutrition. 2006;84(3):483–489. doi: 10.1093/ajcn/84.3.483. [DOI] [PubMed] [Google Scholar]
- 7.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Human Molecular Genetics. 2018;27(20):3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marouli E., Graff M., Medina-Gomez C., Lo K.S., Wood A.R., Kjaer T.R. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542(7640):186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller M.J., Geisler C., Blundell J., Dulloo A., Schutz Y., Krawczak M. The case of GWAS of obesity: does body weight control play by the rules? International Journal of Obesity. 2018;42(8):1395–1405. doi: 10.1038/s41366-018-0081-6. [DOI] [PubMed] [Google Scholar]
- 11.Speakman J.R., Loos R.J.F., O'Rahilly S., Hirschhorn J.N., Allison D.B. GWAS for BMI: a treasure trove of fundamental insights into the genetic basis of obesity. International Journal of Obesity. 2018;42(8):1524–1531. doi: 10.1038/s41366-018-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nature Genetics. 2018;50(11):1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyamdorj R., Pitkaniemi J., Tuomilehto J., Hammar N., Stehouwer C.D., Lam T.H. Ethnic comparison of the association of undiagnosed diabetes with obesity. International Journal of Obesity. 2010;34(2):332–339. doi: 10.1038/ijo.2009.225. [DOI] [PubMed] [Google Scholar]
- 14.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nature Genetics. 2011;43(10):984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattar N., Gill J.M. Type 2 diabetes in migrant south Asians: mechanisms, mitigation, and management. Lancet Diabetes Endocrinology. 2015;3(12):1004–1016. doi: 10.1016/S2213-8587(15)00326-5. [DOI] [PubMed] [Google Scholar]
- 16.Kaminsky Z.A., Tang T., Wang S.C., Ptak C., Oh G.H., Wong A.H. DNA methylation profiles in monozygotic and dizygotic twins. Nature Genetics. 2009;41(2):240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 17.Slomko H., Heo H.J., Einstein F.H. Minireview: epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153(3):1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youngson N.A., Whitelaw E. Transgenerational epigenetic effects. Annual Review of Genomics and Human Genetics. 2008:9233–9257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 19.Portela A., Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 20.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Science Signaling. 2011;4(157):pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 21.Handel A.E., Ebers G.C., Ramagopalan S.V. Epigenetics: molecular mechanisms and implications for disease. Trends in Molecular Medicine. 2010;16(1):7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Saetrom P., Snove O., Jr., Rossi J.J. Epigenetics and microRNAs. Pediatric Research. 2007;61(5 Pt 2):17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- 23.Dolinoy D.C., Huang D., Jirtle R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamson-Reig A., Thyssen S.M., Arany E., Hill D.J. Altered pancreatic morphology in the offspring of pregnant rats given reduced dietary protein is time and gender specific. Journal of Endocrinology. 2006;191(1):83–92. doi: 10.1677/joe.1.06754. [DOI] [PubMed] [Google Scholar]
- 25.Raychaudhuri N., Raychaudhuri S., Thamotharan M., Devaskar S.U. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. Journal of Biological Chemistry. 2008;283(20):13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yajnik C.S., Fall C.H., Coyaji K.J., Hirve S.S., Rao S., Barker D.J. Neonatal anthropometry: the thin-fat Indian baby. The Pune maternal nutrition study. International Journal of Obesity and Related Metabolic Disorders. 2003;27(2):173–180. doi: 10.1038/sj.ijo.802219. [DOI] [PubMed] [Google Scholar]
- 27.Whincup P.H., Kaye S.J., Owen C.G., Huxley R., Cook D.G., Anazawa S. Birth weight and risk of type 2 diabetes: a systematic review. Journal of the American Medical Association. 2008;300(24):2886–2897. doi: 10.1001/jama.2008.886. [DOI] [PubMed] [Google Scholar]
- 28.Einstein F., Thompson R.F., Bhagat T.D., Fazzari M.J., Verma A., Barzilai N. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. Public Library of Science one. 2010;5(1) doi: 10.1371/journal.pone.0008887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feinberg A.P., Irizarry R.A., Fradin D., Aryee M.J., Murakami P., Aspelund T. Personalized epigenomic signatures that are stable over time and covary with body mass index. Science Translational Medicine. 2010;2(49):49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X., Su S., Barnes V.A., De Miguel C., Pollock J., Ownby D. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8(5):522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aissi D., Wahl S. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 33.Demerath E.W., Guan W., Grove M.L., Aslibekyan S., Mendelson M., Zhou Y.H. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Human Molecular Genetics. 2015;24(15):4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers J.C., Loh M., Lehne B., Drong A., Kriebel J., Motta V. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinology. 2015;3(7):526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahl S., Drong A., Lehne B., Loh M., Scott W.R., Kunze S. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayols-Baixeras S., Subirana I., Fernandez-Sanles A., Senti M., Lluis-Ganella C., Marrugat J. DNA methylation and obesity traits: an epigenome-wide association study. The REGICOR study. Epigenetics. 2017;12(10):909–916. doi: 10.1080/15592294.2017.1363951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F., Berg A., Imamura Kawasawa Y., Bixler E.O., Fernandez-Mendoza J., Whitsel E.A. Association between DNA methylation in obesity-related genes and body mass index percentile in adolescents. Scientific Reports. 2019;9(1):2079. doi: 10.1038/s41598-019-38587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott H.R., Shihab H.A., Lockett G.A., Holloway J.W., McRae A.F., Smith G.D. Role of DNA methylation in type 2 diabetes etiology: using genotype as a causal anchor. Diabetes. 2017;66(6):1713–1722. doi: 10.2337/db16-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barajas-Olmos F., Centeno-Cruz F., Zerrweck C., Imaz-Rosshandler I., Martinez-Hernandez A., Cordova E.J. Altered DNA methylation in liver and adipose tissues derived from individuals with obesity and type 2 diabetes. BioMed Central Medical Genetics. 2018;19(1):28. doi: 10.1186/s12881-018-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sala P., de Miranda Torrinhas R.S.M., Fonseca D.C., Ravacci G.R., Waitzberg D.L., Giannella-Neto D. Tissue-specific methylation profile in obese patients with type 2 diabetes before and after Roux-en-Y gastric bypass. Diabetology & Metabolic Syndrome. 2017;9:15. doi: 10.1186/s13098-017-0214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Rodero S., Menendez-Torre E., Fernandez-Bayon G., Morales-Sanchez P., Sanz L., Turienzo E. Altered intragenic DNA methylation of HOOK2 gene in adipose tissue from individuals with obesity and type 2 diabetes. Public Library of Science one. 2017;12(12) doi: 10.1371/journal.pone.0189153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsson E., Jansson P.A., Perfilyev A., Volkov P., Pedersen M., Svensson M.K. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 43.Ribel-Madsen R., Fraga M.F., Jacobsen S., Bork-Jensen J., Lara E., Calvanese V. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. Public Library of Science one. 2012;7(12) doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. European Molecular Biology Organization Journal. 2012;31(6):1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toperoff G., Aran D., Kark J.D., Rosenberg M., Dubnikov T., Nissan B. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Human Molecular Genetics. 2012;21(2):371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hidalgo B., Irvin M.R., Sha J., Zhi D., Aslibekyan S., Absher D. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the genetics of lipid lowering drugs and diet network study. Diabetes. 2014;63(2):801–807. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao F., Wu X., Zhang L., Yuan Y.C., Riggs A.D., Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. Journal of Biological Chemistry. 2007;282(18):13854–13863. doi: 10.1074/jbc.M609446200. [DOI] [PubMed] [Google Scholar]
- 48.Hou C., Zhao M., Li X., Li Y.J., Lin Y., Lu Q.J. Histone H3 acetylation of tumor necrosis factor-alpha and cyclooxygenase-2 in patients with type 2 diabetes. Zhonghua Yixue Zazhi. 2011;91(26):1805–1808. [PubMed] [Google Scholar]
- 49.Paneni F., Costantino S., Battista R., Castello L., Capretti G., Chiandotto S. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circulation: Cardiovascular Genetics. 2015;8(1):150–158. doi: 10.1161/CIRCGENETICS.114.000671. [DOI] [PubMed] [Google Scholar]
- 50.Nie L., Shuai L., Zhu M., Liu P., Xie Z.F., Jiang S. The landscape of histone modifications in a high-fat diet-induced obese (DIO) mouse model. Molecular & Cellular Proteomics. 2017;16(7):1324–1334. doi: 10.1074/mcp.M117.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaikkonen M.U., Lam M.T., Glass C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research. 2011;90(3):430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heneghan H.M., Miller N., McAnena O.J., O'Brien T., Kerin M.J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. Journal of Clinical Endocrinology & Metabolism. 2011;96(5):E846–E850. doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 53.Prats-Puig A., Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Moreno M., Bonet N. Changes in circulating microRNAs are associated with childhood obesity. Journal of Clinical Endocrinology & Metabolism. 2013;98(10):E1655–E1660. doi: 10.1210/jc.2013-1496. [DOI] [PubMed] [Google Scholar]
- 54.Jones A., Danielson K.M., Benton M.C., Ziegler O., Shah R., Stubbs R.S. miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring) 2017;25(10):1734–1744. doi: 10.1002/oby.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallagher I.J., Scheele C., Keller P., Nielsen A.R., Remenyi J., Fischer C.P. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Medicine. 2010;2(2):9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circulation Research. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 57.Kong L., Zhu J., Han W., Jiang X., Xu M., Zhao Y. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetologica. 2011;48(1):61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 58.Karolina D.S., Tavintharan S., Armugam A., Sepramaniam S., Pek S.L., Wong M.T. Circulating miRNA profiles in patients with metabolic syndrome. Journal of Clinical Endocrinology & Metabolism. 2012;97(12):E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 59.Zhang T., Li L., Shang Q., Lv C., Wang C., Su B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochemical and Biophysical Research Communications. 2015;463(1–2):60–63. doi: 10.1016/j.bbrc.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 60.Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Rovira O., Guerra E., Esteve E. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37(5):1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 61.Rong Y., Bao W., Shan Z., Liu J., Yu X., Xia S. Increased microRNA-146a levels in plasma of patients with newly diagnosed type 2 diabetes mellitus. Public Library of Science one. 2013;8(9) doi: 10.1371/journal.pone.0073272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baldeon R.L., Weigelt K., de Wit H., Ozcan B., van Oudenaren A., Sempertegui F. Decreased serum level of miR-146a as sign of chronic inflammation in type 2 diabetic patients. Public Library of Science one. 2014;9(12) doi: 10.1371/journal.pone.0115209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C., Wan S., Yang T., Niu D., Zhang A., Yang C. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Scientific Reports. 2016;6:20032. doi: 10.1038/srep20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y., Gao G., Yang C., Zhou K., Shen B., Liang H. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. International Journal of Molecular Sciences. 2014;15(6):10567–10577. doi: 10.3390/ijms150610567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balasubramanyam M., Aravind S., Gokulakrishnan K., Prabu P., Sathishkumar C., Ranjani H. Impaired miR-146a expression links subclinical inflammation and insulin resistance in type 2 diabetes. Molecular and Cellular Biochemistry. 2011;351(1–2):197–205. doi: 10.1007/s11010-011-0727-3. [DOI] [PubMed] [Google Scholar]
- 66.de Candia P., Spinetti G., Specchia C., Sangalli E., La Sala L., Uccellatore A. A unique plasma microRNA profile defines type 2 diabetes progression. Public Library of Science one. 2017;12(12) doi: 10.1371/journal.pone.0188980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foti D.P., Brunetti A. Editorial: "linking hypoxia to obesity". Frontiers in Endocrinology. 2017;8:34. doi: 10.3389/fendo.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ban J.J., Ruthenborg R.J., Cho K.W., Kim J.W. Regulation of obesity and insulin resistance by hypoxia-inducible factors. Hypoxia (Auckl) 2014:2171–2183. doi: 10.2147/HP.S68771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eltzschig H.K., Carmeliet P. Hypoxia and inflammation. New England Journal of Medicine. 2011;364(7):656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nizet V., Johnson R.S. Interdependence of hypoxic and innate immune responses. Nature Reviews Immunology. 2009;9(9):609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scholz C.C., Taylor C.T. Targeting the HIF pathway in inflammation and immunity. Current Opinion in Pharmacology. 2013;13(4):646–653. doi: 10.1016/j.coph.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 72.Tormos K.V., Chandel N.S. Inter-connection between mitochondria and HIFs. Journal of Cellular and Molecular Medicine. 2010;14(4):795–804. doi: 10.1111/j.1582-4934.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosogai N., Fukuhara A., Oshima K., Miyata Y., Tanaka S., Segawa K. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes. 2007;56(4):901–911. doi: 10.2337/db06-0911. [DOI] [PubMed] [Google Scholar]
- 74.Kabon B., Nagele A., Reddy D., Eagon C., Fleshman J.W., Sessler D.I. Obesity decreases perioperative tissue oxygenation. Anesthesiology. 2004;100(2):274–280. doi: 10.1097/00000542-200402000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trayhurn P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiological Reviews. 2013;93(1):1–21. doi: 10.1152/physrev.00017.2012. [DOI] [PubMed] [Google Scholar]
- 76.Fujisaka S., Usui I., Ikutani M., Aminuddin A., Takikawa A., Tsuneyama K. Adipose tissue hypoxia induces inflammatory M1 polarity of macrophages in an HIF-1alpha-dependent and HIF-1alpha-independent manner in obese mice. Diabetologia. 2013;56(6):1403–1412. doi: 10.1007/s00125-013-2885-1. [DOI] [PubMed] [Google Scholar]
- 77.Pfeiffer S., Krüger J., Maierhofer A., Böttcher Y., Klöting N., Hajj N.E. Hypoxia-inducible factor 3A gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Scientific Reports. 2016;6:27969. doi: 10.1038/srep27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S., Song J., Yang Y., Zhang Y., Wang H., Ma J. HIF3A DNA methylation is associated with childhood obesity and ALT. Public Library of Science one. 2015;10(12) doi: 10.1371/journal.pone.0145944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan H., Lin X., Wu Y., Chen L., Teh A.L., Soh S.E. HIF3A association with adiposity: the story begins before birth. Epigenomics. 2015;7(6):937–950. doi: 10.2217/epi.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463(7281):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Macrae V.E., Davey M.G., McTeir L., Narisawa S., Yadav M.C., Millan J.L. Inhibition of PHOSPHO1 activity results in impaired skeletal mineralization during limb development of the chick. Bone. 2010;46(4):1146–1155. doi: 10.1016/j.bone.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kruit J.K., Wijesekara N., Westwell-Roper C., Vanmierlo T., de Haan W., Bhattacharjee A. Loss of both ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation, and impaired beta-cell function. Diabetes. 2012;61(3):659–664. doi: 10.2337/db11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sturek J.M., Castle J.D., Trace A.P., Page L.C., Castle A.M., Evans-Molina C. An intracellular role for ABCG1-mediated cholesterol transport in the regulated secretory pathway of mouse pancreatic beta cells. Journal of Clinical Investigation. 2010;120(7):2575–2589. doi: 10.1172/JCI41280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Z., Hulver M., McMillan R.P., Cai L., Kershaw E.E., Yu L. Regulation of insulin and leptin signaling by muscle suppressor of cytokine signaling 3 (SOCS3) Public Library of Science one. 2012;7(10) doi: 10.1371/journal.pone.0047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jorgensen S.B., O'Neill H.M., Sylow L., Honeyman J., Hewitt K.A., Palanivel R. Deletion of skeletal muscle SOCS3 prevents insulin resistance in obesity. Diabetes. 2013;62(1):56–64. doi: 10.2337/db12-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haas J.T., Miao J., Chanda D., Wang Y., Zhao E., Haas M.E. Hepatic insulin signaling is required for obesity-dependent expression of SREBP-1c mRNA but not for feeding-dependent expression. Cell Metabolism. 2012;15(6):873–884. doi: 10.1016/j.cmet.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cha-Molstad H., Saxena G., Chen J., Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. Journal of Biological Chemistry. 2009;284(25):16898–16905. doi: 10.1074/jbc.M109.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minn A.H., Hafele C., Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146(5):2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 90.Daneshpajooh M., Bacos K., Bysani M., Bagge A., Ottosson Laakso E., Vikman P. HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia. 2017;60(1):116–125. doi: 10.1007/s00125-016-4113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dayeh T., Volkov P., Salo S., Hall E., Nilsson E., Olsson A.H. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. Public Library of Science Genetics. 2014;10(3) doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jordan S.D., Kruger M., Willmes D.M., Redemann N., Wunderlich F.T., Bronneke H.S. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature Cell Biology. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 93.Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.F., Wythe J.D. miR-126 regulates angiogenic signaling and vascular integrity. Developmental Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang T., Lv C., Li L., Chen S., Liu S., Wang C. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. BioMed Research International. 2013;76:1617. doi: 10.1155/2013/761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stirzaker C., Taberlay P.C., Statham A.L., Clark S.J. Mining cancer methylomes: prospects and challenges. Trends in Genetics. 2014;30(2):75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 96.Jeon J.P., Koh I.U., Choi N.H., Kim B.J., Han B.G., Lee S. Differential DNA methylation of MSI2 and its correlation with diabetic traits. Public Library of Science one. 2017;12(5) doi: 10.1371/journal.pone.0177406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Volkov P., Bacos K., Ofori J.K., Esguerra J.L., Eliasson L., Ronn T. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66(4):1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 98.Suzuki M., Liao W., Wos F., Johnston A.D., DeGrazia J., Ishii J. Whole-genome bisulfite sequencing with improved accuracy and cost. Genome Research. 2018;28(9):1364–1371. doi: 10.1101/gr.232587.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou L., Ng H.K., Drautz-Moses D.I., Schuster S.C., Beck S., Kim C. Systematic evaluation of library preparation methods and sequencing platforms for high-throughput whole genome bisulfite sequencing. Scientific Reports. 2019;9(1):10383. doi: 10.1038/s41598-019-46875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Y., Siejka-Zielinska P., Velikova G., Bi Y., Yuan F., Tomkova M. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nature Biotechnology. 2019;37:424–429. doi: 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- 101.Berdasco M., Esteller M. Clinical epigenetics: seizing opportunities for translation. Nature Reviews Genetics. 2019;20(2):109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 102.Davegardh C., Garcia-Calzon S., Bacos K., Ling C. DNA methylation in the pathogenesis of type 2 diabetes in humans. Molecular Metabolism. 2018:1412–1425. doi: 10.1016/j.molmet.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fu T., Seok S., Choi S., Huang Z., Suino-Powell K., Xu H.E. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Molecular and Cellular Biology. 2014;34(22):4130–4142. doi: 10.1128/MCB.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Agostini M., Knight R.A. miR-34: from bench to bedside. Oncotarget. 2014;5(4):872–881. doi: 10.18632/oncotarget.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Derissen E.J., Beijnen J.H., Schellens J.H. Concise drug review: azacitidine and decitabine. Oncologist. 2013;18(5):619–624. doi: 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]