Abstract
Background
Epigenetic processes control timing and level of gene expression throughout life, during development, differentiation, and aging, and are the link to adapting gene expression profiles to environmental cues. To qualify for the definition of ‘epigenetic’, a change to a gene's activity must be inherited through at least one mitotic division. Epigenetic mechanisms link changes in the environment to adaptions of the genome that do not rely on changes in the DNA sequence. In the past two decades, multiple studies have aimed to identify epigenetic mechanisms, and to define their role in development, differentiation and disease.
Scope of review
In this review, we will focus on the current knowledge of the epigenetic control of pancreatic beta cell maturation and dysfunction and its relationship to the development of islet cell failure in diabetes. Most of the data currently available have been obtained in mice, but we will summarize studies of human data as well. We will focus here on DNA methylation, as this is the most stable epigenetic mark, and least impacted by the variables inherent in islet procurement, isolation, and culture.
Major conclusions
DNA methylation patterns of beta cell are dynamic during maturation and during the diabetic process. In both cases, the changes occur at cell specific regulatory regions such as enhancers, where the methylation profile is cell type specific. Frequently, the differentially methylated regulatory elements are associated with key function genes such as PDX1, NKX6-1 and TCF7L2. During maturation, enhancers tend to become demethylated in association with increased activation of beta cell function genes and increased functionality, as indicated by glucose stimulated insulin secretion. Likewise, the changes to the DNA methylome that are present in pancreatic islets from diabetic donors are enriched in regulatory regions as well.
Keywords: Endocrine pancreas, Beta cells, DNA methylation, Epigenetics, Aging
1. Introduction
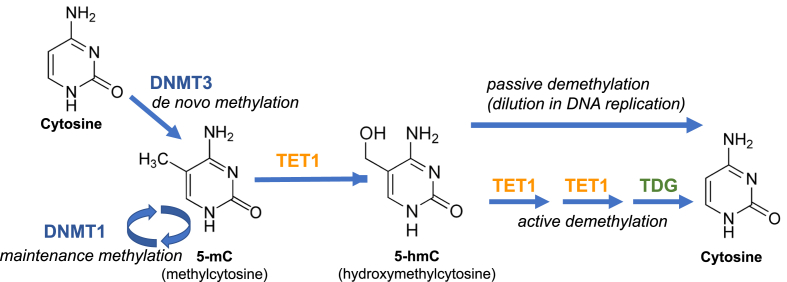
Nearly all the cells in our bodies – with the exception of lymphocytes, which undergo genome rearrangements at the T-cell and B cell receptor genes to produce our repertoire of antigen specific T cells and antibodies, respectively – contain 46 chromosomes whose DNA sequences are faithfully reproduced through cell division. In fact, it is estimated that the somatic mutation rate is less than 1 per 108 base pairs per S-phase. Therefore, as clearly postulated by Waddington in the middle of the 20th century [1], the regulation of gene expression must control development and produce the multitude of cell types and tissues that constitute multicellular organisms. Through work of biochemists, geneticists, and molecular biologists a complex array of mechanisms, such as histone modifications, microRNAs, and DNA methylation, have been identified which interpret the genome in a cell type-specific fashion. While many changes in gene expression occur on a short time scale and do not impact a cell's identity, others are more permanent and, if inherited through at least one cell division, are termed ‘epigenetic’. In this review, we will focus specifically on DNA methylation on CpG dinucleotides as a mark for which epigenetic inheritance through the cell cycle is understood on the molecular level. The cytosine methylation and demethylation system is summarized in Figure 1 and discussed in detail below. For a comprehensive review of all epigenetic factors that can impact the endocrine pancreas the reader is referred to a recent article by Golson and Kaestner in Molecular Metabolism [2].
Figure 1.
The cytosine methylation and demethylation system. Cytosine in the CpG context can be methylated de novo by DNA methyltransferase 3a and 3b (DNMT3) to 5′-methyl cytosine (5 mC). Following copying of the DNA during S-phase of the cell cycle, the now hemi-methylated CpGs are recognized by DNMT1 and restored to the fully methylated state. In specific contexts, likely driven by tissue-specific DNA binding transcription factors, selected 5 mC residues are oxidized by TET enzymes to produce 5hmC (5-hydroxymethyl cytosine), which can be further oxidized to 5 fC (5-formyl cytosine) and 5caC (5-carboxy cytosine), and glycosylated by thymidine DNA glycosylase (TDG), after which the modified base is removed by base excision repair (active demethylation). Alternatively, and possibly quantitatively more impactful, CpG methylation can be removed at selected sites through several rounds of DNA replication once a site has been converted to 5hmC, as this base cannot be recognized by DNMT1 (passive demethylation).
While high levels of DNA methylation at repetitive elements, and low levels of DNA methylation at CpG island containing promoters are largely invariant among cell types, methylation levels at enhancers are strongly cell type-dependent. These cell specific methylation patterns are inherited throughout life and are crucial for stable maintenance of cell identity. Therefore, pathological circumstances that influence normal patterns of DNA methylation may directly affect cell fate, identity and function. We have previously shown that in mice, the final pattern of beta cell specific methylation is achieved gradually though a maturation process that most likely involves functional cues or “training” of the beta cell after birth to accurately and efficiently respond to elevated glucose blood levels by secreting adequate amounts of insulin [3]. The establishment of a mature methylation profile was reflected mainly in demethylation of enhancers highly enriched for binding of beta cell transcription factors, as discussed in detail below.
α- and β-cell dysfunction plays a central role in the progression of Type 2 diabetes (T2D) through multiple mechanisms including partial loss of identity as recently suggested by us and others [4], [5], [6]. However, fundamental questions remain as to the precise nature of these cellular states, their prevalence, level of plasticity and functional outcome. Therefore, understanding the changes in epigenetic players such as DNA methylation which help sustain accurate cell specific gene expression programs throughout life to maintain α- and β-cell identity and fate is key to understanding islet cell plasticity or loss of identity seen in T2D.
2. DNA methylation as a key epigenetic modification
2.1. DNA methylation – general concepts
In mammals, DNA methylation is a key determinant that marks active versus non-active genes [7]. DNA methylation is most common on the cytosine in a CpG dinucleotide context. The human genome contains about 28 million of such CpGs, which is much fewer than would be expected by chance in a 3 billion base-pair genome, indicating that CpGs were selected against in evolution. In addition, the distribution of CpGs within the DNA is not random. Thus, most of the genome has a very sparse distribution of CpGs, while a subset of CpGs are clustered near transcriptional start sites into so-called ‘CpG islands’ (CGIs) [8]. CpG islands display a greater than 50% CpG content over stretches longer than 500 base-pairs [9], [10]. A role for CpG islands in gene regulation was postulated early due to the fact that they are highly enriched near gene promoters and exhibit very low DNA methylation levels. Furthermore, it was established that promoter methylation causes gene silencing [11], [12], providing a direct link between this epigenetic modification and gene activity. In general, DNA methylation is associated with closed chromatin and gene silencing [6]. Critical functions of DNA methylation include the maintenance of genome integrity and the inactivation of one of the X chromosomes in females [13]. These divergent roles are reflected in the global landscape of DNA methylation, where large stretches of repetitive sequences, including those of transposable elements, are fully methylated, while gene promoters are frequently unmethylated.
In contrast to the situation at CpG-rich promoters, in most cells about 85% of cytosines in CpGs are methylated to 5-methylcytosine (5 mC). Methylation is not uniform, in that promoters and tissue-specific enhancers are spared [3], [8], [14]. CpG methylation is the only epigenetic mark for which the inheritance through mitotic cell divisions is understood. This is so because the palindromic nature of the CpG allows for the easy reproduction of the methylated state at any genomic region through DNA replication. When DNA is copied in S-phase, the de novo synthesized DNA strand initially only contains unmodified cytosine. When these are opposite a 5mCpG on the old DNA strand, the DNA methyltransferase 1 enzyme (DNMT1) methylates the newly incorporated cytosine to restore the original state, with S-adenosyl-methionine serving as the methyl donor (see Figure 1).
Originally, it had been thought that because of this replication-dependent re-methylation, all somatic cells in the body would have identical or near-identical DNA methylation patterns, once these are established in early embryogenesis. However, this assumption was not borne out once it became possible to determine DNA methylation patterns genome-wide using both array platforms and DNA sequencing-based assays. When investigators began to compare different cell types and tissues, they noticed that while general hypomethylation at CpG-rich promoters, and high methylation levels at repetitive elements are uniform, DNA methylation at enhancers was strikingly tissue-specific [15], [16], [17]. How could DNA become demethylated? This remained an enigma until the year 2009, when it was discovered that the three enzymes encoded by the Tet loci (‘ten-eleven translocation’; Tet1, Tet2 and Tet3) can catalyze the oxidation of 5-methyl cytosine to 5-hydroxy-methyl cytosine (5hmC) [18], [19], and further to 5-formyl and 5-carboxy-methyl cytosine (Figure 1). This finding led to the realization that these enzymes could be the key in two non-exclusive pathways towards DNA demethylation. De-methylation can either occur via base-excision repair of 5-carboxy-methyl cytosine, or by targeted passive demethylation. The latter occurs if a cell with 5hmC enters the cell cycle, because the aforementioned enzyme DNMT1 does not recognize CpG's containing 5hmC as semi-methylated sites and will thus not methylate the newly synthesized DNA strand at this site (Figure 1). Thus, DNA methylation is not static, although at present it is not clear how the Tet enzymes, which themselves have no DNA sequence specificity, are directed to specific sites during the differentiation of fetal and adult tissues. It seems likely, however, that the DNA binding transcription factors known to maintain mature beta cell identity such as PDX1 and FOXA2 [20], [21], [22] play a role in the recruitment of Tet enzymes to tissue specific enhancers.
2.2. Dynamic DNA methylation during islet cell development and differentiation
In 2015, we published the first study on the dynamics of age-dependent methylome changes in the endocrine pancreas, focusing on the insulin-producing beta cell [3]. This paper summarized a comprehensive and integrative study of the effects of maturation on the beta cells' epigenome using sorted cells from juvenile and old mice. The former group consisted of mice between the ages of 4 and 6 weeks (pre-puberty), the latter of mice between 16 and 20 months of age (post reproductive age). We employed genome-wide base-resolution methylome analysis (WGBS) and integrated these findings with RNAseq transcriptome data and maps of histone modifications and beta cell transcription factors binding sites. When we compared unmethylated regions (‘UMRs’; 5.7% methylated on average) and regions with a low degree of methylation (‘LMRs’; 30% methylated on average) between old and young beta cells we found more than 14,000 chromosomal regions where DNA methylation differed significantly between the two age groups. Remarkable, the total area of the genome with differential DNA methylation spanned more than 13 million base pairs, re-emphasizing the point made above that DNA methylation even within the same developmental lineage or cell type is not static. We discovered that within promoter regions the changes in methylation levels were relatively small (generally up to 10%) whereas distal regions exhibited a change greater than 15% in DNA methylation level, with about a third of the DMRs showing a change of over 50% in methylation with age. Furthermore, regions that lost methylation with age were marked as active enhancers based on enrichment of nucleosomes containing the active histone mark H3K27ac and occupation of beta cell transcription factors such as PDX1 and FOXA2. In contrast, distal DMRs that gained methylation with age were enriched for recognition sequences of early developmental transcription factors such as the SOX and HOX gene families, expression of which is silenced in the postnatal beta cell. Altogether, these data support the notion that chromatin state and the presence of transcription factor binding at distal regulatory elements in the young beta cell target regulatory elements for demethylation with aging. Interestingly, de novo methylated promoters were found to be enriched near genes involved in cell cycle control such as MKi67, Ccnd3, and Plk1, suggesting a possible explanation for the age-dependent decline in the ability of beta cells to proliferate: as the promoters of key cell cycle genes are methylated, they may become refractory to mitogenic signals. In addition, maturation-dependent demethylation occurred in regulatory regions of genes essential for beta cell identity and function, such as the MODY (Maturity Onset Diabetes of the Young) genes, and the ATP-dependent potassium channel genes Kcnj11 and Abcc8. Many of these genes, including Pdx1, Nkx6.1, NeuroD1, Foxa2, and Mnx1 exhibit higher expression in old mice than in four-week old animals. Surprisingly, islets from old mice had improved responsiveness to glucose stimulation as determined by an islet perifusion assay, suggesting that beta cell function might actually improve, not decrease, with beta cell maturity.
These aging-dependent effects on beta cell function and gene expression are not limited to rodent models, but occur in a similar fashion in humans. Thus, Arda and colleagues profiled sorted endocrine and acinar cells from children and adults and found differences in glucose stimulated insulin secretion, as well as higher expression of genes important for hormone secretion in adult islets [23]. In addition, they discovered two transcription factor encoding genes – SIX3 and SIX2 – to be dramatically induced during beta cell aging. Whether these aging-dependent gene expression changes in human islets also correlate with enhancer methylation remains to be determined.
A limited analysis of islet cell type specific DNA methylation was performed by Neiman and colleagues [24]. Remarkably, they found the promoters of the insulin and preproglucagon genes to not be differentially methylated between islet alpha, beta and delta cells. Focusing on sparse CpG sites within a few hundred base-pairs from the transcriptional start sites (neither the insulin nor the pre-proglucagon gene contain CpG islands at their promoters), they found low DNA methylation in all three major hormone-producing cells in mouse and human upstream of the start site. In contrast, CpGs within the first exon of the insulin gene were unmethylated only in beta cells. Similarly, DNA methylation was high in CpGs of the preproglucagon gene in beta cells, while upstream sites were unmethylated in both alpha and beta cells. When they analyzed DNA methylation during mouse fetal development, they observed that promoter methylation of both genes was extremely high at early embryonic stages including the endoderm, which is the precursor of the pancreas, liver and gut. Only after endocrine progenitors, marked by Neurogenin 3, differentiated into early endocrine cells, did rapid demethylation occur. When they extended the DNA methylation analysis to 450,000 CpGs contained on a commercially available array, representing about 2% of the CpGs present in the human genome, they again found that promoter methylation did not differ greatly between alpha and beta cells [24]. In contrast, CpGs differentially methylated between alpha and beta cells were located to putative enhancers, just as they are in the mouse [3]. Given these findings, it appears likely that changing DNA methylation patterns, especially at cell type restricted enhancers, are crucial events in the differentiation of all pancreatic endocrine cell types.
2.3. Islet DNA methylation is altered in type 2 diabetes
Although DNA methylation is relatively stable and inheritable through mitotic cell divisions, it is also reversible as introduced above, and therefore responsive to changes in the environment. Thus, it was shown that the methylation profile of specific genes can be altered as a response to environmental cues such as exercise [25], [26] or changes in hormonal levels, and that these changes can be maintained [27]. Type 2 diabetes mellitus (T2DM) is a metabolic disorder characterized by insufficient beta cell function and peripheral insulin resistance, and is one of the most challenging public health issues today. The world-wide incidence has been estimated at 422 million individuals, resulting in over one million excess deaths and health care costs in the hundreds of billions of dollars each year. Both genetic and epigenetic factors contribute to T2DM, the latter clearly evidenced by the dramatic increase in T2DM incidence over the past two hundred years. T2DM is greatly affected by aging and lifestyle parameters such as diet and physical activity, all likely mediated via epigenetic mechanisms including DNA methylation changes.
Multiple studies have addressed changes to DNA methylation profiles in pancreatic islets in type 2 diabetics, either at specific gene loci, or using array-based assays that capture a small selection of CpGs [28], [29], [30], [31], [32], [33], [34], [35], [36]. Not surprisingly, the number of sites or regions that display differential DNA methylation levels (‘DMRs’) between islets obtained from non-diabetic and type 2 diabetic deceased organ donors is proportional to the number of CpG's analyzed. Thus, in the recent study by Volkov and colleagues, more than 25,000 genomic regions were identified as having differential DNA methylation. As with all epigenomic studies, the number of regions/transcripts/histone modifications reported as altered is dependent on the parameters and ‘cut-offs’ used to identify differences. In the case of DNA methylation analysis by sequencing, an additional important parameter to be considered is sequencing depth. If, for instance, a given CpG is sequenced only to 10-fold coverage, then DNA methylation percentage can be determined only in 10% increments. Thus, frequently only CpGs with a minimum coverage are being considered, limiting the information gained to substantially less than the 28 million CpGs present in the human genome. In the study by Volkov and colleagues, only 692 of the more than 25,000 DMRs identified exhibited an absolute difference in DNA methylation level of greater or equal to 10%, and these are the most likely to result in altered gene activity. Genes of known importance in islet biology included the alpha cell transcription factor ARX and the beta- and delta cell transcription factor PDX1, the latter with a marked increase in DNA methylation in T2DM islets [36]. The increase in methylation at the PDX1 locus correlates with the reduced expression of PDX1 reported by several investigators in islets from T2D mouse models [37]. Notably, however, of the genes associated with these 692 DMRs, only 26 exhibited differential gene expression at the mRNA level, reinforcing the notion that many different epigenetic factors control gene expression in addition to DNA methylation. It should also be emphasized that this and other prior studies employed whole human islets, which differ dramatically from individual to individual in endocrine cell composition and exocrine cell contamination [38], [39], [40]. Thus, any systematic difference in, for instance, alpha cell abundance between type 2 diabetics and controls will skew the DNA methylation profile at the ARX locus and other alpha cell specific genes. Future studies will likely address this caveat using sorted islet cell fractions.
2.4. Multigenerational epigenetic inheritance of beta cell dysfunction
It has been known for more than 25 years that fetal exposure to limited nutrient supply causes intrauterine growth retardation, and that low birth weight causes an increased risk for type 2 diabetes decades later, regardless of genotype of the mother or child [41], [42]. This clear example of an epigenetic effect on metabolic health was studies in mechanistic detail in rodent models of intrauterine growth retardation [43], [44], [45]. Park and colleagues discovered that not only was expression of the key beta cell transcription factor PDX1 reduced persistently in adult rats, but they also determined that this correlated with increased DNA methylation of a CpG island in the proximal promoter of gene [43]. Multiple groups have attempted to reverse these epigenetic changes through the manipulation of methyl donors (S-adenosyl methionine) in the diet or by the administration of beneficial agents such as Exendin-4. Remarkably, administration of Exendin-4, a long acting GLP-1 (glucagon like peptide 1) analog just during the first week of life could reverse repression of Pdx1 which was accompanied by normalization of both histone modifications and DNA methylation status [46].
2.5. Targeting epimutations to alter islet cell function and proliferation
The realization that the epigenome, and in particular CpG methylation, is not only cell type-specific, but also altered in perturbed metabolic states, whether insulin resistance, intrauterine growth retardation, or diabetes, quickly led to attempts to reverse such dysregulation at the epigenetic level to improve metabolic health. Altering DNA methylation levels by changing availability of the methyl donor S-adenosyl methionine is of course a non-specific intervention and fraught with the danger of unintended side effects. Folic acid, a necessary co-factor for the formation of S-adenosyl methionine, is given as a supplement to prevent neural tube defects in children, and is a highly effective intervention in this regard [47]. However, the effects of elevated folate levels on DNA methylation levels and their impact on cancer risk and other diseases are highly complex and controversial [48]. Because of the difficulty of controlling off-target effects of supra-physiological folate levels, researchers have searched for targeted effectors of DNA methylation to produce beneficial ‘epimutations’, and to use it as a research tool to study specific cause and effect relationships.
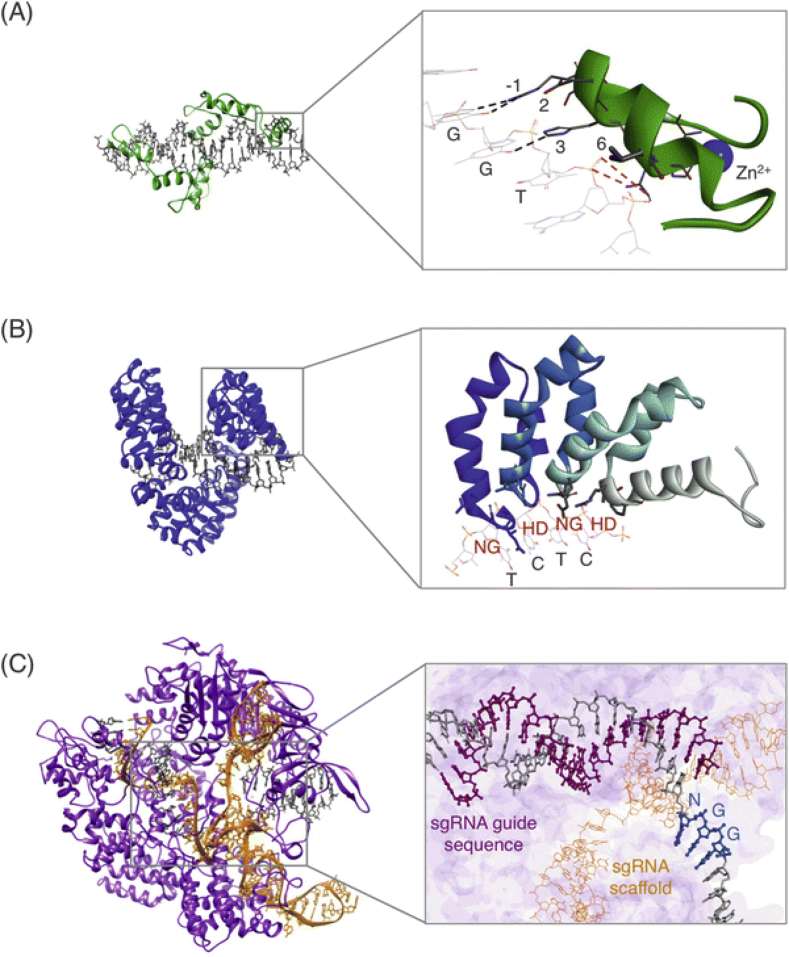
In short succession, three major systems for targeted epimutations were developed (Figure 2). The first system employs zinc finger transcription factor DNA binding moieties to bind to the desired sequence. Repetitive Cys2His2 zinc binding domains consisting of 30 amino acids each bind three DNA base pairs. By combining multiple zinc finger modules, sites of up to 18 base pairs have been targeted, and by adding effector moieties, gene activation, repression or DNA methylation changes have been produced (reviewed in [49]). The second customizable system for epigenetic targeting is based on the ‘transcription activator-like effectors’ or TALEs, which were derived from Xanthomonas bacteria, which are a plant pathogen that hijack the host cells genome to support bacterial growth. TALEs are modular DNA binding proteins, where different 34 amino acid peptides bind to the four bases of DNA. By assembly of these modules in the desired order, DNA binding proteins with exquisite specificity for the target can be designed [49]. Lastly, and used most widely today, modifications of the CRISPR/Cas system that eliminate its DNAse activity have been employed for both targeted gene activation and repression [49].
Figure 2.
Structure of commonly used targeted epigenome modifying systems. (A) A three zinc finger protein (green) in complex with DNA (gray). Inset shows one of the zinc finger modules and its binding to residues −1, 2, 3, and 6 (B) A TALE protein (blue) in complex with DNA (gray). Inset shows TALE repeat-variable di-residues (RVDs) labeled in red and corresponding DNA bases labeled in gray. (C) Nuclease-null dCas9 protein (purple) with designed sgRNA (orange) in complex with DNA (gray). Inset shows the interaction between the sgRNA and DNA. Reprinted with permission from Waryah and colleagues [49].
Recently, TALE-DNMT and TALE-TET1 fusion proteins have been employed to the study of epigenetic misregulation in type 2 diabetic beta cells, and for the induced proliferation of human islets cells. Studying microRNA expression in human islets from type 2 diabetic organ donors, Kameswaran and colleagues had discovered that a cluster of 54 microRNAs encoded at the imprinted DLK1-MEG3 locus is robustly down-regulated in diabetic beta cells [50]. This dramatic change correlated with increased DNA methylation at a known differentially methylated region – in this case meaning differential between the paternally and maternally inherited alleles – termed MEG3-DMR. In order to evaluate causality, we developed a TALE-DNMT fusion protein targeting this element for re-methylation [51]. Following transfection of βTC3 insulinoma cells, we indeed achieved a significant increase in DNA methylation at the locus as predicted. This targeted DNA methylation, or epimutation, reduced gene expression from the locus by four-fold, establishing causality between DNA methylation status and gene expression at this locus [51]. Furthermore, because several of the microRNAs produced from the MEG3 locus target genes that control the sensitivity of cells to apoptotic stimuli and metabolic stress, we evaluated the response of the epimutated beta cells to inflammatory cytokine treatment, and found their sensitivity increased. Thus, the epigenetically dysregulated MEG3 microRNA cluster appears to mediate part of the increased sensitivity to metabolic and apoptotic stress observed in T2DM beta cells.
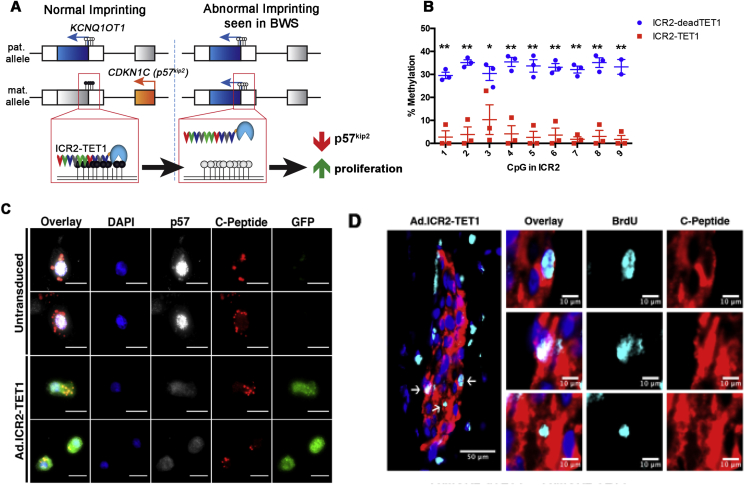
Adult beta cells, whether rodent or human, are largely postmitotic and refractory to mitogenic stimuli. This is in part due to high expression of cell cycle inhibitors such as p16 and p57, the latter of which appears to have a dominant effect, as its loss through mutation or epimutation of the CDKN1C locus results in focal hyperinsulinism characterized by unbridled proliferation of beta cells [52]. Because increasing human beta cell mass – whether ex vivo to increase beta cell mass before islet transplantation for the treatment of type 1 diabetes, or in vivo as a therapeutic approach to T2DM – is a desirable goal, we investigated whether targeted epimutations at the CDKN1C locus is sufficient to induce human beta cell replication [53], [54]. In this case, we targeted the catalytic domain of TET1 to the imprinting control region 2 (ICR2) of the CDKN1C locus to mimic the state of the paternal, and thus CDKN1C silent, allele on the maternal allele (Figure 3A). Even though TET1 only catalyzes cytosine oxidation (see Figure 1), we achieved a large degree of ICR2 demethylation (Figure 3B) and reduced p57 levels in most of the seven human islet preparations manipulated (Figure 3C). The transduced islets were transplanted into immunodeficient mice who were given bromodeoxyuridine, a thymidine analog that can be used to track DNA replication, in the drinking water. When the human islets were recovered three weeks later, four out of five islet preparations exhibited an increase in beta cell replication. An example of BrdU-positive beta cells in the recovered islet graft is shown in Figure 3D. Thus, human beta cells can be forced to adopt a more proliferative state through targeted epimutation, without changing a single base of their genome. Obviously, important safety concerns need to be evaluated before this can be considered in a therapeutic setting, chiefly among them how long the proliferative state lasts, and if insulinomas develop. Nevertheless, this study served as proof-of-principle that epimutations can be employed to produce the desired changes in beta cell properties.
Figure 3.
Targeted epimutation at the CDKN1C locus increases replication of human beta cells. (A) Schematic of the imprinted Chr11p15.5 locus. The ICR2 is methylated (depicted by black circles) at the promoter of lncRNA KCNQ1OT1 on the maternal allele, which correlates with maternal allele-specific expression of CDKN1C/p57kip2. A TALE-TET1 fusion protein was designed to target the ICR2 and remove the methylCpGs at the ICR2 in order to deactivate CDKN1C and increase cell proliferation. (B) Percent methylation of unique CpGs in the ICR2 of human fibroblasts after transduction with either the control ICR2-deadTET1 or ICR2-TET1 adenovirus (*,p < 0.05, **,p < 0.01). (C) Immunocytochemistry of beta cells (identified by C-peptide, red) transduced with the ICR2-TET1 adenovirus. Control beta cells were identified by the absence of GFP staining. Note the absence of nuclear p57kip2 protein in beta cells expressing ICR2-TET1. (D) BrdU+ beta cells in sectioned islet xenografts. C-peptide stained in red, BrdU in blue. Reprinted with permission from [54].
3. Conclusion
Evolution designed a complex array of transcriptional and epigenetic mechanisms to interpret the genome of multicellular organism in a cell type specific and situationally adaptable fashion to enable higher life forms. Among these, DNA methylation stands out as a semi-permanent mark for which the mechanism of inheritance through mitosis – and thus expansion of organ systems – is understood at the molecular level. Over the past ten years it has become apparent that the DNA methylome differs between tissues and cell types, mainly at enhancers, and that its perturbations at minimum correlate with (type 2 diabetes) and sometime cause (Beckwith-Wiedemann syndrome, focal hyperinsulinism) metabolic disease. Further advances in technology will greatly increase the power of methylome analyses and enable base resolution determinations in specific islet cell types, allow for the assessment of de-methylation intermediates with regulatory content such as 5-hydroxy methyl cytosine, and facilitate the targeted manipulation of the epigenome even further. When combined with cell type specific delivery systems, these advances promise a whole new level for precision medicine approaches to diabetes.
FUNDING
This article is part of a supplement entitled ‘Biomarkers of Beta-Cell Health and Dysfunction: Towards Personalised Diabetes Care. Proceedings of the 20th Servier-IGIS Symposium’. The Symposium and the publication of the Supplement have been made possible by an unrestricted educational grant from Institut la Conférence Hippocrate – Servier Group.
Acknowledgements
We thank you our colleagues in the Glaser, Dor, and Kaestner labs for helpful discussions. Related work in our labs was supported through NIH grant UC4-DK104119, the BIRAX Regenerative Medicine Initiative (14BX14NHBG), the Israel Science Foundation (1506/12), and JDRF/Pfizer (2-SRA-2015-5-Q-R). The authors have no financial conflicts of interest to disclose.
Conflict of interest
None declared.
References
- 1.Waddington C.H. George Allen & Unwin Ltd; 1957. The strategy of genes. [Google Scholar]
- 2.Golson M.L., Kaestner K.H. Epigenetics in formation, function, and failure of the endocrine pancreas. Molecular Metabolism. 2017;6(9):1066–1076. doi: 10.1016/j.molmet.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avrahami D., Li C., Zhang J., Schug J., Avrahami R., Rao S. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metabolism. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talchai C., Lin H.V., Kitamura T., Accili D. Genetic and biochemical pathways of beta-cell failure in type 2 diabetes. Diabetes, Obesity and Metabolism. 2009;11:38–45. doi: 10.1111/j.1463-1326.2009.01115.x. [DOI] [PubMed] [Google Scholar]
- 5.Segerstolpe A., Palasantza A., Eliasson P., Andersson E.M., Andreasson A.C., Sun X. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y.J., Schug J., Won K.J., Liu C., Naji A., Avrahami D. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 8.Bird A., Taggart M., Frommer M., Miller O.J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. Journal of Molecular Biology. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 10.Zampieri M., Ciccarone F., Calabrese R., Franceschi C., Burkle A., Caiafa P. Reconfiguration of DNA methylation in aging. Mechanisms of Ageing and Development. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Kass S.U., Goddard J.P., Adams R.L. Inactive chromatin spreads from a focus of methylation. Molecular and Cellular Biology. 1993;13(12):7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keshet I., Yisraeli J., Cedar H. Effect of regional DNA methylation on gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(9):2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancks D.C., Kazazian H.H., Jr. Active human retrotransposons: variation and disease. Current Opinion in Genetics & Development. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergman Y., Cedar H. DNA methylation dynamics in health and disease. Nature Structural & Molecular Biology. 2013;20(3):274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 15.Choi I., Kim R., Lim H.W., Kaestner K.H., Won K.J. 5-hydroxymethylcytosine represses the activity of enhancers in embryonic stem cells: a new epigenetic signature for gene regulation. BioMed Central Genomics. 2014;15:670. doi: 10.1186/1471-2164-15-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L., Zhou R., Kuo Y.-C., Cunniff M., Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nature Communications. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burtscher I., Barkey W., Lickert H. Foxa2-venus fusion reporter mouse line allows live-cell analysis of endoderm-derived organ formation. Genesis. 2013;51(8):596–604. doi: 10.1002/dvg.22404. [DOI] [PubMed] [Google Scholar]
- 21.Doyle M.J., Sussel L. Nkx2.2 regulates beta-cell function in the mature islet. Diabetes. 2007;56(8):1999–2007. doi: 10.2337/db06-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao T., McKenna B., Li C., Reichert M., Nguyen J., Singh T. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metabolism. 2014;19(2):259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arda H.E., Li L., Tsai J., Torre E.A., Rosli Y., Peiris H. Age-dependent pancreatic gene regulation reveals mechanisms governing human beta cell function. Cell Metabolism. 2016;23(5):909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neiman D., Moss J., Hecht M., Magenheim J., Piyanzin S., Shapiro A.M.J. Islet cells share promoter hypomethylation independently of expression, but exhibit cell-type-specific methylation in enhancers. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(51):13525–13530. doi: 10.1073/pnas.1713736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baubec T., Schuebeler D. Genomic patterns and context specific interpretation of DNA methylation. Current Opinion in Genetics & Development. 2014;25:85–92. doi: 10.1016/j.gde.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Rohling M., Herder C., Stemper T., Mussig K. Influence of acute and chronic exercise on glucose uptake. Journal of Diabetes Research. 2016;2016:2868652. doi: 10.1155/2016/2868652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller C.A., Gavin C.F., White J.A., Parrish R.R., Honasoge A., Yancey C.R. Cortical DNA methylation maintains remote memory. Nature Neuroscience. 2010;13(6):664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callinan P.A., Feinberg A.P. The emerging science of epigenomics. Human Molecular Genetics. 2006;15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- 29.Dayeh T., Volkov P., Salo S., Hall E., Nilsson E., Olsson A.H. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. Public Library of Science Genetics. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall E., Dayeh T., Kirkpatrick C.L., Wollheim C.B., Dekker Nitert M., Ling C. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BioMed Central Medical Genetics. 2013;14:76. doi: 10.1186/1471-2350-14-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 32.Ling C., Del Guerra S., Lupi R., Ronn T., Granhall C., Luthman H. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51(4):615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. The European Molecular Biology Organization Journal. 2012;31(6):1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B.T., Dayeh T.A., Kirkpatrick C.L., Taneera J., Kumar R., Groop L. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54(2):360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang B.T., Dayeh T.A., Volkov P.A., Kirkpatrick C.L., Malmgren S., Jing X. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Molecular Endocrinology. 2012;26(7):1203–1212. doi: 10.1210/me.2012-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volkov P., Bacos K., Ofori J.K., Esguerra J.L., Eliasson L., Ronn T. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66(4):1074–1085. doi: 10.2337/db16-0996. [DOI] [PubMed] [Google Scholar]
- 37.Blum B., Roose A.N., Barrandon O., Maehr R., Arvanites A.C., Davidow L.S. Reversal of beta cell de-differentiation by a small molecule inhibitor of the TGFbeta pathway. Elife. 2014;3:e02809. doi: 10.7554/eLife.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirshberg B., Harlan D.M. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. Journal of Histochemistry and Cytochemistry. 2005;53(9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y.J., Golson M.L., Schug J., Traum D., Liu C., Vivek K. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metabolism. 2016;24(4):616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y.J., Traum D., Schug J., Gao L., Liu C., Consortium H. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metabolism. 2019;29(3):769–783 e4. doi: 10.1016/j.cmet.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hales C.N., Barker D.J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35(7):595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 42.Ravelli A.C., van der Meulen J.H., Michels R.P., Osmond C., Barker D.J., Hales C.N. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351(9097):173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 43.Park J.H., Stoffers D.A., Nicholls R.D., Simmons R.A. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. Journal of Clinical Investigation. 2008;118(6):2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmons R.A., Templeton L.J., Gertz S.J. Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes. 2001;50(10):2279–2286. doi: 10.2337/diabetes.50.10.2279. [DOI] [PubMed] [Google Scholar]
- 45.Stoffers D.A., Desai B.M., DeLeon D.D., Simmons R.A. Neonatal exendin-4 prevents the development of diabetes in the intrauterine growth retarded rat. Diabetes. 2003;52(3):734–740. doi: 10.2337/diabetes.52.3.734. [DOI] [PubMed] [Google Scholar]
- 46.Pinney S.E., Santos L.J.J., Han Y., Stoffers D.A., Simmons R.A. Exendin-4 increases histone acetylase activity and reverses epigenetic modifications that silence Pdx1 in the intrauterine growth retarded rat. Diabetologia. 2011;54(10):2606–2614. doi: 10.1007/s00125-011-2250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honein M.A., Paulozzi L.J., Mathews T.J., Erickson J.D., Wong L.Y. Impact of folic acid fortification of the US food supply on the occurrence of neural tube defects. Journal of the American Medical Association. 2001;285(23):2981–2986. doi: 10.1001/jama.285.23.2981. [DOI] [PubMed] [Google Scholar]
- 48.Crider K.S., Yang T.P., Berry R.J., Bailey L.B. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Advances in Nutrition. 2012;3(1):21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waryah C.B., Moses C., Arooj M., Blancafort P. Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing. Methods in Molecular Biology. 2018;1767:19–63. doi: 10.1007/978-1-4939-7774-1_2. [DOI] [PubMed] [Google Scholar]
- 50.Kameswaran V., Bramswig N.C., McKenna L.B., Penn M., Schug J., Hand N.J. Epigenetic regulation of the DLK1-MEG3 MicroRNA cluster in human type 2 diabetic islets. Cell Metabolism. 2014;19(1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kameswaran V., Golson M.L., Ramos-Rodriguez M., Ou K., Wang Y.J., Zhang J. The dysregulation of the DLK1-MEG3 locus in islets from patients with type 2 diabetes is mimicked by targeted epimutation of its promoter with TALE-DNMT constructs. Diabetes. 2018;67(9):1807–1815. doi: 10.2337/db17-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kassem S.A., Ariel I., Thornton P.S., Scheimberg I., Glaser B. Beta-cell proliferation and apoptosis in the developing normal human pancreas and in hyperinsulinism of infancy. Diabetes. 2000;49(8):1325–1333. doi: 10.2337/diabetes.49.8.1325. [DOI] [PubMed] [Google Scholar]
- 53.Avrahami D., Li C., Yu M., Jiao Y., Zhang J., Naji A. Targeting the cell cycle inhibitor p57(Kip2) promotes adult human beta cell replication. Journal of Clinical Investigation. 2014;124(2):670–674. doi: 10.1172/JCI69519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou K., Yu M., Moss N.G., Wang Y.J., Wang A.W., Nguyen S.C. Targeted demethylation at the CDKN1C/p57 locus induces human beta cell replication. Journal of Clinical Investigation. 2019;129(1):209–214. doi: 10.1172/JCI99170. [DOI] [PMC free article] [PubMed] [Google Scholar]