Abstract
Autophagy is a conserved cellular process involving intracellular membrane trafficking and degradation. Pathogens, including hepatitis C virus (HCV), often exploit this process to promote their own survival. The aim of this study was to determine the mechanism by which HCV increases steady-state autophagosome numbers while simultaneously inhibiting flux through the autophagic pathway. Using the lysosomal inhibitor bafilomycin A1, we showed that HCV-induced alterations in autophagy result from a blockage of autophagosome degradation rather than an increase in autophagosome generation. In HCV-infected cells, lysosome function was normal, but a tandem RFP–GFP–LC3 failed to reach the lysosome even under conditions that activate autophagy. Autophagosomes and lysosomes isolated from HCV-infected cells were able to fuse with each other normally in vitro, suggesting that the cellular fusion defect resulted from trafficking rather than an inability of vesicles to fuse. Arl8b is an Arf-like GTPase that specifically localizes to lysosomes and plays a role in autophagic flux through its effect on lysosomal positioning. At basal levels, Arl8b was primarily found in a perinuclear localization and co-localized with LC3-positive autophagosomes. HCV infection increased the level of Arl8b 3-fold and redistributed Arl8b to a more diffuse, peripheral pattern that failed to co-localize with LC3. Knockdown of Arl8b in HCV-infected cells restored autophagosome–lysosome fusion and autophagic flux to levels seen in control cells. Thus, HCV suppresses autophagic flux and increases the steady-state levels of autophagosomes by increasing the expression of Arl8b, which repositions lysosomes and prevents their fusion with autophagosomes.
Keywords: Hepatitis C virus (HCV), autophagy, intracellular trafficking, lysosome, GTPase, Arl8b
Introduction
Macroautophagy (hereafter autophagy) is a process by which organelles, intracellular proteins, or microorganisms are enveloped in a double membrane autophagosome and degraded by fusion with a lysosome (1). Autophagy functions to maintain cellular homeostasis and is especially important during cellular stress. Changes in autophagy pathways frequently result from infection with microorganisms and may serve either to protect the host or to promote the pathogen's life cycle (2).
Hepatitis C virus (HCV)3 is a positive-strand RNA virus with a purely cytosolic life cycle. Numerous studies have shown that HCV modulates autophagy, with the most consistent observation being an induction of LC3-II protein levels and a subsequent increase in the number of autophagosomes present in infected cells (3–8). LC3-II represents the lipidated form of the protein that decorates both sides of the autophagosomal membrane and is used as a marker for active autophagy. It has been suggested that this increase in autophagosome number serves the virus by evading innate immune clearance (6, 9), regulating RNA replication (4, 10–12), or playing a role in virion secretion (5, 13). Increased autophagy might also aid host cell persistence (14, 15). There is still not consensus regarding whether autophagy proceeds to completion with the autophagosome fusing with a lysosome during HCV infection. Long-lived protein degradation, which is a measure of autophagic degradation, was shown to be impaired in JFH-1–infected cells (3). Additionally, GFP-LC3 puncta did not co-localize with LysoTracker, a dye that stains acidic organelles, in JFH-1–infected cells, whereas autophagy induced by nutrient starvation showed a high degree of co-localization between GFP-LC3 and LysoTracker in control cells (3).
The mechanisms by which HCV modulates autophagy are also not fully understood. Endoplasmic reticulum stress activates the unfolded protein response, which in turn activates various autophagy-related genes (8, 11, 16, 17), but the precise steps responsible for autophagosome accumulation are unknown. Other potential mechanisms proposed are that HCV up-regulates Beclin1 to induce initiation of autophagy (18) and then temporally regulates the expression of Rubicon and UVRAG at different time points in infection to control autophagic maturation (19).
Rab and Arf-like GTPases add an additional layer of regulation and play important roles in autophagy, ensuring the appropriate maturation and eventual fusion of autophagosomes, amphisomes, and lysosomes. These GTPases are small guanine nucleotide-binding proteins that control various aspects of intracellular trafficking. In this study, we explore the role of an Arf-like GTPase, Arl8b, during HCV infection. Arl8b (ADP ribosylation factor–like protein 8b) localizes to lysosomes and plays a prominent role in lysosomal positioning (20). Arl8b binds to the kinesin-1 motor protein through its effector SKIP and moves lysosomes in an anterograde fashion toward the cell periphery (21). Lysosomal positioning plays an important role in the final steps of autophagy by governing autophagosome–lysosome fusion. Autophagosomes and lysosomes fuse primarily in the perinuclear region of the cell, and when lysosomes move to the periphery there is a decrease in autophagosome–lysosome fusion (22, 23). These events can be coordinated by Arl8b expression. Korolchuk et al. (24) demonstrated that overexpressing Arl8b could decrease autophagosome–lysosome fusion, whereas knocking down Arl8b had the opposite effect, increasing autophagic flux.
Previous studies have implicated Arl8b in the pathogenesis of microbes, in particular Salmonella typhimurium, by altering intracellular trafficking. During S. typhimurium infection, salmonella-containing vacuoles acquire Arl8b to recruit kinesin and allow for migration of salmonella-containing vacuoles to the periphery for efficient cell-to-cell transfer of the bacteria (25).
In this study, we explore the impact of HCV infection on autophagy, demonstrating that autophagic flux is impaired through failure of autophagosome and lysosome fusion. We determined that the inhibition of fusion was due to a trafficking defect, and we implicated HCV-induced changes in Arl8b expression and localization as part of the mechanism. Overall, we demonstrate that organelle positioning through alteration of GTPases plays a role in viral-induced changes in autophagy and may contribute to the viral life cycle.
Results
HCV infection inhibits autophagic flux
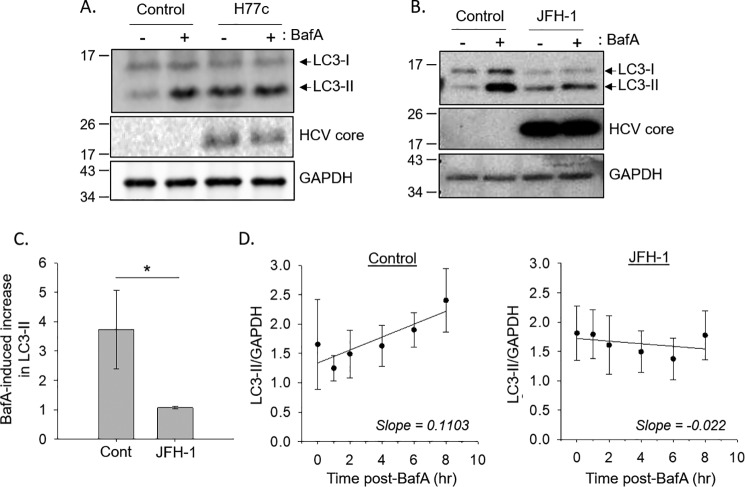
To better understand the effect of HCV on autophagy, we assessed autophagic flux using the inhibitor bafilomycin A1, which prevents the degradation of the autophagosome by inhibiting the vacuolar type H+-ATPase (26). In control cells, treatment with bafilomycin A1 resulted in an ∼4-fold increase in LC3-II levels compared with cells treated with DMSO only (Fig. 1, A and B). In cells bearing the genotype 1 full-length HCV replicon, H77c, the amount of autophagosomes as indicated by the LC3-II level was increased at baseline but did not further increase after bafilomycin A1 treatment (Fig. 1A). We further investigated autophagy flux using direct infection of Huh-7.5 cells with infectious JFH-1 virus. Similar to the replicon cells, JFH-1 infection alone also increased the basal level of LC3-II compared with the uninfected control, and treating infected cells with bafilomycin A1 also did not further increase the levels of LC3-II (Fig. 1, B and C). These data suggest that viral infection prevents lysosomal degradation of the autophagosome. To test this, we treated cells with bafilomycin A1 at different time points up to 8 h. In control cells, there was a statistically significant time-dependent increase in LC3-II levels with the slope of the line reflecting the autophagy initiation rate (Fig. 1D). In JFH-1–infected cells, there was no time-dependent accumulation of LC3-II under the same conditions. Using analysis of variance, there was a significant difference (p = 0.029) between the accumulation of LC3-II in control cells compared with JFH-1–infected cells. This analysis suggests that the viral-induced increase in autophagosomes observed in a cell culture model of HCV infection is likely due to a suppression of autophagosome degradation and not from an increase in autophagy initiation.
Figure 1.
HCV infection inhibits autophagic flux. A, Huh-7.5 cells stably bearing the full-length HCV genotype 1a replicon H77c, or these same cells after the replicon was eliminated by interferon treatment (control), were treated with or without 100 nm bafilomycin A1 (BafA) for 4 h prior to SDS-PAGE analysis. B, Huh-7.5 cells were infected with JFH-1 and treated with or without 100 nm BafA for 4 h prior to SDS-PAGE analysis. C, densitometry analysis of the changes in LC3-II levels, normalized to GAPDH levels, as determined from Western blotting. Quantification of Western blots (n = 5) was done using ImageJ software. The difference between control (Cont) and JFH1 was statistically significant (p = 0.029) as indicated by the Mann–Whitney rank sum test. D, Huh-7.5 cells were treated with 100 nm BafA for the indicated time points, and LC3-II levels were determined by immunoblotting. The plots represent the quantification (n = 5) of the fold increase of LC3-II levels, normalized to GAPDH levels. For control samples, there is a positive correlation between LC3-II levels and length of treatment time by the Spearman rank order correlation test (correlation coefficient = 0.368 and p = 0.0494). There is no significant correlation for the JFH1 samples.
JFH-1 infection does not alter global lysosomal proteolytic activity
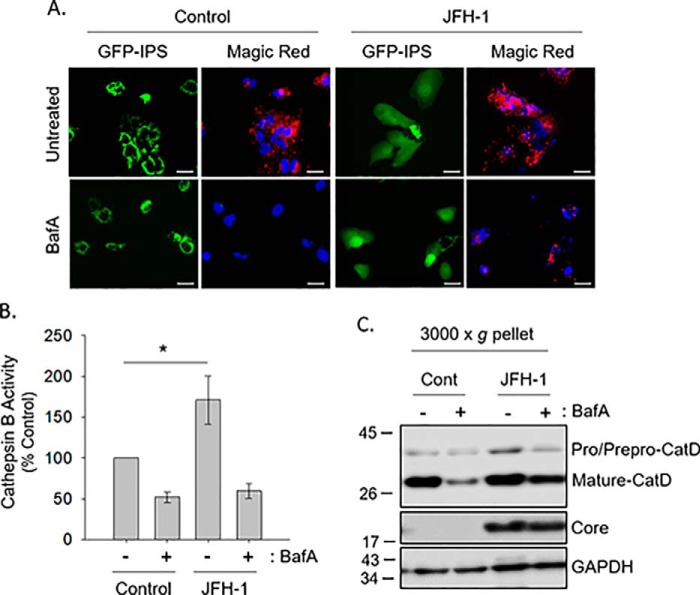
We next asked whether the block in degradation demonstrated by the autophagic flux assay was due to decreased lysosomal integrity and/or function. To measure lysosomal activity, we monitored the proteolytic function of cathepsins during HCV infection. Cathepsin B, a cysteine protease, localizes to lysosomes in its mature form (27). Cathepsin B activity was measured using a substrate, Magic Red (z-arginine-arginine)2, that generates a fluorochrome upon enzymatic cleavage (28). Huh-7.5 cells stably transduced with pEGFP-IPS (MAVS, mitochondrial antiviral-signaling protein) were used to identify HCV-infected cells (29). Under basal conditions, IPS localizes to the mitochondria. After infection, the HCV NS3/4A protease cleaves the EGFP-IPS off the mitochondria, allowing it to appear diffuse throughout the cytoplasm. Using live-cell imaging, uninfected cells displayed red fluorescence, indicating the cleavage of the Magic Red substrate by active cathepsin B (Fig. 2A). Bafilomycin A1 treatment inhibited the generation of the red fluorescence by alkalinizing the lysosome. JFH-1–infected cells showed normal Magic Red cleavage, illustrating that HCV infection does not inhibit cathepsin B activity (Fig. 2, A and B).
Figure 2.
HCV does not alter global lysosomal proteolytic activity. A, hUH-7.5–GFP–IPS cells were treated with 25 nm bafilomycin A1 (BafA) overnight and subsequently exposed to Magic Red cathepsin B. Scale bars represent 20 μm. B, cell lysate cathepsin assay. Huh-7.5 cells were incubated as described for A, and fluorescence measurements were taken as described under “Experimental procedures.” *, p < 0.001 by the Mann–Whitney rank sum test. C, Huh-7.5 cells were treated with 25 nm bafilomycin A1 overnight, and a 3000 × g pellet was assessed by SDS-PAGE.
We also measured the activity of another lysosomal enzyme, cathepsin D. The maturation of this enzyme from its preprocathepsin form to procathepsin D to its mature form is highly dependent on the acidic environment of the lysosome (30). The 28-kDa mature form of cathepsin D was present in equivalent amounts when comparing uninfected and JFH-1–infected cells, indicating efficient enzyme maturation under both conditions (Fig. 2C). Treatment of the cells with bafilomycin A1 reduced the appearance of the mature form of cathepsin D in both uninfected and infected cells. The data thus suggest that altered autophagic flux seen during HCV infection is not due to impaired lysosomal function.
Defects in autophagosome fusion during HCV infection are due to trafficking
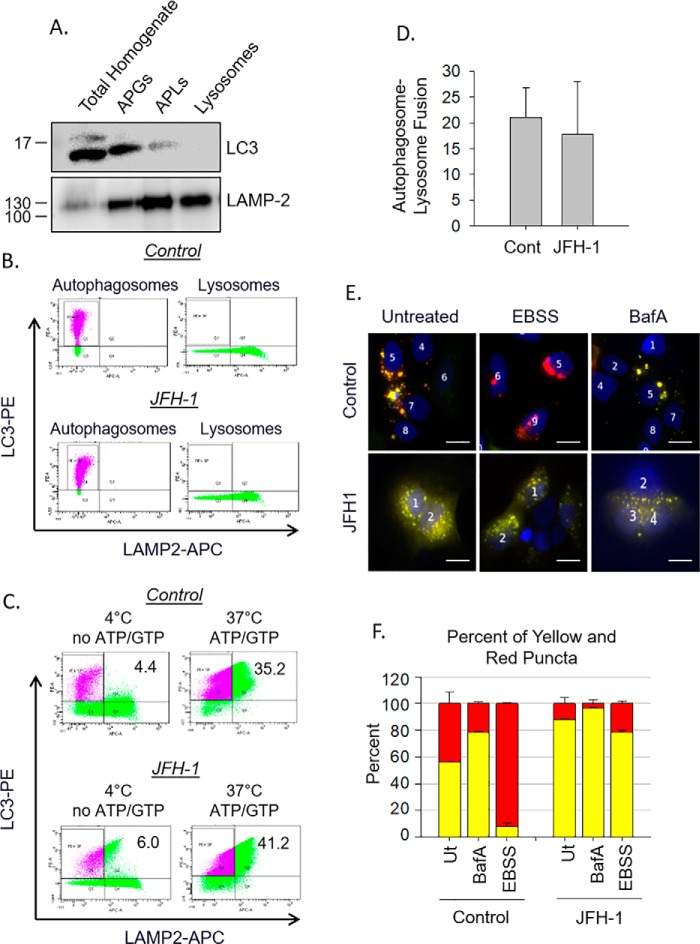
Because HCV infection does not alter global lysosomal proteolytic activity, we reasoned that the HCV-induced defect in autophagosome degradation might be due to a lack of fusion between autophagosomes and lysosomes. A defect in fusion could be due to 1) an altered composition of the autophagosome preventing fusion even if the organelles encounter each other or 2) a defect in trafficking within HCV-infected cells preventing the autophagosomes from contacting lysosomes. To differentiate between these two possibilities, we first assessed the inherent ability of the autophagosome and lysosome to fuse in vitro using a previously established assay (31). Autophagosomes and lysosomes were isolated from Huh-7.5 cells, and the identities of the fractions were confirmed by Western blotting for organelle markers (Fig. 3A). Those same vesicle fractions were separately labeled with fluorescent antibodies for the lysosome (LAMP2-APC) and the autophagosome (LC3-PE) (Fig. 3B) and used for an in vitro fusion assay. The labeled vesicles were incubated at 37 °C to allow fusion to occur (Fig. 3C). Although both the autophagosome and lysosome fractions contain LAMP-2, only the lysosome fraction was labeled with the LAMP2-APC antibody, and thus dually positive vesicles for both APC and PE, upper right quadrant, indicate that fusion had occurred. As a control for no fusion, organelles were either fixed with paraformaldehyde immediately or were held at 4 °C without ATP or GTP in the fusion buffer. Under these circumstances, there were very few dually positive vesicles (Fig. 3C). At 37 °C in the presence of ATP and GTP considerable fusion occurred. There was no difference in autophagosome–lysosome fusion in isolated vesicles from either control or JFH-1–infected cells (Fig. 3, C and D). This suggests that HCV infection does not alter the intrinsic ability of autophagosomes and lysosomes to fuse with each other.
Figure 3.
Defects in autophagosome–lysosome fusion during HCV are due to trafficking. A, organelle fractions isolated from a discontinues Nycodenz gradient were collected and subjected to SDS-PAGE analysis. B, flow cytometry of individual vesicle fractions. Autophagosomes were incubated with anti-LC3-PE antibody, whereas lysosomes were incubated separately with anti-LAMP-2-APC. C, isolated autophagosomes and lysosomes from B were combined, and fusion was assessed as stated under “Experimental procedures.” Fusion is indicated by organelles that are both PE- and APC-positive. Controls for “no fusion” were incubated at 4 °C and had no ATP or GTP added to the reaction. D, the bar graph illustrates the amount of fusion of autophagosomes and lysosomes from control and JFH1-infected cells when compared with their “no fusion” controls. This graph is representative of three independent preparations and flow cytometry experiments. Differences between control and JFH1 were not significant. E, Huh-7.5 cells were transfected with RFP–GFP–LC3 plasmid and treated with either bafilomycin A1 (BafA) or EBSS. Fusion was determined by the loss of GFP fluorescence as indicated under “Experimental procedures.” Scale bars represent 10 μm. F, analysis of E showing the quantification (n = 64–107 cells in three individual experiments) of both yellow and red puncta in each treatment. Cont, control.
To explore whether the fusion defects observed result from alterations in intracellular trafficking, we used the tandem reporter plasmid, RFP–GFP–LC3 (32). Autophagosome formation is marked by yellow fluorescence because of the co-localization of the RFP and GFP signals on the autophagosomal membrane. Autophagosome fusion with a lysosome to form an autophagolysosome causes quenching of the GFP fluorescence, and only the RFP fluorescence is visible because it is more resistant to the acidic environment of the lysosome. If fusion does not occur, autophagosomes will remain yellow. In control, untreated cells, autophagosomes displayed a mixture of red and yellow fluorescence, indicating some basal level of autophagosome–lysosome fusion (Fig. 3E). Nutrient starvation of uninfected cells by incubation in Earle's balanced salt solution (EBSS) activates autophagy and resulted in an increase in red puncta indicating a high degree of autophagosome–lysosome fusion. Treatment with bafilomycin A1, which alkalinizes lysosomes and prevents the quenching of the green fluorescence, caused the puncta to remain yellow (Fig. 3, E and F). Similar to bafilomycin A1 treatment alone, HCV-infected cells displayed more yellow puncta both at baseline and even after cells were treated with EBSS (Fig. 3, E and F). Also of note is that under uninfected conditions, autophagosome–lysosome fusion occurs in a perinuclear location near the mitotic center, whereas in JFH-1–infected cells, the autophagosomes do not localize toward the perinuclear region and are instead peripherally distributed. Collectively, our data indicate that the HCV-induced defect in autophagosome degradation is not due to altered integrity of the autophagosome or the lysosome but is instead a direct result of altered intracellular trafficking.
HCV infection alters the lysosomal Arf-like GTPase, Arl8b
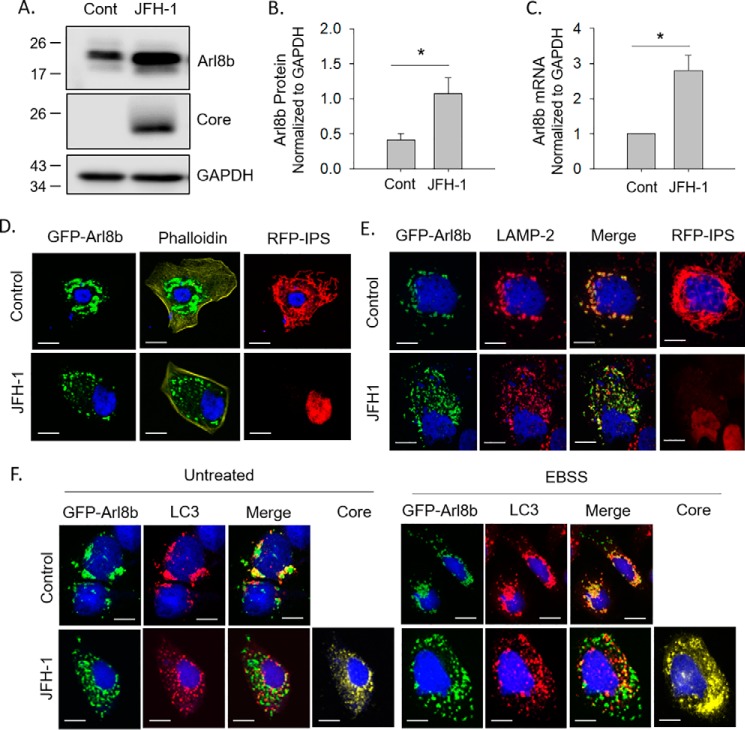
In trying to identify potential proteins involved in autophagosome–lysosome fusion, we examined expression of a number of proteins that had been reported to be involved in lysosomal positioning and autophagic flux. One such protein is the Arf-like GTPase, Arl8b. We found that during HCV infection, both Arl8b protein and mRNA levels are elevated ∼3-fold (Fig. 4, A–C). Next, we examined the localization of Arl8b-positive organelles during infection. We used another Huh-7.5–based reporter cell line that stably expresses an RFP–NLS–IPS fusion protein that is cleavable by HCV NS3 protease. Under basal conditions, the IPS anchor localizes RFP to the mitochondria. After infection, the IPS anchor is cleaved, and RFP–NLS translocates to the nucleus (29). Uninfected and HCV-infected Huh-7.5-RFP–NLS–IPS cells were transfected with a GFP–Arl8b plasmid. Cell boundaries were determined by staining for phalloidin. In uninfected cells, Arl8b localized near the perinuclear region in large, circular structures (Fig. 4D). HCV-infected cells displayed a dramatically altered localization of Arl8b-positive vesicles that were smaller and dispersed throughout the cell periphery (Fig. 4D).
Figure 4.
JFH-1 infection alters the lysosomal Arf-like GTPase, Arl8b. A, Huh-7.5 cells were infected with JFH-1 and assessed for Arl8b expression by Western blotting. B, densitometry analysis of the changes in Arl8b levels, normalized to GAPDH levels, as determined from Western blotting (n = 3). C, quantification of Arl8b mRNA levels normalized to GAPDH. D, Huh-7.5–RFP–IPS cells were transfected with Arl8b–GFP and stained with phalloidin to define cell boundaries. E, overexpression of Arl8b–GFP in Huh-7.5–RFP–IPS cells. The cells were then immunostained for LAMP-2 to identify lysosomes. F, Huh-7.5 cells were transfected with Arl8b–GFP, treated with EBSS, and immunostained for LC3 to identify autophagosomes. Scale bars represent 10 μm. Cont, control.
Arl8b-positive vesicles were located near the perinuclear region and co-localized with LAMP-2, indicating that Arl8b was indeed decorating lysosomes (Fig. 4E). HCV infection did not alter the identity of the Arl8b-positive vesicles because they still co-localized with LAMP-2 (Fig. 4E); however, these Arl8b/LAMP-2 vesicles now distributed throughout the cytoplasm, suggesting that HCV infection resulted in a redistribution of Arl8b-positive lysosomes.
We next assessed the effect of Arl8b on autophagosome positioning. Uninfected and HCV-infected Huh-7.5 cells were transfected with GFP-Arl8b, and the cells were stained for the presence of LC3. In uninfected Huh-7.5 cells, Arl8b-positive vesicles once again were found primarily in the perinuclear region in large puncta and co-localized with LC3-positive vesicles, indicating that the LC3+ autophagosome is fusing with the Arl8b+ lysosome (Fig. 4F). In HCV-infected cells, Arl8b-positive vesicles dispersed into the cellular periphery, and the amount of co-localization with LC3-positive puncta drastically decreased, indicating a complete lack of fusion between autophagosomes and lysosomes. This inhibition of fusion between autophagosomes and lysosomes in HCV-infected cells could not even be overcome by stimulating autophagy with EBSS treatment to simulate starvation. These findings suggest that HCV infection causes Arl8b to be overexpressed within cells, resulting in a repositioning of lysosomes toward the cellular periphery that blocks autophagosome–lysosome fusion.
Impaired autophagic flux in HCV infection is due to an Arl8b-dependent mechanism
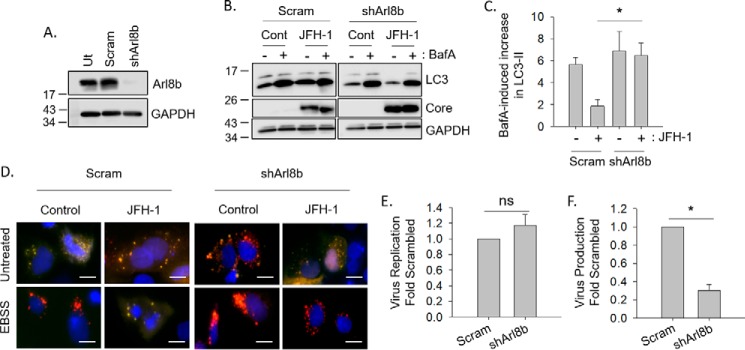
We next determined whether the HCV-induced alteration in autophagosome–lysosome fusion was a consequence of Arl8b-mediated lysosomal repositioning. We first asked whether knocking down Arl8b in HCV-infected cells could alter autophagic flux. By transducing Huh-7.5 cells with a lentivirus expressing an Arl8b shRNA and selecting cells with puromycin, we obtained cells that had a greater than 80% knockdown of Arl8b protein levels (Fig. 5A). Autophagic flux in HCV-infected Arl8b knockdown cells was then measured by treating cells with bafilomycin A1 to block lysosomal clearance of autophagosomes as in Fig. 1. In agreement with our hypothesis, knocking down Arl8b in HCV-infected cells restored autophagic flux to similar levels seen in uninfected cells with almost a 6-fold increase in LC3-II levels after bafilomycin treatment (Fig. 5, B and C). Autophagic flux remained impaired in HCV-infected nontarget shRNA-expressing cells when compared with uninfected cells as was previously shown.
Figure 5.
Impaired autophagic flux in HCV infection is due to an Arl8b-dependent mechanism. A, Huh-7.5 cells stably expressing nontarget shRNA (Scram) or an shRNA to Arl8b assessed by Western blotting. B, cells were infected with JFH-1 and treated with 100 nm bafilomycin A1 (BafA) for 4 h as in Fig. 1. C, densitometry analysis of the changes in LC3 levels after bafilomycin treatment (n = 3). D, nontarget (Scram) or shArl8b Huh-7.5 cells were transfected with RFP–GFP–LC3 plasmid, infected with JFH-1, and treated with EBSS. Fusion was assessed as stated previously. Scale bars represent 10 μm. E, Huh-7.5 cells stably expressing nontarget shRNA (Scram) or shArl8b were infected with the JC1-Luc strain of HCV. Viral replication was assessed in total lysates 3 days postinfection using the Dual-Glo luciferase assay system (n = 4). F, the secretion of infectious virus is decreased when Arl8b is silenced. The cells were infected as in E, and 3 days postinfection, the presence of infectious virus in the culture medium was measured by inoculating virus naïve cells as described under “Experimental procedures” (n = 4). Cont, control.
Next, we examined the effect of Arl8b knockdown on autophagosome–lysosome fusion using the RFP–GFP–LC3 plasmid. As previously seen, JFH-1–infected cells failed to display the normal red puncta after EBSS treatment, and instead the puncta remained yellow, indicating a block in fusion (Figs. 3E and 5D). Arl8b knockdown, however, restored normal fusion in HCV-infected cells as indicated by the presence of red puncta after EBSS treatment (Fig. 5D).
Finally, we assessed the effect of Arl8b knockdown on HCV virus production using a luciferase reporter version of the chimeric JC-1 virus (33). When Arl8b expression was reduced by shRNA, virus secretion decreased 3-fold with no effect on viral replication (Fig. 5, E and F). Taken together, the data suggest that Arl8b up-regulation plays a key role in the HCV-induced autophagic block and is required for efficient virion secretion.
Discussion
It is widely accepted that the autophagy pathway is dramatically altered during HCV infection. HCV infection has been proposed to induce the autophagic response, as measured by increases in LC3-II levels (3–5, 7). However, HCV infection has also been shown to decrease autophagic protein degradation (19, 34). This observation that HCV can increase the number of autophagosomes while simultaneously suppressing autophagic degradation has led to a general confusion regarding whether autophagy goes to completion during HCV infection. In this study, we show that HCV alters the autophagic pathway by suppressing the fusion, and ultimate degradation, of autophagosomes with lysosomes. This finding was seen in both replicon and infection models and with two different HCV genotypes, 2a and 1a.
Through the use of autophagic flux assays as well as vesicular fusion and trafficking assays, we found that the lack of autophagosome degradation in HCV-infected cells was due to a trafficking defect caused by changes in expression of the Arf-like GTPase Arl8b. Effects of Arl8b knockdown were observed for both the JFH-1 strain of the genotype 2a virus as well as the JC-1 strain, a chimeric genotype 2a virus. During infection, Arl8b-dependent repositioning of lysosomes to the periphery serves to impede autophagosome–lysosome fusion, which would normally occur near the perinuclear region.
The data suggest that the main role of Arl8b in the context of HCV infection may be to inhibit autophagy by creating a more peripheral distribution of lysosomes so that autophagosomes and lysosomes do not physically contact each other. However, Arl8b overexpression alone was not sufficient to induce lysosomal repositioning (Fig. 4E, control), indicating that during JFH-1 infection a secondary/additional event must occur to fully reposition the lysosomes. Our lab has also studied the opposing effector protein RILP (Rab-interacting lysosomal protein). RILP links Rab7-containing vesicles to the dynein motor complexes to drive microtubule minus-end-directed transport toward the mitotic center and is thus a key regulator in late endocytic trafficking and the retrograde movement of lysosomes toward the perinuclear region (35, 36). We previously showed that RILP is cleaved specifically during viral infection. This uncouples the Rab7 complex from dynein and promotes outward, kinesin-based trafficking of Rab7-containing vesicles, including lysosomes and multivesicular bodies, toward the cellular periphery (37, 38). Therefore, the peripheral lysosomal repositioning and decreased autophagosome–lysosome fusion observed in the present study likely represent a combination of these two phenomena: the simultaneous loss of dynein binding via RILP cleavage and increased kinesin binding caused by the up-regulation of Arl8b expression.
Multiple reasons exist as to why it might be advantageous for HCV to alter the autophagic pathway. First, autophagy has been shown to play a role in HCV propagation, during both RNA replication as well as protein translation (4, 10, 39, 40). Although there is a consensus that autophagy is important for HCV replication, it is not clear how the autophagic pathway may help HCV replication. If HCV utilizes autophagosomal membranes as part of the replication complexes, then causing stabilization and accumulation of autophagosomes by inhibiting their degradation would provide a source of membranes on which to replicate. Furthermore, if HCV uses the autophagosomal compartments for viral replication and/or assembly, then inhibiting autophagy by repositioning lysosomes and blocking fusion with the autophagosome may also prevent the degradation of not just autophagosomal membranes but also components of the replication complex or viral particles. Another potential advantage for HCV-induced autophagic deregulation would be to increase secretory events ultimately leading to enhanced virion secretion. The simultaneous increase in autophagosome numbers and up-regulation of Arl8b may be a mechanism by which these virion-containing cargo vesicles move toward the cellular membrane.
Pathogens frequently alter various cellular trafficking events as part of their life cycle, whether it be to prevent their destruction by evading contact with lysosomes or to promote their replication by hijacking host organelles. In many cases, cellular GTPases are often targeted. S. typhimurium is a pathogen that bears many similarities to HCV regarding alterations to GTPases. S. typhimurium replicates within vacuoles, which must be prevented from fusing with lysosomes. These vacuoles must also move to the cell periphery as to enable efficient spread of the bacteria. To accomplish this, S. typhimurium not only prevents RILP from binding to Rab7 (41) but also promotes cell-to-cell spread in an Arl8b-dependent manner (25). Viral effects on lysosomal positioning have also been reported. Adenovirus promotes its lifecycle through a direct interaction with dynein. This interaction displaces RILP from the Rab7–dyenin complex, moving the viral components toward the perinuclear region of the cell where viral replication takes place in the nucleus while simultaneously dispersing lysosomes throughout the cell (42).
In conclusion, we have confirmed a role for Arl8b in defective autophagic flux caused by HCV infection. This study adds to our knowledge of how viral infection can modulate GTPases to support their pathogenesis and provides some insight into the importance of organelle positioning in cellular function.
Experimental procedures
Cells and culture conditions
Huh-7.5, Huh-7.5–GFP–IPS, and Huh-7.5–RFP–NLS–IPS cells were obtained from Dr. Charles Rice, Rockefeller Institute. All cell lines were cultured in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose, l-glutamine, and sodium pyruvate, 10% fetal bovine serum, and 1% nonessential amino acids. The cells were maintained at 37 °C with 5% CO2. The H77c genotype 1a HCV replicon cell lines were a gift from Minkyung Yi (University of Texas-Medical Branch) and described previously (43). They were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 μg/ml blasticidin (Invitrogen), and 200 μg/ml Geneticin. Full-length replicon cells were “cured” by culturing with interferon-α (200 units/ml for 4 weeks). The medium used for the culture of cured replicon cell lines contained no Geneticin. The cured cells were used as controls.
Western blotting
Western blotting was performed using anti-LC3B, 1:1000 (Cell Signaling, Beverley, MA); anti-cathepsin D, 1:500 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-GAPDH, 1:1000 (Santa Cruz Biotechnology); anti-core, clone C7-50, 1:1000 (Thermo Scientific); anti-LAMP2 H4B4, 1:2000 (DSHB, University of Iowa); and anti-Arl8b, 1:1000 (Proteintech, Rosemont, IL). Horseradish peroxidase–conjugated secondary antibodies were from Thermo Scientific, and IR-Dye–conjugated secondary antibodies were from LI-COR Biosciences (Lincoln, NE).
HCV infection
JFH-1, a genotype 2a strain, was obtained from Dr. Takaji Wakita (Tokyo, Japan). Huh-7.5 cells were infected with supernatant containing JFH-1 virus at a multiplicity of infection of 1.0 for 24 h. The cells were passaged as necessary, and infection was monitored by immunofluorescence for staining of HCV core protein. JFH-1–infected cells were used in experiments when greater than 70% of the cells were infected.
When indicated, Huh-7.5 cells stably expressing shRNA to Arl8b or nontarget TRC2 were infected with the JC1-Luc strain of HCV (33) (Charles Rice, Rockefeller University) at an multiplicity of infection of 1. After 3 days, extracellular virus was isolated from cell culture supernatants, which were collected and centrifuged at 3,000 × g for 10 min to remove cell debris. The quantification of luciferase reporter activity was used to assay viral titers. 100-μl aliquots of 4-fold dilutions of clarified supernatant cell culture fluids were inoculated onto naïve Huh-7.5 cells seeded 24 h previously in a 96-well plate. After 3 days, the infected cells were washed in PBS, lysed with 20 μl of passive lysis buffer (Promega), and subjected to three rounds of freeze/thaw. The lysates were resuspended by pipetting, and luciferase activity was measured using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's instructions. All luciferase assays were done in triplicate measurements.
Immunofluorescence
Huh-7.5 cells were plated on glass coverslips in 4-well plates. The cells were transfected with Lipofectamine LTX (Thermo Fisher) or Lipofectamine 3000 (Thermo Fisher). ptfLC3 was a gift from Dr. Tamotsu Yoshimori (32) (Addgene, catalog no. 21074), and GFP-Arl8bwt (20) was a gift from Dr. Don J. Mahuran (University of Toronto). For ptfLC3 (RFP–GFP–LC3) experiments, 24 h post-transfection, the cells were treated with either bafilomycin A1 or EBSS for 30 min. The cells were fixed with 3% paraformaldehyde and permeabilized with acetone. For staining with anti-LC3B, the cells were fixed and permeabilized with ice-cold methanol for 10 min. The cells were blocked for 1 h in 1× PBS containing 1% BSA and 1% EDTA at room temperature before being incubated with antibodies. Primary antibodies used were rabbit anti-LC3B, 1:400 (Cell Signaling), mouse anti-LAMP2 H4B4, 1:500 (DSHB), and mouse anti-core clone C7-50, 1:300 (Thermo Fisher). Secondary antibodies used were Alexa Fluor–conjugated donkey anti-mouse or anti-rabbit (Thermo Fisher). Phalloidin-647 (Abcam) was also used. Coverslips were mounted onto slides using Prolong containing DAPI (Thermo Fisher). Images were taken with a Nikon eclipse Ti PFS quantitative fluorescence live-cell and multidimensional imaging system equipped with a digital monochrome Coolsnap HQ2 camera (Roper Scientific, Tucson, AZ) using the MetaMorph software. Confocal images were taken on a Leica TCS SPE confocal configured with a Leica DM550 Q upright microscope.
Autophagy flux assay
The cells were treated with 100 nm bafilomycin A1 for 4 h before being lysed with cell lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton X-100) containing protease inhibitors. The cells treated with 0.1% DMSO were used as a negative control.
Lysosomal enzyme assays
For live-cell imaging, Huh-7.5–GFP–IPS cells were plated on glass-bottomed Microwell dishes (Maktek) at 15,000 cells/dish. Bafilomycin A1 treatment was done at a concentration of 25 nm overnight at 37 °C/5%CO2. The cells were treated with Magic Red cathepsin B reagent (Immunochemistry) at a 1× concentration in HEPES buffer (10 mm HEPES, 133.5 mm NaCl, 2.0 mm CaCl2, 4.0 mm KCl, 1.2 mm MgSO4, 1.2 mm NaH2PO4, 11 mm glucose, pH 7.4) for 1 h at 37 °C with 5%CO2. The nuclei were stained with 1 μg/ml Hoechst stain in HEPES buffer for 20 min at 37 °C with 5% CO2. Live-cell imaging was done using a Nikon eclipse Ti PFS quantitative fluorescence live-cell and multidimensional imaging system equipped with a digital monochrome Coolsnap HQ2 camera (Roper Scientific). Fluorescence images were collected using Metamorph software at wavelengths of 560-nm excitation/607-nm emission for the cathepsin B substrate.
For plate reader assay, Huh-7.5 cells were plated in 96-well plates at 10,000 cells/well with clear bottoms (Grenier Bio-one). The cells were then treated with Magic Red cathepsin B reagent (Immunochemistry) for 2 h at 37 °C/5%CO2. The cells were washed twice with HEPES buffer and lysed with radioimmune precipitation assay buffer for 5 min at 4 °C. Fluorescence measurements were taken at room temperature in a Fluostar Optima plate reader (BMG Labtech, Durham, NC) at excitation/emission wavelengths of 584/620 nm.
To assess cathepsin D processing, Huh-7.5 cells were treated with bafilomycin A1 at a concentration of 25 nm overnight at 37 °C/5% CO2. Cells were trypsinized, washed, and resuspended in 2 ml of homogenization buffer (0.25 m sucrose, 6 mm EDTA, 20 mm HEPES-NaOH, pH 7.4) and homogenized with a Dounce homogenizer with loose-fitting pestle. Homogenate was centrifuged at 3000 × g for 10 min at 4 °C to collect the heavy membrane pellet. The 3000 × g pellet was resuspended in 100 μl of homogenization buffer with protease inhibitors for use in Western blotting.
Subcellular fractionation
Fractionation was carried out as described (31). Briefly, ∼200 million Huh-7.5 cells were collected and ruptured by nitrogen cavitation in a nitrogen bomb (Parr catalog no. 4639) for 7 min at a final pressure of 35 p.s.i. in a volume of 1 ml of 0.25 m sucrose. Disrupted cells were centrifuged at 2000 × g for 5 min at 4 °C to remove intact cells and nuclei. Supernatant containing organelles was centrifuged at 17,000 × g for 12 min at 4 °C to separate organelles (pellet) from cytosol and endoplasmic reticulum fractions (supernatant). The pellet was resuspended in 0.25 m sucrose and mixed with 85.6% Nycodenz (Accurate Chemical) and placed in the bottom of an ultracentrifuge tube. On top of the homogenate, 26% Nycodenz, 24% Nycodenz, 20% Nycodenz, and 15% Nycodenz was layered. Gradients were spun at 104,333 × g for 3 h at 4 °C in a SW41Ti swinging bucket rotor. Autophagosomes were collected from the interface between 15 and 20% Nycodenz, and lysosomes were collected between 24 and 26% Nycodenz. The fractions were washed with 0.25 m sucrose, and autophagosomes and lysosomes were resuspended in 100 μl of 0.25 m sucrose with protease inhibitors for use in the in vitro fusion assay.
In vitro fusion assay and flow cytometry
In vitro fusion assay carried out as described (31). Briefly, isolated autophagosomes were incubated with anti-LC3-PE antibody (Cell Signaling) at a 1:50 dilution, and isolated lysosomes were separately incubated with anti–LAMP2-APC (BD Biosciences) at a 1:20 dilution in 0.25 m sucrose. Antibodies were incubated with organelles for 30 min at room temperature. Organelles were washed with 0.25 m sucrose to remove any unbound antibody. Organelles were resuspended in 5 μl of fusion buffer (10 mm HEPES, pH 7.3, 10 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 0.25 m sucrose) per reaction, and 5 μl of autophagosomes were combined with 5 μl of lysosomes. 5 μl of reaction buffer (0.25 m sucrose, 3 mm ATP, 3 mm GTP, 0.16 mg/ml creatine phosphokinase, 8 mm phosphocreatine, protease inhibitor (Sigma P8340 at 1:100 dilution)) was added to the autophagosome–lysosome mixture, and the fusion reaction was carried out at 37 °C for 30 min. The total volume of the reaction was brought up to 100 μl with 0.25 m sucrose, and 100 μl of 2% paraformaldehyde was added for fixation. Fixed samples were subjected to flow cytometry using a LSR II instrument (BD Biosciences).
Real-time quantitative PCR
RNA was isolated from Huh-7.5 cells using TRI reagent (Ambion) followed by cDNA synthesis using a high-capacity cDNA reverse-transcription kit (Life Technologies). Quantitative PCRs were done using iQ SYBR Green (Bio-Rad) on a CFX96 real-time system on a C1000 Thermal Cycler (Bio-Rad). The following primers were used: 5′-GCGGTATTGCAGAGGAGTCA-3′ (forward primer) and 5′-CCAAGCACTAGCACTGGAA-3′ (reverse primer) for Arl8b and 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward primer) and 5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse primer) for GAPDH.
shRNA-mediated knockdown of Arl8b
MISSION Arl8b shRNA (clone ID NM_018184.2–671s21c1) and TRC2 nontarget shRNA (SHC216) were obtained from Sigma–Aldrich. 293FT cells were transfected with MISSION plasmids, psPax2, and pMDG.2 vectors to produce lentivirus using X-tremeGENE HP transfection reagent (Roche). Lentivirus was collected after 24–48 h. Huh-7.5 cells were transduced with lentivirus and 8 μg/ml Polybrene. The cells were selected with 3 μg/ml puromycin and maintained with 1 μg/ml puromycin.
Statistics
All statistics were performed as stated in the figure legends.
Author contributions
K. N. J.-J. and A. L. W. data curation; K. N. J.-J. and A. L. W. formal analysis; K. N. J.-J., A. L. W., H. K., R. R., and S. A. W. methodology; K. N. J.-J., A. L. W., and S. A. W. writing-original draft; A. L. W., H. K., R. R., and S. A. W. writing-review and editing; S. A. W. conceptualization; S. A. W. supervision; S. A. W. funding acquisition.
Acknowledgments
We thank Dr. MinKyung Yi for providing materials and Dr. Ana Maria Cuervo for technical expertise on the in vitro fusion assay and comments on the manuscript. We acknowledge the Flow Cytometry Core Laboratory, which is sponsored in part by NIGMS, National Institutes of Health COBRE Grant P30 GM103326, and the Confocal Core, which is sponsored by NIGMS, National Institutes of Health COBRE Grant P30 GM122731.
This work was supported by the National Institutes of Health Grant P30 GM118247 and the Lied Pilot Grant Program from the University of Kansas Medical Center Research Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HCV
- hepatitis C virus
- EBSS
- Earle's balanced salt solution
- shRNA
- small hairpin RNA
- IPS
- Interferon beta promoter stimulator protein 1.
References
- 1. Schmid D., and Münz C. (2007) Innate and adaptive immunity through autophagy. Immunity 27, 11–21 10.1016/j.immuni.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim H. J., Lee S., and Jung J. U. (2010) When autophagy meets viruses: a double-edged sword with functions in defense and offense. Semin. Immunopathol. 32, 323–341 10.1007/s00281-010-0226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sir D., Chen W. L., Choi J., Wakita T., Yen T. S., and Ou J. H. (2008) Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48, 1054–1061 10.1002/hep.22464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dreux M., Gastaminza P., Wieland S. F., and Chisari F. V. (2009) The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 106, 14046–14051 10.1073/pnas.0907344106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanida I., Fukasawa M., Ueno T., Kominami E., Wakita T., and Hanada K. (2009) Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5, 937–945 10.4161/auto.5.7.9243 [DOI] [PubMed] [Google Scholar]
- 6. Ke P. Y., and Chen S. S. (2011) Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 121, 37–56 10.1172/JCI41474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ait-Goughoulte M., Kanda T., Meyer K., Ryerse J. S., Ray R. B., and Ray R. (2008) Hepatitis C virus genotype 1a growth and induction of autophagy. J. Virol. 82, 2241–2249 10.1128/JVI.02093-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quan M., Liu S., Li G., Wang Q., Zhang J., Zhang M., Li M., Gao P., Feng S., and Cheng J. (2014) A functional role for NS5ATP9 in the induction of HCV NS5A-mediated autophagy. J. Viral Hepat. 21, 405–415 10.1111/jvh.12155 [DOI] [PubMed] [Google Scholar]
- 9. Shrivastava S., Raychoudhuri A., Steele R., Ray R., and Ray R. B. (2011) Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53, 406–414 10.1002/hep.24073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sir D., Kuo C. F., Tian Y., Liu H. M., Huang E. J., Jung J. U., Machida K., and Ou J. H. (2012) Replication of hepatitis C virus RNA on autophagosomal membranes. J. Biol. Chem. 287, 18036–18043 10.1074/jbc.M111.320085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shinohara Y., Imajo K., Yoneda M., Tomeno W., Ogawa Y., Kirikoshi H., Funakoshi K., Ikeda M., Kato N., Nakajima A., and Saito S. (2013) Unfolded protein response pathways regulate hepatitis C virus replication via modulation of autophagy. Biochem. Biophys. Res. Commun. 432, 326–332 10.1016/j.bbrc.2013.01.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. Y., Wang L., Lee J., and Ou J. J. (2017) Hepatitis C virus induces the localization of lipid rafts to autophagosomes for its RNA replication. J. Virol. 91, e00541–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shrivastava S., Devhare P., Sujijantarat N., Steele R., Kwon Y. C., Ray R., and Ray R. B. (2016) Knockdown of autophagy inhibits infectious hepatitis C virus release by the exosomal pathway. J. Virol. 90, 1387–1396 10.1128/JVI.02383-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vescovo T., Romagnoli A., Perdomo A. B., Corazzari M., Ciccosanti F., Alonzi T., Nardacci R., Ippolito G., Tripodi M., Garcia-Monzon C., Lo Iacono O., Piacentini M., and Fimia G. M. (2012) Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology 142, 644–653.e3 10.1053/j.gastro.2011.11.033 [DOI] [PubMed] [Google Scholar]
- 15. Chandra P. K., Bao L., Song K., Aboulnasr F. M., Baker D. P., Shores N., Wimley W. C., Liu S., Hagedorn C. H., Fuchs S. Y., Wu T., Balart L. A., and Dash S. (2014) HCV infection selectively impairs type I but not type III IFN signaling. Am. J. Pathol. 184, 214–229 10.1016/j.ajpath.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J., Kang R., Huang H., Xi X., Wang B., Wang J., and Zhao Z. (2014) Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression. Autophagy 10, 766–784 10.4161/auto.27954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H., Kang R., Wang J., Luo G., Yang W., and Zhao Z. (2013) Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy 9, 175–195 10.4161/auto.22791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shrivastava S., Bhanja Chowdhury J., Steele R., Ray R., and Ray R. B. (2012) Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J. Virol. 86, 8705–8712 10.1128/JVI.00616-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L., Tian Y., and Ou J. H. (2015) HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS Pathogens 11, e1004764 10.1371/journal.ppat.1004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bagshaw R. D., Callahan J. W., and Mahuran D. J. (2006) The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344, 1186–1191 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- 21. Rosa-Ferreira C., and Munro S. (2011) Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell 21, 1171–1178 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korolchuk V. I., and Rubinsztein D. C. (2011) Regulation of autophagy by lysosomal positioning. Autophagy 7, 927–928 10.4161/auto.7.8.15862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bejarano E., Murray J. W., Wang X., Pampliega O., Yin D., Patel B., Yuste A., Wolkoff A. W., and Cuervo A. M. (2018) Defective recruitment of motor proteins to autophagic compartments contributes to autophagic failure in aging. Aging Cell e12777 10.1111/acel.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., and Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaniuk N. A., Canadien V., Bagshaw R. D., Bakowski M., Braun V., Landekic M., Mitra S., Huang J., Heo W. D., Meyer T., Pelletier L., Andrews-Polymenis H., McClelland M., Pawson T., Grinstein S., et al. (2011) Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell. Microbiol. 13, 1812–1823 10.1111/j.1462-5822.2011.01663.x [DOI] [PubMed] [Google Scholar]
- 26. Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y. (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- 27. Aggarwal N., and Sloane B. F. (2014) Cathepsin B: multiple roles in cancer. Proteomics. Clin. Appl. 8, 427–437 10.1002/prca.201300105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Creasy B. M., Hartmann C. B., White F. K., and McCoy K. L. (2007) New assay using fluorogenic substrates and immunofluorescence staining to measure cysteine cathepsin activity in live cell subpopulations. Cytometry 71, 114–123 [DOI] [PubMed] [Google Scholar]
- 29. Jones C. T., Catanese M. T., Law L. M., Khetani S. R., Syder A. J., Ploss A., Oh T. S., Schoggins J. W., MacDonald M. R., Bhatia S. N., and Rice C. M. (2010) Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28, 167–171 10.1038/nbt.1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zaidi N., Maurer A., Nieke S., and Kalbacher H. (2008) Cathepsin D: a cellular roadmap. Biochem. Biophys. Res. Commun. 376, 5–9 10.1016/j.bbrc.2008.08.099 [DOI] [PubMed] [Google Scholar]
- 31. Koga H., Kaushik S., and Cuervo A. M. (2010) Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 24, 3052–3065 10.1096/fj.09-144519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura S., Noda T., and Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- 33. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., and Rice C. M. (2005) Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- 34. Taguwa S., Kambara H., Fujita N., Noda T., Yoshimori T., Koike K., Moriishi K., and Matsuura Y. (2011) Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J. Virol. 85, 13185–13194 10.1128/JVI.06099-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cantalupo G., Alifano P., Roberti V., Bruni C. B., and Bucci C. (2001) Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683–693 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J. (2001) The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- 37. Adams A., Weinman S. A., and Wozniak A. L. (2018) Caspase-1 regulates cellular trafficking via cleavage of the Rab7 adaptor protein RILP. Biochem. Biophys. Res. Commun. 503, 2619–2624 10.1016/j.bbrc.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wozniak A. L., Long A., Jones-Jamtgaard K. N., and Weinman S. A. (2016) Hepatitis C virus promotes virion secretion through cleavage of the Rab7 adaptor protein RILP. Proc. Natl. Acad. Sci. U.S.A. 113, 12484–12489 10.1073/pnas.1607277113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fahmy A. M., and Labonté P. (2017) The autophagy elongation complex (ATG5–12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci. Rep. 7, 40351 10.1038/srep40351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferraris P., Blanchard E., and Roingeard P. (2010) Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91, 2230–2237 10.1099/vir.0.022186-0 [DOI] [PubMed] [Google Scholar]
- 41. Harrison R. E., Brumell J. H., Khandani A., Bucci C., Scott C. C., Jiang X., Finlay B. B., and Grinstein S. (2004) Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol. Biol. Cell 15, 3146–3154 10.1091/mbc.e04-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scherer J., Yi J., and Vallee R. B. (2014) PKA-dependent dynein switching from lysosomes to adenovirus: a novel form of host-virus competition. J. Cell Biol. 205, 163–177 10.1083/jcb.201307116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wozniak A. L., Griffin S., Rowlands D., Harris M., Yi M., Lemon S. M., and Weinman S. A. (2010) Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathogens 6, e1001087 10.1371/journal.ppat.1001087 [DOI] [PMC free article] [PubMed] [Google Scholar]