Abstract
The outer membrane (OM) of Gram-negative bacteria exhibits unique lipid asymmetry, with lipopolysaccharides (LPS) residing in the outer leaflet and phospholipids (PLs) in the inner leaflet. This asymmetric bilayer protects the bacterium against intrusion of many toxic substances, including antibiotics and detergents, yet allows acquisition of nutrients necessary for growth. To build the OM and ensure its proper function, the cell produces OM constituents in the cytoplasm or inner membrane and transports these components across the aqueous periplasmic space separating the two membranes. Of note, the processes by which the most basic membrane building blocks, i.e. PLs, are shuttled across the cell envelope remain elusive. This review highlights our current understanding (or lack thereof) of bacterial PL trafficking, with a focus on recent developments in the field. We adopt a mechanistic approach and draw parallels and comparisons with well-characterized systems, particularly OM lipoprotein and LPS transport, to illustrate key challenges in intermembrane lipid trafficking. Pathways that transport PLs across the bacterial cell envelope are fundamental to OM biogenesis and homeostasis and are potential molecular targets that could be exploited for antibiotic development.
Keywords: membrane biogenesis, phospholipid, Gram-negative bacteria, outer membrane, lipid trafficking, lipid asymmetry, lipid homeostasis, OmpC–Mla, Tol–Pal
Introduction
Gram-negative bacteria are distinctively characterized by the presence of a complex cell envelope comprising an inner membrane (IM),3 a thin layer of cell wall (or peptidoglycan), and an outer membrane (OM) (1). The IM marks the boundary of the bacterial cytoplasm, and the OM defines a second aqueous compartment known as the periplasm. This double-membrane structure effectively protects the cell from external insults. In particular, the OM restricts the entry of large hydrophobic molecules, in part conferring Gram-negative bacteria intrinsic resistance against many antibiotics and detergents (1).
The OM is an essential lipid bilayer that contains integral proteins (mostly β-barrel OMPs) and peripherally-anchored lipoproteins. It is also highly asymmetric (2–4), whereas the inner leaflet of the OM is composed of typical phospholipids (PLs), the outer leaflet contains tightly-packed lipopolysaccharides (LPS), which impart low fluidity and permeability to the OM (1). This asymmetric arrangement of lipids is critical for proper barrier function. To build a stable and functional OM, the respective components have to be transported from their sites of synthesis at the cytoplasm or IM, across the periplasm, and to the OM (5). Assembly of this second bilayer is extremely challenging not only because of the need for tight coordination between the various transport processes, but also because there is no obvious energy source, such as ATP, in the periplasmic space. Furthermore, all the major components of the OM are strongly amphipathic by nature, which necessitates distinct mechanisms for shielding at least the hydrophobic portions of these molecules from the aqueous environment as they transit the periplasm. In this regard, the transport pathways for OMPs (6), LPS (7), and lipoproteins (8) have been relatively well-characterized. Despite recent advances, however, our understanding of PL transport across the cell envelope is still lacking. In this review, we summarize the current knowledge on possible PL transport pathways that may contribute to OM biogenesis and homeostasis in Gram-negative bacteria. Through discussion of PL trafficking from a mechanistic viewpoint, and in the context of known transport pathways for other lipidated molecules (i.e. lipoproteins and LPS), we hope to highlight outstanding questions in this field and pave a path toward a better understanding of bacterial lipid transport.
Mechanistic models for lipid transport across the periplasm
There are three major steps to be considered for lipid transport between the IM and the OM: (i) release from the first membrane; (ii) transit across the aqueous periplasm; and (iii) insertion into the target membrane. Largely because of the hydrophobic effect, it is most energetically favorable to have the acyl tails of a lipid molecule sequestered in a hydrophobic environment (9). As one can imagine, it would be extremely difficult to pull a lipid molecule out of the membrane and transport it with its acyl tails exposed to aqueous solution; lipid transport across the periplasm therefore does not occur unassisted. A solution to this problem has been proteins that can bind to the acyl tails and shield them from water (Fig. 1). These come in two flavors, those that form soluble lipid–protein complexes as exemplified in lipoprotein transport (8), and those that physically bridge the two membranes to provide a hydrophobic path for lipids moving across the aqueous environment, such as that observed in LPS transport (7). In both of these strategies, the protein-bound lipid molecule is likely in a relatively stable state, thereby facilitating its transfer from the membrane into the aqueous periplasm, and back. External energy input may be required depending on whether it is more energetically favorable to have the lipid in the membrane or bound to transport proteins (Fig. 2).
Figure 1.
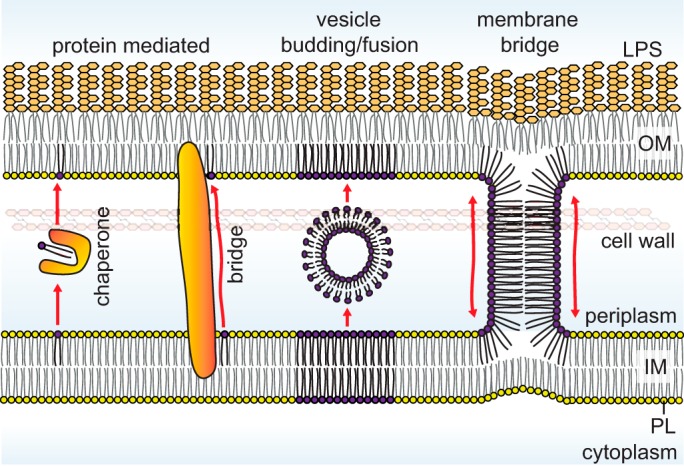
Mechanistic models for lipid trafficking across the periplasm in Gram-negative bacteria. The movement of lipids from the IM to the OM requires shielding of acyl chains from the aqueous periplasmic environment and can involve proteins that act either as chaperones or bridges or may theoretically occur via vesicular transport or hemifusion stalks at sites of membrane juxtaposition.
Figure 2.
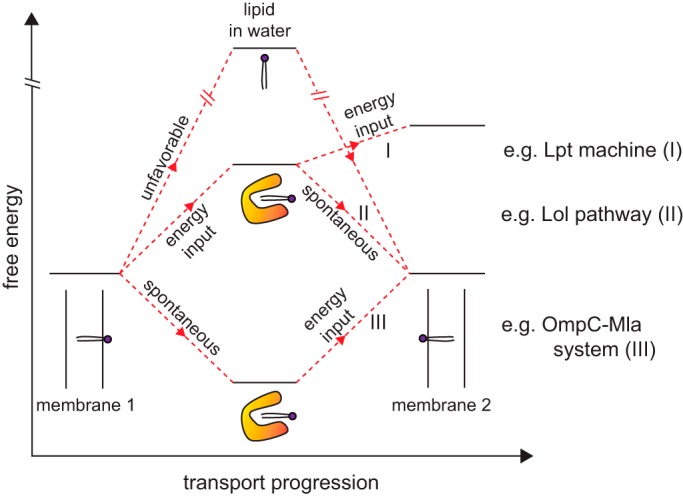
Possible free energy profiles for protein-mediated intermembrane lipid transport. Release of lipids from a membrane (donor) for unassisted diffusion across an aqueous environment to another membrane (recipient) is highly energetically disfavored. In known transport systems, lipid-binding proteins are central to shielding the acyl tails of lipid molecules upon release, giving rise to lipid–protein complexes with energy levels sufficiently close to those of lipids in the membrane environment; this renders lipid transport feasible with or without external energy input, e.g. derived from ATP hydrolysis. Shown here are three different scenarios with distinct energetic requirements at different stages: (I) energy is required to release the lipid molecule from donor membrane as well as to insert it into the recipient membrane, e.g. Lpt machine (Fig. 3); (II) energy is required to release the lipid molecule from the donor membrane only, whereas insertion into recipient membrane is spontaneous, e.g. Lol pathway (Fig. 3); and (III) release from donor membrane is spontaneous (due to high-affinity protein binding) but energy is then required to release the lipid molecule from protein for insertion into recipient membrane, e.g. OmpC–Mla pathway in the context of retrograde transport (Fig. 4). Donor and recipient membranes are labeled as membranes 1 and 2, respectively.
A second possible solution to lipid transport across the periplasm may involve direct exchange of lipids between the two membranes, in ways such that the lipid molecules do not actually leave the membrane environment (Fig. 1). Theoretically, this can occur either via vesicles budding from one membrane and fusing with the target membrane, thereby transferring lipids, or via physical membrane bridges connecting the proximal leaflets of the two membranes, allowing free lipid diffusion. For the former, mechanisms that generate curvature leading to formation of vesicles would be required. For the latter, the two membranes would need to be brought into really-close proximity for hemifusion events to occur. Both pathways would require the assistance of proteins and likely external energy input. At present, there is no evidence for the existence of such transport pathways in bacteria. It has long been suggested that the presence of the peptidoglycan layer and the size of the periplasm (15–20 nm) are not compatible with vesicular transport. Although periplasmic vesicular structures have recently been observed in cryo-tomograms, they were largely found in cells showing signs of envelope stress and may have limited physiological relevance (10). It has also been hypothesized that lipid transport can occur in membrane adhesion zones between the IM and the OM (11). These regions, known as Bayer's patches, could contain proteinaceous or membrane bridges used in lipid trafficking, but they have been highly controversial (12, 13). Recently, intercellular transfer of periplasmic and OM material occurring via transient fusion events between OMs on adjacent cells has been described in Myxococcus sp. (14). The existence of such a pathway for intercellular material exchange suggests that analogous mechanisms may be possible for lipid transport across the periplasm.
Lipoprotein trafficking
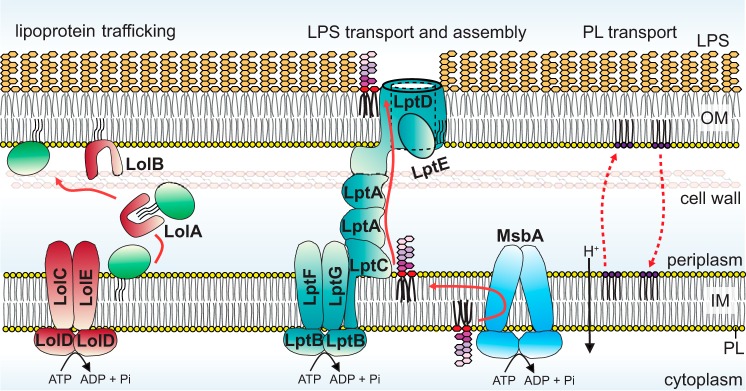
The transport of lipoproteins from the IM to the OM is a classic example for lipid trafficking via soluble lipid–protein intermediates (8). Bacterial lipoproteins are produced with an N-terminal signal peptide that directs secretion across the IM (15). At the periplasmic leaflet of the IM, this signal peptide is processed (16, 17), and the eventual N-terminal cysteine residue is modified with three acyl tails derived from PLs (18–20); this triacyl moiety anchors typically soluble domains of lipoproteins firmly to one leaflet of the bilayer. Lipoproteins destined for the OM are transported across the aqueous periplasm via the Lol pathway, which in Escherichia coli comprises five essential proteins (Fig. 3). The periplasmic protein LolA serves as a lipid chaperone, shielding the acyl tails of lipoproteins from the aqueous environment and shuttling these lipoproteins from the IM to the OM (21). LolCDE constitutes an ATP-binding cassette (ABC) transporter that uses energy from ATP hydrolysis to release lipoproteins from the IM and pass them on to LolA (through LolC) (22, 23). Thus, lipoproteins bound to LolA appear to be in a less stable state compared to when anchored in the IM (Fig. 2). At the OM, LolA hands off the lipoprotein to LolB, itself an OM lipoprotein (Fig. 3) (24). LolA and LolB have similar structures, both containing a large hydrophobic cavity for binding the triacyl moiety of lipoproteins (25). Transfer of lipoprotein from LolA to LolB occurs spontaneously, indicating that the transfer is affinity-driven (26). Finally, LolB inserts the lipoprotein via its acyl tails into the inner leaflet of the OM (27, 28); the mechanism for this step is not known but is also energy-independent, consistent with the idea that the triacyl moiety anchored in the membrane would be the most stable state (Fig. 2).
Figure 3.
Transport pathways of OM lipoproteins and LPS across the cell envelope in Gram-negative bacteria. OM lipoproteins and LPS are transported via a chaperone (Lol pathway) and protein bridge (Lpt machine), respectively. In Lol-mediated transport, ATP hydrolysis is only required to release lipoproteins from the IM. In Lpt-mediated LPS transport, ATP hydrolysis powers LPS transport all the way from the IM to the OM. Bulk PL transport from the IM to the OM depends on the pmf. As opposed to lipoprotein and LPS transport, PL transport across the envelope is bidirectional.
Lipopolysaccharide transport and assembly
LPS transport from the periplasmic side of the IM to the outer leaflet of the OM occurs via a physical protein bridge connecting the two membranes (7). The structure of this glycolipid varies considerably across different Gram-negative species, but in E. coli it typically comprises up to hundreds of sugars anchored to the membrane by six acyl tails (29). LPS is synthesized at the cytoplasmic leaflet of the IM as “rough” LPS (Ra form: ∼10 core sugars linked to lipid A), flipped across the IM by an ABC transporter MsbA (30, 31), and converted to “smooth” LPS via the addition of O-antigen polysaccharides at the periplasmic leaflet of the IM (32). The journey of this completed LPS structure to the cell surface is then mediated by seven essential Lpt proteins (Fig. 3) (7). Here, the IM ABC transporter LptBFGC (33–35) is physically connected to the OM translocon LptDE (36) through interactions with the periplasmic protein LptA (33, 37). The structurally homologous β-jelly roll domains of LptC (38), LptA (39), and the N-terminal domain of LptD (40) interact in a “head–to–tail” fashion to form a protein bridge (41, 42), providing a continuous hydrophobic groove to accommodate the multiple acyl tails of LPS during transit across the periplasm (39). With six acyl tails, E. coli LPS is presumably in its most stable state in a membrane. Therefore, LptBFG harnesses energy derived from ATP hydrolysis to extract LPS from the IM and loads them onto LptC (43–45). Recent in vitro reconstitutions of this system demonstrate that LPS transfer from LptC to LptA, and then to LptD, along the hydrophobic groove also require ATP hydrolysis (46, 47). LptBFG essentially powers direct transport of LPS from the IM all the way to the outer leaflet of the OM (Fig. 2). At the OM, the LptDE translocon assembles incoming LPS into the outer leaflet (Fig. 3). Here, the enormous polysaccharide chain of the LPS molecule likely crosses the OM via the large hydrophilic lumen of the LptD β-barrel domain, which is partially constricted by the lipoprotein LptE (48, 49), whereas its six acyl tails traverse along the side wall of the β-barrel directly into the outer leaflet (40, 50). The placement of LPS onto the cell surface occurs against a concentration gradient and comes at an entropic cost. Strong lateral interactions between LPS molecules may provide part of the driving force for establishing the resulting lipid asymmetry in the OM; however, because ∼106 LPS molecules need to be assembled at the cell surface in every cell cycle (∼20 min for E. coli), it is not surprising that the final step of LPS translocation across the OM also requires ATP hydrolysis at the IM (51).
Phospholipid transport across the cell envelope
Despite being the most basic building block of lipid bilayers, the transport of PLs from the IM to the OM is the least understood. In E. coli, the three major PL species are phosphatidylethanolamine (PE) 75%, phosphatidylglycerol (PG) 20%, and cardiolipin (CL) 5% (52). Some earlier studies suggest that PL compositions of the IM and the OM may be slightly different, with the OM enriched in PE (53, 54). Synthesis of PLs begins with phosphatidic acid (PA), which is converted to cytidine-diphosphate diacylglycerol (CDP-DAG), a common intermediate toward PE and PG (55, 56). CDP-DAG is either converted to phosphatidylserine (PS) before undergoing decarboxylation to give PE (57) or is converted to phosphatidylglycerol phosphate, which undergoes subsequent dephosphorylation to give PG (56, 58). CL is then produced from the condensation of two molecules of PG or one molecule each of PE and PG (59, 60). After their synthesis in the cytoplasmic side of the IM, PLs are presumably flipped across the IM by yet-to-be-identified transporters.
To get to the inner leaflet of the OM where they are predominantly located, PLs need to be extracted from the IM and transported across the aqueous periplasmic space (Fig. 3). As with lipoprotein and LPS transport, this process also requires energy input. Interestingly, even though a minimal requirement for ATP hydrolysis cannot be ruled out, it has been demonstrated that PE translocation from the IM to the OM (anterograde transport) requires the proton motive force (pmf) across the IM (54). Unlike lipoprotein and LPS transport, PL transport has also been shown to be bidirectional (54, 61, 62); PS specifically delivered to the OM, either endogenously via anterograde transport (62) or exogenously by vesicle fusion (61), are transported back to the IM (retrograde transport), where they become processed to give PE. To date, the pathway(s) for anterograde transport of bulk PLs (predominantly PE and PG in E. coli) has not been identified, whereas several systems have been implicated in retrograde PL transport.
Anterograde (IM–to–OM) phospholipid transport
The movement of PLs from the IM to the OM is rapid. It has been demonstrated ∼40 years ago that PE pulse-labeled with radioactivity is translocated to the OM with a t½ of minutes (54). This process depends on the presence of the pmf across the IM. Although no protein has been identified for this transport activity, it has been suggested that PL transport to the OM is fundamentally different from the processes identified for LPS and OM lipoproteins (63). Specifically, lipoproteins destined for the OM can be released from the IM when E. coli spheroplasts are exposed to LolA-containing periplasmic extracts. LPS transport in spheroplasts from the IM to remnants of the OM is also intact. In contrast, PLs are neither released from the IM in the presence of periplasmic extracts nor transported to the OM in spheroplasts. Given that E. coli spheroplasts maintain an intact pmf across the IM, thus allowing study of other known pmf-dependent processes (64–66), these observations suggest that anterograde PL transport is possibly independent of a soluble chaperone or a stable trans-envelope protein bridge.
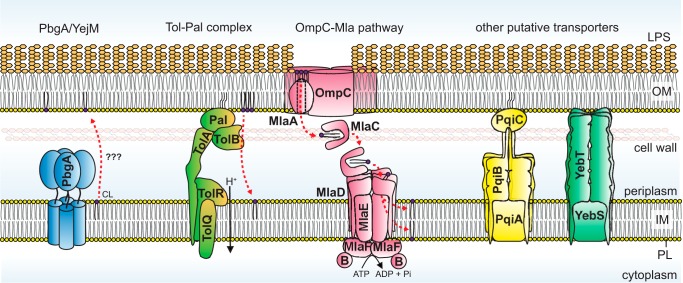
Recently, a couple of studies suggested that an IM protein PbgA/YejM may be involved in CL translocation to the OM (Fig. 4) (67, 68). Using quantitative lipidomics, it was shown that functional PbgA/YejM is required for the enrichment of CL in the OM in Salmonella Typhimurium, which occurs when the stress-response system PhoPQ is activated (67). Curiously, a similar trend of PbgA/YejM-dependent increase of CL was also observed in the IM. Although these results indicate perhaps an overall increase in cellular CL levels, it was thought that this IM protein functions to deliver CL to the OM. The same idea is supported by another study of PbgA/YejM in Shigella flexneri (68), albeit differential detergent extraction and nonquantitative TLC were employed here for membrane separation and lipid analysis, respectively, confounding interpretation. PbgA/YejM is a tetrameric protein with an essential five-helical–bundle transmembrane domain linked at the C terminus to a nonessential periplasmic domain (67, 69). Deletion of the periplasmic domain gives rise to OM permeability defects and reduced LPS levels (69). Why it is required for viability is not known, but overexpression of AcpT, a phosphopantetheinyl transferase, has been shown to suppress lethality in E. coli cells lacking YejM (69). Although the exact role of PbgA/YejM in PL transport still requires thorough investigation, it is likely that this protein plays a critical function in bacterial lipid biology.
Figure 4.
Protein systems implicated in PL transport in Gram-negative bacteria. Mechanism(s) for bulk anterograde PL transport are elusive, but the IM protein PbgA/YejM has been suggested to be involved in the transport of CLs to the OM. Both the Tol–Pal complex and the OmpC–Mla system are important for retrograde transport of PLs from the inner and outer leaflet of the OM, respectively. The directionality of the latter pathway has been a subject of controversy. Pqi and Yeb protein systems are putative PL transporters, but their biological relevant substrate(s) and functions are unknown. For each of these systems, additional experimental evidence is necessary to ascertain direct involvement in and/or directionality of PL transport.
Retrograde (OM–to–IM) phospholipid transport
It is known that PL transport across the cell envelope is bidirectional (54, 61, 62). By monitoring PS–to–PE conversion, a process that only takes place in the IM, it was demonstrated that radioactive PS accumulated in the OM can be translocated back to the IM (61, 62). However, despite making these observations close to 40 years ago, the significance and possible mechanism(s) of retrograde PL transport have not been revealed until recently. Below, we describe two molecular systems implicated in the retrograde transport of PLs (Fig. 4). The Tol–Pal complex is thought to be involved in the movement of bulk PLs presumably from the inner leaflet of the OM to the IM (70), whereas the OmpC–Mla system is believed to maintain OM lipid asymmetry by transporting a small population of PLs that is mislocalized in the outer leaflet of the OM back to the IM (71, 72).
The Tol–Pal complex
The Tol–Pal complex is a trans-envelope system highly conserved in Gram-negative bacteria (Fig. 4) (73, 74). It comprises the TolQRA and TolB–Pal sub-complexes in the IM and the OM, respectively, and these complexes interact with each other in a manner dependent on the pmf (75). The Tol–Pal complex has always been thought to be important for maintaining OM integrity and stability, as mutants are highly sensitive to antibiotics and detergents, leak periplasmic contents, and produce large amounts of OM vesicles (73, 76). However, the exact function of this complex, and how its absence gives rise to the observed phenotypes, was not known. The Tol–Pal complex was also shown to be important for the invagination of the OM during cell division (77, 78).
A recent systematic study by our group has now revealed that the Tol–Pal complex plays an important role in the maintenance of OM lipid homeostasis in E. coli (70). Using steady-state radioactive labeling of cellular lipids, it was demonstrated that cells lacking the Tol–Pal complex maintain WT levels of LPS but accumulate excess PLs in the OM. This molecular defect, which is also observed in S. Typhimurium (79), likely destabilizes the OM and can potentially account for the OM phenotypes described above. It was further established that PL buildup in the OM in tol–pal mutants is due to defects in retrograde PL transport. Here, by monitoring the turnover of pulse-labeled radioactive anionic PLs (PG/CL) in the OM through processes that occur only in the IM, it was shown that a functional Tol–Pal complex is required for efficient transport of bulk PLs from the OM back to the IM. This assay, which couples PL transport to turnover, is similar to those previously employed to demonstrate retrograde transport in cells (62). The Tol–Pal complex thus represents the first molecular machine implicated in bulk PL transport.
Even so, it is still not clear whether the Tol–Pal complex directly mediates PL transport between the two membranes. One possibility is that the Tol–Pal complex indirectly affects the function of a true PL transport system. Cells lacking the Tol–Pal complex exhibit delayed invagination of the OM during division (77); the resulting wider distance between the two membranes (around the division site) may somehow give rise to nonspecific effects on other processes, including retrograde PL transport. Alternatively, the Tol–Pal complex may physically move PLs, which could potentially happen in three ways. First, parts of the complex might bind PLs. However, PL-binding activities for various periplasmic domains of the complex have not been detected in vitro.4 Second, the complex may interact with a PL-binding protein, a yet-to-be-identified component of this system. Third, the Tol–Pal complex may bring the two membranes in close proximity to allow hemifusion and thus lipid diffusion to occur (i.e. Bayer's patch/bridge model). Each of these speculative transport mechanisms needs to account for the proposed directionality of transport and may require energy input. In this regard, the TolQRA complex is homologous to the ExbBD–TonB (80, 81), MotAB (80, 82), and AglQRS (83) systems, which transduce energy from the pmf for the generation of force involved in siderophore uptake, flagella motility, and cell gliding, respectively. How this force is utilized in the Tol–Pal complex to maintain OM lipid homeostasis requires further investigation.
The OmpC–Mla system
The OmpC–Mla system has been shown to play an important role in the maintenance of OM lipid asymmetry in Gram-negative bacteria (71, 72). It comprises the OmpC–MlaA complex at the OM, a periplasmic protein MlaC, and an ABC transporter MlaFEDB at the IM (Fig. 4). Removing any member of the OmpC–Mla system results in aberrant accumulation of PLs at the outer leaflet of the OM in E. coli, thereby disrupting lipid asymmetry; this system is believed to remove the mislocalized PLs from the OM and transport them back to the IM (71, 72). Transport between the two membranes occurs via a soluble PL–protein intermediate. MlaC has been crystallized with a PL bound, revealing a hydrophobic pocket that shields both acyl tails of the PL molecule from the aqueous periplasm (84). In addition, MlaC has been shown to interact with both OM and IM complexes in vitro (84) and in vivo (85). Although removing MlaC does not cause observable defects in retrograde PL transport in cells, it was recently demonstrated that overproduction of MlaC together with the MlaFEDB complex is able to partially rescue defects in PL transport in cells lacking the Tol–Pal complex (70). Therefore, the OmpC–Mla system does in fact transport PLs in a retrograde fashion, even though it may only be important to remove a small subset of PLs from the OM, particularly those mislocalized to the outer leaflet.
The OM lipoprotein MlaA forms complexes with trimeric porins, including OmpC and OmpF, in the OM of E. coli, but it appears that only the OmpC–MlaA complex is important for the maintenance of lipid asymmetry (72). This complex is proposed to extract PLs from the outer leaflet of the OM presumably without external energy input. Despite being a lipoprotein, recent structural and biochemical studies revealed that MlaA is really an integral membrane protein that binds porin trimers within the bilayer, at one or more of its dimeric interfaces (86, 87). Interestingly, MlaA forms a hydrophilic channel across the OM, likely providing a path for PL translocation across the OM. In this context, even though a single amino acid change gives rise to OM lipid asymmetry defects, the exact function of OmpC within the complex remains unclear (87). Based on the architecture of MlaA, it has been proposed that PLs extracted from the outer leaflet of the OM do not enter the inner leaflet but are delivered directly to MlaC in the periplasm in an energy-independent fashion. Such a pathway circumvents the need to work against a PL concentration gradient at the OM; however, it necessitates that PLs in the outer leaflet of the OM are in a less stable state compared with those bound to MlaC. In support of this idea, molecular dynamic simulations have shown that the size of the hydrophobic pocket of MlaC can change with PL occupancy (88), suggesting MlaC may have conformational flexibility that possibly allows it to maximize its affinity for PLs via an induced fit mechanism. High affinity binding of PLs by MlaC is also corroborated by the observation that MlaC does not spontaneously transfer PLs to the second lipid-binding protein in this system MlaD in vitro (85).
PLs are delivered from MlaC into the IM via the MlaFEDB complex, which is an ABC transporter (89). MlaF and MlaE represent the nucleotide-binding domain and transmembrane domain of the transporter, respectively. Biochemical characterization of the complex elucidated functions of auxiliary proteins MlaB and MlaD (89). MlaB is important for the assembly of the transporter and its ATP hydrolytic activity, whereas MlaD forms stable hexamers that bind PLs in vitro (84, 89). The crystal structure of the periplasmic domain of MlaD reveals six protomers organized in a donut-shaped architecture containing a central hydrophobic pore (84), presumably for interactions with the acyl tails of PLs. MlaD also interacts directly with MlaC in cells (85), indicating that MlaC transfers PLs to MlaD within the MlaFEDB complex. Here, because MlaC has high affinity for PLs (85), the energy derived from ATP hydrolysis in the complex may then be required to release the bound PLs and/or activate transfer to MlaD and then into the IM. Whether these PLs get flipped back to the cytoplasmic leaflet of the IM is not known.
There has in fact been some controversy regarding the directionality of lipid transport for the OmpC–Mla system. ABC transporters found in chloroplasts (TGD2) and mycobacteria (Mce1/4 complexes) but homologous to the MlaFEDB complex are known to be involved in PA and fatty acid/cholesterol uptake, respectively (90, 91). In E. coli, the OmpC–Mla system was thus initially annotated to function in retrograde PL transport, especially given that overexpressing OM phospholipase PldA rescues asymmetry defects in the OM of ompC–mla mutant strains (71, 72); in this scenario, if outer leaflet PLs in the OM cannot be removed (by transport), they can be degraded instead. More recently, evolution experiments revealed that removing the Mla system in Acinetobacter baumannii improves growth and restores OM barrier function in strains that do not make lipooligosaccharides (LOS) (92); this makes sense in the context of retrograde PL transport given that cells would require sufficient PLs in the outer leaflet of the OM when no LOS is made. Although these studies inferred function solely from genetic interactions, it has also been shown that overexpressing the mlaFEDCB operon partially rescues retrograde PL transport defects in tol–pal mutants (70). Therefore, both genetics and biochemical data support a role of the OmpC–Mla system in retrograde PL transport. Interestingly, there have also been some recent observations supporting anterograde PL transport. A separate group characterizing mla mutants in A. baumannii found that these strains have severely compromised OM function due to reduced PL content in the OM. By monitoring the rate of appearance of newly-synthesized PLs relative to existing PLs in the IMs and OMs using MS, the authors propose a role for Mla proteins in anterograde PL transport (93). Unfortunately, because existing PL levels are supposedly lower in the OMs of mla mutants, it may not be straightforward to infer about overall changes in PL transport from the rates of change of new versus existing PLs in these strains (as compared with that in WT cells). In vitro reconstitution experiments also suggest a role for the OmpC–Mla system in anterograde transport (94). It has been shown that the complete MlaFEDB complex may transfer PLs spontaneously to MlaC in vitro. However, proper inactive enzyme controls were lacking. Furthermore, this in vitro reaction did not appear to be dependent on or modulated by ATP hydrolysis, which is quite puzzling indeed for an ABC transporter. Overall, additional studies are definitely needed to provide more clarity to this problem.
Other putative lipid transporters
The periplasmic domain of MlaD contains the mammalian cell entry (MCE) domain, which is widely conserved in proteobacteria and actinomycetes (95). Based on homology, two other MCE domain proteins have been described in E. coli, and they have recently been proposed to also be involved in lipid transport (Fig. 4) (95, 96). PqiB and YebT contain three and seven MCE domains, respectively, and both have been co-purified with PLs (84). Structural characterization of these proteins revealed that they form hexameric assemblies via their MCE domains, giving rise to structures that can span the entire periplasmic space between the IM and the OM (84), potentially facilitating transport of lipid substrates or other hydrophobic molecules. Even though cells lacking both proteins have some perturbations to the OM (95, 96), the true functions of these proteins are not yet clear.
OM lipid homeostasis via retrograde PL transport
The processes of lipid transport are inherent for the synthesis and maintenance of the OM in Gram-negative bacteria. Anterograde PL transport is essential for OM biogenesis. In contrast, the role(s) of retrograde PL transport pathways is less clear until recently, where they have been implicated in OM lipid homeostasis (70). The current model (proposed by our group) suggests cells transport more PLs than required to the OM to fill up spaces that may arise from changes in the OM; this process ensures that the bilayer is always complete. To maintain OM stability, however, excess PLs must then be continuously removed, particularly via retrograde transport to the IM in a manner dependent on the Tol–Pal complex. This model makes sense in the context of unidirectional (IM–to–OM) transport of LPS, β-barrel proteins, and lipoproteins and directly alleviates the need for fine control over the levels of these components in the OM.
Lipid homeostasis in the OM also impacts structural organization such as lipid asymmetry. It is perhaps logical to assume that both bulk anterograde and retrograde PL transport delivers PLs to and removes them from the inner leaflet of the OM, respectively. In the absence of the Tol–Pal complex, the cell would accumulate a large excess of PLs in the inner leaflet of the OM, which generates instability in the bilayer. This presumably allows PLs to flip across the OM, leading to subsequent accumulation in the outer leaflet and thus loss of lipid asymmetry (70). Even in a WT cell (with an intact Tol–Pal complex), new OM needs to be continuously synthesized for growth and division. As such, it is conceivable that anterograde PL transport would still be faster than retrograde transport; this scenario may inevitably lead to a slight buildup of PLs in the inner leaflet of the OM and subsequently the outer leaflet. Besides employing PL-degrading enzymes (PldA (97) and PagP (98)) to correct such perturbations in OM lipid asymmetry, retrograde PL transport mediated by the OmpC–Mla system additionally contributes to OM homeostasis by removing this small amount of PLs that ended up aberrantly in the outer leaflet of the OM (71, 72).
Conclusions and outlook
There are still huge gaps in our knowledge of OM biogenesis. Although significant advances have been made in understanding OM lipoprotein trafficking, LPS transport and assembly, and β-barrel protein folding over the past 2 decades, we have made little progress in deciphering PL transport across the cell envelope. This contrasts with the major advances made toward elucidating nonvesicular PL transport pathways within single organelles (i.e. chloroplast (90) and mitochondria (99)) or between separate ones (i.e. membrane contact sites (100)) in eukaryotic cells. The obvious major mystery in Gram-negative bacteria cell envelope biology is anterograde PL transport. Why these systems have remained elusive may be due to the lack of targeted genetic approaches to discover factors. One component of the β-barrel protein-folding machine (BamB) was identified in a chemical genetic selection as a mutation that rescues OM leakiness in an lptD mutant (101), which has now been shown to be a β-barrel assembly defect (102, 103). One may therefore argue that a genetic selection targeted at correcting OM lipid dyshomeostasis, such as those found in tol–pal strains (70) or gain–of–function mlaA mutants (104), could instead lead to new information regarding PL transport across the cell envelope. Genetic screens based specifically on direct detection of defects in OM lipid homeostasis and/or lipid asymmetry may also aid in the identification of PL transport pathways. It is especially intriguing what the (main) anterograde PL transport system could be, given that the process differs from OM lipoprotein and LPS transport in its requirement for energy derived from the pmf (54).
The functions of various proposed PL transport systems should also be thoroughly investigated. It is not yet clear whether the Tol–Pal complex directly mediates retrograde PL transport nor is it known whether PbgA/YejM, PqiB, and YebT are in fact lipid transporters. The transport directionality of the OmpC–Mla system also requires further clarification. In general, functional assignment of PL transport systems should include both cell-based and in vitro assays monitoring PL movement between the two membranes. The cell-based PL transport assays are clearly nontrivial (even in model organisms like E. coli), and extra caution should be taken to ensure proper separation of and minimal lipid mixing between IM and OM fragments during analysis. In this regard, current studies, including ours, employ IM and OM integral membrane proteins and LPS (very different properties from PLs) as molecular markers in membrane separation, which cannot truly inform the extent of PL mixing, if any. Therefore, methods that may influence the propensity of PL mixing between membranes should be avoided. Specifically, removal of the cell wall layer and divalent cations by use of lysozyme (2) and EDTA (106), respectively, is known to disrupt lipid asymmetry in the OM, and it could have other unexpected effects on membrane stability and thus PL mixing between membranes. In addition, cell lysis by sonication or French press at high pressures (>12,000 p.s.i.), albeit after spheroplasting, has been shown to cause significant membrane mixing or cross-contamination (107). Careful evaluation of protocols for cell lysis and/or membrane separation would go a long way to ensure more robust and reliable tracking of PL movement across the cell envelope. Beyond these intricate cell-based assays, in vitro reconstitutions of each putative PL transport system would ultimately be required for definitive assignment of function. In vitro approaches to study lipid transfer between two membranes are likely highly challenging yet feasible, as exemplified by the recent elegant demonstration of intermembrane LPS transport using purified Lpt proteins and artificial membranes (46).
Gram-negative bacteria are intrinsically resistant to many clinically-used antibiotics due in part to the OM permeability barrier. The OM is also essential for bacterial growth and therefore is a great molecular target for novel antibiotic intervention. To this end, small molecule inhibitors against the Lol (108, 109) and MsbA/Lpt pathways (105, 110) have already been identified. Continued efforts in deciphering PL-trafficking processes will ultimately yield new targets that can be exploited for combating Gram-negative bacterial pathogens.
Acknowledgements
S.-S.C. would like to thank the ASBMB for the 2019 Walter A. Shaw Young Investigator Award, and the JBC editorial board for the invitation to write this article. S.-S.C. is deeply indebted to his mentors Daniel Kahne (Harvard U) and Jonathan Beckwith (Harvard Medical Sch) for providing exceptional training, friendship and support throughout his career. He is also very grateful to postdocs and students of the Chng group past and present for their commitment, patience and hard work. Finally, S.-S.C. would like to acknowledge his award nominator Jean-Francois Collet (U catholique de Louvain) and referees Hiroshi Nikaido (UC Berkeley), Tracy Palmer (Newcastle U) and M. Stephen Trent (U Georgia Atlanta) for their generosity and support.
This work was supported by Singapore Ministry of Health National Medical Research Council under its Cooperative Basic Research Grant NMRC/CBRG/0072/2014 and its Open Fund Individual Research Grant MOH-000145 (both to S.-S. C.). The authors declare that they have no conflicts of interest with the contents of this article.
A. Z. H. Tan and S.-S. Chng, unpublished observations.
- IM
- inner membrane
- OM
- outer membrane
- OMP
- outer membrane protein
- PL
- phospholipid
- LPS
- lipopolysaccharide
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PA
- phosphatidic acid
- CL
- cardiolipin
- CDP-DAG
- cytidine-diphosphate diacylglycerol
- LOS
- lipooligosaccharide
- pmf
- proton motive force
- ABC
- ATP-binding cassette
- MCE
- mammalian cell entry.
References
- 1. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mühlradt P. F., and Golecki J. R. (1975) Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur. J. Biochem. 51, 343–352 10.1111/j.1432-1033.1975.tb03934.x [DOI] [PubMed] [Google Scholar]
- 3. Kamio Y., and Nikaido H. (1976) Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 10.1021/bi00657a012 [DOI] [PubMed] [Google Scholar]
- 4. Funahara Y., and Nikaido H. (1980) Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J. Bacteriol. 141, 1463–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Konovalova A., Kahne D. E., and Silhavy T. J. (2017) Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 10.1146/annurev-micro-090816-093754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hagan C. L., Silhavy T. J., and Kahne D. (2011) β-barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 10.1146/annurev-biochem-061408-144611 [DOI] [PubMed] [Google Scholar]
- 7. Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., and Kahne D. (2016) Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 10.1038/nrmicro.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okuda S., and Tokuda H. (2011) Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259 10.1146/annurev-micro-090110-102859 [DOI] [PubMed] [Google Scholar]
- 9. Tanford C. (1978) The hydrophobic effect and the organization of living matter. Science 200, 1012–1018 10.1126/science.653353 [DOI] [PubMed] [Google Scholar]
- 10. Dobro M. J., Oikonomou C. M., Piper A., Cohen J., Guo K., Jensen T., Tadayon J., Donermeyer J., Park Y., Solis B. A., Kjær A., Jewett A. I., McDowall A. W., Chen S., Chang Y. W., et al. (2017) Uncharacterized bacterial structures revealed by electron cryotomography. J. Bacteriol. 199, e00100–17 10.1128/JB.00100-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bayer M. E. (1968) Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 53, 395–404 10.1099/00221287-53-3-395 [DOI] [PubMed] [Google Scholar]
- 12. Kellenberger E. (1990) The “Bayer bridges” confronted with results from improved electron microscopy methods. Mol. Microbiol. 4, 697–705 10.1111/j.1365-2958.1990.tb00640.x [DOI] [PubMed] [Google Scholar]
- 13. Bayer M. E. (1991) Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 107, 268–280 10.1016/1047-8477(91)90052-X [DOI] [PubMed] [Google Scholar]
- 14. Ducret A., Fleuchot B., Bergam P., and Mignot T. (2013) Direct live imaging of cell–cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. Elife 2, e00868 10.7554/eLife.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kudva R., Denks K., Kuhn P., Vogt A., Müller M., and Koch H. G. (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534 10.1016/j.resmic.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 16. Hussain M., Ichihara S., and Mizushima S. (1982) Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257, 5177–5182 [PubMed] [Google Scholar]
- 17. Tokunaga M., Tokunaga H., and Wu H. C. (1982) Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc. Natl. Acad. Sci. U.S.A. 79, 2255–2259 10.1073/pnas.79.7.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sankaran K., and Wu H. C. (1994) Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 19. Jackowski S., and Rock C. O. (1986) Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J. Biol. Chem. 261, 11328–11333 [PubMed] [Google Scholar]
- 20. Gupta S. D., and Wu H. C. (1991) Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiol. Lett. 62, 37–41 [DOI] [PubMed] [Google Scholar]
- 21. Matsuyama S., Tajima T., and Tokuda H. (1995) A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14, 3365–3372 10.1002/j.1460-2075.1995.tb07342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yakushi T., Yokota N., Matsuyama S., and Tokuda H. (1998) LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J. Biol. Chem. 273, 32576–32581 10.1074/jbc.273.49.32576 [DOI] [PubMed] [Google Scholar]
- 23. Yakushi T., Masuda K., Narita S., Matsuyama S., and Tokuda H. (2000) A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 10.1038/35008635 [DOI] [PubMed] [Google Scholar]
- 24. Matsuyama Si, Yokota N., and Tokuda H. (1997) A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 10.1093/emboj/16.23.6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., and Miki K. (2003) Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209 10.1093/emboj/cdg324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okuda S., and Tokuda H. (2009) Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl. Acad. Sci. U.S.A. 106, 5877–5882 10.1073/pnas.0900896106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsukahara J., Mukaiyama K., Okuda S., Narita S., and Tokuda H. (2009) Dissection of LolB function: lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504 10.1111/j.1742-4658.2009.07156.x [DOI] [PubMed] [Google Scholar]
- 28. Hayashi Y., Tsurumizu R., Tsukahara J., Takeda K., Narita S., Mori M., Miki K., and Tokuda H. (2014) Roles of the protruding loop of factor B essential for the localization of lipoproteins (LolB) in the anchoring of bacterial triacylated proteins to the outer membrane. J. Biol. Chem. 289, 10530–10539 10.1074/jbc.M113.539270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raetz C. R., and Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doerrler W. T., Gibbons H. S., and Raetz C. R. (2004) MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109 10.1074/jbc.M408106200 [DOI] [PubMed] [Google Scholar]
- 31. Mi W., Li Y., Yoon S. H., Ernst R. K., Walz T., and Liao M. (2017) Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 10.1038/nature23649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitfield C., and Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 10.1146/annurev-biochem-060713-035600 [DOI] [PubMed] [Google Scholar]
- 33. Sperandeo P., Cescutti R., Villa R., Di Benedetto C., Candia D., Dehò G., and Polissi A. (2007) Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253 10.1128/JB.01126-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sperandeo P., Lau F. K., Carpentieri A., De Castro C., Molinaro A., Dehò G., Silhavy T. J., and Polissi A. (2008) Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 10.1128/JB.00270-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruiz N., Gronenberg L. S., Kahne D., and Silhavy T. J. (2008) Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 10.1073/pnas.0801196105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu T., McCandlish A. C., Gronenberg L. S., Chng S.-S., Silhavy T. J., and Kahne D. (2006) Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 10.1073/pnas.0604744103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chng S.-S., Gronenberg L. S., and Kahne D. (2010) Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 10.1021/bi100493e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran A. X., Dong C., and Whitfield C. (2010) Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 10.1074/jbc.M110.144709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suits M. D., Sperandeo P., Dehò G., Polissi A., and Jia Z. (2008) Novel structure of conversed Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380, 476–488 10.1016/j.jmb.2008.04.045 [DOI] [PubMed] [Google Scholar]
- 40. Qiao S., Luo Q., Zhao Y., Zhang X. C., and Huang Y. (2014) Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 10.1038/nature13484 [DOI] [PubMed] [Google Scholar]
- 41. Freinkman E., Okuda S., Ruiz N., and Kahne D. (2012) Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806 10.1021/bi300592c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Villa R., Martorana A. M., Okuda S., Gourlay L. J., Nardini M., Sperandeo P., Dehò G., Bolognesi M., Kahne D., and Polissi A. (2013) The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 10.1128/JB.02057-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narita S., and Tokuda H. (2009) Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164 10.1016/j.febslet.2009.05.051 [DOI] [PubMed] [Google Scholar]
- 44. Li Y., Orlando B. J., and Liao M. (2019) Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 10.1038/s41586-019-1025-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Owens T. W., Taylor R. J., Pahil K. S., Bertani B. R., Ruiz N., Kruse A. C., and Kahne D. (2019) Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 10.1038/s41586-019-1039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okuda S., Freinkman E., and Kahne D. (2012) Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 10.1126/science.1228984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sherman D. J., Xie R., Taylor R. J., George A. H., Okuda S., Foster P. J., Needleman D. J., and Kahne D. (2018) Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 10.1126/science.aar1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chng S.-S., Ruiz N., Chimalakonda G., Silhavy T. J., and Kahne D. (2010) Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368 10.1073/pnas.0912872107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Freinkman E., Chng S.-S., and Kahne D. (2011) The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 108, 2486–2491 10.1073/pnas.1015617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong H., Xiang Q., Gu Y., Wang Z., Paterson N. G., Stansfeld P. J., He C., Zhang Y., Wang W., and Dong C. (2014) Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 10.1038/nature13464 [DOI] [PubMed] [Google Scholar]
- 51. Xie R., Taylor R. J., and Kahne D. (2018) Outer membrane translocon communicates with inner membrane ATPase to stop lipopolysaccharide transport. J. Am. Chem. Soc. 140, 12691–12694 10.1021/jacs.8b07656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raetz C. R., and Dowhan W. (1990) Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 265, 1235–1238 [PubMed] [Google Scholar]
- 53. Osborn M. J., Gander J. E., Parisi E., and Carson J. (1972) Mechanism of assembly of the outer membrane of Salmonella typhimurium: isolation and characterization of the cytoplasmic and outer membrane. J. Biol. Chem. 247, 3962–3972 [PubMed] [Google Scholar]
- 54. Donohue-Rolfe A. M., and Schaechter M. (1980) Translocation of phospholipids from the inner to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 77, 1867–1871 10.1073/pnas.77.4.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanfer J., and Kennedy E. P. (1963) Metabolism and function of bacterial lipids. I. Metabolism of phospholipids in Escherichia coli B. J. Biol. Chem. 238, 2919–2922 [PubMed] [Google Scholar]
- 56. Kanfer J., and Kennedy E. P. (1964) Metabolism and function of bacterial lipids. II. Biosynthesis of phospholipids in Escherichia coli. J. Biol. Chem. 239, 1720–1736 [PubMed] [Google Scholar]
- 57. Kanfer J. N., and Kennedy E. P. (1962) Synthesis of phosphatidylserine by Escherichia coli. J. Biol. Chem. 237, PC270–PC271 [PubMed] [Google Scholar]
- 58. Lu Y. H., Guan Z., Zhao J., and Raetz C. R. (2011) Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 286, 5506–5518 10.1074/jbc.M110.199265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hirschberg C. B., and Kennedy E. P. (1972) Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 69, 648–651 10.1073/pnas.69.3.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan B. K., Bogdanov M., Zhao J., Dowhan W., Raetz C. R., and Guan Z. (2012) Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 10.1073/pnas.1212797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jones N. C., and Osborn M. J. (1977) Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J. Biol. Chem. 252, 7405–7412 [PubMed] [Google Scholar]
- 62. Langley K. E., Hawrot E., and Kennedy E. P. (1982) Membrane assembly: movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. J Bacteriol. 152, 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tefsen B., Geurtsen J., Beckers F., Tommassen J., and de Cock H. (2005) Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 280, 4504–4509 10.1074/jbc.M409259200 [DOI] [PubMed] [Google Scholar]
- 64. Harms N., Koningstein G., Dontje W., Muller M., Oudega B., Luirink J., and de Cock H. (2001) The early interaction of the outer membrane protein PhoE with the periplasmic chaperon Skp occurs at the cytoplasmic membrane. J. Biol. Chem. 276, 18804–18811 10.1074/jbc.M011194200 [DOI] [PubMed] [Google Scholar]
- 65. Ollis A. A., and Postle K. (2012) ExbD mutants define initial stages in TonB energization. J. Mol. Biol. 415, 237–247 10.1016/j.jmb.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rubino F. A., Kumar S., Ruiz N., Walker S., and Kahne D. E. (2018) Membrane potential is required for MurJ function. J. Am. Chem. Soc. 140, 4481–4484 10.1021/jacs.8b00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dalebroux Z. D., Edrozo M. B., Pfuetzner R. A., Ressl S., Kulasekara B. R., Blanc M. P., and Miller S. I. (2015) Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451 10.1016/j.chom.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rossi R. M., Yum L., Agaisse H., and Payne S. M. (2017) Cardiolipin synthesis and outer membrane localization are required for Shigella flexneri virulence. MBio 8, e01199–17 10.1128/mBio.01199-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. De Lay N. R., and Cronan J. E. (2008) Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 178, 1327–1337 10.1534/genetics.107.081836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shrivastava R., Jiang X., and Chng S.-S. (2017) Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol. 106, 395–408 10.1111/mmi.13772 [DOI] [PubMed] [Google Scholar]
- 71. Malinverni J. C., and Silhavy T. J. (2009) An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014 10.1073/pnas.0903229106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chong Z. S., Woo W. F., and Chng S.-S. (2015) Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol. 98, 1133–1146 10.1111/mmi.13202 [DOI] [PubMed] [Google Scholar]
- 73. Lloubès R., Cascales E., Walburger A., Bouveret E., Lazdunski C., Bernadac A., and Journet L. (2001) The Tol–Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529 10.1016/S0923-2508(01)01226-8 [DOI] [PubMed] [Google Scholar]
- 74. Sturgis J. N. (2001) Organisation and evolution of the tol-pal gene cluster. J. Mol. Microbiol. Biotechnol. 3, 113–122 [PubMed] [Google Scholar]
- 75. Cascales E., Gavioli M., Sturgis J. N., and Lloubès R. (2000) Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38, 904–915 10.1046/j.1365-2958.2000.02190.x [DOI] [PubMed] [Google Scholar]
- 76. Bernadac A., Gavioli M., Lazzaroni J. C., Raina S., and Lloubès R. (1998) Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180, 4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gerding M. A., Ogata Y., Pecora N. D., Niki H., and de Boer P. A. (2007) The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025 10.1111/j.1365-2958.2006.05571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Petiti M., Serrano B., Faure L., Lloubes R., Mignot T., and Duché D. (2019) Tol energy-driven localization of pal and anchoring to the peptidoglycan promote outer membrane constriction. J. Mol. Biol. 431, 3275–3288 10.1016/j.jmb.2019.05.039 [DOI] [PubMed] [Google Scholar]
- 79. Masilamani R., Cian M. B., and Dalebroux Z. D. (2018) Salmonella Tol–Pal reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect. Immun. 86, e00173–18 10.1128/IAI.00173-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cascales E., Lloubès R., and Sturgis J. N. (2001) The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA–MotB. Mol. Microbiol. 42, 795–807 10.1046/j.1365-2958.2001.02673.x [DOI] [PubMed] [Google Scholar]
- 81. Celia H., Noinaj N., Zakharov S. D., Bordignon E., Botos I., Santamaria M., Barnard T. J., Cramer W. A., Lloubes R., and Buchanan S. K. (2016) Structural insight into the role of the Ton complex in energy transduction. Nature 538, 60–65 10.1038/nature19757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thormann K. M., and Paulick A. (2010) Tuning the flagellar motor. Microbiology 156, 1275–1283 10.1099/mic.0.029595-0 [DOI] [PubMed] [Google Scholar]
- 83. Faure L. M., Fiche J. B., Espinosa L., Ducret A., Anantharaman V., Luciano J., Lhospice S., Islam S. T., Tréguier J., Sotes M., Kuru E., Van Nieuwenhze M. S., Brun Y. V., Théodoly O., Aravind L., et al. (2016) The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539, 530–535 10.1038/nature20121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ekiert D. C., Bhabha G., Isom G. L., Greenan G., Ovchinnikov S., Henderson I. R., Cox J. S., and Vale R. D. (2017) Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285.e17 10.1016/j.cell.2017.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ercan B., Low W. Y., Liu X., and Chng S.-S. (2019) Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC–Mla system. Biochemistry 58, 114–119 10.1021/acs.biochem.8b00897 [DOI] [PubMed] [Google Scholar]
- 86. Abellón-Ruiz J., Kaptan S. S., Baslé A., Claudi B., Bumann D., Kleinekathöfer U., and van den Berg B. (2017) Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 10.1038/s41564-017-0046-x [DOI] [PubMed] [Google Scholar]
- 87. Yeow J., Tan K. W., Holdbrook D. A., Chong Z. S., Marzinek J. K., Bond P. J., and Chng S.-S. (2018) The architecture of the OmpC–MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J. Biol. Chem. 293, 11325–11340 10.1074/jbc.RA118.002441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang Y. M., Miao Y., Munguia J., Lin L., Nizet V., and McCammon J. A. (2016) Molecular dynamic study of MlaC protein in Gram-negative bacteria: conformational flexibility, solvent effect and protein-phospholipid binding. Protein Sci. 25, 1430–1437 10.1002/pro.2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thong S., Ercan B., Torta F., Fong Z. Y., Wong H. Y., Wenk M. R., and Chng S.-S. (2016) Defining key roles for auxillary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife 5, e19042 10.7554/eLife.19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91 10.1146/annurev.cellbio.042308.113414 [DOI] [PubMed] [Google Scholar]
- 91. Casali N., and Riley L. W. (2007) A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60 10.1186/1471-2164-8-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Powers M. J., and Trent M. S. (2018) Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl. Acad. Sci. U.S.A. 115, E8518–E8527 10.1073/pnas.1806714115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kamischke C., Fan J., Bergeron J., Kulasekara H. D., Dalebroux Z. D., Burrell A., Kollman J. M., and Miller S. I. (2019) The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 8, e40171 10.7554/eLife.40171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hughes G. W., Hall S. C. L., Laxton C. S., Sridhar P., Mahadi A. H., Hatton C., Piggot T. J., Wotherspoon P. J., Leney A. C., Ward D. G., Jamshad M., Spana V., Cadby I. T., Harding C., Isom G. L., et al. (2018) Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 2018 10.1038/s41564-019-0481-y [DOI] [PubMed] [Google Scholar]
- 95. Isom G. L., Davies N. J., Chong Z. S., Bryant J. A., Jamshad M., Sharif M., Cunningham A. F., Knowles T. J., Chng S. S., Cole J. A., and Henderson I. R. (2017) MCE domain proteins: conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci. Rep. 7, 8608 10.1038/s41598-017-09111-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nakayama T., and Zhang-Akiyama Q. M. (2017) pqiABC and yebST, Putative mce operons of Escherichia coli, encode transport pathways and contribute to membrane integrity. J. Bacteriol. 199, e00606–16 10.1128/JB.00606-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dekker N. (2000) Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35, 711–717 10.1046/j.1365-2958.2000.01775.x [DOI] [PubMed] [Google Scholar]
- 98. Bishop R. E. (2005) The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912 10.1111/j.1365-2958.2005.04711.x [DOI] [PubMed] [Google Scholar]
- 99. Tatsuta T., and Langer T. (2017) Intramitochondrial phospholipid trafficking. Biochim. Biophys. Acta 1862, 81–89 10.1016/j.bbalip.2016.08.006 [DOI] [PubMed] [Google Scholar]
- 100. Cockcroft S., and Raghu P. (2018) Phospholipid transport protein function at organelle contact sites. Curr. Opin. Cell Biol. 53, 52–60 10.1016/j.ceb.2018.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Eggert U. S., Ruiz N., Falcone B. V., Branstrom A. A., Goldman R. C., Silhavy T. J., and Kahne D. (2001) Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 294, 361–364 10.1126/science.1063611 [DOI] [PubMed] [Google Scholar]
- 102. Chng S.-S., Xue M., Garner R. A., Kadokura H., Boyd D., Beckwith J., and Kahne D. (2012) Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide transport. Science 337, 1665–1668 10.1126/science.1227215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lee J., Xue M., Wzorek J. S., Wu T., Grabowicz M., Gronenberg L. S., Sutterlin H. A., Davis R. M., Ruiz N., Silhavy T. J., and Kahne D. (2016) Characterization of a stalled complex on the β-barrel assembly machine. Proc. Natl. Acad. Sci. U.S.A. 113, 8717–8722 10.1073/pnas.1604100113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sutterlin H. A., Shi H., May K. L., Miguel A., Khare S., Huang K. C., and Silhavy T. J. (2016) Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 10.1073/pnas.1601375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang G., Baidin V., Pahil K. S., Moison E., Tomasek D., Ramadoss N. S., Chatterjee A. K., McNamara C. W., Young T. S., Schultz P. G., Meredith T. C., and Kahne D. (2018) Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proc. Natl. Acad. Sci. U.S.A. 115, 6834–6839 10.1073/pnas.1804670115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jia W., El Zoeiby A., Petruzziello T. N., Jayabalasingham B., Seyedirashti S., and Bishop R. E. (2004) Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem. 279, 44966–44975 10.1074/jbc.M404963200 [DOI] [PubMed] [Google Scholar]
- 107. De Leij L., and Witholt B. (1977) Structural heterogeneity of the cytoplasmic and outer membranes of Escherichia coli. Biochim. Biophys. Acta 471, 92–104 10.1016/0005-2736(77)90396-0 [DOI] [PubMed] [Google Scholar]
- 108. McLeod S. M., Fleming P. R., MacCormack K., McLaughlin R. E., Whiteaker J. D., Narita S., Mori M., Tokuda H., and Miller A. A. (2015) Small-molecule inhibitors of Gram-negative lipoprotein trafficking discovered by phenotypic screening. J. Bacteriol. 197, 1075–1082 10.1128/JB.02352-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nickerson N. N., Jao C. C., Xu Y., Quinn J., Skippington E., Alexander M. K., Miu A., Skelton N., Hankins J. V., Lopez M. S., Koth C. M., Rutherford S., and Nishiyama M. (2018) A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob. Agents Chemother. 62, e02151–17 10.1128/AAC.02151-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Srinivas N., Jetter P., Ueberbacher B. J., Werneburg M., Zerbe K., Steinmann J., Van der Meijden B., Bernardini F., Lederer A., Dias R. L., Misson P. E., Henze H., Zumbrunn J., Gombert F. O., Obrecht D., et al. (2010) Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 10.1126/science.1182749 [DOI] [PubMed] [Google Scholar]