SUMMARY
Specialized microenvironments, called niches, control adult stem cell proliferation and differentiation. The brain lateral ventricular (LV) neurogenic niche is generated from distinct postnatal radial glial progenitors (pRGPs), giving rise to adult neural stem cells (NSCs) and niche ependymal cells (ECs). Cellular-intrinsic programs govern stem versus supporting cell maturation during adult niche assembly, but how they are differentially initiated within a similar microenvironment remains unknown. Using chemical approaches, we discovered that EGFR signaling powerfully inhibits EC differentiation by suppressing multiciliogenesis. We found that EC pRGPs actively terminated EGF activation through receptor redistribution away from CSF-contacting apical domains and that randomized EGFR membrane targeting blocked EC differentiation. Mechanistically, we uncovered spatiotemporal interactions between EGFR and endocytic adaptor protein Numb. Ca2+-dependent basolateral targeting of Numb is necessary and sufficient for proper EGFR redistribution. These results reveal a previously unknown cellular mechanism for neighboring progenitors to differentially engage environmental signals, initiating adult stem cell niche assembly.
Graphical Abstract
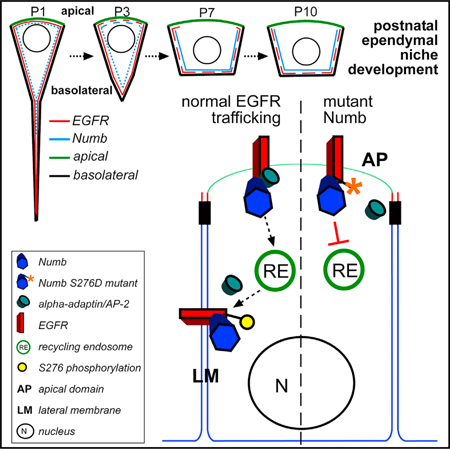
In Brief
Constructing adult stem niches from postnatal progenitors requires differential cellular interpretations of similar microenvironmental signals. Abdi et al. show that in an EGF-rich environment needed for adult neural stem cell proliferation, a subpopulation of postnatal progenitors downregulates EGFR signaling via targeted receptor trafficking, promoting multiciliated ependymal niche cell differentiation.
INTRODUCTION
Adult stem cells are integral components of normal tissue homeostasis, and their dysfunctions can contribute to human disease (Gage and Temple, 2013; Gonzales and Fuchs, 2017; Tomasetti and Vogelstein, 2015). To balance proliferation and differentiation, adult stem cells often reside in dedicated microenvironments called niches, which are cellular complexes composed of tissue-restricted stem cells interacting with neighboring cells (Gehart and Clevers, 2019; Hogan et al., 2014; Ihrie and Alvarez-Buylla, 2011). Although much is known about the cellular identities of stem cell niche components, the principles governing their differentiation from specified precursors into three-dimensional (3D) environments in adult tissues remain unclear. Understanding these principles will be important for tissue regeneration strategies that use in vivo and ex vivo platforms for engineering cellular transplants (Barrilleaux et al., 2006; Kim et al., 2012; Pacelli et al., 2017).
In the adult rodent brain, the lateral ventricular (LV) neurogenic region supports continuous new neuron production throughout life (Bjornsson et al., 2015; Lim and Alvarez-Buylla, 2016). This subependymal zone (SEZ) and/or subventricular zone (SVZ) niche containing adult neural stem cells (NSCs) is constructed within the first 2 weeks after birth from embryonically specified radial glial progenitors (Paez-Gonzalez et al., 2011) and serves as an excellent model system to study mechanisms regulating adult stem cell niche assembly. For the SEZ niche, differentiation of at least 2 types of postnatal radial glial progenitors (pRGPs) is required for its functional assembly at the LV surface: (1) pRGPs that retain proliferative capacity to become future adult NSCs (Fuentealba et al., 2015) and (2) pRGPs that differentiate into neighboring niche ependymal cells (ECs) (Paez-Gonzalez et al., 2011; Spassky et al., 2005). Although little is known about the steps governing transition of pRGPs into adult GFAP+ SEZ NSCs, differentiation of niche ECs from pRGPs requires timely and coordinated activation of numerous transcription factors, including Mcidas, Myb, and Foxj1 (Spassky and Meunier, 2017). This transcriptional cascade results in EC morphological specialization, including basal body duplication and multiciliogenesis (Stubbs et al., 2012). Proper EC differentiation is critical assembling SEZ niches and sustaining adult neurogenesis (Paez-Gonzalez et al., 2011), as well as preventing hydrocephalus (Abdi et al., 2018; Del Bigio, 2010; Tissir et al., 2010).
The cerebrospinal fluid (CSF), containing various signaling molecules and growth factors during development (Dani and Lehtinen, 2016), contacts the apical membranes of both pRGP populations during postnatal SEZ niche development. Because extracellular environments are temporally and contextually similar for pRGPs destined to becoming adult NSCs and niche ECs, it has generally been assumed that cell-intrinsic programs differentially drive the differentiation of pRGP subpopulations during SEZ niche development. Given the comparable local microenvironments along the LV wall, it remains unclear how specified niche EC progenitors initiate their differentiation while acting on similar extracellular cues as adult NSC progenitors. Although embryonically specified progenitors mature postnatally into adult SEZ niche ECs and NSCs (Ortiz-Alvarez et al., 2019; Redmond et al., 2019), EC progenitor differentiation can be specifically tracked and readily studied via Foxj1 expression (Paez-Gonzalez et al., 2011), unlike postnatal NSC progenitors, which at present cannot be easily identified. Using a chemical screen to uncover signaling pathways controlling pRGP differentiation into SEZ niche ECs, we uncovered a previously unknown cellular mechanism for neighboring radial glial progenitors to differentially act on the same extracellular cues, enabling distinct downstream programs in EC progenitors for assembling the adult LV neurogenic niche.
RESULTS AND DISCUSSION
EGF Signaling Inhibits pRGP Differentiation into SEZ Niche ECs
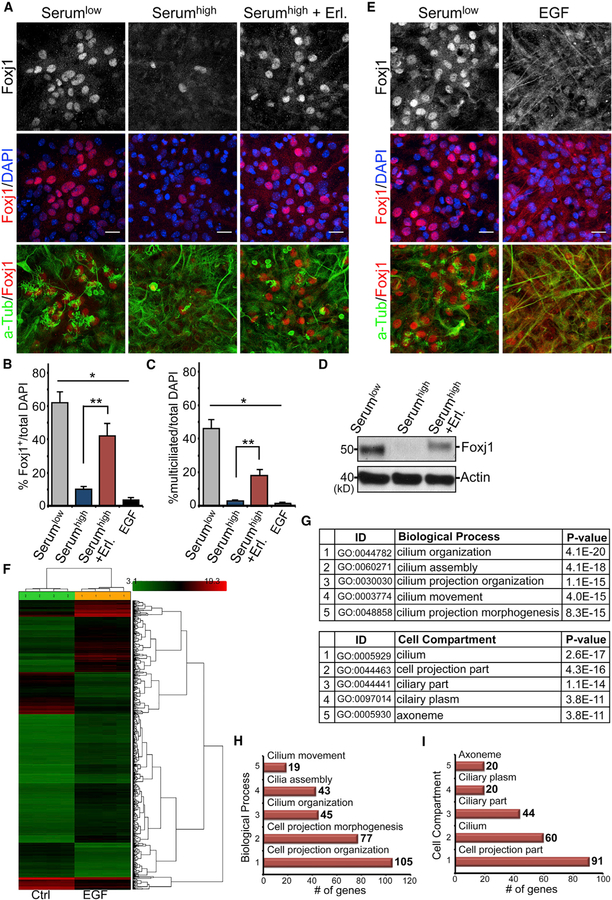
To interrogate molecular signals controlling the initiation of SEZ niche EC differentiation, we first grew postnatal day 0 (P0) LV progenitors using our established EC culture assay. We showed previously that this culture can efficiently differentiate into ECs following a reduction of media serum concentration from 10% to 2% (Paez-Gonzalez et al., 2011). We wondered whether reducing certain factors within the serum is necessary to initiate niche EC differentiation from specified pRGPs. To test this, we used chemical inhibitors of fibroblast growth factor (FGF), epidermal growth factor (EGF), platelet-derived growth factor (PDGF), or transforming growth factor β (TGF-β) pathways in EC cultures incubated in 10% serum and screened for their potential to initiate EC differentiation. Although FGF, PDGF, and TGF-β pathway inhibitors showed no significant effects, we found that epidermal growth factor receptor (EGFR)-specific inhibitor (Erlotinib) enabled Foxj1+ EC differentiation from pRGPs while incubated in 10% serum (Figures 1A–1D and S1A). To confirm that EGFR activity can directly block the initiation of niche EC differentiation, upon initiating EC differentiation in 2% serum, we added EGF to the culturing media. This effectively prevented EC Foxj1 expression and multiciliogenesis (Figures 1B–1E). Chemical inhibition of MEK1/2 (signaling effector kinases downstream of activated EGFR) was also sufficient to initiate Foxj1 expression and EC differentiation under 10% serum condition (Figures S1A and S1B). Immunohistochemical (IHC) staining for phospho-EGFR (pEGFR, the Y1068-activated form of the receptor) on differentiating EC cultures with the addition of EGF showed strong cellular enrichment compared with controls (Figure S1C). Cultures harvested from P0 FOXJ1-GFP reporter mice (Ostrowski et al., 2003) showed a lack of GFP expression following EGF addition during EC differentiation (compared with robust GFP upregulation in control condition), suggesting lack of foxj1 transcriptional activation instead of protein stability (Abdi et al., 2018) (Figure S1C).
Figure 1. EGF Inhibition of Ependymal Differentiation.
(A) IHC images of EC cultures grown in differentiation media (Serumlow), media containing 10% serum (Serumhigh), or media containing both 10% serum and Erlotinib (Serumhigh + Erl.). Samples were labeled with antibodies to Foxj1, acetylated tubulin (a-Tub), and DAPI. Scale bar: 20 µm.
(B) IHC images of EC cultures grown in differentiation media or differentiation media + EGF (EGF) and labeled with antibodies to Foxj1, a-Tub, and DAPI. Scale bar: 20 µm.
(C) Quantifications of Foxj1+ cells as the percentage of total DAPI-labeled cells. *p < 0.0001, one-way ANOVA; **p < 0.0064, Wilcoxon two-sample test; n = 5; mean ± SEM.
(D) Quantifications of multiciliated cells (visualized by a-Tub staining) as the percentage of total DAPI-labeled cells. *p < 0.0001, one-way ANOVA; **p < 0.0064, Wilcoxon two-sample test; n = 5; mean ± SEM.
(E) Western blot analysis of EC cultures grown in differentiation media (Serumlow), 10% serum (Serumhigh), or 10% serum + Erlotinib (Serumhigh + Erl.). Actin is the loading control.
(F) Transcriptome heatmap, with genes changing at least 2-fold between EGF-treated (EGF) and untreated (Ctrl) cultures. Log2 expression values were row normalized using Z scores and clustered using hierarchical clustering.
(G–I) Enriched GO terms (G) ranked by the percentage of genes located in each library. The number of genes identified for biological process (H) and cell compartment (I) is indicated for each bar graph.
See also Figure S1.
To evaluate EGF’s strong effects on inhibiting niche EC differentiation from pRGPs, we performed transcriptome analyses on differentiating EC cultures in low serum with or without EGF addition. Microarray analyses showed that expression levels of ~3,200 genes were altered when comparing EGF-treated with untreated conditions (Figure 1F). Gene Ontology (GO) analysis identified the main biological process difference as being related to motile cilium assembly, with the top five terms including cilium organization, assembly, movement, and multicilia formation (Figures 1G–1I, S1D, and S1E). Key factors known to be required for multiciliated cell differentiation (Brooks and Wallingford, 2014; Marshall and Kintner, 2008), including Mcidas, Gemc1, Myb, and Foxj1, were repressed by the addition of EGF to the culturing media (Figure S1F). Additional downstream genes, such as DNAH6, DNAH9, Kif9, and Kif27 (Choksi et al., 2014), were uniformly downregulated by at least 5-fold (Figure S1G). STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) protein-network analyses diagramming gene associations within our dataset specifically indicated EGF’s strong effects on attenuating the initiation of EC differentiation from pRGPs (Figure S1H). Although the upstream control of multiciliogenesis transcriptional regulator Foxj1 remains poorly understood, these results suggest that reduced MEK activation in EC progenitors may induce Foxj1 expression, resulting in multiciliogenesis initiation. However, it remains possible that downregulation of MEK activity directly control aspects of multiciliogenesis gene induction.
EGFR Localization and Downregulation during Postnatal Ependymal Niche Development
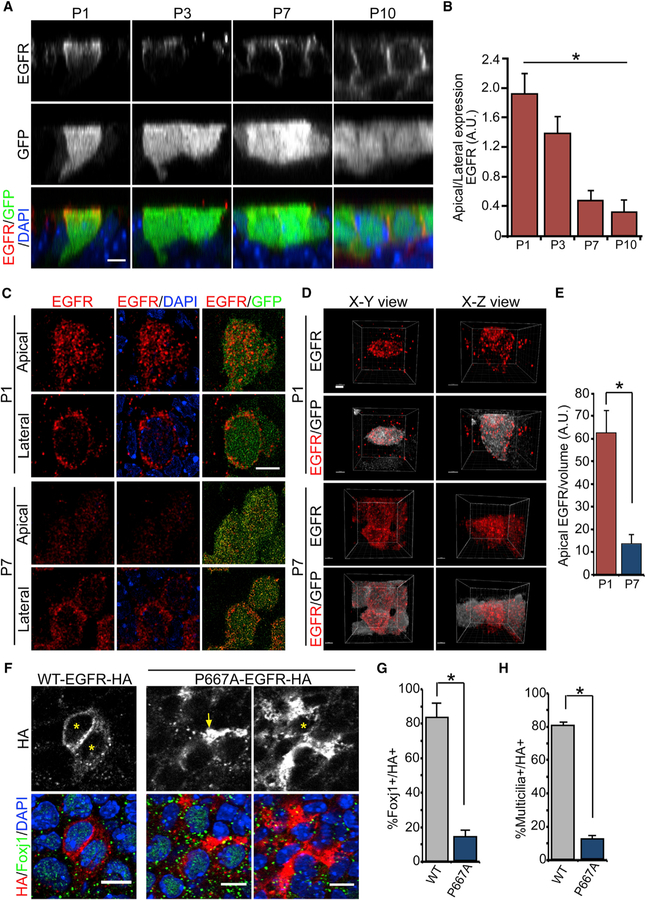
Our ependymal culture results suggested that terminating developmental EGF signaling after birth is required for initiating niche EC differentiation from pRGPs. Because EGF is known to be present in the CSF contacting the apical surfaces of differentiating ECs (Cieślak et al., 1986; Doetsch et al., 2002), we next wondered whether EGFR expression may be dynamically regulated during this important developmental time window. To track pRGPs fated to become niche ECs, we imaged FOXJ1-GFP-expressing cells along the LV neurogenic niche in P1, P3, P7, and P10 animals. Although FOXJ1-GFP can mislabel some neural progenitors postnatally (Abdi et al., 2018; Ostrowski et al., 2003), it remains a highly efficient reporter for ECs during their differentiation. At P1, LV germinal matrix GFP+ cells retained a radial glial-like morphology with smaller apical surface areas and single basal processes (Figure S2A). By P3, these GFP+ cells showed apical surface expansion while retaining their basal processes (Figure S2A). This apical surface expansion continued during EC differentiation at P7 and P10, with concurrent disappearance of basal processes from GFP+ cells (Figure S2A). Using orthogonal imaging of GFP+ cells on the x-z plane, at P1 we found EGFR expression in GFP+ pRGPs at their apical domain, as well as intracellularly (Figure 2A). By P3, EGFR was mostly intracellular, with decreased apical surface expression, while some cells began to show EGFR enrichment at their basolateral membranes (Figure 2A). At P7, we observed EGFR concentrating at the lateral domains of differentiating GFP+ ECs, which became mainly basolateral by P10 (Figure 2A). Quantifications of the apically to laterally localized EGFR ratio confirmed redistribution during postnatal EC maturation (Figure 2B). To quantify EGFR redistribution in ependymal pRGPs, we turned to super-resolution stimulated emission depletion (STED) microscopy (Klar and Hell, 1999). We detected distinct EGFR+ vesicular particles on the apical and lateral membrane domains of developing P1 FOXJ1-GFP+ progenitors (Figure 2C). Using Imaris 3D reconstructing of STED microscopic images and volumetric analysis of EGFR apical density, we were able to calculate the average numbers of EGFR particles per GFP+ cell at the apical domain in P1 and P7 animals, showing a significant reduction in the total numbers of apical EGFR particles from P1 to P7 (Figures 2D and 2E).
Figure 2. Apical to Basolateral Ependymal EGFR Localization.
(A) IHC images in x-z views of LV whole mounts from FOXJ1-GFP+ animals, with antibodies to EGFR, GFP, and DAPI. Scale bar: 10 µm.
(B) Quantification of the apical/basolateral EGFR expression ratio in FOXJ1-GFP+ pRGPs. *p < 0.0001, one-way ANOVA, n = 10 cells for each group, mean ± SEM.
(C) STED super-resolution images from the P1 and P7 LV surface labeled with EGFR and GFP antibodies. Single apical and lateral optical sections are shown. Scale bar: 5 µm.
(D) 3D rendering of STED super-resolution images from (C), with EGFR in red and GFP in white for clarity. Scale bars: 2 µm.
(E) Quantifications of apical EGFR particles as the fraction of the volume measured. *p < 0.001, Student’s t test, n = 10, mean ± SEM.
(F) IHC images from P14 LV whole mounts injected P1 with either WT-EGFR-HA or P667A-EGFR-HA lentivirus and labeled with anti-Foxj1/HA antibodies and DAPI. The asterisk indicates the cell body of HA+ cells quantified; note apically expressed P667A-EGFR-HA (arrow, left panels) from the deeper section of the same cell in right panels (*). Scale bars: 10 µm.
(G) Quantifications of Foxj1+ cells per total HA+ cells for each construct. *p < 0.0286, Wilcoxon two-sample test, n = 4 animals, mean ± SEM.
(H) Quantifications of multicilia+ cells per total HA+ cells for each construct. *p < 0.0286, Wilcoxon two-sample test, n = 4 animals, mean ± SEM.
See also Figures S2 and S3.
To determine whether this postnatal downregulation of EGFR from EC progenitors corresponded to receptor signaling activity, we performed IHC staining for activated pEGFR on LV whole mounts from P3 and P7 FOXJ1-GFP animals. Consistently, although we detected pEGFR signals on the apical domains of GFP+ pRGPs at P3, they were absent by P7 (Figure S2B). The presence of nearby pEGFR+ (but GFP-negative) cells within the developing neurogenic niche at P7 suggested that differentiating ECs were able to terminate their EGFR activity within a microenvironment that is activating EGFR in other cell types (Figure S2B). In the P35 mature SEZ niche, tamoxifen induction of nestion-CreERtm4; R26R-tdTomato; FOXJ1-GFP animals at P14 labels both niche ECs and NSCs (Figure S2C). Although as expected we readily detected EGFR IHC staining in tdTomato+ SEZ niche astrocytes at P35, we did not find EGFR expression in neighboring GFP+ niche ECs (Figure S2C).
Because the dynamic redistribution of EGFR from apical to basolateral cellular domains during EC development had not been described previously, we next wanted to examine whether this is functionally required for EC differentiation. We used a well-characterized EGFR mutation, located within the juxtamembrane domain (EGFR-P667A), resulting in receptor misaccumulation at the cellular apical domains, in addition to basolateral targeting (He et al., 2002). We generated hemagglutinin (HA)-tagged wild type (WT)-EGFR and P667A-EGFR lentiviral constructs, and injected them into P1 animal LVs. To minimize overexpression of EGFR, we limited in vivo viral infection to 1%–5% of differentiating pRGPs. IHC staining of LV whole mounts 3 days after injection confirmed that pRGPs can efficiently express WT-EGFR-HA and P667A-EGFR-HA constructs (Figures S2D and S2E). While WT-EGFR-HA was redistributed temporally from apical to lateral domains similar to endogenously expressed EGFR (Figures S2D and S2F), P667A-EGFR-HA was retained apically concurrent with a block in pRGP transition to EC morphology (Figures S2E and S2F). We next performed LV lentiviral injections in P1 FOXJ1-GFP+ mice. IHC staining of P14 LV whole mounts readily showed control WT-EGFR-HA construct expression in GFP+ ECs (Figure S3A). Foxj1 and acetylated tubulin antibody IHC co-staining confirmed that WT-EGFR-HA-expressing cells were able to properly differentiate into ECs (Figures 2F–2H and S3B). Although most control WT-EGFR-HA expression co-localized with ECs, we also detected as expected its expression in neighboring SEZ astrocytes from targeting P1 pRGPs (Figures S3A). In contrast to controls, most mutant P667A-EGFR-HA protein expression co-localized with GFAP+ SEZ astrocytes (Figures 2F and S3C). It was difficult to detect P667A-EGFR-HA expression in GFP+ Foxj1+-multiciliated cells (Figures 2F–2H, S3A, and S3B). Quantifications of control and mutant EGFR construct expressions in Foxj1+/multiciliated cells over multiple experiments revealed significant reductions of their cellular co-localization with the mutant receptor (Figures 2G and 2H), consistent with our earlier in vitro data showing that persistent EGFR activation can effectively block pRGP differentiation into niche ECs. Furthermore, our results showed that instead of simply regulating receptor expression, selective protein redistribution is a key step in terminating EGFR activation and allowing postnatal niche EC differentiation to proceed.
EGFR Activation Defects in Numb/Numblike Mutant Ependymal Niche Progenitors
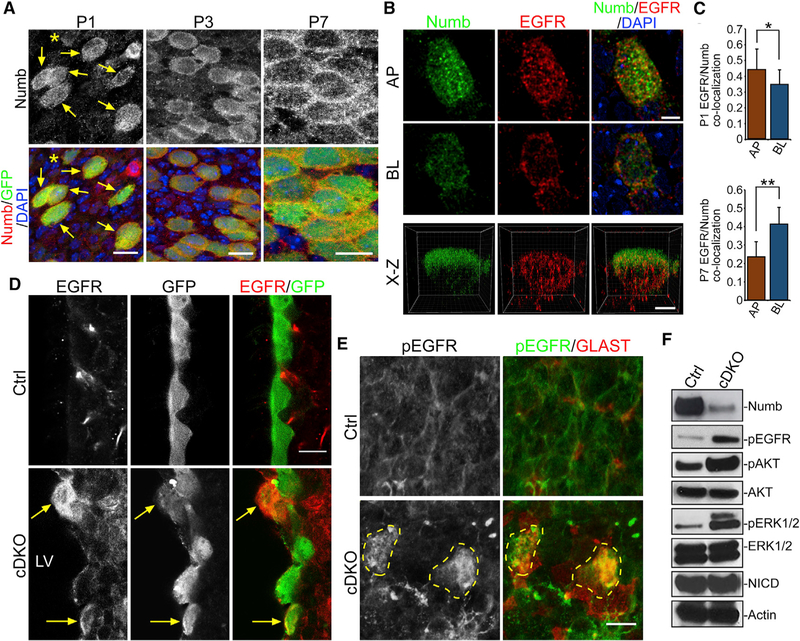
We next wondered which molecular mechanisms control EGFR redistribution in pRGP to downregulate extracellular growth factor signaling. The endocytic adaptor protein Numb was first identified as a key mediator of cellular asymmetric division (Rhyu et al., 1994; Roegiers and Jan, 2004; Uemura et al., 1989). Subsequent experiments have shown that Numb can interact with plasma membrane receptors to control their recycling and degradation (Gulino et al., 2010). At P1, Numb IHC staining on LV whole mounts from FOXJ1-GFP reporter animals showed strong intracellular distribution in GFP+ ependymal pRGPs (Figure 3A). Between P3 to P7, Numb became localized to lateral cellular domains in GFP+ cells during niche EC differentiation (Figure 3A). STED super-resolution imaging of differentiating EC pRGPs showed EGFR and Numb co-localizing to vesicular structures (Figures 3B, 3C, and S4A). At P1, there was robust EGFR and Numb co-localization at apical and basolateral domains, which became more basolateral by P7 (Figures 3B, 3C, and S4A). Upregulation of Numb protein in FOXJ1-GFP+ pRGPs consistently coincided with EGFR expression (Figure S4B).
Figure 3. EGFR Activation Defects in Numb/Numblike Mutants.
(A) IHC images of LV whole mounts from FOXJ1-GFP+ mice labeled with antibodies to Numb/GFP and DAPI. At P1, most GFP+ cells strongly expressed Numb (arrows), while GFP-dim cells showed lower Numb levels (*). Scale bar: 10 µm.
(B) STED super-resolution images of LV whole mounts from P1 FOXJ1-GFP+ animals labeled with antibodies to Numb, EGFR, and DAPI. AP, apical domain; BL, basolateral domain. x-z view, optical section from longest x-y axis. Scale bar: 5 µm.
(C) Quantifications of Numb/EGFR co-localization at AP or BL domains from P1 (top graph) or P7 (bottom graph) animals. Co-localization, average Pearson’s coefficient. *p < 0.03, **p < 0.0001, Student’s t test, n = 12 cells, mean ± SEM.
(D) IHC images of LV neurogenic niche coronal sections from P14 control (Ctrl) or FOXJ1-Cre; Nbflox/flox; NblKO/KO; CAG-GFP (cDKO) animals labeled with antibodies to GFP and EGFR. Note the cDKO GFP+ cells with high-level EGFR and AP localization (arrows).
(E) IHC images of LV whole mounts from P14 Ctrl or cDKO animals, labeled with antibodies to pEGFR and GLAST and showing mutant cells strongly expressing pEGFR (dashed circles). Scale bar: 20 µm.
(F) Western blot analysis of LV whole mounts from P14 Ctrl or cDKO animals. Actin is the loading control.
See also Figure S4.
To determine whether this Numb protein redistribution was responsible for EGFR trafficking, we deleted Numb and its closely related homolog Numblike (Nbl) in EC progenitors by inter-crossing FOXJ1-Cre; NumbF/+; NblKO/+ and NumbF/+; NblKO/+ animals to generate FOXJ1-Cre; NumbF/+; NblKO/KO (control) and FOXJ1-Cre; NumbF/F; NblKO/KO (conditional double knockout [cDKO]) mice. The CAG-GFP Cre-dependent reporter line was also crossed into the genetic background to label FOXJ1-Cre-targeted cells. Numb cDKO mice developed significant ventriculomegaly, glial scaring, and reduced multicilia coverage on the LV wall postnatally (Figures S4B–S4G) and did not survive past 5 weeks of age. IHC staining of P14 control versus cDKO ventricular sections showed that although, as expected, we did not detect EGFR expression in GFP+ ECs in control samples, there was strong EGFR expression in GFP+ cells in cDKO samples (Figure 3D). Furthermore, IHC staining of P14 LV whole mounts detected aberrant pEGFR on the apical membrane of cDKO mutant cells (Figure 3E). LV whole-mount western blot analyses comparing P14 control and cDKO animals showed that although total AKT and ERK1/2 protein levels were equivalent, pAKT, pERK1/2, and pEGFR levels (activated forms) were all significantly increased in cDKO samples (Figure 3F), confirming abnormal EGFR activation. Quantifications of GFP+ cells comparing P10 cDKO with control LV whole mounts showed cDKO cells had significantly increased GLAST expression and reduced apical surface diameter, indicative of pRGP to EC maturation defects (Figures S4F and S4G). There was phenotypic variability in cDKO GFP+ cells postnatally, and we believe this is caused by our timing of conditional Numb deletion and Numb protein perdurance. Using onset of foxj1 gene transcription to express Cre for Numb deletion also initiates the Foxj1-driven multiciliogenesis program in Cre-targeted cells. Ideally, one would need an EC progenitor-specific Cre driver that turns on postnatally before Foxj1-initiated multiciliogenesis, but to our knowledge such a driver line has not been identified.
Endocytic Adaptor Protein Numb Regulates EGFR Trafficking in Ependymal Progenitors
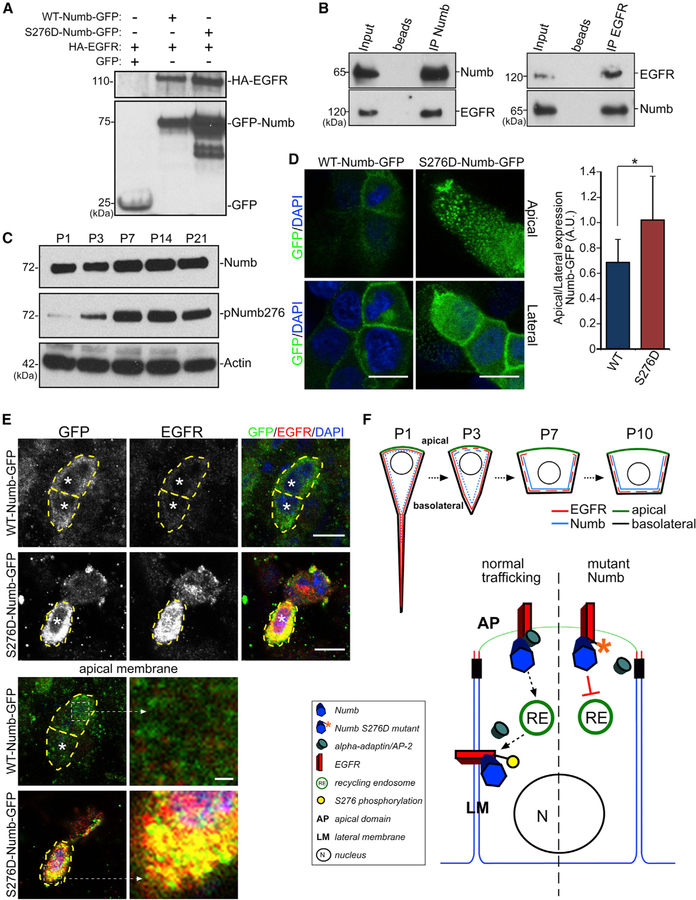
To examine potential biochemical interactions between Numb and EGFR, we first preformed immunoprecipitation (IP) experiments using tagged protein expression constructs. Co-transfections of HA-EGFR and Numb-GFP into HEK293 cells, followed by protein lysate IP with anti-GFP antibody, resulted in a robust pull-down of HA-EGFR (Figure 4A). As a control, co-transfections of HA-EGFR and GFP followed by anti-GFP IP did not pull down HA-EGFR (Figure 4A). IP experiments on acutely isolated P3 LV whole mounts showed efficient co-precipitation of EGFR and Numb proteins (Figure 4B). EGFR is internalized and recycled from the plasma membrane in a clathrin-dependent manner through activity of AP-2 protein containing complexes (Tomas et al., 2014). Numb has been shown to interact with AP-2 and clathrin-coated pits via α-adaptin protein (Berdnik et al., 2002). Moreover, Numb becomes decoupled from the recycling endocytic machinery following Ca2+-dependent phosphorylation at serine 276 (S276) by calmodulin-dependent protein kinase (CaMK), inhibiting its interactions with AP-2 and clathrin-mediated endocytosis (Smith et al., 2007; Tokumitsu et al., 2005, 2006). Thus, EGFR may be interacting with Numb at the apical membrane, internalized, and recycled, where Numb S276 phosphorylation at the basolateral membrane accumulates EGFR/Numb by decoupling from the endocytic machinery. Western blot analyses of postnatal LV wall samples revealed that S276 phospho-Numb (pNumb) levels increased from nearly undetectable levels at P1 to high expression by P7, while there was only modest increase in total Numb protein concurrently (Figure 4C), consistent with temporal dynamics of Numb lateralization in ependymal progenitors (Figure 3A).
Figure 4. EGFR Trafficking via Numb Phosphorylation.
(A) IP from HEK293 cell lysates co-transfected with constructs as indicated (+) and probed with antibodies to HA (EGFR, upper blot) and GFP (Numb, lower blot).
(B) IP from P3 LV whole mounts using anti-Numb or anti-EGFR antibodies. Blots were then probed with anti-Numb and anti-EGFR antibodies.
(C) Western blot analyses of LV walls of indicated ages and blotted for Numb and pNumb276. Actin is the loading control.
(D) IHC images of MDCK cells expressing either WT-Numb-GFP or S276D-Numb-GFP stained with anti-GFP antibody and DAPI. Single optical section views are from cellular apical surfaces (apical view, top row) or below the surface showing lateral membrane domains (lateral view, bottom row). Scale bars: 10 mm. The ratio of GFP fluorescent intensity at apical versus lateral domains for WT-Numb-GFP and S276D-Numb-GFP is shown. *p < 0.001, Student’s t test, n = 10, mean ± SEM.
(E) IHC images of P7 LV whole mounts expressing WT-Numb-GFP or S276D-Numb-GFP (from P1 lentiviral infection) labeled with anti-GFP, EGFR antibodies, and DAPI. Note high-level EGFR expression in S276D-Numb-GFP-expressing cells, but not WT-Numb-GFP-expressing cells (*). Right panels: single optical plane of the apical membrane, with enlarged areas showing high-level EGFR localization with S276D-Numb-GFP (dashed boxes). Scale bars: 10 µm; inset 1 µm.
(F) Schematic illustrations showing EGFR/Numb localizations during postnatal ependymal maturation (top) and the putative molecular pathway of EGFR redistribution (bottom).
Although it remains unclear which signaling cascades are at the basolateral membranes during ependymal niche maturation that resulted in increased Numb accumulation instead of continued endocytic recycling, we reasoned that constitutive S276 phosphorylation will prevent Numb from recycling EGFR from the apical membrane through the normal endocytic pathway. To test whether Numb directly regulates trafficking of EGFR, we generated a phosphomimetic version of Numb converting S276 to aspartic acid (S276D), which would continuously inhibit its interaction with the endocytic AP-2 protein complex. IP experiment showed that similar to WT-Numb-GFP, S276D-Numb-GFP mutant protein can physically interact with EGFR (Figure 4A). To confirm protein expression and localization, we first expressed WT-Numb-GFP or S276D-Numb-GFP in Madin-Darby canine kidney (MDCK) cells. While as expected WT-Numb-GFP protein was basolaterally localized, the S276D-Numb-GFP mutant protein also showed significant apical membrane localization (Figure 4D). We next injected lentiviral constructs expressing WT-Numb-GFP or S276D-Numb-GFP proteins into the LVs of P1 animals and stained LV whole mounts for EGFR and GFP expression at P7. We found that EGFR was dramatically upregulated in S276D-Numb-GFP-expressing cells with EGFR localized both apically and intracellularly, in contrast to control WT-Numb-GFP-expressing cells with mainly basolateral EGFR (Figure 4E). We observed considerable co-localization between EGFR and S276D-Numb-GFP both apically and intracellularly (Figure 4E). Together with the genetic mutant phenotype described earlier, these results demonstrated that Numb critically controls EGFR localization during EC differentiation, thereby downregulating their EGF-mediated cellular activation.
Despite its functional importance in sustaining the proliferative capacity of adult NSCs in health and disease, principles governing postnatal construction of the LV neurogenic niche from pRGPs remain poorly understood. Here we show a previously undescribed cellular pathway for postnatal development of the LV ependymal niche. EGFR signaling plays important roles in the expansion and migration of neuroprogenitors during development, but it was unclear how neighboring pRGPs along the LV surface switch from proliferation to a differentiation program postnatally during SEZ niche assembly. While we did not observe obvious EGFR localization changes in Foxj1-negative pRGPs during this early postnatal period, we found that Foxj1+ EC progenitors accomplished timely suppression of EGFR activity through a receptor trafficking step from their apical to basolateral membrane domains, away from ligand-induced activation. This cellular program enabled EC progenitors to efficiently switch from proliferation to differentiation, without waiting for receptor turnover, while the microenvironment continues to support EGF-mediated neurogenic proliferation.
Mechanistically, we identified physical interactions between EGFR and endocytic adaptor protein Numb. Originally described in Drosophila as a critical regulator of progenitor asymmetric cell division, mammalian Numb and Nbl also play important roles during neural development (Li et al., 2003; Petersen et al., 2002; Shen et al., 2002). Numb has been shown to biochemically interact with clathrin-coated vesicles, AP-2, and EHD/RME-1 proteins via aspargine-proline-phenylalanine (NPF) and aspartate-proline-phenylalanine (DPF) interaction motifs (Santolini et al., 2000; Smith et al., 2004). These protein-protein interactions between the plasma membrane and the intracellular endocytic compartments enable Numb to recycle TrkB receptors at the leading edge of migrating neuroblasts (Zhou et al., 2011), as well as metabotropic glutamate receptor 1 in Purkinje cells (Zhou et al., 2015). While we uncovered Numb/EGFR interactions in ependymal pRGPs, it is likely that Numb has cargos beside EGFR in ependymal pRGPs during postnatal development.
What are the possible upstream signals triggering Numb trafficking of EGFR from the apical membrane of ependymal pRGPs? In epithelial cells, Numb is targeted basolaterally and recycles from endocytic to plasma membrane domains via Ca2+-dependent signaling (Smith et al., 2007). Here we show highly specific Numb basolateral trafficking in pRGPs during EC development, temporally and functionally important for EGFR targeting and downregulation of its activity. While a conserved function for Numb in mammalian asymmetric cell division during neural development remains debated, it is exciting that Numb’s endocytic trafficking function in this context allowed differential engagement of a key environmental stimulus between neighboring pRGPs. Because we observed increased Numb phosphorylation at the CaMK-dependent S276 site postnatally, and because CaMK is activated by increased cytosolic [Ca2+] (Wayman et al., 2008), these results predict that increased intracellular [Ca2+] is a likely initiator of Numb basolateral localization developmentally. It is unknown whether increased [Ca2+] in pRGPs originates from extracellular sources or intracellular stores, and future research in these areas should illuminate cellular principles controlling adult stem cell niche assembly from progenitors.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chay T. Kuo (Chay.kuo@duke.edu). Plasmids generated in this study will be deposited to Addgene.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Knockout and transgenic mice
Mice were housed (up to 5 animals per cage) in a controlled with 12 h/12 h light/dark cycle at Duke University. Mice were fed a standard chow diet and provided ad libitum access to food and water throughout the study. All mouse experiments were performed according to an approved protocol by the Institutional Animal Care and Use Committee at Duke University. Both male and female mice were used throughout the study. No significant effect of sex was observed during analysis. Age of animals used is described in figure legends. The following mouse lines were used: FOXJ1-GFP (Ostrowski et al., 2003); nestin-CreERtm4 (Kuo et al., 2006); FOXJ1-Cre (Zhang et al., 2007); numbflox/+ (Zhong et al., 2000); numblikeKO/+ (Petersen et al., 2002); CAG-EGFP reporter (Kawamoto et al., 2000); R26R-tdTomato (Arenkiel et al., 2011).
Primary ependymal cultures
Primary ependymal cultures were generated from postnatal radial glial progenitors harvested from P0-P1 neonatal mouse pups as previously described (Abdi et al., 2018). Briefly, lateral ventricular walls were dissected, triturated in growth media (DMEM-High Glucose4.5 g/L (GIBCO)/DMEM-F12 1:1 mixture, 10% FBS (Hyclone), and 1% Pen/Strep) and plated in 24 well plates coated with Poly-D-Lysine (Sigma), then incubated under normal cell culture conditions in growth media. After cell culture reached 90%–100% confluence (~3–4 days after plating), media were switched and kept in differentiation media conditions (DMEM-High Glucose 4.5 g/L (GIBCO), 2% FBS (Hyclone), 1% L-glutamine, and 1% Pen/Strep 2% FBS).
Cell lines
293T17 and MDCK cell lines were acquired from ATCC and were cultured according to their recommended conditions (see Key Resources Table).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
Antibodies | ||
| Mouse monoclonal anti-Acetylated-tubulin | Sigma | Cat#- T7451; RRID:AB_609894 |
| Rabbit polyclonal anti-Foxj1 | Sigma | Cat#- HPA005714; RRID:AB_1078902 |
| Mouse monoclonal anti-GLAST | Miltenyi Biotech | Cat#- 130-095-822; RRID:AB_10829302 |
| Mouse monoclonal anti-HA-tag | Genescript | Cat#- A01244-100; RRID:AB_1289306 |
| Chicken polyclonal anti-HA-tag | Aves | Cat#- ET-HA100; RRID:AB_2313511 |
| Rabbit polyclonal anti-EGFR | Genetex | Cat#- GTX100448; RRID:AB_1950176 |
| Rabbit polyclonal anti-pEGFR | Abcam | Cat#- ab5644; RRID:AB_305012 |
| Rabbit polyclonal anti-AKT | Cell Signaling | Cat#- 4691T; RRID:AB_915783 |
| Rabbit polyclonal anti-pAKT | Cell Signaling | Cat#- 4060T; RRID:AB_2315049 |
| Rabbit polyclonal anti-ERK | Cell Signaling | Cat#- 9102S; RRID:AB_330744 |
| Rabbit polyclonal anti-pERK | Cell Signaling | Cat#- 9101S; RRID:AB_331646 |
| Goat polyclonal anti-Numb | Abcam | Cat#- ab4147; RRID:AB_304320 |
| Rabbit polyclonal anti-Notch | Cell Signaling | Cat#- 4147S; RRID:AB_2153348 |
| Mouse monoclonal anti-S100b | Sigma | Cat#- SAB1402349; RRID:AB_10640343 |
| Chicken polyclonal anti-GFP chicken | Aves | Cat#- GFP1020; RRID:AB_10000240 |
| Chicken polyclonal anti-GFAP | Aves | Cat#- GFAP; RRID:AB_2313547 |
| Mouse monoclonal anti-GFAP | Sigma | Cat#- G3893; RRID:AB_477010 |
| Rabbit polyclonal anti-γ-Tubulin | Sigma | Cat#- T5192; RRID:AB_261690 |
|
Bacterial and Virus Strains | ||
| Lentivirus WT-Numb-GFP | This paper | N/A |
| Lentivirus S276S-Numb-GFP | This paper | N/A |
| Lentivirus WT-EGFR-HA | This paper | N/A |
| Lentivirus P67A-EGFR-HA | This paper | N/A |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| Erlotinib | Selleckchem | S7786 |
| Mek Inhibitor | Selleckchem | S1036 |
| EGF | Invitrogen | PHG0311 |
|
Critical Commercial Assays | ||
| Q5 site directed mutagenesis kit | New England Biolabs | E0554S |
| Superscript Vilo cDNA synthesis kit | Invitrogen | 11754050 |
|
Deposited Data | ||
| Ependymal microarray data | This paper | GEO: GSE124518 |
|
Experimental Models: Cell Lines | ||
| 293T17 | ATCC | CRL-11268 |
| MDCK | ATCC | PTA-6500 |
|
Experimental Models: Organisms/Strains | ||
| FOXJ1-GFP | Ostrowski et al., 2003 | N/A |
| nestin-CreERtm4 | Kuo et al., 2006 | N/A |
| FOXJ1-Cre | Zhang et al., 2007 | N/A |
| numblikeKO/+ | Petersen et al., 2002 | N/A |
| CAG-EGFP | Kawamoto et al., 2000 | N/A |
| numbflox/+ | Zhong et al., 2000 | N/A |
| R26R-tdTomato | Arenkiel et al., 2011 | N/A |
|
Oligonucleotides | ||
| Refer to Table S1 | N/A | |
|
Recombinant DNA | ||
| PWPXLD-WT-Numb-GFP | This paper | N/A |
| PWPXLD-S276D-Numb-GFP | This paper | N/A |
| PWPXLD-WT-EGFR-HA | This paper | N/A |
| PWPXLD-P667-EGFR-HA | This paper | N/A |
METHOD DETAILS
Staining of primary ependymal cultures
ECs were grown from primary postnatal radial glial progenitors harvested from P0-P1 neonatal mouse pups as previously described (Abdi et al., 2018). EGF (20 ng/ml, Invitrogen) was added to 2% serum differentiation media for the treatment condition. For immunofluorescence staining, cell cultures were grown on 12 mm glass coverslips coated with Poly-D-Lysine, washed once in PBS, fixed in either 2% or 4% paraformaldehyde solution. Fixed cells were permeabilized in PBS containing 0.1% Triton X-100 (PBST), and blocked using 10% donkey serum in PBST. Primary antibody solutions were incubated overnight at 4°C, and conjugated secondary antibodies were incubated for 3 hours at 4°C. DAPI was incubated for 20 minutes at room temperature.
Immunohistochemistry and imaging
IHC of brain coronal sections and lateral ventricular wall wholemounts were prepared as previously described (Abdi et al., 2018). Briefly, samples fixed directly in paraformaldehyde solution (4% or 2%) followed by permeabilization in 0.1% PBST solution. Donkey serum at 10% in PBST was used to block tissue sections and wholemounts prior to immunolabeling. Confocal images were acquired on Leica TCS SP5 confocal microscope, with control and experimental samples imaged under identical settings. Super-resolution images were acquired using a Leica DMi8 motorized inverted microscope with STED and confocal capability. Deconvolution was carried out using the Huygens deconvolution software. Primary antibodies solutions were incubated overnight at 4°C with tissue, and fluorescently conjugated secondary antibodies were incubated for 2 hours at room temperature. All antibodies are listed in the Key Resources Table.
Biochemistry and immunoprecipitation
Cell cultures or tissue samples were lysed and prepared for immunoblotting as previously described (Abdi and Bennett, 2008). Lysates were resolved on Tris-Glycine SDS-PAGE gels (Bio-rad) and transferred onto PVDF membranes (Bio-rad). Antibodies were diluted in PBS containing 0.1% Triton X-100 with 2% BSA, followed by overnight incubation at 4°C. Detection was accomplished through secondary antibodies conjugated to horseradish peroxidase (Cell Signaling) and treated with enhanced chemiluminescence. HEK293 cells were grown on 24-well culture dishes. 24 hours after plating, HEK293 cells were transfected with equal amounts of plasmids for WT-Numb-GFP, S276D-Numb-GFP, and WT-EGFR-HA. 24 hours after transfection, media was replaced with low serum media then harvested 24 hours later. Immunoprecipitations were performed on HEK293 cells lysed in buffer containing 0.02 M HEPES Buffer, 0.15 M NaCl, 1% Triton X-100, sodium orthovanadate (Sigma), protease inhibitor cocktail (Roche), 10 mM NaF. Supernatants were collected from lysates spun at 15K rpm for 20 minutes, and primary antibodies incubated either 4 hours at room temperature or overnight at 4°C. Secondary antibodies conjugated to agarose beads were incubated for 2 hours at room temperature. Beads were washed 5 times in lysis buffer, harvested in 5X PAGE, processed for SDS-PAGE analysis and western blotting. All antibodies are listed in the Key Resources Table.
Transcriptome analyses
Affymetrix Gene Chip microarray data underwent strict quality control processing using the simpleaffy package in the Bioconductor2 suite from the R statistical programming environment. Log-scale Robust Multiarray Analysis (RMA) from the affy package was used to normalize the data and eliminate systematic differences across the arrays. Differential expression of genes across the conditions was identified from a moderated test statistic employed by the limma package. The False Discovery Rate (FDR) method was used to correct for multiple hypothesis testing. Gene set enrichment analysis was performed to identify differentially regulated pathways and gene ontology terms for each of the comparisons performed. Heatmap of the differentially expressed genes (FDR ≤ 0.05) for each < Ctrl > versus < EGF > comparison. Gene expression has been z-score normalized and the samples and genes are clustered by correlation distance with complete linkage. The heatmap shows all genes that were identified as significant in at least one of the comparisons. Heatmaps only include genes that had an FDR ≤ 0.05 and a log2FC > 5 or < −5. Hierarchical clustering plot of all of the samples based on expression of all genes on the arrays were shown. The samples are clustered using a correlation distance with complete linkage. Samples are colored by their condition.
Quantitative PCR
Total RNA was extracted using RNA easy mini Kit (QIAGEN), followed by cDNA preparation using Superscript Vilo cDNA synthesis kit (Invitrogen). Quantitative PCR analyses were performed using SYBR Green as previously described (Abdi et al., 2018). All primers are listed in Table S1.
DNA constructs and lentiviral preparation
The WT-Numb-GFP, cloned into peGFP plasmid, was a gift from Jane McGlade (Smith et al., 2007). The S276D mutation was generated in WT-EGFR-GFP using site directed mutagenesis (Q5, New England Biosciences), sequenced to confirm no additional mutations were generated. WT-Numb-GFP and S276D-Numb-GFP were cloned into PWXLD plasmid to generate lentiviruses. The EGFR-HA construct contains a c-terminal 3x-HA tag and was cloned into pEGFP by replacing the EGFP cassette. WT-EGFR-HA and P667A-EGFR-HA were cloned into PWXLD plasmid to generate lentiviruses. Lentiviruses were produced in 293T17 cells through triple-transfection of transfer plasmids pWPXLD, psPAX2, and pMD2.G. Lentivirus-containing media was collected, filtered through 0.45 micron membranes. To concentrate lentiviruses, media was spun for 2 hours at 25K rpm using swinging bucket rotor in Beckman Coulter ultracentrifuge. Numb and EGFR mutagenesis was performed by site-directed mutagenesis using the New England Biolabs Q5 site directed mutagenesis system according to the manufacturer’s instructions. All DNAs sequenced to verify that only the desired mutations were present.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical methods were used to pre-determine samples sizes, but they are similar to those reported elsewhere (Benner et al., 2013; Kuo et al., 2006; Paez-Gonzalez et al., 2011). For representative images and western blots, the results were shown to be reproducible by at least five separate experiments. Blocking of experimental design was assigned by animal genotype and was not randomized. Depending on sample size, unpaired Student’s t test (≥R10) or Wilcoxon two-sample test (≥R5) were used for statistical comparisons between two datasets. One-way ANOVA was used to compare more than two datasets. Graphs are presented as mean ± SEM and P values are presented in the figure legends. Statistical significance was set at a P value < 0.05.
Supplementary Material
Highlights.
EGFR downregulation promotes postnatal radial glial differentiation into ependyma
Apical EGFR redistributes to ependymal basolateral domains, limiting its activation
Endocytic adaptor Numb traffics ependymal EGFR during adult neurogenic niche assembly
Progenitors differentially act on the same extracellular cues via receptor redistribution
ACKNOWLEDGMENTS
We thank J. McGlade (University of Toronto) for providing the WT-Numb-GFP construct, D. Holtzman (Washington University) for the FOXJ1-Cre line, Duke Sequencing & Genomic Technologies for help with transcriptome analyses, B. Hogan and P. Tata (Duke University) for comments on the manuscript, and D. Fromme for technical assistance throughout. This work was supported by NIH grants R01NS078192, R01NS096096, and R01MH105416 and by the March of Dimes.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.056.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
Microarray data are available from the GEO database, accession number GSE124518. All other remaining data are available within the main article and supplemental files.
REFERENCES
- Abdi KM, and Bennett V (2008). Adducin promotes micrometer-scale organization of beta2-spectrin in lateral membranes of bronchial epithelial cells. Mol. Biol. Cell 19, 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdi K, Lai CH, Paez-Gonzalez P, Lay M, Pyun J, and Kuo CT (2018). Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat. Commun 9, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Hasegawa H, Yi JJ, Larsen RS, Wallace ML, Philpot BD, Wang F, and Ehlers MD (2011). Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PLoS ONE 6, e29423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrilleaux B, Phinney DG, Prockop DJ, and O’Connor KC (2006). Review: ex vivo engineering of living tissues with adult stem cells. Tissue Eng 12, 3007–3019. [DOI] [PubMed] [Google Scholar]
- Benner EJ, Luciano D, Jo R, Abdi K, Paez-Gonzalez P, Sheng H, Warner DS, Liu C, Eroglu C, and Kuo CT (2013). Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature 497, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Török T, González-Gaitán M, and Knoblich JA (2002). The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221–231. [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, Apostolopoulou M, Tian Y, and Temple S (2015). It takes a village: constructing the neurogenic niche. Dev. Cell 32, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks ER, and Wallingford JB (2014). Multiciliated cells. Curr. Biol 24, R973–R982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Lauter G, Swoboda P, and Roy S (2014). Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141, 1427–1441. [DOI] [PubMed] [Google Scholar]
- Cieślak D, Szulc-Kuberska J, Stepień H, and Klimek A (1986). Epidermal growth factor in human cerebrospinal fluid: reduced levels in amyotrophic lateral sclerosis. J. Neurol 233, 376–377. [DOI] [PubMed] [Google Scholar]
- Dani N, and Lehtinen MK (2016). CSF Makes Waves in the Neural Stem Cell Niche. Cell Stem Cell 19, 565–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bigio MR (2010). Ependymal cells: biology and pathology. Acta Neuropathol 119, 55–73. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Verdugo JM, Caille I, Alvarez-Buylla A, Chao MV, and Casaccia-Bonnefil P (2002). Lack of the cell-cycle inhibitor p27Kip1 results in selective increase of transit-amplifying cells for adult neurogenesis. J. Neurosci 22, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A (2015). Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, and Temple S (2013). Neural stem cells: generating and regenerating the brain. Neuron 80, 588–601. [DOI] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol 16, 19–34. [DOI] [PubMed] [Google Scholar]
- Gonzales KAU, and Fuchs E (2017). Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev. Cell 43, 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulino A, Di Marcotullio L, and Screpanti I (2010). The multiple functions of Numb. Exp. Cell Res 316, 900–906. [DOI] [PubMed] [Google Scholar]
- He C, Hobert M, Friend L, and Carlin C (2002). The epidermal growth factor receptor juxtamembrane domain has multiple basolateral plasma membrane localization determinants, including a dominant signal with a polyproline core. J. Biol. Chem 277, 38284–38293. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. (2014). Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, and Alvarez-Buylla A (2011). Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron 70, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, and Miyazaki J (2000). A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett 470, 263–268. [DOI] [PubMed] [Google Scholar]
- Kim H, Cooke MJ, and Shoichet MS (2012). Creating permissive microenvironments for stem cell transplantation into the central nervous system. Trends Biotechnol 30, 55–63. [DOI] [PubMed] [Google Scholar]
- Klar TA, and Hell SW (1999). Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett 24, 954–956. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Mirzadeh Z, Soriano-Navarro M, Rasin M, Wang D, Shen J, Sestan N, Garcia-Verdugo J, Alvarez-Buylla A, Jan LY, and Jan YN (2006). Postnatal deletion of Numb/Numblike reveals repair and remodeling capacity in the subventricular neurogenic niche. Cell 127, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, and Jan YN (2003). Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 40, 1105–1118. [DOI] [PubMed] [Google Scholar]
- Lim DA, and Alvarez-Buylla A (2016). The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb. Perspect. Biol 8, a018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, and Kintner C (2008). Cilia orientation and the fluid mechanics of development. Curr. Opin. Cell Biol 20, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Alvarez G, Daclin M, Shihavuddin A, Lansade P, Fortoul A, Faucourt M, Clavreul S, Lalioti ME, Taraviras S, Hippenmeyer S, et al. (2019). Adult Neural Stem Cells and Multiciliated Ependymal Cells Share a Common Lineage Regulated by the Geminin Family Members. Neuron 102, 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski LE, Hutchins JR, Zakel K, and O’Neal WK (2003). Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther 8, 637–645. [DOI] [PubMed] [Google Scholar]
- Pacelli S, Basu S, Whitlow J, Chakravarti A, Acosta F, Varshney A, Modaresi S, Berkland C, and Paul A (2017). Strategies to develop endogenous stem cell-recruiting bioactive materials for tissue repair and regeneration. Adv. Drug Deliv. Rev 120, 50–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, and Kuo CT (2011). Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron 71, 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, and Zhong W (2002). Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934. [DOI] [PubMed] [Google Scholar]
- Redmond SA, Figueres-Onate M, Obernier K, Nascimento MA, Parraguez JI, López-Mascaraque L, Fuentealba LC, and Alvarez-Buylla A (2019). Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell Rep 27, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, and Jan YN (1994). Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491. [DOI] [PubMed] [Google Scholar]
- Roegiers F, and Jan YN (2004). Asymmetric cell division. Curr. Opin. Cell Biol 16, 195–205. [DOI] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, and Di Fiore PP (2000). Numb is an endocytic protein. J. Cell Biol 151, 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, and Temple S (2002). Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853. [DOI] [PubMed] [Google Scholar]
- Smith CA, Dho SE, Donaldson J, Tepass U, and McGlade CJ (2004). The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell 15, 3698–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. (2007). aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J 26, 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, and Meunier A (2017). The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol 18, 423–436. [DOI] [PubMed] [Google Scholar]
- Spassky N, Merkle FT, Flames N, Tramontin AD, García-Verdugo JM, and Alvarez-Buylla A (2005). Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci 25, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, and Kintner C (2012). Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol 14, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Qu Y, Montcouquiol M, Zhou L, Komatsu K, Shi D, Fujimori T, Labeau J, Tyteca D, Courtoy P, et al. (2010). Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat. Neurosci 13, 700–707. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Inuzuka H, Sueyoshi Y, Yokokura S, Ichimura T, Nozaki N, and Kobayashi R (2005). Phosphorylation of Numb family proteins. Possible involvement of Ca2+/calmodulin-dependent protein kinases. J. Biol. Chem 280, 35108–35118. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Hatano N, Yokokura S, Sueyoshi Y, Nozaki N, and Kobayashi R (2006). Phosphorylation of Numb regulates its interaction with the clathrin-associated adaptor AP-2. FEBS Lett 580, 5797–5801. [DOI] [PubMed] [Google Scholar]
- Tomas A, Futter CE, and Eden ER (2014). EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 24, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, and Vogelstein B (2015). Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, and Jan YN (1989). numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva AJ, and Soderling TR (2008). Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron 59, 914–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, and Holtzman MJ (2007). A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am. J. Respir. Cell Mol. Biol 36, 515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Jiang MM, Schonemann MD, Meneses JJ, Pedersen RA, Jan LY, and Jan YN (2000). Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl. Acad. Sci. USA 97, 6844–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xiao H, and Wang H (2011). Developmental changes of TrkB signaling in response to exogenous brain-derived neurotrophic factor in primary cortical neurons. J. Neurochem 119, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Yang D, Wang DJ, Xie YJ, Zhou JH, Zhou L, Huang H, Han S, Shao CY, Li HS, et al. (2015). Numb deficiency in cerebellar Purkinje cells impairs synaptic expression of metabotropic glutamate receptor and motor coordination. Proc. Natl. Acad. Sci. USA 112, 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.