Abstract
Imaging of rod photoreceptor outer-segment disc membranes by atomic force microscopy and cryo-electron tomography has revealed that the visual pigment rhodopsin, a prototypical class A G protein–coupled receptor (GPCR), can organize as rows of dimers. GPCR dimerization and oligomerization offer possibilities for allosteric regulation of GPCR activity, but the detailed structures and mechanism remain elusive. In this investigation, we made use of the high rhodopsin density in the native disc membranes and of a bifunctional cross-linker that preserves the native rhodopsin arrangement by covalently tethering rhodopsins via Lys residue side chains. We purified cross-linked rhodopsin dimers and reconstituted them into nanodiscs for cryo-EM analysis. We present cryo-EM structures of the cross-linked rhodopsin dimer as well as a rhodopsin dimer reconstituted into nanodiscs from purified monomers. We demonstrate the presence of a preferential 2-fold symmetrical dimerization interface mediated by transmembrane helix 1 and the cytoplasmic helix 8 of rhodopsin. We confirmed this dimer interface by double electron–electron resonance measurements of spin-labeled rhodopsin. We propose that this interface and the arrangement of two protomers is a prerequisite for the formation of the observed rows of dimers. We anticipate that the approach outlined here could be extended to other GPCRs or membrane receptors to better understand specific receptor dimerization mechanisms.
Keywords: G protein-coupled receptor (GPCR), receptor, rhodopsin, retinoid-binding protein, retina, cryo-electron microscopy (cryo-EM), rod outer segment, transmembrane helix 1 (TM1), helix 8 (H8), cell signaling, double electron-electron resonance (DEER), rhodopsin dimerization, transducin
Introduction
G protein–coupled receptors (GPCRs)6 are the largest and most conserved family of membrane receptors in higher eukaryotes; they respond to and transduce sensory, chemotactic, hormonal, and neuronal signals that encompass most of the physiological processes in development and disease (1–3). Rhodopsin-like (or class A) GPCRs consist of a hydrophobic core of seven transmembrane α-helices (TM1–TM7) that interact to form a helix bundle (4, 5), an extracellular N-terminal region bearing N-linked glycosylation sites, and a C-terminal region that starts after TM7. TM7 is typically followed by a short cytoplasmic helix 8 (H8) that is often tethered by palmitoylated Cys residues and runs parallel to the membrane. Class A GPCRs are further divided into groups associated with a particular ligand specificity, such as the amines, peptides, cannabinoids, odorants, and retinals, with the prototypical light-sensing rhodopsin being the best-characterized (1, 6).
A body of growing biophysical, pharmacological, and genetic evidence indicates that many additional class A GPCRs other than rhodopsin (7, 8) can form concentration-dependent dimers and oligomers, which could be mediated by transient hydrophobic interactions between TM α-helical domains and other integrations of the extracellular domains (e.g. neurotensin 1 (9, 10), D1/D2-dopamine (11–13), β1- and β2-adrenergic (AR) (14–16), and C5a (17) receptors). From studies with a multitude of class A GPCRs, it seems true that the symmetrical intradimeric interface involves TM1 and H8 or TM4–6 (11, 12, 15, 17–19). Adding to the complexity in GPCR dimerization is its effect with respect to downstream signaling (20, 21). Although monomeric GPCRs can efficiently couple to the heterotrimeric G protein and arrestin in vitro (22–24), conformational changes at the intradimeric GPCR interface can transmit additional information to enhance or weaken the particular downstream signaling activity in vivo (10, 25–27). Further, some GPCR dimers have been shown to signal through intermolecular cooperation (e.g. luteinizing hormone receptor) such that binding-deficient and signaling-deficient forms of a receptor can re-establish normal signaling through functional complementation in vivo (28).
The recent cryo-EM structure of the metabotropic glutamate receptor subtype 5 confirmed that class C GPCRs are obligate dimers that are commonly linked by disulfide bonds or coiled-coil interactions between the two protomers in the case of GABAB receptors (29, 30). Higher-order complexes are essential for cellular trafficking and signaling of these receptors (31). However, it has yet to be concluded whether there is a common dimeric interface that is employed by most GPCR dimers or if there are very distinct dimerization/oligomerization strategies specific to a particular native environment (32). Higher-order organization of some class A GPCRs in native tissues is critical to their signaling properties, regulation, and allosteric pharmacology as, for example, observed for the muscarinic M2 receptor (33).
Although it had been demonstrated that a single rhodopsin is sufficient for activation of its G protein transducin (Gt), the high density of rhodopsin (24,000–30,000 molecules/μm2) (34, 35) in the rod disc membranes suggests that dimers—measured to be present already at low concentrations when reconstituted in liposomes (36) or when expressed as opsin apoprotein in mammalian cells (37, 38)—form another active structural unit. Previous atomic force microscopy, cryo-EM, and lateral diffusion studies showed that the native organization of rhodopsin in isolated mouse rod outer segment (ROS) disc membranes can even form paracrystalline areas, depending on poorly defined factors (7, 8, 39, 40). These results were corroborated, as rhodopsin could be extracted as dimers or as entire rows of dimers with mild detergents (41). With peptide-interfering assays, we determined that TM1, TM2, TM4, and TM5 are all significant for rhodopsin dimerization or oligomerization (42). Structural evidence of rhodopsin dimerization emanating from crystallography studies of photoactivated rhodopsin were suggestive of an interaction interface involving TM1, TM2, and H8 (43–47). In this report, we present a cryo-EM structure and complementary biophysical characterization of the rhodopsin dimer with the primary intradimeric interface formed by TM1 and H8 of each rhodopsin as well as a secondary interdimeric contact that is formed when dimers oligomerize.
Results
Characterization of rhodopsin dimers in nanodiscs
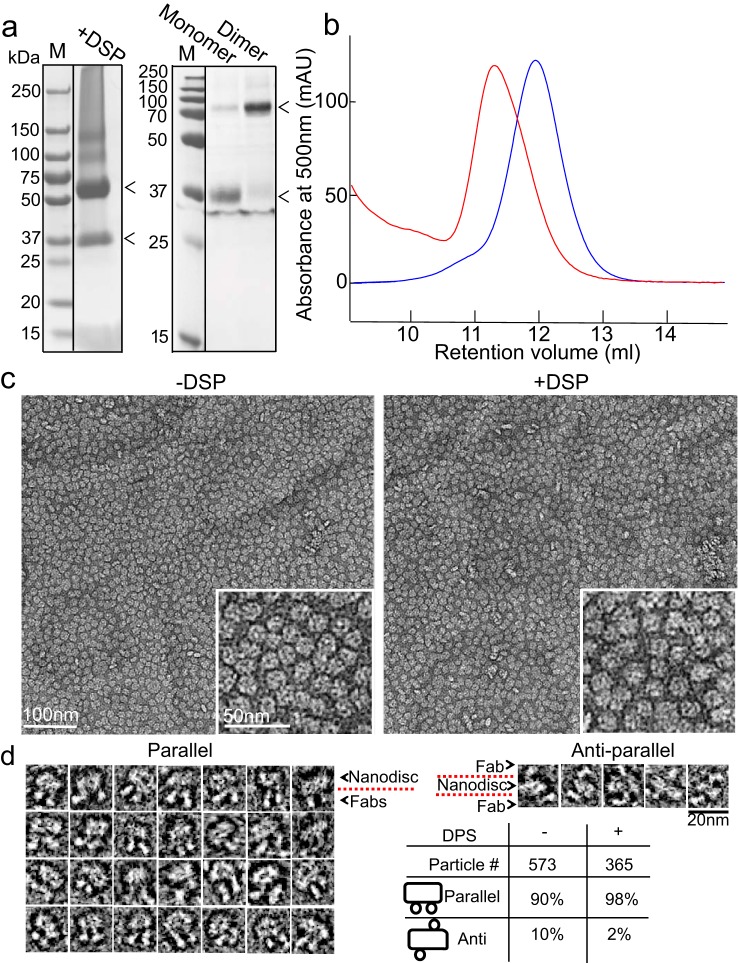
We generated nanodiscs contain rhodopsin dimer with the MSP1E3D1 protein. This belt protein, which is known to produce a disc diameter of ∼130 Å (48), should be able to properly accommodate a dimeric rhodopsin structure (∼100 Å in the widest dimension). Following rhodopsin solubilization from disc membranes in 3% n-octylglucoside (OG) and fractionation by gel filtration, we reconstituted the dimer into nanodiscs as described previously (at a 1:1:80 ratio of rhodopsin/MSP1E3D1/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids) (48, 49). To identify the native rhodopsin dimerization interface, we also incubated disc membranes with a homobifunctional cross-linker with a length of 12 Å prior to solubilization that can conjugate proteins through the ϵ-amino group of Lys residues (dithiobis(succinimidyl)propionate (DSP) cross-linker; Fig. S1, a and b). Following solubilization and gel filtration purification, cross-linked rhodopsin dimers were similarly incorporated into nanodiscs at a 1:1:80 ratio of rhodopsin/MSP1E3D1/POPC lipids.
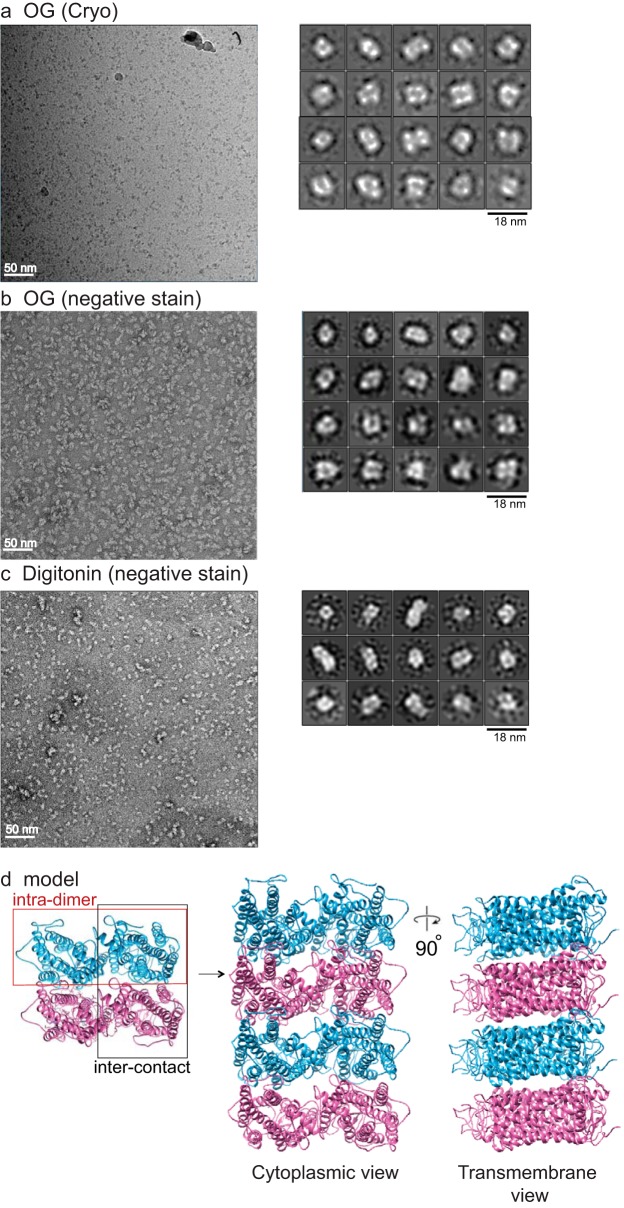
After overnight incubation with BioBeads resin for detergent removal, we observed a relatively pure dimer nanodisc population that was well-separated from monomer-containing nanodiscs by gel filtration (Fig. 1 (a and b) and Fig. S1 (c and d)). The ratio of different forms could be deduced from the chromatography profile (Fig. S1c). Negatively stained transmission EM (TEM) micrographs of rhodopsin dimers with or without cross-linking showed that rhodopsin dimer nanodiscs were homogeneous in size (Fig. 1c). To determine the orientation of the rhodopsin dimers following reconstitution, we incubated rhodopsin dimer nanodiscs with the 1D4-Fab generated by papain cleavage of the monoclonal rhodopsin 1D4 antibody, at a 1:10–15 ratio of rhodopsin dimer nanodisc/Fab overnight and quantified them (Fig. 1d). The rhodopsin dimers in nanodiscs are mostly parallel, with the in vitro reconstituted rhodopsin dimers at a 90:10 ratio of parallel/anti-parallel and in disc membrane cross-linked rhodopsin dimers at a 98:2 ratio of parallel/anti-parallel. From our study, as well as others, it appears that rhodopsin has a propensity to form parallel dimers when reconstituted into a lipid environment in vitro (18). Therefore, we could obtain parallel rhodopsin dimers in nanodiscs either by reconstitution in vitro or by cross-linking in disc membranes for structure determination by cryo-EM.
Figure 1.
Formation of rhodopsin dimers in nanodiscs and characterization by 1D4 Fab. a, Coomassie-stained gels show that by cross-linking, primarily dimers were obtained after solubilization, before (left) or after (right) gel filtration (Fig. S1c). The arrowheads point to the positions of rhodopsin dimer and monomer on the gel. b, gel filtration of rhodopsin dimer-containing nanodiscs (red) and monomer-containing nanodiscs (blue) showing distinct peaks. c, negatively stained micrographs show that nanodiscs with in vitro reconstituted or in disc membranes cross-linked (with DSP) rhodopsin dimer are uniform in size. d, 1D4 Fab quantification of dimer orientation shows that both in disc membranes cross-linked dimers and the in vitro reconstituted dimers form parallel dimers.
Rhodopsin dimers in nanodiscs are functional
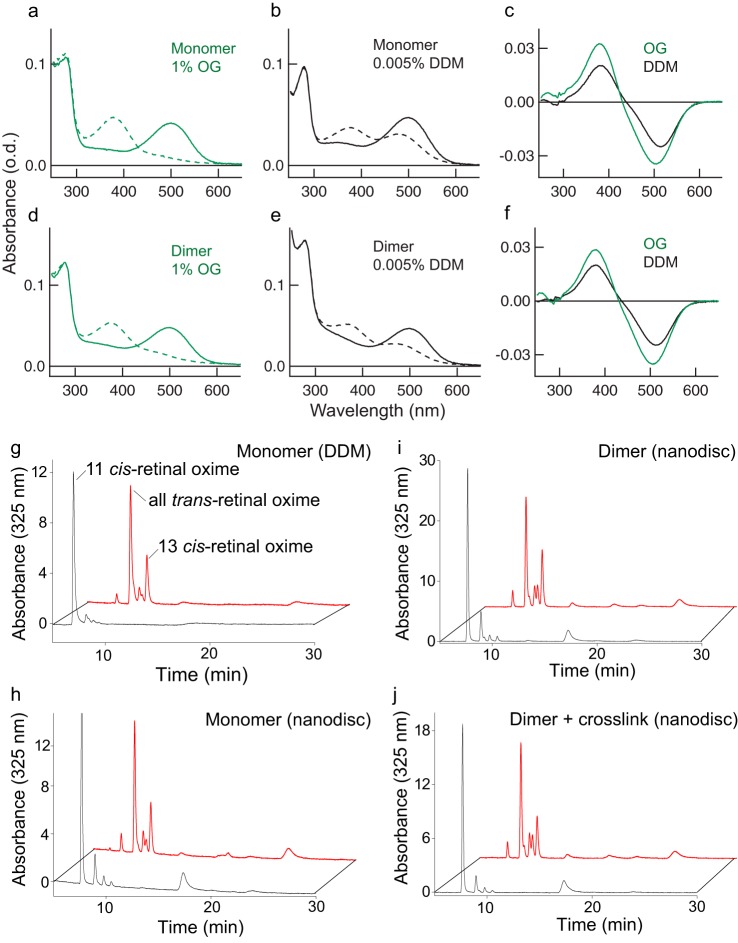
We conducted light activation and retinoid analyses to show that the cross-linked rhodopsin dimer retains photochemical activity after extraction from disc membranes and insertion into nanodiscs (Fig. 2). We first showed that following cross-linking in disc membranes and gel filtration, both rhodopsin dimer and monomer fractions could transition to Meta-II (λmax of 380 nm) after light activation in various detergents that include OG and n-dodecyl-β-d-maltoside (DDM) (Fig. 2, a–f). However, due to the presence of the DSP cross-linker on multiple Lys residues of the cytoplasmic loops, G protein–binding activity of the cross-linked rhodopsin monomer or dimer was abolished. This was tested with a high-affinity peptide derived from the C terminus of the Gtα-subunit that was previously co-crystallized with the active receptor (43) (Fig. S2). Next, when rhodopsin dimers with or without cross-linking were incorporated into nanodiscs, the binding of the Gtα-subunit peptide was abolished (cross-linked dimer) or reduced (no cross-link), as the Meta-II transition upon light activation was hindered by the physical constraint of the nanodiscs, and therefore more of the Meta-I state was observed in both cases (Fig. S2). Reduced binding of Gt to rhodopsin dimers in nanodiscs has been observed previously (48). We then performed retinoid analyses with HPLC that separates 11-cis-retinal and all-trans-retinal to show that rhodopsin monomers and dimers with or without cross-linking in the nanodiscs undergo retinal isomerization under light stimulation (Fig. 2, g–j). Compared with the dark state, upon light activation, the 11-cis peak is replaced by the all-trans peak in all cases.
Figure 2.
Functional activity of rhodopsin dimer by Meta-II transition and retinoid analyses. Shown are UV-visible spectra of DSP-cross-linked rhodopsin monomer (a and b) and dimer (d and e) fractionated by gel filtration and diluted with 1.0% OG (a and d) or 0.005% DDM (b and e). Dark- and light-activated rhodopsin spectra are indicated as solid lines and dashed lines, respectively. Difference spectra (after activation − before activation) for monomer (c) and dimer (f) are shown to expand the light-dependent formation of active Meta-II (380 nm) from dark rhodopsin (500 nm). g–j, retinoid analysis shows normal 11-cis-retinal to all-trans-retinal transition upon light activation; some 13-cis-retinal is observed as a by-product in all cases. The retinals in monomer in DDM (g) or in nanodiscs (h) and dimer in nanodiscs without cross-link (i) or with cross-link (j) were converted to corresponding retinal oximes and analyzed by HPLC monitoring the UV absorbance at 325 nm. Compared with the dark state (black line), light activation (red line) shows a shift from 11-cis-retinal to all-trans-retinal.
Cryo-EM of rhodopsin dimer cross-linked in disc membranes
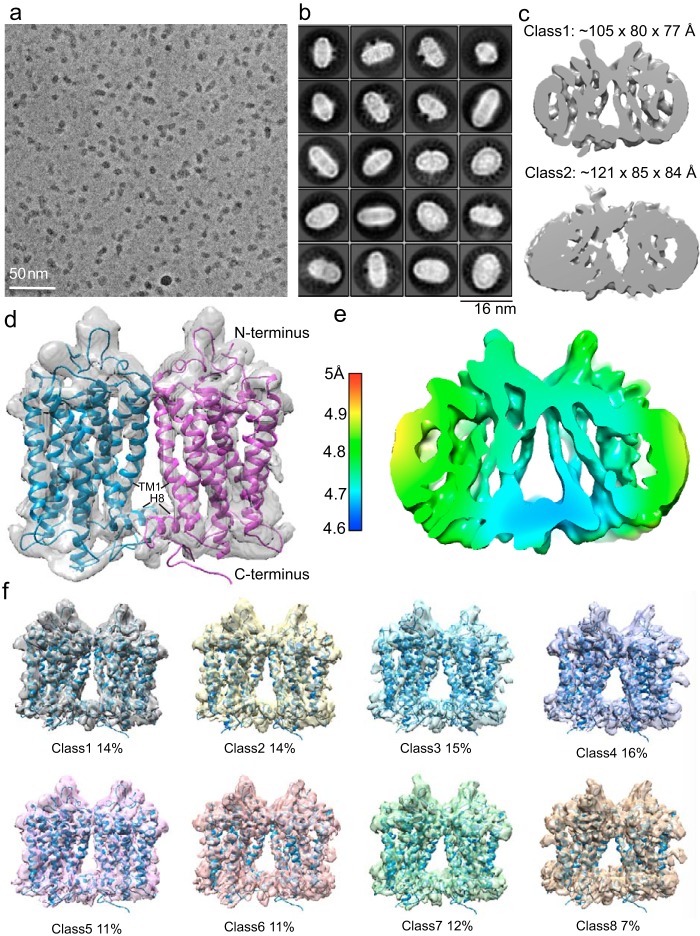
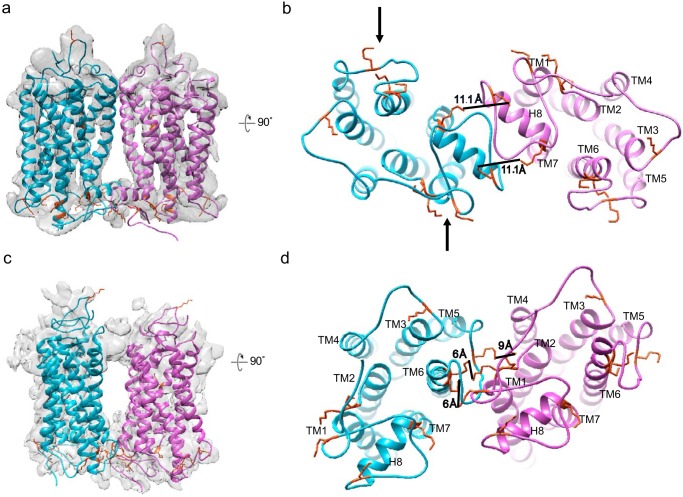
Images of rhodopsin dimer (∼70 kDa) in nanodiscs were recorded at 300 kV with a Titan Krios equipped with a Bioquantum energy filter, a K2 direct electron detector, and a Volta phase plate (VPP) that improves image contrast (Fig. S3) (50–52). For the cross-linked dimer sample, picked particles were subjected to 2D classification in Relion 2, resulting in many class averages with recognizable secondary structure elements (Fig. 3, a and b), and 762,000 particles were selected for 3D classifications (53). Classification into three classes was performed with imposed C2 symmetry, resulting in the most populated class (class 1) containing 434,000 particles (57% of all aligned particles in 2D classification) (Fig. 3c and Fig. S4a). The refinement (C2 symmetry) for class 1 dimer reached a resolution of ∼4.7 Å, as determined by the gold-standard Fourier shell correlation (FSC) = 0.143 criterion between the independently refined two half-sets (53) (Fig. S4, b and c). The angular distribution indicated complete coverage of all particle orientations (Fig. S4d). Rigid-body fitting allowed the docking of two rhodopsin crystal structures (PDB entry 1U19) (54) into the refined cryo-EM density of class 1, yielding a good visual fit (Fig. 3d); the intradimeric interface is clearly observed to be mediated by TM1 and H8. Local resolution estimation showed that the TMs were resolved to a higher resolution and are more rigid than the intraluminal domains (Fig. 3e). Further 3D classification (C2 symmetry) on class 1 into eight classes did not identify additional intradimeric interfaces other than TM1 and H8 (Fig. 3f). Our finding is supported by studies including cysteine cross-link/MS in disc membranes (55) and in vivo retinal peptide competition assays (56), both of which suggested TM1 and H8 at the dimer interface. Our intradimeric interface is estimated to include a surface area of >300 Å2 created mainly by hydrophobic residues (55), suggesting that it could be weak and reversible in the native ROS disc membrane.
Figure 3.
Cryo-EM analysis of the rhodopsin dimer cross-linked in native disc membranes and then reconstituted into nanodiscs. a, representative cryo-EM micrograph imaged at 300 kV with the VPP in combination with a K2 detector at a pixel size of 0.8 Å and a total dose of 50 e−/Å2. b, Relion 2D classification; classes are aligned to the protein density and are ranked by population. c, 762,000 particles were selected for 3D classifications; the two main refined classes have different dimensions. d, upon the removal of nanodisc EM density, docking of rhodopsin (PDB entry 1U19) into the rhodopsin EM density shows that the intradimeric interface is mediated by TM1 and H8 of each rhodopsin. e, local resolution estimation. f, class 1 particles were further classified by a second round of 3D classification (C2 symmetry) for eight classes; all analyses showed TM1 and H8 at the intradimeric interface, and no additional dimer configuration was identified. Segmented views with the docking of rhodopsin (PDB entry 1U19) are shown.
3D classification also was performed on the 762,000 cross-linked particles without imposing any symmetry; two major classes (class 1 and class 2) were identified to contain 318,000 particles (42% of all aligned particles) and 373,000 particles (49% of all aligned particles), respectively (Fig. S5a). These two classes have visibly different dimensions, suggesting distinct dimer arrangements and, therefore, different nanodisc sizes (Fig. 3c and Fig. S5a). Class 1 particles led to a refined (C1 symmetry) cryo-EM density map that resembles Fig. 3d, allowing the same docking of two rhodopsin molecules (PDB entry 1U19) interfaced by TM1 and H8 (Fig. S5b) with the angular distribution covering all particle orientations (Fig. S5c). Further asymmetric 3D classifications of class 1 into three classes did not identify additional intradimeric interfaces (Fig. S5d). In all 3D refinement runs (C1 or C2 symmetry), local refinement searches including a mask around the dimer were performed after the initial global 3D refinement without masking.
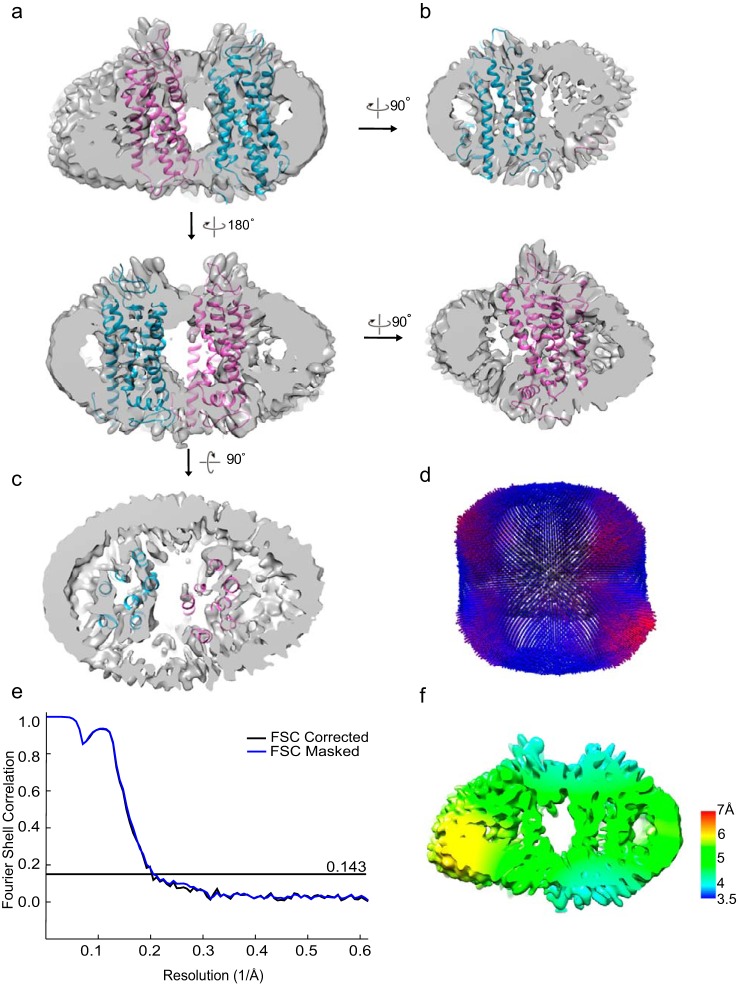
The class 2 dimer from the asymmetric 3D classification was further classified and refined to 4.5–6 Å (C1 symmetry); 213,000 particles eventually revealed an interface formed by TM1 and TM2 of one rhodopsin and the cytoplasmic portion between TM5 and TM6 of the adjacent rhodopsin (Figs. 3c, 4, and 5). We suggest that this hints at the interdimeric contact that occurs when the paracrystalline track of rhodopsin dimer rows forms, previously observed by atomic force microscopy and by cryo-electron tomography (7, 8). This contact interface is likely flexible, and due to constraints by the small nanodiscs, the observed arrangement of the two protomers is distorted by a few degrees, which prevents modeling into rows. We modeled the positions of Lys residues that can mediate cross-linking formation in both class 1 and class 2 EM densities to show that, given the flexibility of the cytoplasmic loops, multiple Lys residues are available within the allowed DSP cross-linker distance of 12 Å to stabilize the dimeric interfaces (Fig. 5).
Figure 4.
An interdimeric contact is observed in the cross-linked rhodopsin dimer cryo-EM data set. a–c, from 3D classification (C1 symmetry) class 2, a refined density map (C1 symmetry) with 213,000 particles was obtained. Cross-sectional views of the rhodopsin dimer with the docking of rhodopsin (PDB entry 1U19) are shown. d, angular distribution of the refined structure shows no strong bias in particle orientation for the reconstruction. e, Relion post-processing with the FSC = 0.143 criterion. f, local resolution estimation.
Figure 5.
Modeling of Lys ϵ-amino groups as cross-link sites in the intradimeric interface and the interdimeric contact. A total of nine exposed Lys residues on the cytoplasmic side of rhodopsin are available for chemical cross-linking by DSP. Black arrows point to Lys residues that can be linked to DSP and thus block Gt interaction. Shown are side (a) and cytoplasmic (b) views of the intradimeric interface with rhodopsin crystal structure (PDB entry 1U19) docked into the EM densities. Lys-311 and Lys-325 on both rhodopsin molecules allow two cross-links to form. The cross-sectional area taken up by intradimer is 84 × 47 Å = 3948 Å2. Shown are side (c) and cytoplasmic (d) views of the interdimeric contact; Lys-245/248/231 on one rhodopsin can cross-link with Lys-66/67 on the adjacent rhodopsin based on the 12-Å length of the DSP linker. The cross-sectional area of the interdimer is 87 × 64 Å = 5568 Å2. The MSP1E3D1 nanodisc is relatively small for the observed large cross-sectional area and could result in some distortion of the rhodopsin arrangement.
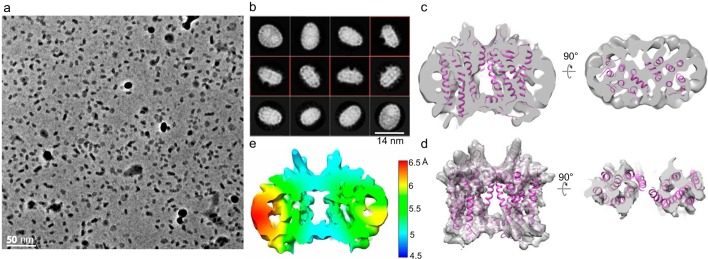
Further corroborating the existence of the intra- and interdimeric rhodopsin interfaces in the retina, rhodopsin tetramers of 160 kDa (Fig. 1a and Fig. S1 (c and d)) were purified similarly upon cross-linking in disc membranes, followed by solubilization with 2% OG or 0.1% digitonin for gel filtration fractionation. Because of their larger sizes, tetramers cannot be incorporated into MSP1E3D1 nanodiscs for structural determination by cryo-EM. Nevertheless, from negatively stained EM and cryo-EM images of the detergent-solubilized samples, the majority of the 2D classes form a square-shaped configuration rather than linear, L-shaped, or T-shaped configurations (Fig. 6, a–c). This indicates that via the intradimer interface at TM1 and H8, the basic unit forms and that multiple units can arrange in parallel, given some flexibility of the observed interdimeric contact, to eventually form rows of dimers. A model of the oligomeric organization of rhodopsin is presented in Fig. 6d.
Figure 6.
Cross-linked rhodopsin tetramer structure and model of rhodopsin oligomers. a, rhodopsin cross-linked in disc membranes was solubilized in 2% OG, and tetramers were isolated by gel filtration, vitrified, and imaged by cryo-EM with a Tecnai F20. Particles (∼65,000) were autopicked and analyzed for 2D classification by Relion 2. Most of the tetramer classes have a square shape rather than linear, T, or L shape. A representative micrograph is shown to the left. Cross-linked tetramers solubilized in 2% OG (b) or in 0.1% digitonin (c) were negatively stained and imaged with a Tecnai F20, and 35,000 particles from each condition were autopicked and analyzed by Relion 2. Most of the tetramer classes have a square shape. A representative micrograph is shown to the left. Some 2D classes for hexamer are also seen with 0.1% digitonin solubilization. Circle, represents mask diameter at 180 Å. d, model of rhodopsin oligomer. A pair of rhodopsin molecules with the same color forms the intradimeric interface, whereas two adjacent rhodopsin molecules with different colors form the interdimeric contact. The organization of the tetramer and oligomer (cytoplasmic and transmembrane views) is displayed.
Cryo-EM of rhodopsin dimer reconstituted in vitro
Based on the cryo-EM structure of the cross-linked dimers, we anticipated that there also exists a substantial propensity for the rhodopsin's TM1 and H8 to induce dimerization in vitro and therefore imaged the in vitro reconstituted rhodopsin dimer in nanodiscs without cross-linking at a 1:1:80 ratio of rhodopsin/MSP1E3D1/POPC lipid (48, 49) (Fig. 7). Approximately 2100 images were taken at 1.35 Å/pixel size with a total dose of 50 e−/Å2 (Fig. 7a). From the 2D classification of autopicked particles, 96,000 particles (∼15%) aligned to the protein density rather than to the nanodiscs (Fig. 7b), indicating that these particles retain the unperturbed dimeric interface under surface tension introduced during freezing. 3D refinement with C1 or C2 symmetry generated a map with the same geometry as the cross-linked intradimeric structure in Fig. 3d (Fig. 7 (c and d) and Fig. S6a) at resolutions in the 6–7 Å range (Fig. 7e and Fig. S6 (b–d)) and angular distributions covering all orientations (Fig. S6, e and f). Only one 3D map was obtained from the cryo-EM data set with the intradimeric interface formed by TM1 and H8 (Fig. 7 (c and d) and Fig. S6a).
Figure 7.
Cryo-EM analysis of the in vitro reconstituted rhodopsin dimer in nanodiscs. a, representative cryo-EM micrograph imaged at 300 kV using the VPP in combination with a K2 detector at a pixel size of 1.35 Å and a total dose of 50 e−/Å2. b, Relion 2D classification; classes are ranked by population; classes with the transmembrane view are boxed in red. c, 3D refinement (C2 symmetry) of 96,000 particles aligned to protein densities shows that the intradimeric interface is mediated by TM1 and H8; two rhodopsin molecules (PDB entry 1U19) are docked, with cross-sectional views shown from the side (left) and bottom (right). d, docking of rhodopsin (PDB entry 1U19) into the EM density after nanodisc removal in Chimera; cross-sectional view shown to the right. e, local resolution estimate (C2 symmetry).
Double electron–electron resonance (DEER) of rhodopsin dimer reconstituted in vitro
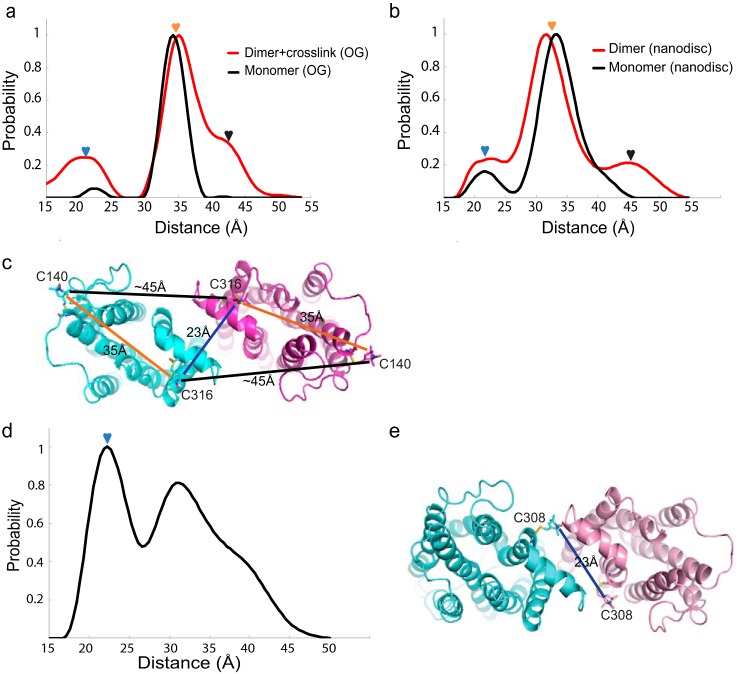
DEER measurements were also performed on the rhodopsin dimer cross-linked in disc membranes as well as the in vitro reconstituted dimer samples with rhodopsin extracted from disc membranes (Fig. 8 (a–c) and Fig. S7 (a–d)). Native rhodopsin contains Cys-140 and Cys-316 to which nitroxide spin labels can be attached. An intramolecular spin–spin distance of ∼35 Å was observed (Fig. 8, a–c). Our DEER distances revealed marked similarity between the dimer cross-linked in disc membranes and the in vitro reconstituted dimers, both showing additional signals at ∼23 and ∼45 Å relative to controls (Fig. 8, a and b). This supports the concept that the preferential rhodopsin dimerization interface formed by TM1 and H8 can be reconstituted in vitro (Fig. 8c). Furthermore, we reconstituted a mutant rhodopsin, which carries a single reactive Cys-308, as dimers into nanodiscs. DEER revealed a main peak at ∼23 Å, again predicting the existence of the intradimeric interface formed by TM1/H8 (Fig. 8d and Fig. S7 (e and f)). A schematic of dimer configuration with the observed distance between the spin-labeled Cys-308 as evidenced from DEER is shown in Fig. 8e.
Figure 8.
DEER distance distributions of spin-labeled rhodopsin dimer. a, rhodopsin monomer (black) with Cys-140 and Cys-316 spin-labeled in 3% OG showed a single peak centered at ∼35 Å. In disc membranes, cross-linked rhodopsin dimer (red) in 3% OG showed additional peaks at ∼23 and ∼45 Å. b, rhodopsin monomer reconstituted into nanodiscs (black) with Cys-140 and Cys-316 spin-labeled showed a peak centered ∼35 Å. In vitro reconstituted rhodopsin dimer in nanodiscs (red) showed additional peaks at ∼23 and ∼45 Å. c, schematics of the rhodopsin dimer configuration predicted by the DEER measurement, with the distances between Cys-140 and Cys-316 color-labeled. The nitroxide label is modeled into the Cys-140 and Cys-316 sites of rhodopsin. d, rhodopsin mutant spin-labeled at site 308 and in vitro reconstituted as dimer in nanodisc showed additional intradimeric DEER distance at ∼23 Å (blue arrow); the width of the peak suggests flexibility of the intradimeric interface. e, schematic of the dimer configuration predicted by the DEER measurement, with the distance from Cys-308 labeled. The nitroxide label is modeled into the Cys-308 site of the rhodopsin mutant.
Comparison of the intradimeric structure with known class A GPCR crystal and cryo-EM structures
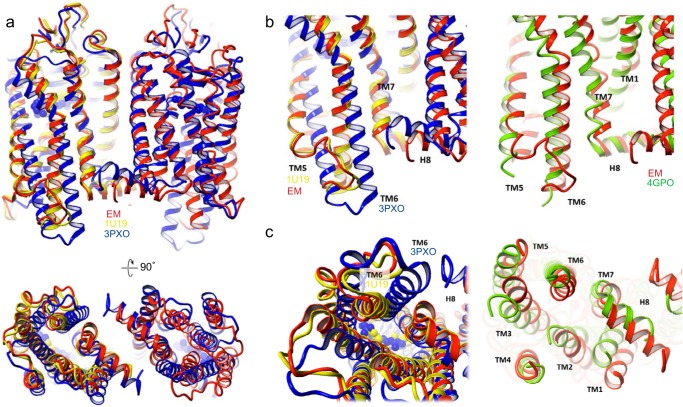
We superimposed our intradimeric structure with three other class A GPCR crystal structures that include the monomeric rhodopsin (PDB entry 1U19), activated rhodopsin (PDB entry 3PXO), and inactive β1-AR (PDB entry 4GPO) dimers (Fig. 9). β1-AR was chosen for comparison as an example of an inactive class A GPCR dimer obtained from crystallization (57). As expected, the topology of the cryo-EM rhodopsin dimer (red) most resembles inactive rhodopsin (yellow) and then the inactive β1-AR dimer (green) and least the active rhodopsin dimer (blue) (Fig. 9). Side and cytoplasmic views of TM5 and TM6 are enlarged for clarity (Fig. 9, b and c), and the activated rhodopsin (blue) showed a rotational tilt of TM6, which is the critical conformational change that is clearly different from the cryo-EM dimer (red) in the dark state.
Figure 9.
Comparison of rhodopsin cryo-EM structure with known GPCR class A crystal structures. Overlay of EM rhodopsin dimer structure (red) with crystal structures of dark state rhodopsin monomer (yellow, PDB entry 1U19), active opsin dimer (blue, PDB entry 3PXO), and inactive β1-AR dimer (green, PDB entry 4GPO). a, top, side view; bottom, cytoplasmic view. b and c, enlarged side view and cytoplasmic view of TM5/TM6 regions that change upon rhodopsin activation. Left, EM rhodopsin dimer superimposed with inactive rhodopsin (PDB entry 1U19) and activated opsin (PDB entry 3PXO). Right, EM rhodopsin dimer superimposed with β1-AR (PDB entry 4GPO).
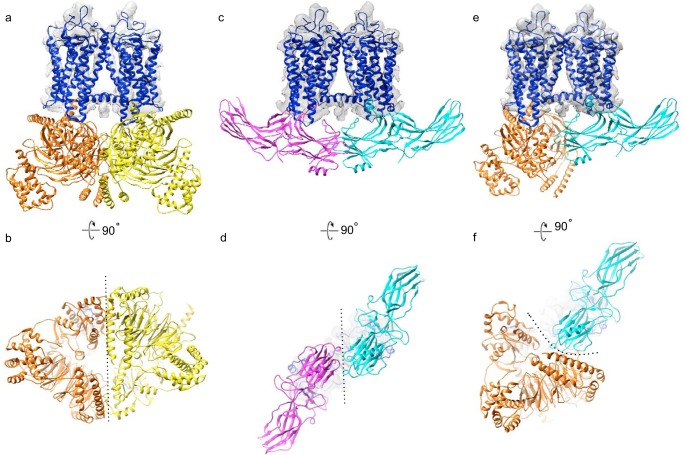
When superimposing active rhodopsin from the recent cryo-EM structure of the rhodopsin-heterotrimeric G protein (Gi) complex (58) onto the current dimer map, the intradimeric interface neither interferes nor enhances the interaction between one rhodopsin and one Gi (Fig. 10, a and b). Considering that Gi forms a relatively flexible complex, as analyzed by molecular dynamics simulations (59), the rhodopsin dimer may be able to accommodate the binding of two Gi proteins simultaneously without creating a clash between the Gi proteins. Additionally, when overlaying the crystal structure of the rhodopsin–arrestin complex (60) with the present EM structure, the rhodopsin dimer also may be able to accommodate two arrestin molecules (Fig. 10, c and d). Furthermore, with overlaying one rhodopsin bound to Gi and one rhodopsin bound to arrestin with the present EM structure, it appears that both Gi and arrestin could also be accommodated simultaneously (Fig. 10, e and f).
Figure 10.
Overlay of rhodopsin–Gi and rhodopsin–arrestin complex structures with rhodopsin docked into intradimeric EM densities (Fig. 3d). Shown are a side view (a) and cytoplasmic view (b) of activated rhodopsin (blue); and two Gαβγ complex: one in yellow and one in orange. Two Gi complexes are separated by a dotted line. Shown are a side view (c) and cytoplasmic view (d) of activated rhodopsin (blue); and two arrestin: one in magenta and one in cyan, separated by a dotted line. Shown are a side view (e) and cytoplasmic view (f) of activated rhodopsin (blue), a G complex in orange, and the arrestin in cyan. A dotted line separates arrestin from the Gi complex.
Discussion
GPCRs mediate an incredible array of physiological processes in the human body. An increased mechanistic understanding of their activity has led to the recognition of GPCRs as major pharmacological targets. Recent findings also indicate that GPCR homo- or heterodimerization is important for drug screening and design (21, 32). We expect that the approach outlined in this study can be extended to other GPCRs or membrane receptors to allow for a better understanding of their specific dimerization mechanisms.
Studies by fluorescence or bioluminescence resonance energy transfer, peptide competition, mutagenesis, and cross-linking methods have suggested various dimerization interfaces for numerous GPCR dimers (14, 18). Different interfaces for dimerization of class A GPCRs have been predicted (e.g. β2-AR, TM1/TM2/H8 (61); opsin, TM1/TM2/H8 (43–46); CXCR4, TM4–6 (62); opioid, TM1 and TM4–6 (63, 64); C5a chemotactic receptor 1, TM4/TM5 (65); and smoothened, TM4/TM5 (66)) by crystallographic methods but do not demonstrate the state of dimerization in the native membrane. To our knowledge, the structural study presented here is the first to resolve the dimerization/tetramerization interfaces of a class A GPCR extracted from its native membrane. The intradimeric interface formed by TM1 and H8 could have several implications in the biogenesis and function of rhodopsin in vivo; we, however, favor the idea that this arrangement occurs primarily due to the high-density packing of rhodopsin in the rod photoreceptors.
Genetic studies have shown that doubling the amount of rhodopsin molecules in the retina will double the size of ROS (67), whereas reducing the expression of rhodopsin by half roughly reduces the volume of ROS by half, and yet paracrystalline arrays can form in both cases (35). Collectively, these studies indicate that in vivo rhodopsin density and their contacts are relatively stable. The nanodisc system employed in our study nicely mimics the native ROS disc membranes not only as a lipid environment, but also with an imposed physical constraint. The fact that we detected similar dimerization interfaces for rhodopsin cross-linked in disc membranes or reconstituted in vitro is good evidence that TM1 and H8 provide the structural basis for rhodopsin dimerization. Further, our interdimeric structure points at a likely more flexible contact between TM1 and TM2 of one rhodopsin and TM5 and TM6 of a second rhodopsin, which is also observed when rhodopsin is modeled into rows of dimers (Figs. 5 and 6).
Rhodopsin dimerization can also play a role in receptor maturation, trafficking to disc membranes, and recycling. It is well-known that a class C GPCR, such as the GABAB receptor, exists as a heterodimer and that dimerization is needed to mask the ER retention signal for the maturation of the receptors to the cell membrane (68–70). Our intradimeric interface with TM1 and H8 is in agreement with the findings of a peptide competition study that showed this dimeric form to be important for the proper insertion of rhodopsin into the disc membrane (56); the injection of peptide targeting TM1 and H8 in murine retina in the study disrupted both rhodopsin dimerization and ROS maturation. Collectively, findings indicate that the intradimeric interface of TM1 and H8 of rhodopsin is likely necessary for its targeting and maturation to ROS disc membranes.
Experimental procedures
Expression and purification of membrane scaffold proteins
A pET-28a vector containing the MSP1E3D1 gene construct (Addgene, Watertown, MA) was transformed into Escherichia coli BL21Gold (DE3) cells for growth and expression. The cell cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (BioShop, Burlington, Canada) overnight at 28 °C and were harvested and resuspended in lysis buffer (20 mm Na3PO4, 1% Triton X-100, 1 mm PMSF, pH 7.4). After sonication, the lysate was cleared by centrifugation at 43,399 × g for 30 min in a JA-25.50 rotor (Beckman Coulter, Brea, CA) and was loaded onto an Ni2+-nitrilotriacetic acid–agarose resin (Qiagen, Hilden, Germany). The column was then washed sequentially with the following buffers: 1) 40 mm Tris, 300 mm NaCl, 1% Triton X-100, pH 8.0; 2) 40 mm Tris, 300 mm NaCl, 50 mm cholate, 20 mm imidazole, pH 8.0; and 3) 40 mm Tris, 300 mm NaCl, 50 mm imidazole, pH 8.0. Last, MSP1E3D1 scaffold was eluted with 40 mm Tris, 300 mm NaCl, 300 mm imidazole, pH 8.0, and buffer-exchanged using a PD-10 column (GE Healthcare) into a buffer with 40 mm Tris, 150 mm NaCl, 0.5 mm EDTA, 30 mm sodium cholate, pH 7.4.
Purification and cross-linking of rhodopsin dimer from bovine retinas
All procedures related to rhodopsin were carried out under dim red light (>650 nm) to keep rhodopsin in the dark state. Rhodopsin-containing disc membranes were purified from dark-adapted retinas as described previously (41). For chemical cross-linking with DSP (Thermo Fisher Scientific), disc membranes (1 mg/ml) were incubated with DSP at a 125-fold molar excess to rhodopsin in buffer (100 mm Na2HPO4, pH 8, 150 mm NaCl) for 30 min at 4 °C (41), followed by quenching with 100 mm Tris-Cl, pH 8, for 15 min. The cross-linked sample was washed twice with buffer (100 mm Na2HPO4, pH 8, 150 mm NaCl) and pelleted by ultracentrifugation (104,300 × g) for 5 min at 4 °C in an Optima TLX ultracentrifuge (Beckman Coulter).
OG and DDM were purchased from Glycon Biochemicals (Luckenwalde, Germany). Disc membranes with or without cross-linking were solubilized in 3% OG at 4 °C overnight, and the lysate was cleared by ultracentrifugation (104,300 × g) for 25 min at 4 °C in an Optima TLX (Beckman Coulter) centrifuge. The sample was then concentrated with a 30-kDa cut-off Amicon Ultra centrifugal filter device (Sigma-Aldrich) and fractionated into dimer or monomer with 1–2% OG in buffer (100 mm Na2HPO4, pH 8, 150 mm NaCl) by two rounds of gel filtration, using a Superdex 200 Increase 10/300 GL column (GE Healthcare) running at 0.5 ml/min at 25 °C with Akta Pure (GE Healthcare). Absorbance was continuously monitored at 280 and 500 nm.
Construction of rhodopsin nanodiscs
POPC lipid stock was obtained from Avanti Polar Lipids. The lipid was evaporated to remove its chloroform solvent and was then dissolved in 100 mm sodium cholate (Sigma-Aldrich) for a final 50 mm stock. For the preparation of dimer- or monomer-containing nanodiscs, purified rhodopsin (50–100 μm) after gel filtration was mixed with MSP1E3D1 (50–100 μm) and POPC/cholate at a 1:1:80 ratio of rhodopsin/MSP1E3D1/POPC (dimer) or at a 0.1:1:80 ratio of rhodopsin/MSP1E3D1/POPC (monomer) in 1% OG and 14 mm sodium cholate, as described previously (48, 49). After 15 min of incubation at 4 °C, BioBeads SM-2 resin (Bio-Rad) was added, which was prewashed with methanol followed by water. The dimer-containing nanodiscs were incubated at 25 °C for 45 min and then at 4 °C overnight to remove residual OG; 4 °C incubation was omitted for the preparation of monomer-containing nanodiscs. The next day, BioBeads were removed, and the sample was purified by gel filtration with a Superdex 200 Increase 10/300 GL column running at 0.5 ml/min at 25 °C in buffer (20 mm Tris-Cl, pH 8, 150 mm NaCl). Absorbance was continuously monitored at 280 and 500 nm.
Characterization of rhodopsin dimer in nanodiscs using a rhodopsin 1D4 antibody
For the determination of the orientation of rhodopsin in the reconstituted and cross-linked dimers in nanodiscs, rhodopsin-containing nanodiscs were incubated with 1D4 Fab fragment generated by papain digestion of the rhodopsin 1D4 mAb, at a 1:10–15 nanodisc/Fab ratio at 4 °C overnight. For papain digestion, 10 mg of rhodopsin 1D4 antibody (6.68 mg/ml; University of British Columbia) was incubated with 2 mg of papain (Sigma-Aldrich) in 10 ml of buffer (10 mm Na2HPO4, 137 mm NaCl, 2.7 mm KCl, 1.8 mm KH2PO4, 20 mm EDTA, pH 7.4) and 20 mm cystine for 2 h at 37 °C. Completion of the digestion was monitored by nonreducing PAGE. The digest was then concentrated and buffer-exchanged in a PD-10 column with 20 mm Tris-Cl, pH 8. To purify 1D4 Fab, the digest was passed through 5 ml of DEAE-Sepharose fast flow resin (GE Healthcare), as both the Fc fragment and papain bind to DEAE. Fab was collected in the flowthrough. The rhodopsin-containing nanodisc-1D4-Fab complex was then examined and quantified using negatively stained EM.
UV-visible spectroscopy
UV-visible spectroscopy of rhodopsin with or without nanodiscs was performed using a Cary 60 spectrophotometer with a single-cell Peltier assembly (Agilent Technologies, Santa Clara, CA). An optical cuvette with one compartment (width, 2 mm; light path, 1 cm) was used for recording the spectra of the samples. The sample temperature was kept at 15 °C, and the dark spectra were recorded after incubation for 10 min. Subsequently, light-activated spectra were recorded after photobleaching in the cuvette by >500-nm yellow light from a Fiber-Lite MI-150 illuminator (Dolan-Jenner, Boxborough, MA) that had been passed through a glass infrared cutoff filter. The difference spectra were calculated by subtracting dark spectra from light-activated spectra. Rhodopsin was quantified by the change in absorbance at 500 nm (40,600 m−1 cm−1) upon complete bleaching in the presence of hydroxylamine.
Retinoids analysis
Samples of reconstituted rhodopsin in nanodiscs (50 μg) with or without 1-min white light photobleaching on ice were dissolved in 800 μl of buffer (10 mm Na3PO4, pH 8.0, 50% methanol (v/v), 100 mm hydroxylamine). The resulting mixture was extracted with 400 μl of hexanes, and 100 μl of the extract were injected into a normal-phase HPLC column (Agilent Sil, 5 μm, 4.6 × 250 mm; Agilent Technologies) with 10% ethyl acetate in hexanes as eluent at a flow rate of 1.4 ml/min. Retinoids were detected by monitoring absorbance at 325 nm. 11-cis-Retinal oxime is known to have an elution time of ∼7 min, and all-trans-retinal oxime has an elution time of ∼8 min.
Sample preparation for negatively stained EM and cryo-EM screenings
Copper grids (Electron Microscopy Sciences, Hatfield, PA) for negative stain were coated with a continuous carbon film support. The nanodisc sample (3 μl) of 0.01–0.03 mg/ml concentration was incubated on a carbon-coated copper grid after low discharging at 15 mA for 30 s. The sample was then stained in 2% (w/v) uranyl acetate (Electron Microscopy Sciences) by dipping the grid onto four droplets of stain for 10–20 s each, followed by blotting with a filter paper.
For the screening, images were manually acquired using a FEI Tecnai F20 transmission electron microscope operating at 200 kV acceleration voltage equipped with a Tietz TVIPS 4000 × 4000 CMOS camera, using a defocus range of −1 to −3 μm at ×60,000 magnification (1.6 Å/pixel) (Cleveland Center for Membrane and Structure Biology, Case Western Reserve University, Cleveland, OH). Data analysis was performed by Relion 2 (53).
For cryo-EM, Quantifoil® R 1.2/1.3 holey carbon grids (Electron Microscopy Sciences) were glow-discharged at 15–30 mA for 30 s prior to sample application. Concentrated sample (1 mg/ml; 3.5 μl) was applied on the grid in the Vitrobot chamber (Vitrobot Mark IV, Thermo Fisher Scientific). The Vitrobot chamber was set to 100% humidity at 4 °C. The sample was blotted for 2.5 s and then plunged into liquid ethane cooled by liquid nitrogen. All sample preparation procedures were done in dim red light (>650 nm) to keep rhodopsin in the dark state.
Cryo-EM analysis of cross-linked and reconstituted rhodopsin dimers in nanodiscs
For the cross-linked dimer nanodisc sample, 5500 cryo-EM micrographs were acquired using a Titan Krios (Thermo Fisher Scientific) operated at 300 kV acceleration voltage, equipped with the VPP, a Gatan Bioquantum energy filter, and a K2 Summit direct electron detector (Heidelberg, Germany). Data were collected automatically using the SerialEM (71) software package in counting mode at a target defocus of 300–1000 nm, with a pixel size of 0.8 Å and a total electron dose of 50 e−/Å2 fractioned into 50 movie frames. Stacks of image frames were aligned using Motioncorr2 (72). Contrast transfer function parameters were estimated using Gctf (73), and micrographs where the theoretical CTF fit did not agree with the signal beyond 5 Å were discarded (Fig. S3). Automated particle picking was performed in Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/)7 and resulted in a total of ∼1.7 million particles. The picked particles were binned three times and subjected to 2D classification in Relion 2 (53), and 762,000 particles were selected for 3D classification distributed in three classes with or without imposing C2 symmetry. A second round of 3D classification for eight classes (C2 symmetry) or three classes (C1 symmetry) was done on unbinned class 1 particles to examine for additional dimer configurations. After global refinement (with or without imposing C2 symmetry) on the unbinned particles, local refinement searches with additional masks around the dimer were performed. Postprocessing was done in Relion 2 with a soft mask around the nanodisc to estimate the resolution using the gold-standard FSC = 0.143 criterion. Details of the cryo-TEM data collection have been provided in Table S1.
For the reconstituted dimer nanodisc sample, 2100 cryo-EM micrographs were acquired using a 300-kV Titan Krios equipped with the VPP, a Gatan Bioquantum, and a K2 detector (Martinsried, Germany). Data were collected using a pixel size of 1.35 Å. Data analysis was done similarly as above with imposed C1 or C2 symmetry. Details of the cryo-TEM data collection have been provided in Table S2.
Model building and refinement
For modeling of the rhodopsin crystal structure into the cryo-EM electron densities, the Chimera software package was used for the initial rigid body docking. Nanodisc cryo-EM density far away from the rhodopsin density was removed in UCSF Chimera by segmentation (74).
The cryo-EM map of rhodopsin dimer was put into an artificial crystal lattice to calculate its structure factors using the phenix.map_to_structure_factors script in the Phenix program (75). The 3D density map was sharpened by applying a negative B-factor of 201.8 Å (54) as determined with the phenix.auto_sharpen script (75) and by manual intervention. Two monomers of bovine rhodopsin (54) were placed in the sharpened 3D density map using rigid body fitting in Chimera (74). The model fitting was then improved manually by adjusting the main-chain fitting in Coot 0.8.8 (76). The N-terminal β-hairpin and helix H8 were used as references during the backbone tracing and for the calculation of the dimer interface. The initial model was then refined by rigid body refinement of individual chains in the PHENIX program (75), where the amplitudes and phases of the structural factors were used as pseudo-experimental diffraction data for the model refinement. Multiple rounds of Phenix real-space refinement (75) and REFMAC version 5.8 (77) refinement against the overall map at a resolution of 4.7 Å were used to improve the dimer model. Each round of refinement was followed by manual model building and adjustments with Coot 0.8.8 (76). The stereochemical quality of the final rhodopsin dimer model was assessed with the Molprobity (78) and wwPDB validation servers (79). Details of the cryo-TEM data collection and structural refinement statistics have been provided in Table S1. Protein coordinates and cryo-EM density maps were deposited in the PDB and EMDB with accession codes 6OFJ and EMD-20047, respectively.
Purification of Cys-308 rhodopsin mutant from HEK293 cells
Expression, purification, and spin labeling of bovine rhodopsin mutants were performed as described previously with minor modifications (80). For the present study, single rhodopsin cysteine mutant M308C in a C140S/C316S base mutant was produced in stable HEK293S GnTI− cell cultures. The mutant was eluted from a 1D4 antibody resin in a buffer containing 10 mm Bistris propane, 0.03% DDM at 4 °C using a peptide with sequence identity to the last 18 amino acids of rhodopsin's C terminus. The rhodopsin mutant showed WT UV-visible absorption characteristics consistent with previous reports (59).
DEER measurement and analysis
Spin-labeling was carried out during solubilization of rhodopsin from disc membranes or after nanodisc formation for the mutant Cys-308 rhodopsin, with nitroxide label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)methanethiosulfonate (Toronto Research Chemicals, Toronto, Canada) at a 2-fold molar excess of the receptor for 3–12 h at 4 °C.
For DEER measurements, spin-labeled rhodopsin in 3% OG or in nanodiscs (generally 50–100 μm) was flash-frozen within quartz capillaries (1.5-mm inner diameter and 1.8-mm outer diameter) (VitroCom, Boonton, NJ). After freezing, the capillaries were loaded into an EN 5107D2 resonator, and Q-band measurements were performed at 80 K on a Bruker Elexsys 580 spectrometer with a Super Q-FTu Bridge (Bruker, Billerica, MA). For the four-pulse DEER experiment used, a 32-ns π-pump pulse was applied to the low-field peak of the nitroxide field-swept spectrum, and the observer π/2 (16 ns) and π (32 ns) pulses were positioned 17.8 gauss upfield, which corresponds to the nitroxide center line. Distance distributions were obtained from the raw dipolar evolution data using the LabVIEW (National Instruments, Austin, TX) program “LongDistances.” Nitroxide labels were modeled at sites Cys-140 and Cys-316 or at Cys-308 in the dimer model using the Discovery Studio (BioVIA) software package.
Author contributions
K. P. and O. P. E. supervised the project; D. Y. Z., S. G., O. P. E.. and K. P. wrote the manuscript; all authors edited the manuscript. D. Y. Z. and T. M. performed sample preparations and spectroscopic characterizations; D. Y. Z., M. P., and S. G. performed cryo-EM analyses; D. Y. Z., M. P., and S. G. acquired cryo-EM data with support from J. M., J. P., and W. B.; D. Y. Z. and J. Z. performed retinoid analyses; N. V. E. and D. Y. Z performed DEER measurements; S. G. performed protein modeling and refinement. P. M. and S. F. performed molecular dynamics simulations.
Supplementary Material
Acknowledgments
We thank Dr. Beata Jastrzebska (Case Western Reserve University) for involvement in designing the cross-linking experiments and Dr. Maryam Khoshouei (Novartis Corp.) for helpful discussions. The authors wish to acknowledge Allergan for their generous support of research at the Center for Translational Vision Research (CTVR) at Gavin Herbert Eye Institute, University of CA, Irvine.
This work was supported in part by National Institutes of Health Grants EY R24024864 and R01EY027283 (to K. P.), a grant from the Alcon Research Institute (ARI) (to K. P.), a grant from Research to Prevent Blindness (to the Department of Ophthalmology at the University of California, Irvine, CA), and a grant from the Canada Excellence Research Chairs program (to O. P. E.). This research is based on work supported by the Canadian Institute for Advanced Research (CIFAR) Program in Molecular Architecture of Life through a catalyst grant. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S7.
The atomic coordinates and structure factors (code 6OFJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Cryo-EM density maps for this work were deposited in the Electron Microscopy Data Bank under the accession code EMD-20047.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GPCR
- G protein–coupled receptor
- DEER
- double electron–electron resonance
- DDM
- n-dodecyl-β-d-maltoside
- DSP
- dithiobis(succinimidyl)propionate
- OG
- n-octyl-β-d-glucoside
- ROS
- rod outer segment
- TM
- transmembrane α-helix
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- H8
- helix 8
- AR
- adrenergic receptor
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- TEM
- transmission EM
- VPP
- Volta phase plate
- 2D and 3D
- two- and three-dimensional, respectively
- FSC
- Fourier shell correlation
- PDB
- Protein Data Bank.
References
- 1. Salon J. A., Lodowski D. T., and Palczewski K. (2011) The significance of G protein-coupled receptor crystallography for drug discovery. Pharmacol. Rev. 63, 901–937 10.1124/pr.110.003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reiter E., Ahn S., Shukla A. K., and Lefkowitz R. J. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 10.1146/annurev.pharmtox.010909.105800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rohrer D. K., and Kobilka B. K. (1998) G protein-coupled receptors: functional and mechanistic insights through altered gene expression. Physiol. Rev. 78, 35–52 10.1152/physrev.1998.78.1.35 [DOI] [PubMed] [Google Scholar]
- 4. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., and Miyano M. (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- 5. Palczewski K. (2006) G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 10.1146/annurev.biochem.75.103004.142743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 10.1021/cr4003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., and Palczewski K. (2003) Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature 421, 127–128 10.1038/421127a [DOI] [PubMed] [Google Scholar]
- 8. Gunkel M., Schöneberg J., Alkhaldi W., Irsen S., Noé F., Kaupp U. B., and Al-Amoudi A. (2015) Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure 23, 628–638 10.1016/j.str.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 9. Harding P. J., Attrill H., Boehringer J., Ross S., Wadhams G. H., Smith E., Armitage J. P., and Watts A. (2009) Constitutive dimerization of the G-protein coupled receptor, neurotensin receptor 1, reconstituted into phospholipid bilayers. Biophys. J. 96, 964–973 10.1016/j.bpj.2008.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dijkman P. M., Castell O. K., Goddard A. D., Munoz-Garcia J. C., de Graaf C., Wallace M. I., and Watts A. (2018) Dynamic tuneable G protein-coupled receptor monomer-dimer populations. Nat. Commun. 9, 1710 10.1038/s41467-018-03727-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo W., Urizar E., Kralikova M., Mobarec J. C., Shi L., Filizola M., and Javitch J. A. (2008) Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 27, 2293–2304 10.1038/emboj.2008.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo W., Shi L., Filizola M., Weinstein H., and Javitch J. A. (2005) Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. Sci. U.S.A. 102, 17495–17500 10.1073/pnas.0508950102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S. P., So C. H., Rashid A. J., Varghese G., Cheng R., Lança A. J., O'Dowd B. F., and George S. R. (2004) Dopamine D1 and D2 receptor co-activation generates a novel phospholipase C-mediated calcium signal. J. Biol. Chem. 279, 35671–35678 10.1074/jbc.M401923200 [DOI] [PubMed] [Google Scholar]
- 14. Dorsch S., Klotz K. N., Engelhardt S., Lohse M. J., and Bünemann M. (2009) Analysis of receptor oligomerization by FRAP microscopy. Nat. Methods 6, 225–230 10.1038/nmeth.1304 [DOI] [PubMed] [Google Scholar]
- 15. Hebert T. E., Moffett S., Morello J. P., Loisel T. P., Bichet D. G., Barret C., and Bouvier M. (1996) A peptide derived from a β2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J. Biol. Chem. 271, 16384–16392 10.1074/jbc.271.27.16384 [DOI] [PubMed] [Google Scholar]
- 16. Angers S., Salahpour A., Joly E., Hilairet S., Chelsky D., Dennis M., and Bouvier M. (2000) Detection of β 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U.S.A. 97, 3684–3689 10.1073/pnas.060590697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klco J. M., Lassere T. B., and Baranski T. J. (2003) C5a receptor oligomerization. I. Disulfide trapping reveals oligomers and potential contact surfaces in a G protein-coupled receptor. J. Biol. Chem. 278, 35345–35353 10.1074/jbc.M305606200 [DOI] [PubMed] [Google Scholar]
- 18. Kota P., Reeves P. J., Rajbhandary U. L., and Khorana H. G. (2006) Opsin is present as dimers in COS1 cells: identification of amino acids at the dimeric interface. Proc. Natl. Acad. Sci. U.S.A. 103, 3054–3059 10.1073/pnas.0510982103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berthouze M., Rivail L., Lucas A., Ayoub M. A., Russo O., Sicsic S., Fischmeister R., Berque-Bestel I., Jockers R., and Lezoualc'h F. (2007) Two transmembrane Cys residues are involved in 5-HT4 receptor dimerization. Biochem. Biophys. Res. Commun. 356, 642–647 10.1016/j.bbrc.2007.03.030 [DOI] [PubMed] [Google Scholar]
- 20. Ferré S., Casadó V., Devi L. A., Filizola M., Jockers R., Lohse M. J., Milligan G., Pin J. P., and Guitart X. (2014) G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol. Rev. 66, 413–434 10.1124/pr.113.008052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith N. J., and Milligan G. (2010) Allostery at G protein-coupled receptor homo- and heteromers: uncharted pharmacological landscapes. Pharmacol. Rev. 62, 701–725 10.1124/pr.110.002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whorton M. R., Bokoch M. P., Rasmussen S. G., Huang B., Zare R. N., Kobilka B., and Sunahara R. K. (2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 104, 7682–7687 10.1073/pnas.0611448104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsukamoto H., Sinha A., DeWitt M., and Farrens D. L. (2010) Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J. Mol. Biol. 399, 501–511 10.1016/j.jmb.2010.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ernst O. P., Gramse V., Kolbe M., Hofmann K. P., and Heck M. (2007) Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. U.S.A. 104, 10859–10864 10.1073/pnas.0701967104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrick-Davis K., Grinde E., Harrigan T. J., and Mazurkiewicz J. E. (2005) Inhibition of serotonin 5-hydroxytryptamine2c receptor function through heterodimerization: receptor dimers bind two molecules of ligand and one G-protein. J. Biol. Chem. 280, 40144–40151 10.1074/jbc.M507396200 [DOI] [PubMed] [Google Scholar]
- 26. Banères J. L., and Parello J. (2003) Structure-based analysis of GPCR function: evidence for a novel pentameric assembly between the dimeric leukotriene B4 receptor BLT1 and the G-protein. J. Mol. Biol. 329, 815–829 10.1016/S0022-2836(03)00439-X [DOI] [PubMed] [Google Scholar]
- 27. Pellissier L. P., Barthet G., Gaven F., Cassier E., Trinquet E., Pin J. P., Marin P., Dumuis A., Bockaert J., Banères J. L., and Claeysen S. (2011) G protein activation by serotonin type 4 receptor dimers: evidence that turning on two protomers is more efficient. J. Biol. Chem. 286, 9985–9997 10.1074/jbc.M110.201939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rivero-Müller A., Chou Y. Y., Ji I., Lajic S., Hanyaloglu A. C., Jonas K., Rahman N., Ji T. H., and Huhtaniemi I. (2010) Rescue of defective G protein-coupled receptor function in vivo by intermolecular cooperation. Proc. Natl. Acad. Sci. U.S.A. 107, 2319–2324 10.1073/pnas.0906695106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koehl A., Hu H., Feng D., Sun B., Zhang Y., Robertson M. J., Chu M., Kobilka T. S., Laeremans T., Steyaert J., Tarrasch J., Dutta S., Fonseca R., Weis W. I., Mathiesen J. M., et al. (2019) Structural insights into the activation of metabotropic glutamate receptors. Nature 567, E10 10.1038/s41586-019-0983-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pin J. P., Kniazeff J., Binet V., Liu J., Maurel D., Galvez T., Duthey B., Havlickova M., Blahos J., Prézeau L., and Rondard P. (2004) Activation mechanism of the heterodimeric GABA(B) receptor. Biochem. Pharmacol. 68, 1565–1572 10.1016/j.bcp.2004.06.035 [DOI] [PubMed] [Google Scholar]
- 31. Pin J. P., and Bettler B. (2016) Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68 10.1038/nature20566 [DOI] [PubMed] [Google Scholar]
- 32. Bouvier M. (2001) Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2, 274–286 10.1038/35067575 [DOI] [PubMed] [Google Scholar]
- 33. Redka D. S., Morizumi T., Elmslie G., Paranthaman P., Shivnaraine R. V., Ellis J., Ernst O. P., and Wells J. W. (2014) Coupling of G proteins to reconstituted monomers and tetramers of the M2 muscarinic receptor. J. Biol. Chem. 289, 24347–24365 10.1074/jbc.M114.559294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang Y., Fotiadis D., Filipek S., Saperstein D. A., Palczewski K., and Engel A. (2003) Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J. Biol. Chem. 278, 21655–21662 10.1074/jbc.M302536200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang Y., Fotiadis D., Maeda T., Maeda A., Modzelewska A., Filipek S., Saperstein D. A., Engel A., and Palczewski K. (2004) Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. J. Biol. Chem. 279, 48189–48196 10.1074/jbc.M408362200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mansoor S. E., Palczewski K., and Farrens D. L. (2006) Rhodopsin self-associates in asolectin liposomes. Proc. Natl. Acad. Sci. U.S.A. 103, 3060–3065 10.1073/pnas.0511010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Comar W. D., Schubert S. M., Jastrzebska B., Palczewski K., and Smith A. W. (2014) Time-resolved fluorescence spectroscopy measures clustering and mobility of a G protein-coupled receptor opsin in live cell membranes. J. Am. Chem. Soc. 136, 8342–8349 10.1021/ja501948w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mishra A. K., Gragg M., Stoneman M. R., Biener G., Oliver J. A., Miszta P., Filipek S., Raicu V., and Park P. S. (2016) Quaternary structures of opsin in live cells revealed by FRET spectrometry. Biochem. J. 473, 3819–3836 10.1042/BCJ20160422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suda K., Filipek S., Palczewski K., Engel A., and Fotiadis D. (2004) The supramolecular structure of the GPCR rhodopsin in solution and native disc membranes. Mol. Membr. Biol. 21, 435–446 10.1080/09687860400020291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Govardovskii V. I., Korenyak D. A., Shukolyukov S. A., and Zueva L. V. (2009) Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Mol. Vis. 15, 1717–1729 [PMC free article] [PubMed] [Google Scholar]
- 41. Jastrzebska B., Fotiadis D., Jang G. F., Stenkamp R. E., Engel A., and Palczewski K. (2006) Functional and structural characterization of rhodopsin oligomers. J. Biol. Chem. 281, 11917–11922 10.1074/jbc.M600422200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jastrzebska B., Chen Y., Orban T., Jin H., Hofmann L., and Palczewski K. (2015) Disruption of rhodopsin dimerization with synthetic peptides targeting an interaction interface. J. Biol. Chem. 290, 25728–25744 10.1074/jbc.M115.662684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., and Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 10.1038/nature09789 [DOI] [PubMed] [Google Scholar]
- 44. Salom D., Lodowski D. T., Stenkamp R. E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J. A., and Palczewski K. (2006) Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128 10.1073/pnas.0608022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruprecht J. J., Mielke T., Vogel R., Villa C., and Schertler G. F. (2004) Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620 10.1038/sj.emboj.7600374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., and Schertler G. F. (2011) The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471, 656–660 10.1038/nature09795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., and Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 10.1038/nature07063 [DOI] [PubMed] [Google Scholar]
- 48. Bayburt T. H., Leitz A. J., Xie G., Oprian D. D., and Sligar S. G. (2007) Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 282, 14875–14881 10.1074/jbc.M701433200 [DOI] [PubMed] [Google Scholar]
- 49. Banerjee S., Huber T., and Sakmar T. P. (2008) Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 377, 1067–1081 10.1016/j.jmb.2008.01.066 [DOI] [PubMed] [Google Scholar]
- 50. Danev R., Tegunov D., and Baumeister W. (2017) Using the Volta phase plate with defocus for cryo-EM single particle analysis. Elife 6, e23006 10.7554/eLife.23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khoshouei M., Radjainia M., Phillips A. J., Gerrard J. A., Mitra A. K., Plitzko J. M., Baumeister W., and Danev R. (2016) Volta phase plate cryo-EM of the small protein complex Prx3. Nat. Commun. 7, 10534 10.1038/ncomms10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khoshouei M., Radjainia M., Baumeister W., and Danev R. (2017) Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Nat. Commun. 8, 16099 10.1038/ncomms16099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kimanius D., Forsberg B. O., Scheres S. H., and Lindahl E. (2016) Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., and Buss V. (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 342, 571–583 10.1016/j.jmb.2004.07.044 [DOI] [PubMed] [Google Scholar]
- 55. Knepp A. M., Periole X., Marrink S. J., Sakmar T. P., and Huber T. (2012) Rhodopsin forms a dimer with cytoplasmic helix 8 contacts in native membranes. Biochemistry 51, 1819–1821 10.1021/bi3001598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang T., Cao L. H., Kumar S., Enemchukwu N. O., Zhang N., Lambert A., Zhao X., Jones A., Wang S., Dennis E. M., Fnu A., Ham S., Rainier J., Yau K. W., and Fu Y. (2016) Dimerization of visual pigments in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 9093–9098 10.1073/pnas.1609018113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang J., Chen S., Zhang J. J., and Huang X. Y. (2013) Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 20, 419–425 10.1038/nsmb.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang Y., Kuybeda O., de Waal P. W., Mukherjee S., Van Eps N., Dutka P., Zhou X. E., Bartesaghi A., Erramilli S., Morizumi T., Gu X., Yin Y., Liu P., Jiang Y., Meng X., et al. (2018) Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 10.1038/s41586-018-0215-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Van Eps N., Altenbach C., Caro L. N., Latorraca N. R., Hollingsworth S. A., Dror R. O., Ernst O. P., and Hubbell W. L. (2018) Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc. Natl. Acad. Sci. U.S.A. 115, 2383–2388 10.1073/pnas.1721896115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kang Y., Zhou X. E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T. A., Yefanov O., Han G. W., Xu Q., de Waal P. W., Ke J., Tan M. H., Zhang C., et al. (2015) Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 10.1038/nature14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., and Stevens R. C. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 10.1126/science.1150577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., and Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 10.1126/science.1194396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Mathiesen J. M., Sunahara R. K., Pardo L., Weis W. I., Kobilka B. K., and Granier S. (2012) Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 10.1038/nature10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu H., Wacker D., Mileni M., Katritch V., Han G. W., Vardy E., Liu W., Thompson A. A., Huang X. P., Carroll F. I., Mascarella S. W., Westkaemper R. B., Mosier P. D., Roth B. L., Cherezov V., and Stevens R. C. (2012) Structure of the human κ-opioid receptor in complex with JDTic. Nature 485, 327–332 10.1038/nature10939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Robertson N., Rappas M., Doré A. S., Brown J., Bottegoni G., Koglin M., Cansfield J., Jazayeri A., Cooke R. M., and Marshall F. H. (2018) Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 553, 111–114 10.1038/nature25025 [DOI] [PubMed] [Google Scholar]
- 66. Wang C., Wu H., Katritch V., Han G. W., Huang X. P., Liu W., Siu F. Y., Roth B. L., Cherezov V., and Stevens R. C. (2013) Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 10.1038/nature12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wen X. H., Shen L., Brush R. S., Michaud N., Al-Ubaidi M. R., Gurevich V. V., Hamm H. E., Lem J., Dibenedetto E., Anderson R. E., and Makino C. L. (2009) Overexpression of rhodopsin alters the structure and photoresponse of rod photoreceptors. Biophys. J. 96, 939–950 10.1016/j.bpj.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. White J. H., Wise A., Main M. J., Green A., Fraser N. J., Disney G. H., Barnes A. A., Emson P., Foord S. M., and Marshall F. H. (1998) Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature 396, 679–682 10.1038/25354 [DOI] [PubMed] [Google Scholar]
- 69. Margeta-Mitrovic M., Jan Y. N., and Jan L. Y. (2000) A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27, 97–106 10.1016/S0896-6273(00)00012-X [DOI] [PubMed] [Google Scholar]
- 70. Jones K. A., Borowsky B., Tamm J. A., Craig D. A., Durkin M. M., Dai M., Yao W. J., Johnson M., Gunwaldsen C., Huang L. Y., Tang C., Shen Q., Salon J. A., Morse K., Laz T., et al. (1998) GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature 396, 674–679 10.1038/25348 [DOI] [PubMed] [Google Scholar]
- 71. Mastronarde D. N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 72. Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., and Agard D. A. (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang K. (2016) Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 75. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 77. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 78. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berman H., Henrick K., and Nakamura H. (2003) Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980 10.1038/nsb1203-980 [DOI] [PubMed] [Google Scholar]
- 80. Caro L. N., Li Z., Balo A. R., Van Eps N., Rini J. M., and Ernst O. P. (2015) Rapid and facile recombinant expression of bovine rhodopsin in HEK293S GnTI(−) cells using a PiggyBac inducible system. Methods Enzymol. 556, 307–330 10.1016/bs.mie.2015.01.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.