Abstract
Members of a large family of Ankyrin Repeat Domain (ANKRD) proteins regulate numerous cellular processes by binding to specific protein targets and modulating their activity, stability, and other properties. The same ANKRD protein may interact with different targets and regulate distinct cellular pathways. The mechanisms responsible for switches in the ANKRDs' behavior are often unknown. We show that cells' metabolic state can markedly alter interactions of an ANKRD protein with its target and the functional outcomes of this interaction. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 in vesicle-like structures. Upon nutrient limitation, ANKRD9 loses its vesicular pattern and assembles with IMPDH2 into rodlike filaments, in which IMPDH2 is stable. Inhibition of IMPDH2 activity with ribavirin favors ANKRD9 binding to IMPDH2 rods. The formation of ANKRD9/IMPDH2 rods is reversed by guanosine, which restores ANKRD9 associations with the vesicle-like structures. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicle-to-rods transition as well as binding and regulation of IMPDH2. Oppositely to overexpression, ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Taken together, the results suggest that a guanosine-dependent metabolic switch determines the mode of ANKRD9 action toward IMPDH2.
Keywords: ankyrin, nucleotide, cell metabolism, complex, protein-protein interaction, ANKRD9, GTP, IMPDH2, nucleotide, ankyrin repeat domain, rod, cytoophidia
Introduction
Ankyrin Repeat Domains proteins (ANKRD)2 form a large and fascinating family of proteins that regulate numerous cellular pathways. The activities of ANKRDs involve protein complex assembly, interactions with cytoskeleton, posttranslational modification, regulation of protein degradation, and many others (1–8). Structural basis of interactions between ANKRDs and their targets is becoming clearer (1); however, little is known about regulation of these interactions in cells. In some cases, the same ANKRD binds to different protein targets and modulates distinct cellular processes. These different forms of action and the factors that control ANKRD behavior remain poorly understood.
ANKRD9 (ankyrin domain containing protein 9) is an ANKRD protein implicated in a broad spectrum of cellular processes, including lipid metabolism (9), copper homeostasis (10), and cell proliferation (11). Recent genome-wide association studies have uncovered the potential role for ANKRD9 in hypertension (12) and myocardial repolarization (13). Despite this accumulating evidence of ANKRD9's physiologic and clinical significance, information about functional properties and mechanisms of ANKRD9 is very limited. Analysis of protein sequence predicted ANKRD9 to be a 35-kDa protein with at least one ankyrin repeat. In a large MS screen of human interactome, ANKRD9 was copurified with inosine monophosphate dehydrogenase 2 (IMPDH2) (14), the rate-limiting enzyme in the GTP biosynthetic pathway. Recent studies confirmed this interaction and found that overexpression of ANKRD9 decreases IMPDH2 levels by stimulating interactions with the ubiquitin ligase complex and causing protein degradation (11).
These findings were difficult to reconcile with the original report, which demonstrated that in cells ANKRD9 formed vesicle-like structures (9) and, therefore, is presumably spatially separated from IMPDH2, which is a cytosolic enzyme. ANKRD9 mRNA levels are under nutritional control (9) and, therefore, it is possible that ANKRD9 protein has different localization and functions under different metabolic conditions. Our studies have tested this hypothesis. We have found that the intracellular location of ANKRD9 and its regulation of IMPDH2 depend on availability of metabolites (guanosine) as well as a tertiary structure of IMPDH2. Taken together, our results suggest a mechanism through which ANKRD9 modulates the intracellular properties of IMPDH2 in response to metabolic changes.
Results
In cells, ANKRD9 adopts two distinct forms
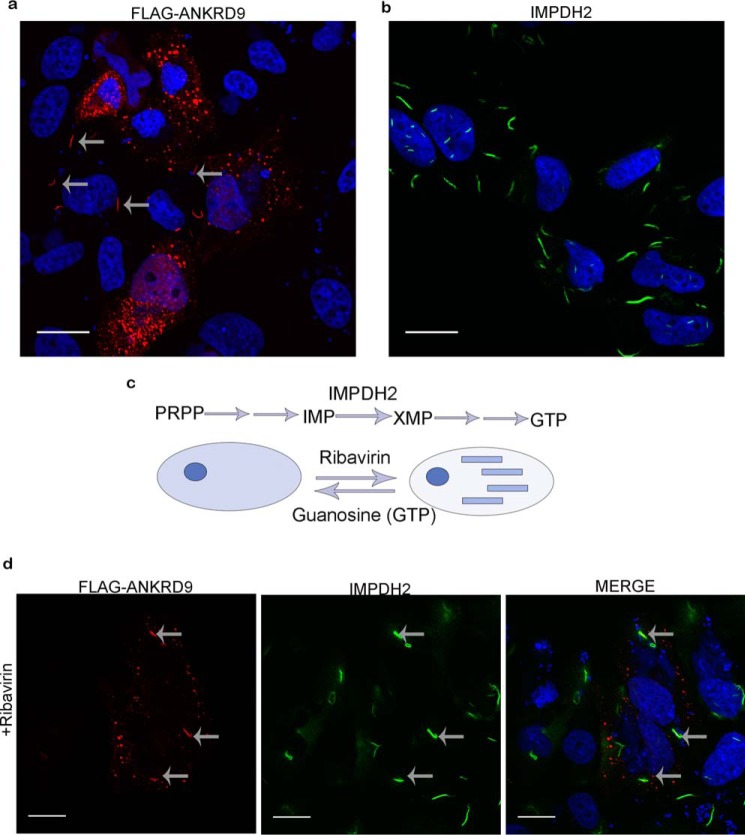
This work has started with an observation that expression of a FLAG-tagged ANKRD9 in HeLa cells produced two very different intracellular patterns. In the majority of cells, ANKRD9 formed vesicle-like structures (Fig. 1a), in agreement with a previous report (9). In other cells, ANKRD9 was present in distinct 3- to 5-micron “rodlike” structures (Fig. 1a, arrows). Although “vesicles” were numerous and distributed throughout the cell, typically only few rods (1–3 per cells) were formed in the cytoplasm and around the nucleus. The appearance of the rods was intriguing, because IMPDH2 (an established target of ANKRD9) is known to assemble into rodlike filaments (also called cytoophidia) in response to various metabolic perturbations (15–18, 20–22).
Figure 1.
ANKRD9 adopts two distinct forms. a, HeLa cells were transfected with FLAG-ANKRD9 expressing plasmid and immunostained for the FLAG-tag. FLAG-ANKRD9 (red) shows a vesicle-like and rodlike pattern (gray arrows); cell nuclei are visualized with DAPI (blue); n = 3 of independent experiments. b, IMPDH2 forms rods when inhibited with ribavirin. HeLa cells were treated with 10 mm ribavirin for 1 h then stained for IMPDH2 (green) and DAPI (blue); n = 2. c, IMPDH2 mediates the rate-limiting step (conversion of inosine monophosphate (IMP) to xanthine monophosphate (XMP)) in a pathway beginning with phosphoribosyl pyrophosphate (PRPP) and eventually leading to GTP. IMPDH2 transitions from a diffuse cystolic form (left) to rods (right) when inhibited with ribavirin; this transition is reversed with guanosine. d, HeLa cells were treated with 10 μm ribavirin for 1 h then transfected with FLAG-ANKRD9 and immunostained for FLAG (red) and IMPDH2 (green). Gray arrows point to rods containing both IMPDH2 and ANKRD9. Scale bar, 20 μm.
Normally, in HeLa cells IMPDH2 shows a diffuse cytosolic pattern. Formation of IMPDH2 rods can be rapidly induced by incubating cells with ribavirin, a potent cell-permeable inhibitor of IMPDH2 (Fig. 1, b and c). Consequently, to test whether the recombinant ANKRD9 associates with IMPDH2 in rods, we treated cells with ribavirin for 1 h prior to ANKRD9 expression. Under these conditions, ANKRD9 colocalized with the IMPDH2 rods and was also detected in vesicle-like structures in the same cells (Fig. 1d). The overlap between ANKRD9 and IMPDH2 in rods was complete, i.e. no ANKRD9 rods without IMPDH2 were detected. Thus, in addition to the “basal” vesicle-like form, ANKRD9 interacts with inactive IMPDH2 in rodlike filaments.
ANKRD9 forms rods upon nutrient limitation
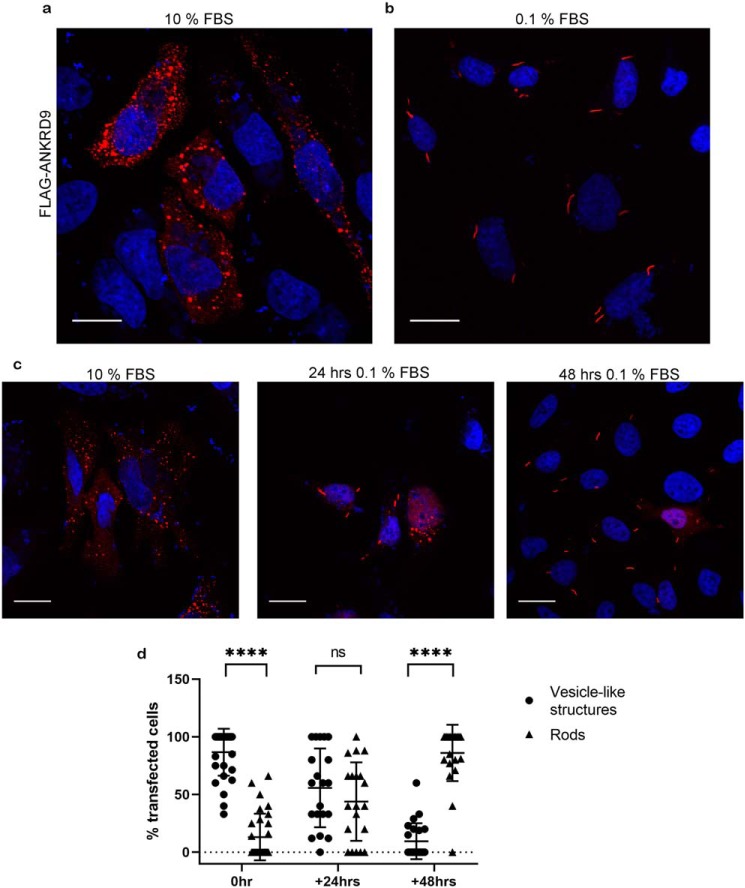
These results were interesting because they demonstrated the previously unknown ability of ANKRD9 to assemble with IMPDH2 in large macromolecular complexes. To better understand the physiologic trigger for the ANKRD9-IMPDH2 assembly we considered that ANKRD9 mRNA levels respond to starving/refeeding (i.e. are nutrient dependent) (9) and that IMPDH2 forms rods when cells are starved of certain metabolites (15, 20). Consequently, we tested whether nutrient deprivation would cause transition of ANKRD9 from vesicle-like structures to rods. Cells transfected with ANKRD9 plasmid were grown in low serum (0.1% FBS) for 24 and 48 h and immunostained for ANKRD9 and IMPDH2. After 48 h of nutrient limitation all ANKRD9 was found in rods (compare Fig. 2, a and b). The transition to rods was time dependent (Fig. 2c); i.e. after a shorter nutrient limitation (24 h) ANKRD9 was detected in both vesicle-like structures and in rods (Fig. 2, c and d).
Figure 2.
ANKRD9 forms rods upon nutrient depletion. a, HeLa cells were transfected with FLAG-ANKRD9 (red) as in Fig. 1a and incubated in the presence of 10% FBS. n = 3 independent experiments. b, HeLa cells were transfected with FLAG-ANKRD9 and incubated in the presence of 0.1% FBS for 48 h. Rods are indicated by gray arrows; nuclei are stained with DAPI (blue). n = 3 independent experiments. c, representative images of each time point; rods appear in the majority of cells after 48 h in 0.1% FBS media. For 24 h, n = 3. Scale bar, 20 μm. d, percentage of cells containing ANKRD9 in vesicle-like structures (black filled circles) or rods (black filled triangles) after overnight transfection (control), followed by 24 or 48 h in starvation (0.1% FBS) media. The total number of analyzed cells is as follows: n = 137 cells (control, 0 h of 0.1% FBS), n = 149 cells (24 h of 0.1% FBS), n = 131 cells (48 h of 0.1% FBS). Error bars represent S.D. Each individual point represents at least 5 cells. ****, p value < 0.0001., unpaired t test; ns, non-significant.
To further verify interactions between ANKRD9 and IMPDH2 in rods we attempted co-immunoprecipitation but neither protein could bind to beads under standard immunoprecipitation conditions, presumably because of rods' large size and/or dimensions. Consequently, we homogenized cells under nondenaturing conditions and fractionated cell homogenate using differential centrifugation. Analysis of fractions revealed presence of both IMPDH2- and ANKRD9-FLAG2 in pellets, which typically contain heavy organelles such as nuclei and mitochondria (Fig. S1). No soluble IMPDH2 and FLAG-ANKRD9 was detected in agreement with the confocal imaging data.
Formation of ANKRD9 rods is caused by depletion of IMPDH2-dependent metabolites
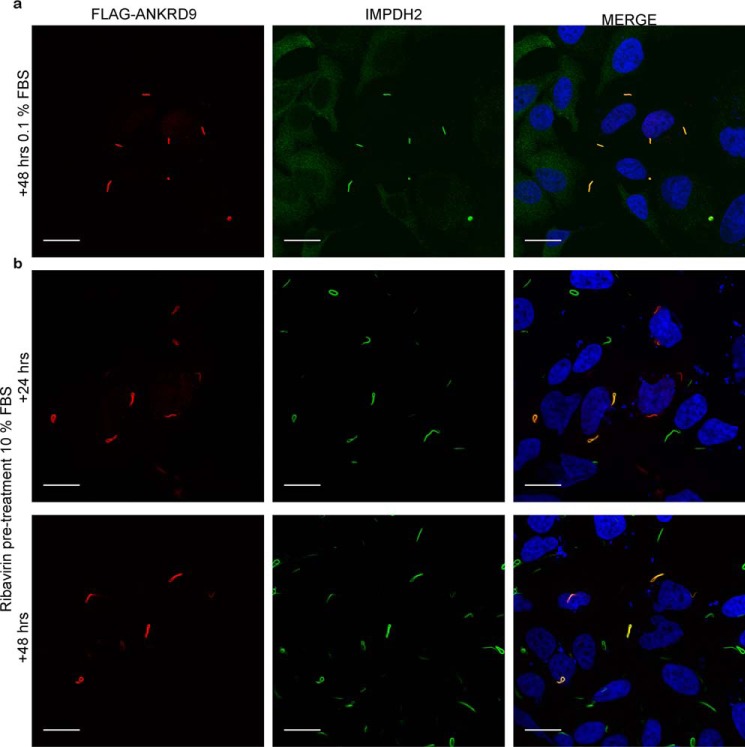
Brief treatment of cells with ribavirin and a prolonged nutrient depletion both cause IMPDH2 inhibition and the ANKRD9/IMPDH2 assembly. In the first case, the vesicle-to-rod transition of ANKRD9 was partial, whereas in the second case it was complete. One important difference between these two conditions is the duration of IMPDH2 inactivation. Partial effect of brief treatment with ribavirin on ANKRD9 pattern suggested that the inhibition of IMPDH2 per se could be insufficient to trigger loss of ANKRD9 association with the vesicle-like structures and that the depletion of IMPDH2-dependent metabolite(s) may be needed for complete conversion of ANKRD9 into a rod-bound form. To test this hypothesis, we added ribavirin to cells prior to ANKRD9 expression, as previously, and then allowed cells to grow for 24 and 48 h under basal conditions (i.e. in normal serum) in the presence of ribavirin (Fig. 3). After 24 h, ANRKD9 fully transitioned to rods (Fig. 3b). The rods contained both ANKRD9 and IMPDH2 and were very similar to those induced by 48 h of nutrient limitation (compare Fig. 3, a and b). The result suggests that the prolonged incubation with the IMPDH2 inhibitor, which depletes cells of IMPDH2-dependent metabolites, mimics serum starvation conditions.
Figure 3.
Formation of ANRKD9 rods is caused by depletion of IMPDH2 metabolites. a, HeLa cells were transfected with FLAG-ANKRD9, placed in 0.1% FBS for 48 h and immunostained for FLAG and IMPDH2. n = 3 independent experiments. b, HeLa cells were treated with 10 μm ribavirin for 1 h then transfected with FLAG-ANRKD9 and kept in 10% FBS for 24 and 48 h. Cells were immunostained with FLAG (red) and IMPDH2 (green). Two independent experiments. Merged images show overlap between ANKRD9 and IMPDH2 rods. Scale bar, 20 μm.
To test for specificity of metabolite depletion, we considered that ANKRD9 function was important for copper homeostasis and tested whether limiting copper levels in cells had effects similar to limiting IMPDH2-dependent metabolites. Prolonged incubation with copper chelators did not cause rod formation (Fig. S2), i.e. ANKRD9 responds to specific metabolic changes.
Guanosine reverses the ANKRD9 relocalization from vesicle-like structures to rods
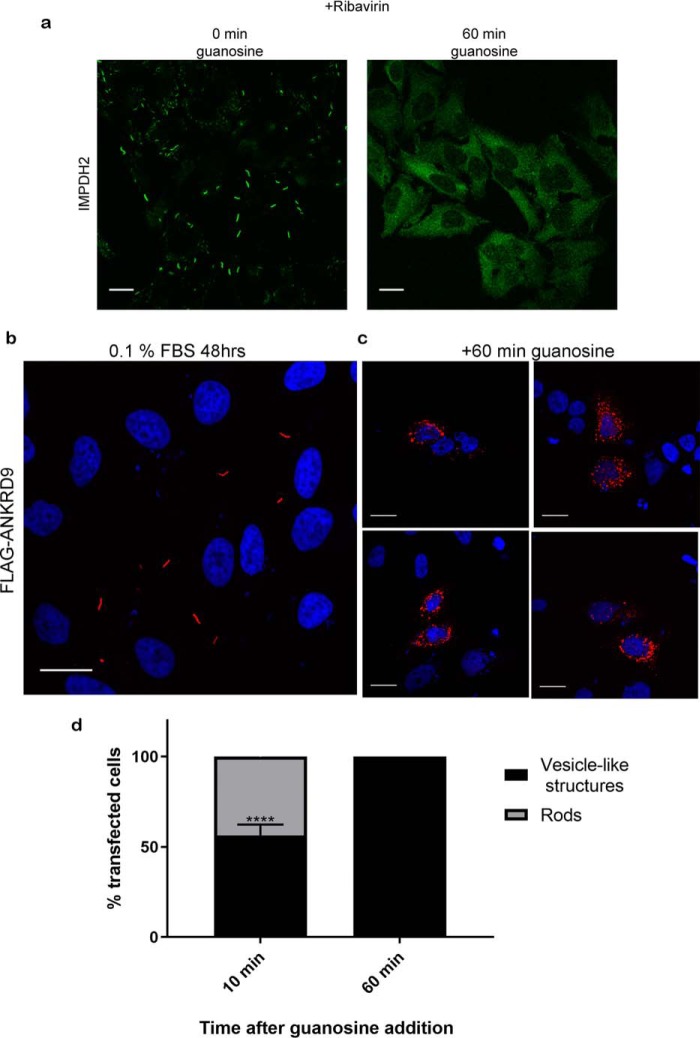
To examine further whether the vesicle-to-rods transition of ANKRD9 represents a regulatory response to metabolic state of the cell, we tested whether ANKRD9 transition to rods can be reversed. First, cells were grown under nutrient-limiting conditions for 24 h to induce partial rod formation, then washed and placed into the basal growth medium for 24 h. This treatment decreased the abundance of rods and caused appearance of ANKRD9 in vesicle-like structures, but did not fully restore the vesicular pattern of ANKRD9 (Fig. S2). We then considered that the depletion of guanine pools was previously suggested to cause IMPDH2 macro-assembly and that the formation of IMPDH2 rods can be reversed by addition of guanosine (Fig. 4a) (21, 23). Consequently, we tested whether the ANKRD9/IMPDH2 rods, formed under conditions of nutrient limitation, were affected by guanosine. To this end, we expressed ANKRD9, induced rod formation by incubating cells for 48 h in low serum, and then added 100 μm guanosine for 1 h (this concentration was previously shown to cause disassembly of IMPDH2 rods (Fig. 4) (23). Addition of guanosine completely reversed the ANKRD9 interactions with IMPDH2 and caused uniform transition of ANKRD9 back to vesicles (compare Fig. 4, b and c). This effect was rapid; the vesicle-like pattern of ANKRD9 was already observed at 10 min following addition of guanosine (Fig. 4, c and d). Thus, the disassembly of ANKRD9/IMPDH2 rods occurs on a much faster timescale than rod formation, perhaps reflecting addition of a large excess of guanosine. Guanosine-dependent change in ANKRD9 pattern also provides an indication that the cellular behavior of ANKRD9 may be controlled by the same factors that regulate IMPDH2 macro-assembly, specifically, lower guanine pools.
Figure 4.
ANKRD9 transition to rods is reversible with guanosine addition. a, left, IMPDH2 rod formation is reversed with guanosine addition. HeLa cells were treated for 1 h with 10 μm ribavirin to induce IMPDH2 rod formation and immunostained for IMPDH2 (green); right, HeLa cells were treated with 10 μm ribavirin for 1 h to induce rods formation, then 100 μm guanosine was added for 60 min and cells were immunostained for IMPDH2. n = 3 for each condition. Scale bar, 20 μm. b, control, FLAG-ANKRD9 forms rods following incubation of transfected HeLa cells in 0.1% FBS for 48 h. c, the same treatment, followed by incubation with 100 μm guanosine for 1 h. The ANKRD9 rods disappear and ANKRD9 shows a vesicle-like pattern. n = 3. Scale bar, 20 μm. d, percentage of cells with ANKRD9 vesicle-like structures or rods at 10 and 60 min after addition of guanosine. n = 3 for each time point. Error bar represents S.D. ****, p value < 0.0001., unpaired t test. Scale bar, 20 μm.
ANKRD9 expression is associated with longer and fewer IMPDH2 rods
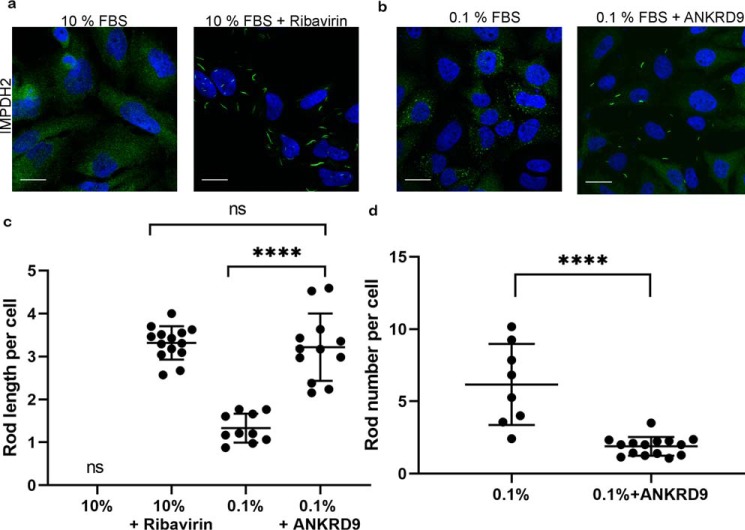
Purified IMPDH2 can assemble into rods in vitro in the absence of any other proteins (24–29). Therefore, the role of ANKRD9 in IMPDH2 rods is likely to be regulatory and involve modulation of rod properties. It was shown previously that IMPDH2 rods were dynamic and could change their length over time (30). Consequently, to clarify the functional significance of ANKRD9/IMPDH2 macro-assembly, we compared the length and numbers of IMPDH2 rods in cells with either endogenous or overexpressed ANKRD9 under conditions of nutrient limitation. Ribavirin treatment was used as a control for rod formation. In nutrient-depleted HeLa cells (that have endogenous ANKRD9), the length of IMPDH2 rods was about 1.5 microns, whereas in HeLa cells overexpressing FLAG-ANKRD9 the length of IMPDH2 rods was significantly increased (by about 2.5-fold) to 3–5 microns (Fig. 5, a and b). The length of IMPDH2/ANKRD9 rods is similar to IMPDH2 rods formed in the presence of ribavirin (Fig. 5c). This result further suggests that under conditions of nutrient limitation ANRKD9 binds to an inhibited IMPDH2. An increased length of IMPDH2 rods also suggested that excess ANKRD9 stabilized the IMPDH2 rods favoring longer structures. Indeed, we found that without ANKRD9 overexpression the number of IMPDH2 rods per cell was about six, whereas significant increase in rod length upon ANKRD9 overexpression was associated with a decreased number of rods, to 1–2 per cell (Fig. 5d).
Figure 5.
Higher ANKRD9 abundance is associated with longer IMPDH2 rods. a, immunostaining of IMPDH2 in HeLa cells under basal conditions (10% FBS) shows an expected diffuse pattern (left panel). Treatment of these cells with 10 μm ribavirin for 1 h in the absence of recombinant ANKRD9 triggers formation of IMPDH2 rods (right panel). n = 3 for both conditions. Scale bar, 20 μm. b, in the absence of recombinant ANKRD9, nutrient limitation (0.1% FBS for 48 h) causes appearance of short IMPDH2 rods (left panel). Expression of recombinant FLAG-ANKRD9 significantly increases the length of IMPDH2 rods (right panel). n = 3 independent experiments. c, quantification of IMPDH2 rod length in different treatment conditions. The total number of cells analyzed is as follows: n = 150 cells (10% FBS + ribavirin), n = 73 cells (0.1% FBS), and n = 156 cells (0.1% FBS + ANKRD9 expression). Statistical analysis was done using an unpaired t test. ****, p < 0.0001. Error bars represent S.D. Each point is the average rod length of at least 5 cells. d, quantification of the number of rods per cell under nutrient limitation in the absence and presence of recombinant ANKRD9. unpaired t test. ****, p < 0.0001. Error bars represent S.D. Each point is the average rod length of at least 5 cells.
A conserved CysCys motif is required for ANKRD9 transition between two cellular forms
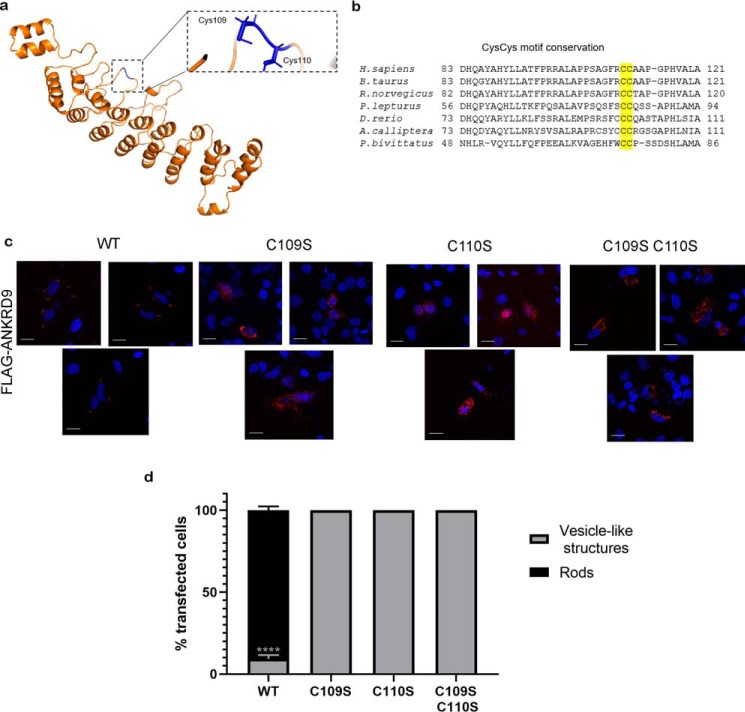
ANKRD9 has not been characterized biochemically and its structure is unknown. Consequently, to better understand the biochemical basis of ANKRD9/IMPDH2 interactions, we predicted the structure of ANKRD9 using the Robetta server (Fig. 6a). The molecular model of ANKRD9 shows nine helix-turn-helix repeats assembled into a concaved structure. To identify amino acid residues important for ANKRD9 function, we searched for highly conserved residues that are located at the protein surface and therefore are available for protein-protein interaction. ANKRD9 is highly conserved, but the sequence alignment of ANKRD9 orthologs from multiple species identified several invariant residues in the loop regions of ANKRD9 (Fig. 6a). Two conserved vicinal cysteines Cys109Cys110 (Fig. 6b) were of particular interest, because cysteines are susceptible to various modifications and hence can contribute to observed changes in ANKRD9 intracellular behavior.
Figure 6.
A conserved CysCys motif in ANKRD9 is required for transition from vesicle-like structures to rods. a, structural model of ANKRD9 with conserved Cys residues highlighted in blue; the boxed area shows the position of Cys109 and Cys110 in the loop. b, multiple sequence alignment of the Cys109Cys110-containing region illustrates conservation of this motif. c, HeLa cells were transfected with WT ANKRD9 (control) or indicated ANKRD9 mutants and placed in 0. 1% FBS-containing medium for 48 h. Unlike WT, none of the mutants formed rods under these conditions. n = 3 for each condition. Scale bar, 20 μm. d, quantification of ANKRD9 mutant rod formation where vesicle-like structures are in black and rods are in gray. Unlike WT, none of the mutants formed rods under these conditions. n = 3. ****, p < 0.0001 unpaired t test. Error bar represents S.D.
To directly test the role of Cys109Cys110 motif in rod formation we generated a series of mutants, in which Cys109 and Cys110 were converted to serine individually or together. The C109S, C110S, and C109S/C110S mutants were expressed in HeLa cells and their ability to form rods was analyzed under nutrient-limiting conditions. Unlike WT ANKRD9, which shows complete transition to rods after 48 h in 0.1% FBS, neither mutant lost its vesicle-like pattern nor formed rods under these conditions (Fig. 6, c and d). The expression levels of ANKRD9 mutants did not differ significantly from the WT (Fig. 7c). These results suggest that although Cys109 and Cys110 do not play an important role in stabilizing ANKRD9 in the vesicle-like structures, they are required for assembly with IMPDH2 under nutrient limitation. The inability of the Cys109Cys110 mutants to lose vesicular patterns and transition to rods may reflect the role of this motif in sensing the metabolite depletion (see “Discussion”). This effect is specific for Cys109/Cys110 mutants because mutation of another surface-exposed conserved residue Arg125 to Ala yielded a very different phenotype. The R125A variant was expressed normally; however, unlike Cys mutants it lost the vesicle-like pattern and became diffuse (Fig. S3).
Figure 7.
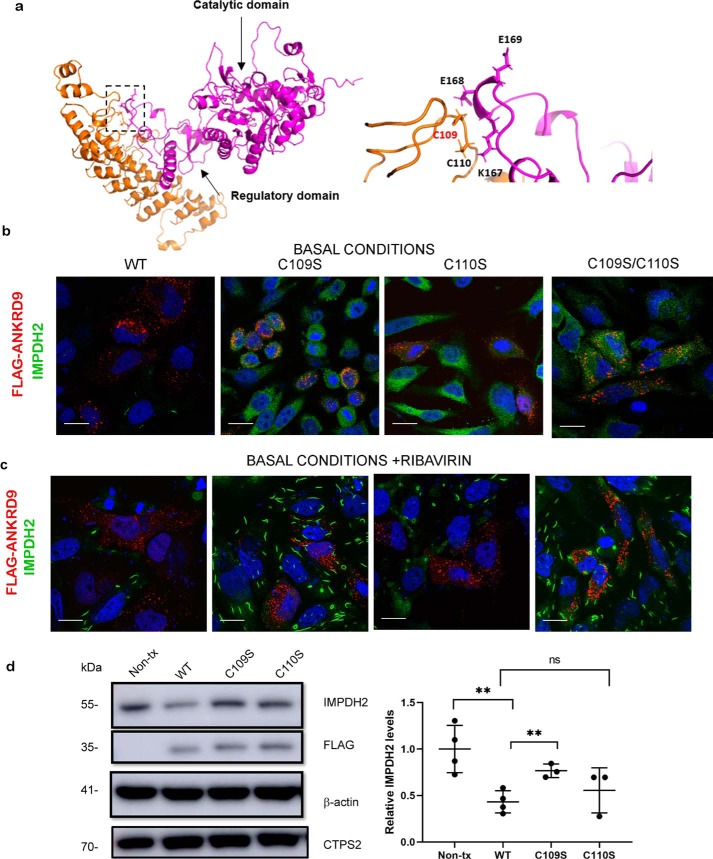
The Cys109Cys110 motif contributes to ANKRD9-IMPDH2 interaction. a, left, docking model of ANKRD9 and IMPDH2 (PDB ID 1NF7, see Fig. S7). IMPDH2 catalytic and regulatory domains are indicated by the arrows; the predicted interaction site is indicated by the box. Right, the magnified view of the predicted interaction site between ANKRD9 (orange) and IMPDH2 (purple). The CysCys motif in ANKRD9 interacts with Lys167, Glu168 and Glu169 of IMPDH2. b, Cys mutants do not facilitate IMPDH2 degradation. HeLa cells were transfected with WT ANKRD9 and indicated mutants under basal conditions (10% FBS) and immunostained for FLAG and IMPDH2. Cells expressing WT ANKRD9 showed significantly reduced IMPDH2 staining whereas cells with the Cys mutants did not. n = 3 independent experiments for each condition. Scale bar, 20 μm. c, Cys mutants of ANKRD9 do not form rods with IMPDH2. HeLa cells were transfected with the indicated constructs and subsequently treated with 10 μm ribavirin for 1 h prior to immunostaining for FLAG and IMPDH2. n = 3. Scale bar is 20 μm. d, left, the WT and ANKRD9 mutants were expressed in HeLa cells under basal conditions (10% FBS) and cell lysates, were separated and immunoblotted for FLAG, IMPDH2, CTPS2, and β-actin used as a loading control. WT ANKRD9 but not the C109S mutant significantly decreases IMPDH2 abundance, the C110S mutant is intermediate between the two and similar to WT. Right, densitometry of IMPDH2 intensity after WT ANKRD9 or mutant expression in HeLa cells. n = 3. ****, p value < 0.0001., unpaired t test.
Cys109Cys110 motif contributes to ANKRD9-IMPDH2 interaction
To determine whether the Cys109/Cys110 residues were involved in binding and regulation of IMPDH2, we first explored the possible structural interactions between ANKRD9 and IMPDH2 by docking our Robetta model and the known crystal structure of IMPDH2 (Fig. 7a, left panel). The docking model revealed favorable interactions between the ANKRD9 loop containing the Cys109Cys110 motif with the regulatory domain of IMPDH2 (Fig. 7a, right panel) (see Fig. S4 for additional information). Interestingly, the IMPDH2 residues involved in predicted interactions with ANKRD9 are located near the known nucleotide-binding site of IMPDH2 (Fig. S5) (25), which may explain sensitivity of ANKRD9/IMPDH2 assembly to guanosine.
To directly verify these interactions and their consequences, we took advantage of the report that appeared when this work was ongoing (11). This published study suggested a role for elevated ANKRD9 in proteasome-mediated degradation of IMPDH2. Consequently, we expressed the WT ANKRD9 and ANKRD9 mutants in HeLa cells under basal conditions and compared their effect on IMPDH2 abundance. Although the intracellular localization of WT and mutants in vesicular structures was very similar, their effects on IMPDH2 levels were markedly different.
Expression of WT ANKRD9 was associated with a marked decrease of IMPDH2 protein (Fig. 7, b and d). This effect was specific for IMPDH2, as overexpression of ANKRD9 did not influence levels of cytidine triphosphate synthetase 2 (CTPS2) (Fig. 7d and Fig. S6), another metabolic enzyme known to form rodlike structures (31–35). Use of proteasome inhibitor MG132 after transfection of cells with WT ANKRD9 partially restored IMPDH2 levels (Fig. S6) supporting the proposed role for ANKRD9 in facilitating proteasomal degradation of IMPDH2.
In contrast to WT ANKRD9, the C109S and C109S/C110S mutants did not significantly affect IMPDH2 levels (Fig. 7b). Treatment with 10 μm ribavirin for 1 h confirmed the difference in IMPDH2 abundance between cells expressing WT and mutant ANKRD9s. IMPDH2 rods were absent in cells expressing WT ANKRD9 mutants (Fig. 7c) reflecting the marked decrease in IMPDH2 abundance, but present in cells with C109S and C109S/C110S variants. The effects of C110S mutant were less pronounced that those of the C109S, C109S/C110S mutants and were closer to WT ANKRD9 (Fig. 7d). This result suggested that Cys109 plays the primary role in interactions with IMPDH2. This conclusion was confirmed by Western blotting (Fig. 7d). Taken together, these results suggest that under basal conditions excess ANKRD9 negatively affects IMPDH2 abundance. Under nutrient-deprivation conditions ANKRD9 loses the ability to form vesicle-like structures but retains ability to bind to IMPDH2 in rods. In rods, ANKRD9-dependent degradation of IMPDH2 is blocked, presumably by the tertiary structure of rods. The Cys109Cys110 motif plays an important role in both nutrient sensing and binding/regulation of IMPDH2.
ANKRD9 knockdown increases IMPDH2 levels and inhibits rod formation under nutrient-limiting conditions
The above experiments provided evidence for the impact of elevated ANKRD9 on IMPDH2. To better understand how variation in ANKRD9 abundance affects IMPDH2, we decreased levels of ANKRD9 in HEK293A cells using siRNA. HEK293 cells were chosen because they endogenously express ANKRD9 at higher levels than HeLa cells, facilitating more accurate measurements of siRNA-mediated down-regulation. HEK293 cells also show IMPDH2 assembly in rods upon serum starvation and the recombinant FLAG-ANKRD9 has the same vesicle-like pattern under basal conditions as in HeLa cells (Fig. S7).
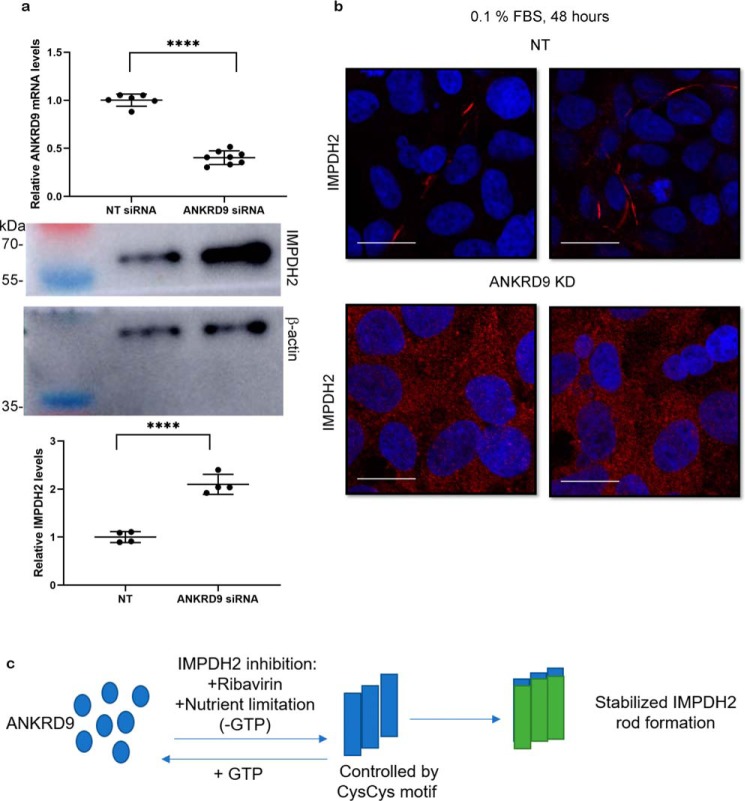
The siRNA-mediated knockdown reduced ANKRD9 mRNA levels by about 60% (Fig. 8a, upper panel); the protein levels could not be measured because of low sensitivity of available antibodies. The cells were then examined under basal and low serum conditions. ANKRD9 down-regulation increased IMPDH2 expression levels (Fig. 8a, lower panel) in agreement with previous findings (11). Despite up-regulation, IMPDH2 did not form rods in response to nutrient limitation in cells with down-regulated ANKRD9 (Fig. 8b). This finding suggests that ANKRD9 serves to stabilize IMPDH2 rods when nutrients (presumably GTP pools) are depleted (Fig. 8c).
Figure 8.
ANKRD9 knockdown increases IMPDH2 expression and reduces rod formation under nutrient limiting conditions. a, top, HEK293A cells were transfected with nontargeted (NT) siRNA and ANKRD9 siRNA, and ANKRD9 mRNA levels were quantified. n = 6 for NT siRNA and n = 8 for ANKRD9 siRNA; Bottom: IMPDH2 levels in corresponding cells were analyzed by Western blotting with β-actin used as a loading control (lower panels). n = 3 for each condition. Decrease in ANKRD9 levels is associated with higher IMPDH2 abundance. ****, p < 0.0001 (unpaired t test). Error bar represents S.D. b, ANKRD9 down-regulation diminishes IMPDH2 rod formation under nutrient limitation (0.1% FBS). HEK293A cells were transfected with the indicated siRNAs and placed in 0.1% FBS for 48 h and immunostained for IMPDH2. IMPDH2 rods were apparent in control cells; no IMPDH2 rods are visible in cells treated with ANKRD9 siRNA. n = 3 for each condition. Scale bar, 20 μm. c, cartoon showing that ANKRD9 forms rods with inhibited IMPDH2 when GTP pools are lowered and stabilizes rods.
Discussion
ANKRD9 is a highly conserved protein found in all vertebrates. The cellular and physiologic functions of ANKRD9 are poorly understood, and the limited available data paint a complex and confusing picture. The first report, published in 2009, found ANKRD9 mRNA levels to be responsive to changes in lipid metabolism and nutritional availability, whereas the second report, published in 2018, provided evidence that ANKRD9 could be involved in proteasomal degradation of IMPDH2, a nucleotide processing enzyme. Our laboratory previously identified ANKRD9 as a novel regulator of copper homeostasis based on specific elevation of cellular copper in response to ANKRD9 knockdown (9–11). These ANKRD9-infuenced processes have little in common; to modulate these distinct pathways ANKRD9 is likely to interact with more than one cellular target and these interactions can be spatially, temporarily, and metabolically controlled. Here we show that the recombinant ANKRD9 is associated with at least two distinct intracellular structures: the vesicle-like puncta and cytosolic rods (Fig. 8c). The presence of ANKRD9 in these structures depends on a metabolic state of the cell, and changes in protein levels of either endogenous or recombinant ANKRD9 have distinct consequences for IMPDH2 abundance and macro-assembly. We have generated the molecular model for ANKRD9 and identified cysteine residues required for changes in ANKRD9 intracellular localization and activity toward IMPDH2.
The transition of ANKRD9 to a rodlike form occurs when cells are grown in a medium with low serum. Although low serum conditions can be used to block cell cycle, we suggest that ANKRD9 senses nutrient depletion rather than inhibition of the cell cycle progression. Our reasoning is the following. First, the formation of rods takes longer than the inhibition of cell cycle. Second, prolonged inhibition of IMPDH2 with ribavirin accelerates formation of ANKRD9 rods, suggesting that the transition of ANKRD9 from vesicle-like structures to rods is triggered by IMPDH2-dependent nucleotide misbalance (compare Fig. 3, a and b). Third, addition of guanosine rapidly disassembles ANKRD9 rods, suggesting that in low serum cellular guanine pools are lowered (Fig. 4).
The assembly into rods and rings is a characteristic property of IMPDH2 that has been extensively studied (17, 21, 22). This macro-assembly occurs in response to various triggers, including IMPDH2 inhibition, signaling during T-cell activation, or depletion of certain amino acids. IMPDH2 has propensity for self-assembly, and no proteins with enzymatic functions have been found to interact with IMPDH2 rods. GTP pools are critical for IMPDH rods formation because guanosine, but no other nucleosides, reverse the macro-assembly (16, 21, 23, 30). We found that guanosine also disassembles the ANKRD9/IMPDH2 complex and, significantly, restores vesicular pattern for ANKRD9. This observation suggests that the ANKRD9 may sense levels of nucleotide pools and respond by changing its location/association with IMPDH2. In fact, the predictive algorithms suggest presence of a nucleotide-binding site in the vicinity of Cys109/Cys110. Further studies are needed to directly test whether ANKRD9 is a nucleotide-binding protein and whether it binds to specific vesicles or self-aggregates into structures with a vesicle-like appearance.
GTP and ATP bind to the regulatory domain of IMPDH2 and influence not only macro-assembly but also IMDPH2 activity (25, 26, 36, 37). In our docking model, ANKRD9 interacts near these nucleotide-binding sites. This close proximity suggests that ANKRD9/IMPDH2 interaction in rods may influence nucleotide binding to IMPDH2. Overall, our working model is the following. ANKRD9 senses nucleotide levels either through direct binding or interactions with GTP/GDP binding protein(s) in vesicular structures. As nutrients deplete, nucleotide binding is lost and ANKRD9 transitions to rods where it binds to the GTP-less IMPDH2. The C109C110 mutations lock ANKRD9 in a nucleotide-bound state so that even in low serum conditions vesicular-like ANKRD9 is retained. The details of this model and the specific role of Cys109/Cys110 in regulation of IMPDH2 require further testing. Specifically, available evidence strongly suggests that ANKRD9 interacts with IMPDH2 (Refs. 11, 14 and our study), however, it remains to be determined whether these interactions are always direct (as was shown in Ref. 14) or, in rods, may involve other partners.
We show that ANKRD9 binds to the inhibited IMPDH2 and increases the length of IMPDH2 rods. The significance of IMPDH2 rods has been a subject of great interest and debate in the literature, because the appearance of the rods is not linked to either IMPDH2 abundance or activity (both active and inhibited IMPDH2 form rods). Rods are readily formed in nutrient-starved cells as well as in rapidly proliferating cells, which may also have low surplus of nutrients. Our data suggest that under these circumstances macro-assembly into rods may protect IMPDH2 from ANKRD9-mediated degradation, which may otherwise occur when ANKRD9 loses association with vesicles in response to nutrient deprivation. ANKRD9 binding to IMPDH2 rods appears to stabilize them. In other words, the metabolic state of the cell determines whether ANKRD9 destroys or protects IMPDH2.
CTPS2 also forms rods and rings. The CTPS2 rods are distinct from IMPDH2 rods although their assembly could be regulated by similar factors such as nucleotide metabolites (16). Previous proteome screen (6) did not find CTPS2 among ANKRD9-binding partners (only IMPDH1 and IMPDH2 were detected) and in our studies we observed no effect of ANKRD9 on CTPS2, suggesting that the ANKRD9-IMPDH2 assembly and regulation is specific.
Comparison of the predicted ANKRD9 structure to structures of other better characterized ANKRD-containing proteins using 3D protein Blast (http://3d-blast.life.nctu.edu.tw/)3 (39, 40) show that three-dimensional fold of ANKRD9 resembles that of other ANKRDs including the ankyrin-binding domain of phospholipase iPLA2b, gankyrin tankyrase. The homology to the latter enzyme is particularly interesting. Tankyrase mediates poly-ADP–ribosylation of various proteins, marking them for degradation via ubiquitin ligase pathway, and regulates mitosis, genome integrity, and cell signaling. As ANKRD9, tankyrase regulates protein abundance in various cellular locations; β-catenin in the cytosol and GLUT4 transporter in vesicles (38). It is tempting to speculate that coupling sensitivity to metabolites with changes in intracellular localization could be a common mechanistic feature of ANKRD-containing proteins with a structural similarity to ANKRD9.
Experimental procedures
Cell lines
HeLa cells and HEK293 cells were maintained in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin-streptomycin, and 1% nonessential amino acids and passaged every 2–3 days, when cells were 80% confluent in T75 cm2 flasks.
Site-directed mutagenesis in ANKRD9
An N-terminally FLAG-tagged ANKRD9 construct was generated using cDNA of human ANKRD9 as a template. The construct was cloned into a pcDNA3.1 vector containing blasticidin and ampicillin resistance. Mutations were introduced using the QuikChange XL Mutagenesis kit. Primers were purchased from Integrative DNA Technologies. Presence of correct mutations and the lack of unwanted mutations were verified by sequencing the coding region. Primers used for mutagenesis were: C109S forward primer: 5′-AGCCGCGCAGCTGCGGAAGCCGG-3′; C109S reverse primer: 5′-CCGGCTTCCGCAGCTGCGCGGCT-3′; C110S forward primer: 5′-GGGAGCCGCGCTGCAGCGGAAGC-3′; C110S reverse primer: 5′-GCTTCCGCTGCAGCGCGGCTCCC-3′; C109S/C110S forward primer: 5′-GGGAGCCGCGCTGCTGCGGAAGCCGG-3′; C109S/C110S reverse primer: 5′-CCGGCTTCCGCAGCAGCGCGGCTCCC-3′. R125A forward primer: 5′-GCAGAATGCCCACGGCGGCGTTGTAGCGCACTG-3′; and reverse primer: 5′-CAGTGCGCTACAACGCCGTGGGCATTCTGC-3′.
Immunofluorescence microscopy
Cells were transfected with 1 μg FLAG-ANKRD9 or FLAG-ANKRD9 mutants using 2% Turbofect in 200 μl Opti-MEM in 2 ml of 45,000 cells/ml in 12-well plates for 16 h and processed for immunofluorescence as follows. HeLa cells were washed with PBS then fixed with 4% formaldehyde for 15 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 for 10 min with mild shaking. Cells were then blocked with 1% BSA, 1% gelatin in PBS overnight at 4 °C. The next day cells were incubated with primary antibody for 1 h at room temperature in a humidified chamber. Mouse anti-FLAG (catalogue no. F1804; Millipore-Sigma) was used at a 1:500 dilution, rabbit anti-IMPDH2 (catalogue no. ab75790; Abcam), and rabbit anti-CTPS2 (catalogue no. ab235109; Abcam) were used at 1:300 dilution. Cells were then washed with PBS once, twice with PBS + 0.1% Tween, once with PBS, and then incubated with fluorescent secondary antibody in the dark for 1 h. Donkey anti-mouse Alexa Fluor 555 (catalogue no. A31570, Thermo Fisher) and donkey anti-rabbit Alexa Fluor 488 (catalogue no. A21202, Thermo Fisher) were used at 1:500 dilution. Donkey anti-rabbit Alexa Fluor 555 (catalogue no. ab150070, Abcam) was used in Fig. 8b. Coverslips were then washed as with the primary antibodies with a final wash in water to remove excess salts. Coverslips were then mounted with Fluoromount and DAPI at 50% each. Images were taken with an Olympus FV3000RS confocal microscope and processed using Fiji. Images in Figs. S1 and S5 were taken with a Zeiss LSM 5 Pascal confocal microscope and processed using Image J.
Quantifying rod length
For rod length, images were processed in Fiji, as described previously (35). Briefly, images were first deconvoluted to 8-bit. The image threshold, or the amount of contrast in an image, was set to between 2.5 and 3.5%. This range was set using the default and black and white settings in Fiji and allowed visualization of rod structures above background for accurate rod length measurements. Lastly, the “Analyze particles” application was used where objects with the length below 0.5 microns were excluded to only quantify rod structures which are typically 1 micron and above. Circularity, which denotes whether or not the particle analyzed is a circle, was set to the lowest value 0.01; as in this report we were interested in the ANKRD9-dependent length of IMPDH2 rods and not rings. Rod lengths were determined for all cells in a given image then the DAPI channel was used to determine rod number per cell in the image. At least 10 images in random locations containing at least 10 cells were used for quantification in each condition. Statistical analysis was performed using unpaired t tests.
Structural modeling
First, a crystal structure of IMPDH2 (PDB ID 1NF7) was used to refill all missed regions of the structure. This was done using the Molecular Operating Environment (MOE) software, version 2018.01 (CCG, Montreal, Canada), the full amino acid sequence of IMPDH2 and the Homology Model application of Protein module, using Amber10: EHT force field with a Reaction Field (R-Field). A model with best root mean square deviation (RMSD) and contact energy was chosen. See Fig. S4 for additional details.
Docking of IMPDH2 and ANKRD9 was conducted with the Dock application of the Compute module in MOE. A number of docking configurations were generated and scored. The score was calculated as a combination of free energy of binding including, among others contributions, solvation and entropy terms, and targeted regions interaction score that was calculated as a number of heavy atoms from the conserved regions of six sites in protein ANKRD9 contacting heavy atoms of protein IMPDH2. See Fig. S3 for additional details.
siRNA knockdown of ANKRD9
siGENOME SMART pool constructs against human ANKRD9 (catalogue no. M-015551-01-0005) was used in HEK293A cells. Wet-reverse transfection was performed as follows: Cells were grown to ∼60% confluency in complete medium without antibiotics. DharmaFECT at 2.5% (catalogue no. T-2001-002, Dharmacon) and Opti-MEM were mixed with 20 nm siRNA and Opti-MEM, added to wells and incubated for 30 min. 1.37 × 105 cells/ml cells were then added and incubated with siRNA plus DharmaFECT at 37 ° for 60 h. cOmplete medium without antibiotics was added for 12 h for a total of 72 h incubation. Cells were then processed for RT-qPCR immunostaining for IMPDH2 rod formation.
Real-time quantitative PCR (RT-qPCR)
RNA isolation and mRNA quantitation were performed as described previously (19). Briefly, RNA isolation was done with the RNeasy Kit (Qiagen), cDNA was generated and RT-PCR was done with the One-Step SYBR Green kit (Applied Biosystems) on an ABI 7500 Sequence Detection System (Applied Biosystems). S18 was used for normalization for ΔΔCt analysis. Primers used for qPCR were as follows: S18 forward: 5′-TTCTGGCCAACGGTCTAGACAAC-3′; S18 reverse: 5′-CCAGTGGTCTTGGTGTGCTGA-3′; ANKRD9 forward: 5′-CTGGTCACCGCCATCTCT-3′ ANKRD9 reverse: 5′-CTAGCCTTTGCCAGTGAGGT-3′.
Western blotting
HeLa cells were transfected with FLAG-ANKRD9 plasmid, protein was expressed for 16 h and cells were lysed with 1× RIPA buffer in PBS. Lysates were cleared with centrifugation at 9000 × g for 10 min. Supernatants were separated using Laemmli gel electrophoresis, and proteins were transferred to PVDF membrane for Western blotting. The following antibodies were used: Mouse anti-FLAG, dilution 1:10,000; rabbit anti-IMPDH2, 1:10,000; rabbit anti-CTPS2, 1:10,000; mouse anti-β actin (catalogue no. NB600–501, Novus, 1:10,000). Membranes were blocked for 1 h in 5% milk in PBS at room temperature and incubated with primary antibody in 1% milk in PBS-T. Membranes were washed the next day in PBS-T and incubated with HRP-conjugated secondary antibody, dilution 1:10,000, for 1 h at room temperature and imaged with chemiluminescence.
For fractionation, HeLa cells were transfected with the FLAG-Tag ANKRD9 and rods were formed using low serum conditions as described above. Cells were collected, pelleted, and resuspended with 50 mm MOPS, pH 7.5, 150 mm NaCl, 20% glycerol, 0.1% n-dodecyl-β-d-maltoside (DDM) and homogenized with a Dounce homogenizer. The homogenate was fractionated at the following speeds/times: 500 × g for 5 min to pellet the nuclear-enriched fraction, 6000 × g to pellet the mitochondria-enriched fraction, and 10,000 × g to pellet the plasma membrane fraction. Samples from the supernatant and pellet were used for immunoblotting as above. Rabbit anti-FLAG antibody (Catalogue no. F7425, Sigma) was used at 1:10,000 dilution.
Statistical analysis
GraphPad Prism 6 was used for all statistical analyses.
Author contributions
D. H. and S. L. conceptualization; D. H., N. M. H., I. F. T., and S. L. resources; D. H. data curation; D. H., V. L. K., and S. L. formal analysis; D. H., H. E. P., N. M. H., I. F. T., and S. L. supervision; D. H. and S. L. funding acquisition; D. H., V. L. K., N. M. H., and E. R. G. investigation; D. H., V. L. K., and E. R. G. visualization; D. H., H. E. P., N. M. H., I. F. T., and S. L. methodology; D. H. writing-original draft; D. H. and S. L. project administration; D. H., H. E. P., N. M. H., I. F. T., and S. L. writing-review and editing; N. M. H., I. F. T., and S. L. validation.
Supplementary Material
Acknowledgments
We thank Dennis Chang for performing initial experiments that confirmed distinct ANKRD9 intracellular patterns and Samuel Jayakanthan for help with fractionation experiments. The Robetta server was used for the protein structure prediction.
This work was supported by National Institutes of Health Grants R01DK071865 and 2R56DK071865 (to S. L.). This work was also supported by Training Grant T32 GM007445 (to D. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ANKRD
- ankyrin repeat domains
- IMPDH2
- inosine monophosphate dehydrogenase 2
- CTPS2
- cytidine triphosphate synthetase 2
- qPCR
- quantitative PCR.
References
- 1. Islam Z., Nagampalli R. S. K., Fatima M. T., and Ashraf G. M. (2018) New paradigm in ankyrin repeats: Beyond protein-protein interaction module. Int. J. Biol. Macromol. 109, 1164–1173 10.1016/j.ijbiomac.2017.11.101 [DOI] [PubMed] [Google Scholar]
- 2. Li J., Mahajan A., and Tsai M. D. (2006) Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178 10.1021/bi062188q [DOI] [PubMed] [Google Scholar]
- 3. Sedgwick S. G., and Smerdon S. J. (1999) The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 10.1016/S0968-0004(99)01426-7 [DOI] [PubMed] [Google Scholar]
- 4. Mosavi L. K., Minor D. L. Jr., and Peng Z.-Y. (2002) Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. U.S.A. 99, 16029–16034 10.1073/pnas.252537899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inada H., Procko E., Sotomayor M., and Gaudet R. (2012) Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry 51, 6195–6206 10.1021/bi300279b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi N., Hamada-Nakahara S., Itoh Y., Takemura K., Shimada A., Ueda Y., Kitamata M., Matsuoka R., Hanawa-Suetsugu K., Senju Y., Mori M. X., Kiyonaka S., Kohda D., Kitao A., Mori Y., and Suetsugu S. (2014) TRPV4 channel activity is modulated by direct interaction of the ankyrin domain to PI(4,5)P2. Nat. Commun. 5, 4994 10.1038/ncomms5994 [DOI] [PubMed] [Google Scholar]
- 7. Michel F., Soler-Lopez M., Petosa C., Cramer P., Siebenlist U., and Müller C. W. (2001) Crystal structure of the ankyrin repeat domain of Bcl-3: A unique member of the Ikb protein family. EMBO J. 20, 6180–6190 10.1093/emboj/20.22.6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H.-L., Fan S.-S., Pang M., Liu Y.-H., Guo M., Liang J.-B., Zhang J.-L., Yu B.-F., Guo R., Xie J., and Zheng G.-P. (2015) The ankyrin repeat domain 49 (ANKRD49) augments autophagy of serum-starved GC-1 cells through the NF-κB pathway. PLoS One 10, e0128551 10.1371/journal.pone.0128551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X., Newkirk R. F., Carre W., Ghose P., Igobudia B., Townsel J. G., and Cogburn L. A. (2009) Regulation of ANKRD9 expression by lipid metabolic perturbations. BMB Rep. 42, 568–573 10.5483/BMBRep.2009.42.9.568 [DOI] [PubMed] [Google Scholar]
- 10. Malinouski M., Hasan N. M., Zhang Y., Seravalli J., Lin J., Avanesov A., Lutsenko S., and Gladyshev V. N. (2014) Genome-wide RNAi ionomics screen reveals new genes and regulation of human trace element metabolism. Nat. Commun. 5, 3301 10.1038/ncomms4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee Y., Lim B., Lee S. W., Lee W. R., Kim Y. I., Kim M., Ju H., Kim M. Y., Kang S.-J., Song J.-J., Lee J. E., and Kang C. (2018) ANKRD9 is associated with tumor suppression as a substrate receptor subunit of ubiquitin ligase. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3145–3153 10.1016/j.bbadis.2018.07.001 [DOI] [PubMed] [Google Scholar]
- 12. Chiang K. M., Yang H. C., and Pan W. H. (2018) A two-stage whole-genome gene expression association study of young-onset hypertension in Han Chinese population of Taiwan. Sci. Rep. 8, 1800 10.1038/s41598-018-19520-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arking D. E., Pulit S. L., Crotti L., van der Harst P., Munroe P. B., Koopmann T. T., Sotoodehnia N., Rossin E. J., Morley M., Wang X., Johnson A. D., Lundby A., Gudbjartsson D. F., Noseworthy P. A., Eijgelsheim M., et al. (2014) Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet. 46, 826–836 10.1038/ng.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex Network: A systematic exploration of the human interactome. Cell 162, 425–440 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calise S. J., Carcamo W. C., Krueger C., Yin J. D., Purich D. L., and Chan E. K. L. (2014) Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell Mol. Life Sci. 71, 2963–2973 10.1007/s00018-014-1567-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duong-Ly K. C., Kuo Y.-M., Johnson M. C., Cote J. M., Kollman J. M., Soboloff J., Rall G. F., Andrews A. J., and Peterson J. R. (2018) T cell activation triggers reversible inosine-5′-monophosphate dehydrogenase assembly. J. Cell Sci. 131, jcs223289 10.1242/jcs.223289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keppeke G. D., Calise S. J., Chan E. K. L., and Andrade L. E. C. (2015) Assembly of IMPDH2-based, CTPS-based, and mixed rod/ring structures is dependent on cell type and conditions of induction. J. Genet. Genomics 42, 287–299 10.1016/j.jgg.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 18. Keppeke G. D., Andrade L. E. C., Grieshaber S. S., and Chan E. K. L. (2015) Microinjection of specific anti-IMPDH2 antibodies induces disassembly of cytoplasmic rods/rings that are primarily stationary and stable structures. Cell Biosci. 5, 1 10.1186/2045-3701-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang H., Ralle M., Wolfgang M. J., Dhawan N., Burkhead J. L., Rodriguez S., Kaplan J. H., Wong G. W., Haughey N., and Lutsenko S. (2018) Copper-dependent amino oxidase 3 governs selection of metabolic fuels in adipocytes. PLoS Biol. 16, e2006519 10.1371/journal.pbio.2006519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calise S. J., Purich D. L., Nguyen T., Saleem D. A., Krueger C., Yin J. D., and Chan E. K. L. (2016) 'Rod and ring' formation from IMP dehydrogenase is regulated through the one-carbon metabolic pathway. J. Cell Sci. 129, 3042–3052 10.1242/jcs.183400 [DOI] [PubMed] [Google Scholar]
- 21. Keppeke G. D., Chang C. C., Peng M., Chen L.-Y., Lin W.-C., Pai L.-M., Andrade L. E. C., Sung L.-Y., and Liu J.-L. (2018) IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div. 13, 5 10.1186/s13008-018-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang C.-C., Lin W.-C., Pai L.-M., Lee H.-S., Wu S.-C., Ding S.-T., Liu J.-L., and Sung L.-Y. (2015) Cytoophidium assembly reflects upregulation of IMPDH activity. J. Cell Sci. 128, 3550–3555 10.1242/jcs.175265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ji Y., Gu J., Makhov A. M., Griffith J. D., and Mitchell B. S. (2006) Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J. Biol. Chem. 281, 206–212 10.1074/jbc.M507056200 [DOI] [PubMed] [Google Scholar]
- 24. Anthony S. A., Burrell A. L., Johnson M. C., Duong-Ly K., Kuo C. Y.-M., Simonet J. C., Michener P., Andrews A., Kollman J. M., and Peterson J. R. (2017) Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell 28, 2600–2608 10.1091/mbc.e17-04-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buey R. M., Ledesma-Amaro R., Velazquez-Campoy A., Balsera M., Chagoyen M., de Pereda J. M., and Revuelta J. L. (2015) Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 6, 8923 10.1038/ncomms9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buey R. M., Fernández-Justel D., Marcos-Alcalde Í., Winter G., Gómez-Puertas P., de Pereda J. M., Luis Revuelta J. (2017) A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 7, 2648 10.1038/s41598-017-02805-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Labesse G., Alexandre T., Vaupré L., Salard-Arnaud I., Him J. L., Raynal B., Bron P., and Munier-Lehmann H. (2013) MgATP regulates allostery and fiber formation in IMPDHs. Structure 21, 975–985 10.1016/j.str.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 28. Aughey G. N., and Liu J.-L. (2015) Metabolic regulation via enzyme filamentation. Crit. Rev. Biochem. Mol. Biol. 51, 282–293 10.3109/10409238.2016.1172555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Connell J. D., Zhao A., Ellington A. D., and Marcotte E. M. (2012) Dynamic reorganization of metabolic enzymes into intracellular bodies. Ann. Rev. Cell Dev. Biol. 28, 89–111 10.1146/annurev-cellbio-101011-155841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thomas E. C., Gunter J. H., Webster J. A., Schieber N. L., Oorschot V., Parton R. G., and Whitehead J. P. (2012) Different characteristics and nucleotide binding properties of inosine monophosphate dehydrogenase (IMPDH) isoforms. PLoS One 7, e51096 10.1371/journal.pone.0051096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carcamo W. C., Satoh M., Kasahara H., Terada N., Hamazaki T., Chan J. Y. F., Yao B., Tamayo S., Covini G., von Mühlen C. A., and Chan E. K. L. (2011) Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One 6, e29690 10.1371/journal.pone.0029690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo K.-M., Chang C.-C., Shen Q.-J., Sung L.-Y., and Liu J.-L. (2014) CTP synthase forms cytoophidia in the cytoplasm and nucleus. Exp. Cell Res. 323, 242–253 10.1016/j.yexcr.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 33. Liu J.-L. (2010) Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genomics 37, 281–296 10.1016/S1673-8527(09)60046-1 [DOI] [PubMed] [Google Scholar]
- 34. Lynch E. M., Kicks D. R., Shepherd M., Endrizzi J. A., Maker A., Hansen J. M., Barry R. M., Gitai Z., Baldwin E. P., and Kollman J. M. (2017) Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol. 24, 507–514 10.1038/nsmb.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noree C., Monfort E., Shiau A. K., and Wilhelm J. E. (2014) Common regulatory control of CTP synthase enzyme activity and filament formation. Mol. Biol. Cell 25, 2282–2290 10.1091/mbc.e14-04-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., and Hardie D. G. (2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 113, 274–284 10.1172/JCI19874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Labesse G., Alexandre T., Gelin M., Haouz A., and Munier-Lehmann H. (2015) Crystallographic studies of two variants of Pseudomonas aeruginosa IMPDH with impaired allosteric regulation. Acta Crystallogr. D Biol. Crystallogr. 71, 1890–1899 10.1107/S1399004715013115 [DOI] [PubMed] [Google Scholar]
- 38. Su Z., Deshpande V., James D. E., and Stöckli J. (2018) Tankyrase modulates insulin sensitivity in skeletal muscle cells by regulating the stability of GLUT4 vesicle proteins. J. Biol. Chem. 293, 8578–8587 10.1074/jbc.RA117.001058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tung C.-H., Huang J.-W., and Yang J.-M. (2007) Kappa-alpha plot derived structural alphabet and BLOSUM-like substitution matrix for fast protein structure database search. Genome Biol. 8, R31.1–R31.16 10.1186/gb-2007-8-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang J.-M. and Tung C.-H. (2006) Protein structure database search and evolutionary classification. Nucleic Acids Res. 34, 3646–3659 10.1093/nar/gkl395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.