Abstract
Hyperdiploidy with greater than 50 chromosomes is usually associated with favorable prognosis in pediatric acute lymphoblastic leukemia (ALL), whereas hypodiploidy with ≤43 chromosomes is associated with extremely poor prognosis. Sometimes, hypodiploidy is “masked” and patients do not have a karyotypically visible clone with ≤43 chromosomes. Instead, their abnormal karyotypes contain 50–78 or more chromosomes from doubling of previously hypodiploid cells. When the hypodiploid and doubled hyperdiploid clones are both present, patients can be identified by traditional test methods [karyotype, DNA Index (DI), fluorescence in situ hybridization (FISH)], but the incidence of masked hypodiploid cases in which only the doubled clone is visible is unknown. We analyzed 7013 patients with B-ALL enrolled in COG AALL03B1 (2003–2011) for whom chromosome studies were available. Of 115 patients with hypodiploidy (25–39 chromosomes), karyotypes of 40 showed only the hypodiploid clone, 47 showed mosaicism with both hypodiploid and hyperdiploid (doubled) karyotypes, and 28 with masked hypodiploidy showed only a hyperdiploid (doubled) clone. Unique karyotypic signatures were identified, and widespread loss of heterozygosity (LOH) was seen in the microsatellite panel for all patients with masked hypodiploidy. An increased awareness of the unusual karyotypic profile associated with a doubled hypodiploid clone and coordinated use of DI, FISH, and LOH studies when indicated can identify patients with masked hypodiploidy and allow appropriate treatment selection.
Keywords: hypodiploid, near-haploid, low-hypodiploid, B-ALL, cytogenetics, doubling
Introduction
Over the last 30 years, outcomes of children with acute lymphoblastic leukemia (ALL) have steadily improved, with current 5-year overall survival (OS) rates above 90% (1–5). Key to this improvement has been the use of risk-adapted therapy, and the presence or absence of certain blast cell karyotypic abnormalities are very important indicators of prognosis (6). Hyperdiploidy with more than 50 chromosomes, which is found in approximately 25% of children with ALL, is usually associated with a favorable prognosis (7). In contrast, hypodiploidy with 43 or fewer chromosomes occurs in only 1–2% of children with ALL and is associated with an extremely poor prognosis (8–12).
Three distinct subgroups of hypodiploidy are recognized: near-haploidy (24–31 chromosomes), low hypodiploidy (32–39 chromosomes), and high hypodiploidy (40–43 chromosomes). Recent studies have provided new insights into the genomic features of different hypodiploid ALL subgroups (13). Remarkably, almost all children with low-hypodiploid ALL harbor TP53 mutations in leukemia cells and approximately 50% also harbor them in the germline, suggesting that low-hypodiploid ALL is often a manifestation of Li-Fraumeni syndrome.
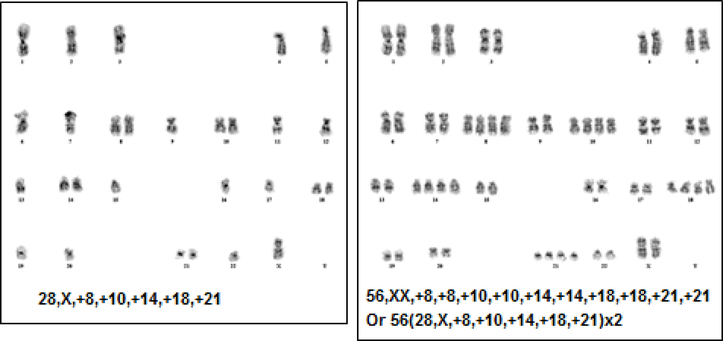
It is common for leukemia cells with near-haploid or low-hypodiploid chromosome numbers to undergo an exact or near-exact doubling of the hypodiploid clone, which results in a clone with a modal chromosome number in the hyperdiploid range (50–78 or more) and might be misconstrued as a favorable prognostic feature (Figure 1). Previous studies of hypodiploidy have shown that cases with a hypodiploid doubling have been mosaic with both hypodiploid and hyperdiploid (doubled) clones visible by standard cytogenetics, DI, and/or FISH. In this study, we report the frequency of hypodiploidy among patients registered in the Children’s Oncology Group (COG) study AALL03B1 and highlight a previously unrecognized subgroup with “masked” hypodiploidy characterized by the presence of only a single hyperdiploid (doubled) clone that accounts for approximately 25% of patients with hypodiploid ALL. We also describe the characteristic patterns of chromosome gain and loss that allow the identification of patients with suspected masked hypodiploidy, and demonstrate how this can be confirmed by testing for loss of heterozygosity (LOH).
Figure 1.
Mosaicism for a near haploid clone and an exact “doubled” copy of the near haploid clone. The doubled clone is often mistaken as a typical hyperdiploid cell. Note the “hyperdiploidy by tetrasomy” pattern.
Patients and Methods
Patients
Between 29 December 2003, and 6 September 2011, a total of 7759 children and young adults <31 years of age with newly diagnosed precursor B-cell ALL (B-ALL) were enrolled in the COG AALL03B1 () risk classification and biology study. In AALL03B1, a standard battery of cytogenetic and FISH studies, DI determination, and early disease response assessment were performed, and these data were used in conjunction with clinical features to determine risk and allocate patients to post-induction therapy.
Chromosome and FISH analyses
Chromosome analyses of bone marrow specimens at the time of diagnosis were performed in COG-approved local laboratories throughout the study. FISH analysis using probes for the centromeres of chromosomes 4, 10, and 17 was performed throughout the course of the study. This analysis was initially done in COG reference or approved local (2003–2006) laboratories and later (2007–2011) exclusively in COG-approved local laboratories. Presence of ETV6–RUNX1, BCR-ABL1, and KMT2A (infants only) rearrangements was determined by reverse-transcription (RT) PCR or FISH in 1 of 2 central reference laboratories (2003–2006) or by FISH in COG-approved local laboratories (2007–2011). All karyotype and FISH results from approved local laboratories were centrally reviewed in real time by AJC or NAH. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature [ISCN (1995)] (14). As in the Harrison CJ et al. 2004 study (11), the following modification was used: all hypodiploid karyotypes, regardless of modal number, were described as gains to the haploid chromosome complement (1n) rather than losses from the diploid one (2n).
Microsatellite panel analysis
For patient samples that required microsatellite panel analysis, DNA was isolated from diagnostic bone marrow or involved peripheral blood samples as well as from uninvolved post-induction peripheral blood by using the DNeasy kit (Qiagen). Lymphoblast and germline DNA were amplified using the Powerplex® 16 (Promega) microsatellite panel kit, which includes 15 polymorphic short tandem repeat polymorphisms from 13 different chromosomes and the amelogenin locus for gender confirmation. Allelic patterns for informative loci between tumor and normal samples were compared to identify LOH in tumor samples. LOH patterns were compared with the karyotype of lymphoblasts. Chromosomes that were disomic in a hypodiploid clone were tetrasomic in the doubled clone and showed no LOH. Chromosomes that were monosomic in the hypodiploid clone were usually present in only 2 copies in the doubled clone and showed LOH in the microsatellite panel. Allele height was carefully considered when assessing trisomies for the presence or absence of LOH.
DNA ploidy analysis by flow cytometry
To determine DNA ploidy level by calculating the DI, single-cell suspensions of samples were prepared. Nucleated cell count was performed, followed by preparation of Giemsa-stained smears and cytospin slides for morphologic evaluation and verification of the presence and integrity of lymphoblasts as well as estimation of blast percentage in each sample. The Beckman Coulter DNA-Prep Reagents kit was used for cell lysis and DNA staining. Cells were lysed by adding 100 μL of DNA PREP LPR and vortexed for 8 sec. Then, 2 mL of propidium iodide was added for DNA staining and the mixture was vortexed for 10 sec. Specimens were incubated at room temperature for 15 min and analyzed on the Beckman Coulter FC500 cytometer. A sample of normal whole blood stained for DNA was used as a control. A gating strategy was designed to negate doublets or cellular aggregates. Samples were run for 30 min or until 10,000 events were collected in the gate of interest. Data were analyzed by using Multicycle Analysis software (Phoenix Flow Systems). Results were interpreted by a pathologist, using well-established guidelines (15,16). Interpreted data were correlated with morphologic findings from Giemsa-stained smears and cytospin slides. For some difficult cases, interpreted data were also correlated with cytogenetic findings.
Therapy
Patients received therapy in one of several COG studies for B-ALL: AALL0331 () for standard-risk patients (age 1–9.99 years and initial white blood cell count [WBC] <50,000/μL), AALL0232 ()(17) for high-risk patients (age ≥10 years and/or initial WBC ≥50,000/μL), or AALL0031 (18–20) or AALL0622 () for very high-risk patients [presence of adverse features such as induction failure, t(9;22), KMT2A (MLL) rearrangement with poor early response, or hypodiploidy(≤43 chromosomes)].
Results
Distribution of patients with hypodiploidy by modal chromosome number
Of 7013 patients with B-ALL for whom chromosome studies were successfully performed, 121 (1.72%) were classified as hypodiploid with ≤43 chromosomes (Table 1). Of them, 65 had near-haploidy, 50 had low hypodiploidy, and 6 had high hypodiploidy. Because of the limited number and karyotypic heterogeneity of patients with high hypodiploidy, this subgroup was not further evaluated. Thus, 115 patients underwent further analysis.
Table 1.
Distribution of hypodiploid (≤43 chromosomes) patients by modal chromosome number
| Modal chromosome number | Number of patients |
|---|---|
| 24–31 | 65 |
| 32–39 | 50 |
| 40–43 | 6 |
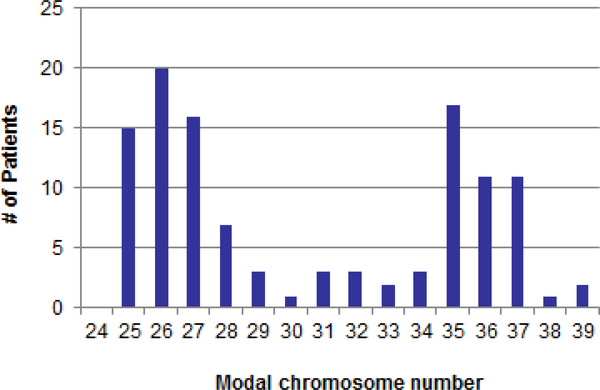
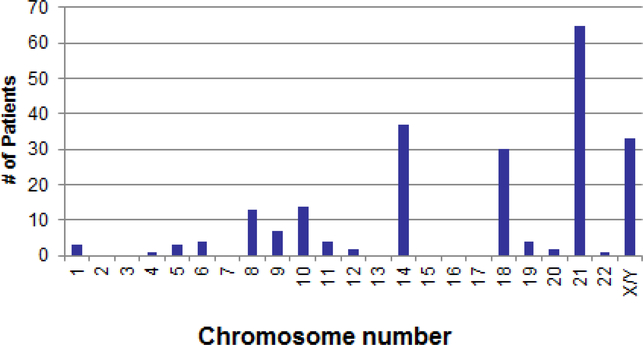
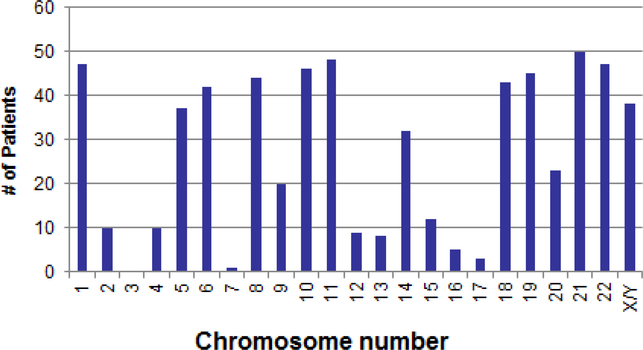
There were no patients with clones containing 23 or 24 chromosomes, suggesting that true haploidy is extremely rare. Figure 2 shows the distribution of 115 patients with hypodiploidy by modal chromosome numbers 25–39. The distribution was bimodal, with peaks at modal chromosomal numbers 25–27 for those with near-haploidy and at 35–37 for those with low hypodiploidy. Most of the chromosomal abnormalities were numerical in nature and involved loss of an entire chromosome. Structural chromosomal abnormalities occurred in 7 (11%) patients with near-haploidy and 17 (34%) with low hypodiploidy. Chromosome loss or retention in patients with near-haploidy or low hypodiploidy followed a pattern. Patients with near-haploidy were most likely to remain disomic for chromosomes 14, 18, 21, and X/Y (Figure 3a), whereas those with low hypodiploidy were most likely to show disomy for chromosomes 1, 5, 6, 8, 10, 11, 14, 18, 19, 21, 22, and X/Y (Figure 3b). None of the 115 patients had a clone with loss of both homologs of a chromosome pair. Of note, chromosome 21 was retained in the disomic state in all 115 patients with hypodiploidy. Table 2 gives karyotypes for the 115 patients.
Figure 2.
Distribution of hypodiploid cases by modal chromosome number
Figure 3a.
Chromosomes retained (disomic) in 65 near-haploid patients (modal chromosome # 24–31)
Figure 3b.
Chromosomes retained (disomic) in 50 low hypodiploid patients (modal chromosome # 32–39)
Table 2.
Karyotypes for the 115 patients with hypodiploidy
| COG # | Modal # | Karyotype | Category* |
|---|---|---|---|
| 1 | 25 | 25,X,+18,+21[9]/50,idemx2[5]/46,XY[6] | Hypo/Doubled |
| 2 | 25 | 25,X,+X,+21[17]/50,idemx2[2]/46,XX[1] | Hypo/Doubled |
| 3 | 25 | 25,X,+Y,+21[1]/50,idemx2[19] | Hypo/Doubled |
| 4 | 25 | 50(25,X,+Y,+21)x2[17]/52,idem,+17,+18[4] | Doubled |
| 5 | 25 | 53(25,X,+14,+21)x2,+X,+9,add(11)(q24),add(17)(q25),+mar[12]/46,XX[9] | Doubled |
| 6 | 25 | 25,X,+Y,+21[10]/50,idemx2[1] | Hypo/Doubled |
| 7 | 25 | 25,X,+Y,+21[8]/46,XY[13] | Hypo |
| 8 | 25 | 25,X,+18,+21,add(21)(p11.2)[11]/50,idemx2[3]/46,XX[7] | Hypo/Doubled |
| 9 | 25 | 25,X,+14,+21[6]/50,idemx2[7]/46,XX[7] | Hypo/Doubled |
| 10 | 25 | 25,X,+14,+21[8]/46,XY[14] | Hypo |
| 11 | 25 | 50(25,X,+X,+21)x2[11]/46,XX[9] | Doubled |
| 12 | 25 | 25,X,+14,+21[9]/46,XY[13] | Hypo |
| 13 | 25 | 25,X,+18,+21[6]/50,idemx2[14]/46,XY[0] | Hypo/Doubled |
| 14 | 25 | 50(25,X,+14,+21)x2[11]/46,XX[9] | Doubled |
| 15 | 25 | 25,X,+18,+21[4]/50,idemx2[7]/46,XY[11] | Hypo/Doubled |
| 16 | 26 | 52(26,X,+10,+18,+21)x2[20]/46,XY[0] | Doubled |
| 17 | 26 | 26,X,+Y,+14,+21[6]/46,XY[13] | Hypo |
| 18 | 26 | 26,X,+Y,t(6;14)(q13;q32),+14,+21[6]/27,idem,+18[2]/52,idemx2[9]/46,XY[4] | Hypo/Doubled |
| 19 | 26 | 52(26,X,+Y,+14,+21)x2[4]/46,XY[17] | Doubled |
| 20 | 26 | 26,X,+X,+14,+21[12]/46,XX[12] | Hypo |
| 21 | 26 | 26,X,+Y,+14,+21[3]/52,idemx2[3]/46,XY[12] | Hypo/Doubled |
| 22 | 26 | 26,X,+14,+18,+21[19]/46,XX[1] | Hypo |
| 23 | 26 | 26,X,+X,+14,+21[14]/46,XX[6] | Hypo |
| 24 | 26 | 26,X,+18,+21,+21[4]/52,idemx2[1]/49,idemx2,+14,−18,−18,−21,−21[cp4]/49,idemx2,der(12)t(1;12)(q21;p13),+14,−18,−18,del(18)(q12),−21,−21[cp3]/46,XY[1] | Hypo/Doubled |
| 25 | 26 | 52(26,X,+Y,+10,+21)x2[cp39]/46,XY[20] | Doubled |
| 26 | 26 | 26,X,+10,+18,+21[8]/26,idem,−10,+r(10)[3]/46,XY[7] | Hypo |
| 27 | 26 | 26,X,+Y,+14,+21[13]/52,idemx2[1]/46,XY[6] | Hypo/Doubled |
| 28 | 26 | 26,X,+Y,+14,+21[9]/52–54,idemx2,+9[cp5]/46,XY[7] | Hypo/Doubled |
| 29 | 26 | 52(26,X,+Y,+9,+21)x2[6]/51,idem,−9[7]/46,XY[7] | Doubled |
| 30 | 26 | 26,X,+Y,+14,+21[5]/46,XY[15] | Hypo |
| 31 | 26 | 52(26,X,+Y,+14,+21)x2,add(1)(p36)[cp11]/46,XY[9] | Doubled |
| 32 | 26 | 26,X,+Y,+14,+21[5]/52,idemx2[15] | Hypo/Doubled |
| 33 | 26 | 26,X,+X,+14,+21[5]/46,XX[25] | Hypo |
| 34 | 26 | 26,X,+Y,+14,+21[16]/46,XY[4] | Hypo |
| 35 | 26 | 51(26,X,+Y,+8,+21)x2,−Y[3]/51,idem,−Y,+21[4]/50,idem,−Y,del(14)(q22q24)[cp2]/46,XY[11] | Doubled |
| 36 | 27 | 27,X,+X,+10,+18,+21[6]/54,idemx2[7]/46,XX[15] | Hypo/Doubled |
| 37 | 27 | 27,X,+Y,+8,+10,+21[5]/52,idemx2,−Y,−8[3]/46,XY[14] | Hypo/Doubled |
| 38 | 27 | 27,X,+Y,add(2)(p11.2),+9,add(9)(p13),+14,add(14)(q13),+21[11]/46,XY[9] | Hypo |
| 39 | 27 | 27,X,+X,+8,+18,+21[11]/54,idemx2[7]/46,XX[2] | Hypo/Doubled |
| 40 | 27 | 27,X,+Y,+10,+18,+21[7]/54,idemx2[4]/46,XY[9] | Hypo/Doubled |
| 41 | 27 | 27,X,+8,+14,+18,+21[14]/28,idem,+mar[2]/46,XX[4] | Hypo |
| 42 | 27 | 27,X,+Y,+9,+18,+21[1]/47,XYYc[2] | Hypo |
| 43 | 27 | 27,X,+X,+8,+10,+21[5]/54,idemx2[14]/46,XX[1] | Hypo/Doubled |
| 44 | 27 | 55(27,X,+Y,+14,+18,+21)x2,+10[2]/46,XY[18] | Doubled |
| 45 | 27 | 55(27,X,+Y,+14,+18,+21)x2,+mar1[2]/56,idem,+mar2[3]/46,XY[9] | Doubled |
| 46 | 27 | 54(27,X,+X,+14,+21,+r)x2[6]/46,XX[27] | Doubled |
| 47 | 27 | 27,X,+Y,+8,+18,+21[4]/46,XY[20] | Hypo |
| 48 | 27 | 55(27,X,+Y,+4,+14,+21)x2,+9[10]/46,XY[6] | Doubled |
| 49 | 27 | 54(27,X,+8,+14,+18,del(18)(q21),+21)x2[13]/54,idem,−14,+15[cp4] | Doubled |
| 50 | 27 | 54(27,X,+Y,+9,+14,+21)x2[9]/53,idem,−9[12] | Doubled |
| 51 | 27 | 27,X,+Y,+14,+18,+21[12]/54,idemx2[4]/46,XY[5] | Hypo/Doubled |
| 52 | 28 | 28,X,+Y,+9,+14,+18,+21[5]/54,idemx2,−9,−18[18]/46,XY[4] | Hypo/Doubled |
| 53 | 28 | 28,X,+Y,+6,+10,+18,+21[3]/56,idemx2[17] | Hypo/Doubled |
| 54 | 28 | 57(28,X,+X,+10,+14,+18,+21)x2,+6[24]/46,XX[8] | Doubled |
| 55 | 28 | 28,X,+Y,+6,+10,+18,+21[24]/46,XY[7] | Hypo |
| 56 | 28 | 56(28,X,+Y,+8,+10,+18,+21)x2[10]/46,XY[10] | Doubled |
| 57 | 28 | 28,X,+Y,+9,del(9)(p13),+14,+18,+21[10]/56,idemx2[2]/46,XY[8] | Hypo/Doubled |
| 58 | 28 | 28,X,+Y,+8,+14,+18,+21[18]/46,XY[2] | Hypo |
| 59 | 29 | 29,X,+X,+1,+14,+18,+?20,+21[cp2]/46,XX[36] | Hypo |
| 60 | 29 | 65(29,X,+Y,+11,+12,+14,+19,+21)x2,+1,+2,+4,+6,+10,+18,+22[cp15]/46,XY[5] | Doubled |
| 61 | 29 | 29,X,+X,+8,+10,+14,+18,+21[10]/58,idemx2[5]/74,idemx2,+1,+1,+2,+4,+5,+6,+10,+10,+11,+12,+15,+17,+19,+20,+21,+21[2] | Hypo/Doubled |
| 62 | 30 | 64(30,X,+1,+5,+6,+8,+18,+19,+21)x2,+10,+11,+14,+22[14]/46,XY[12] | Doubled |
| 63 | 31 | 31,X,+8,+10,+11,+12,+14,+18,+21,+22[3]/62,idemx2[8]/46,XX[2] | Hypo/Doubled |
| 64 | 31 | 31,X,+Y,+5,+8,+9,add(9)(p13),der(10)t(9;10)(p13;p15),+11,+14,+19,+21[6]/62,idemx2,−5,−add(9)(p13)x2,+der(9)t(9;13)(p13;q22)inv(9)(q13q34)x2,+10,+10,+10,+10,− der(10)t(9;10)x2,−11,−14,+18[5]/72~74,idemx2,+X,+Y,+1,+2,+3,+9,− add(9)(p13)x2,+der(9)t(9;13)(p13;q22)inv(9)(q13q34)x2, +10,+10,+10,+10,−der(10)t(9;10)x2,+16,+18,+18,+21,+22[cp4]/46,XY[6] | Hypo/Doubled |
| 65 | 31 | 65(31,X,+1,+5,+6,+8,+11,+19,+20,+21)x2,+9,+14,+22[5]/46,XX[15] | Doubled |
| 66 | 32 | 32,X,+4,+5,+8,+10,+12,+13,+20,+21,+22[cp9]/46,XY[21] | Hypo |
| 67 | 32 | 32,add(X)(q28),+1,+6,+8,+10,+14,+18,+19,+21,+22[9]/64,idemx2[cp5]/31,idem,+X,-add(X),−19,add(22)(p11.2[2] | Hypo/Doubled |
| 68 | 32 | 32,X,+1,+5,del(5)(q31q35),+6,+8,+10,+11,+18,+21,+22[cp6]/64,idemx2,−del(5),−6,+14,−18,+19,+19[cp4]/46,XX[20] | Hypo/Doubled |
| 69 | 33 | 73(33,X,+Y,+1,+6,+8,+11,+15,+18,+19,+20,+21)x2,+2,+4,+5,+9,+10,+14,+22[cp17]/46,XY[4] | Doubled |
| 70 | 33 | 33,X,+X,+1,+5,+6,+10,+11,+18,+19,+21,+22[7]/46,XX[13] | Hypo |
| 71 | 34 | 67(34,X,+X,+1,+6,+8,+10,+11,+14,+18,+19,+21,+22)x2,+5,−10,−11,+13,−14[cp5]/46,XX[15] | Doubled |
| 72 | 34 | 34,X,+Y,+5,+8,+10,+11,+14,+16,+18,+20,+21,+22[cp3]/61~67,idemx2[cp2]/46,XY[15] | Hypo/Doubled |
| 73 | 34 | 61(34,X,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+21)x2,−1,−5,−6,−8,−9,−10,−14[3]/46,XX[17] | Doubled |
| 74 | 35 | 62(35,X,+X,+5,+6,+10,+11,+13,+15,+16,+19,+20,+21,+22)x2,−X,−6,−10,−13,−15,−16,−19,−20[cp2]/46,XX[16] | Doubled |
| 75 | 35 | 35,X,+1,+4,+5,+6,+8,+9,+10,+11,+18,+19,+21,+22[cp13]/46,XY[7] | Hypo |
| 76 | 35 | 67(35,X,+Y,+1,+2,+6,+11,+12,+14,+15,+18,+19,+21,+22)x2,−Y,−2,add(13)(p11.2),−14,−15,−21,+22,+mar[cp6]/46,XY[15] | Doubled |
| 77 | 35 | 35,X+X,+1,+5,+6,+8,+10,+11,+14,+18,+19,+21,+22[11]/46,XX[18] | Hypo |
| 78 | 35 | 35,X,+Y,+1,+5,+8,+9,+10,+11,+18,+19,+20,+21,+22[3]/46,XY[16] | Hypo |
| 79 | 35 | 35,X,+1,+2,del(2)(q31),+6,+8,+10,+11,+12,+14,+18,+19,+21,+22[3]/61~62,idemx2,−1,+2,−del(2)(q31)x2,+5,−8,−8,−10,add(10)(q24),−11,−12[cp6]/67~71,idemx2,der(1)t(1;11)(q42;q14),−del(2)(q31),+3,+5,add(10)(q24)x2,+17,der(19)t(11;19)(q14;p13)x2,+21,+21[cp10]/46,XX[3] | Hypo/Doubled |
| 80 | 35 | 35,X,+X,+1,+5,+6,+8,+10,+11,+14,+15,+19,+21,+mar[cp10]/64,idemx2,−5,−11,−14,−15,−19,−mar[cp16]/46,XX[4] | Hypo/Doubled |
| 81 | 35 | 35,X,+X,+1,+5,+6,+8,+10,+11,+18,+19,+21,+22,+mar[5]/75,idemx2,+2,+2,+16,+16,+r[8]/46,XX[22] | Hypo/Doubled |
| 82 | 35 | 35,X,+Y,+1,+5,+6,+8,+9,add(9)(q32),+10,+11,add(13)(q32),+14,+19,+21,+22[9]/46,XY[10] | Hypo |
| 83 | 35 | 35,X,+Y,+1,+4,+5,+6,+8,der(9)add(9)(p13)add(9)(q22),+10,+11,del(13)(q12q22),+13,+19,+21,+22[3]/46,XY[4] | Hypo |
| 84 | 35 | 35,X,+1,+2,+6,+8,+10,+11,+12,+14,+18,+19,+21,+22[7]/46,XX[12] | Hypo |
| 85 | 35 | 35,X,+1,+5,+6,+8,+10,der(10)t(7;10)(q11.2;q24),+11,+12,+14,+18,+19,+21,+mar[cp4]/61,idemx2,−1,−5,−6,−11,i(13)(q10),−14,−18,−19,−marx2[cp7]/61,idemx2,−1,−5,−6,−11,i(13)(q10),−14,−18,−19,+21,add(21)(q22),−22,−marx2[cp3]/62,idemx2,−1,−5,−6,−11,der(13)t(1;13)(q21;p11.2),−18,der(18)t(1;18)(p22;q21),−19,−marx2[cp3]/86,idemx2,+X,+1,+2,+3,+3,+4,+7,+10,+der(10)t(7;10),+12,+13,+13,+15,+17,+20,+21,+22,−marx2[cp3]/46,XY[7] | Hypo/Doubled |
| 86 | 35 | 70(35,X,+X,+1,+5,+6,+8,+11,+14,+18,+19,+20,+21,+22)x2[6]/46,XX[14] | Doubled |
| 87 | 35 | 35,X,+X,+1,+5,+6,+8,+10,+11,+14,+18,+19,+21,+22[7]/35,idem,del(6)(q22)[4]/46,XX[9] | Hypo |
| 88 | 35 | 35,X,+X,+1,+2,+6,+10,+11,+12,+14,+18,+19,+21,+22[10]/46,XX[10] | Hypo |
| 89 | 35 | 35,X,+1,+5,+8,+9,+10,+11,+14,+18,+19,+21,+22,+mar[3]/64,idemx2,−5,−10,−10,−11,−19,−mar[11]/46,XY[13] | Hypo/Doubled |
| 90 | 35 | 35,X,+Y,+1,+5,+6,+8,+10,+11,+14,+18,+19,+21,+22[6]/70,idemx2[1]/46,XY[18] | Hypo/Doubled |
| 91 | 36 | 36,X,+X,+1,+5,+6,+8,+10,+11,+14,+15,+18,+19,+21,+22[9]/46,XX[12] | Hypo |
| 92 | 36 | 73(36,X,+Y,+1,+5,+6,+8,+9,+10,+14,+18,+19,+20,+21,+22)x2,−X,+14,+mar[cp31]/46,XY[1] | Doubled |
| 93 | 36 | 36,X,+X,+1,+5,+8,+10,+11,+12,+13,+16,+19,+20,+21,+22[10]/46,XX[5] | Hypo |
| 94 | 36 | 36,X,+Y,+1,+4,+6,+8,+10,+11,+15,+17,+18,+19,+21,+22[5]/46,XY[17] | Hypo |
| 95 | 36 | 36,X,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+20,+21,+22[5]/72,idemx2[1]/46,XX[21] | Hypo/Doubled |
| 96 | 36 | 36,X,+Y,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+21,+22[8]/72,idemx2[1]/62,idemx2,−X,−Y,−5,−6,−8,−9,−10,−11,−14,−22[4] | Hypo/Doubled |
| 97 | 36 | 36,X,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+20,+21,+22[21]/46,XX[22] | Hypo |
| 98 | 36 | 36,X,+Y,+1,+2,+5,+6,+10,+11,+16,+18,+19,+20,+21,+22[5]/70,idemx2,−10,−16[5]/46,XY[10] | Hypo/Doubled |
| 99 | 36 | 36,X,+X,+1,+5,+6,+8,+10,+11,+14,+18,+19,+20,+21,+22[6]/71,idemx2,−14[cp11]/46,XX[8] | Hypo/Doubled |
| 100 | 36 | 36,X,+Y,+1,+2,+6,+8,+10,+11,+12,+14,+18,+19,+21,+22[6]/68,idemx2,−Y,−1,−2,+3,−11,−21[8]/63,idemx2,−Y,−1,−2,−11,−12,−14,−18,−19,−21[4]/46,XY[2] | Hypo/Doubled |
| 101 | 36 | 36,X,+Y,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+21,+22[12]/46,XY[10] | Hypo |
| 102 | 37 | 37,X,+X,+1,+4,+5,+6,+8,+9,+10,+11,+18,+19,+20,+21,+22[3]/72,idemx2,−9,−9[5]/46,XX[12] | Hypo/Doubled |
| 103 | 37 | 37,X,+Y,+1,+4,+8,+11,+15,+17,+18,+19,+20,+21,+22,+2mar[6]/46,XY[20] | Hypo |
| 104 | 37 | 37,X,+X,+1,+5,+6,dup(7)(q22q36),+8,+9,+10,+11,+14,+18,+19,+20,+21,+22[4]/74,idemx2[5]/72,idemx2,−9,−9[3]/46,XX[4] | Hypo/Doubled |
| 105 | 37 | 37,X,+Y,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+20,+21,+22[26]/74,idemx2[2]/46,XY[26] | Hypo/Doubled |
| 106 | 37 | 37,X,+1,+2,add(2)(q33),+4,+6,+8,+10,+11,+12,+13,+14,+18,+19,+21,+22[2]/62,idemx2,−1,−2,−add(2),−4,−6,−10,−11,−12,−13,−14,−18,−19[7]/46,XY[9] | Hypo/Doubled |
| 107 | 37 | 37,X,+Y,+1,+5,+6,+8,+9,del(9)(p10),+10,add(10)(q11),+11,+14,+18,+19,+20,+21,+22[1]/69,idemx2,−8,−10,−11,−14,−19[20]/46,XY[18] | Hypo/Doubled |
| 108 | 37 | 37,X,+Y,+1,+5,+6,+8,+9,+10,+11,+14,+18,+19,+20,+21,+22[5]/74,idemx2[cp2] | Hypo/Doubled |
| 109 | 37 | 37,X,+Y,+1,+6,+7,+8,+9,+10,+11,add(11)(q25),+15,+18,+19,+20,+21,+22[2]/46,XY[17] | Hypo |
| 110 | 37 | 37,X,+X,+1,+4,+5,+6,+8,+9,+10,+11,+14,+18,+19,+21,+22[4]/46,XX[18] | Hypo |
| 111 | 37 | 37,X,+X,+1,+2,+6,+8,+9,+10,+11,+14,+15,+20,+21,+22,+mar[3]/46,XX[17] | Hypo |
| 112 | 37 | 37,X,+X,+1,+2,+4,+5,+9,+10,+11,+14,+15,+17,+18,+21,+22[cp5]/46,XX[20] | Hypo |
| 113 | 38 | 38,X,+Y,+1,+2,+5,+6,+9,+10,+11,+13,+15,+18,+19,+20,+21,+22[6]/46,XY[14] | Hypo |
| 114 | 39 | 39,X,+Y,+1,+4,+5,+6,+8,+10,+11,+13,+15,+16,+18,+19,+20,+21,+22[8]/64,idemx2,−Y,−4,−6,−11,−13,−13,−15,−15,−16,−16,−19,−19,−20,−20[3] | Hypo/Doubled |
| 115 | 39 | 39,X,+Y,+1,+5,+6,+8,+9,+10,+11,+13,+14,+15,+18,+19,+20,+21,+22[7]/72,idemx2,−6,−9,−9,−10,−10,−15[cp9]/72,idemx2,−6,−8,−9,−9,−10,−15[cp4] | Hypo/Doubled |
Partially and completely masked hypodiploidy
Of the 115 patients with hypodiploidy, karyotypes of 39 (18 with near-haploidy and 21 with low hypodiploidy) showed only the hypodiploid clone. Karyotypes of 48 (26 with near-haploidy and 22 with low hypodiploidy) showed mosaicism, with both hypodiploid karyotypes and hyperdiploid karyotypes that had arisen due to an exact or near-exact doubling of the hypodiploid clone. Karyotypes of the remaining 28 patients (21 with near-haploidy and 7 with low hypodiploidy) showed only a hyperdiploid (doubled) clone, which had the effect of “masking” the true karyotypic nature of the clone (Table 3). However, these doubled karyotypes demonstrated a unique pattern of aneuploidy characterized primarily by tetrasomy rather than trisomy, which distinguished them from a “typical” hyperdiploid case with trisomies and an equivalent number of chromosomes (Figure 1). For 12 of 28 patients with only a hyperdiploid (doubled) clone, the hypodiploidy was considered to be only partially masked, because there was evidence of a small population of hypodiploid cells by DI and/or FISH analyses. For the 16 remaining patients, the hypodiploidy was completely masked, with no evidence of a detectable level of hypodiploid cells by either DI or FISH analysis.
Table 3.
Distribution of clone types among hypodiploid patients
| Hypodiploid clone only | Both hypo & hyperdiploid (doubled) clones | Hyperdiploid (doubled) clone only | |
|---|---|---|---|
| near-haploid | 18 (28%) | 26 (40%) | 21 (32%) |
| low-hypodiploid | 21 (42%) | 22 (44%) | 7 (14%) |
| total | 39 (34%) | 48 (42%) | 28 (24%) |
Loss of heterozygosity in tumor and germline DNA for patients with completely masked hypodiploidy
Tumor and germline DNA in the 16 patients exhibiting completely masked hypodiploidy, which was identified solely on the basis of the unusual hyperdiploid karyotype with tetrasomic chromosomes, were evaluated for LOH by using a microsatellite panel that tested 15 distinct loci on 13 different chromosomes. In all 16 patients, LOH on multiple different chromosomes confirmed that the hyperdiploid cells had arisen by doubling of a hypodiploid clone. For patients in whom masked hypodiploidy was confirmed, the following nomenclature was used for description: the presumed hypodiploid karyotype was written within parentheses, prefaced by the total number of chromosomes, and with ×2 following the parentheses (Table 2).
Discussion
In previously published studies, outcomes of children with B-ALL who have hypodiploidy in leukemic clones are very poor (OS ≤ 40%) (8–12). Consequently, it is important to identify all patients with hypodiploidy at the time of diagnosis for timely administration of intense consolidation and continuation therapy. Notably, in our study, we identify a subset of children whose leukemia is characterized as near-haploid or low hypodiploid, but the only visible leukemic clone contains chromosome numbers that would normally be considered hyperdiploid. Since hyperdiploidy with >50 chromosomes is usually associated with excellent prognosis, it is very important for cytogeneticists and treating physicians to distinguish true hyperdiploidy associated with favorable prognosis from the “hyperdiploidy” that results from doubling of a near-haploid or low-hypodiploid clone. There are no studies in the literature that address outcomes for patients who display only a masked doubled hypodiploid clone. It has been reported (12) that there is no difference in outcome for patients who were mosaic for a doubled clone and a hypodiploid clone versus those who displayed only a hypodiploid clone. We expect that patients showing only the doubled clone will fare no differently than those with a detectable hypodiploid clone but this remains to be proven.
The tendency of hypodiploid clones to undergo exact or near-exact doubling was first reported in 1990 (8) and has been confirmed in several subsequent series of children with ALL (9–12). A near-haploid or low-hypodiploid clone usually contains only 1 or 2 copies of each chromosome; therefore, if the clone undergoes exact doubling, the new clone will contain 2 or 4 copies of each chromosome (Figure 1). This is in contrast to the typical hyperdiploidy associated with favorable risk, wherein most of the aneuploidy occurs in the form of trisomies. In effect, the doubling of a hypodiploid clone will result in “hyperdiploidy by tetrasomy,” in contrast to the “hyperdiploidy by trisomy” that characterizes true hyperdiploidy associated with favorable risk. Further, the apparent “disomy” seen in the doubled hypodiploid clone is actually isodisomy and will be associated with LOH for all loci on that chromosome.
Doubled near-haploid karyotypes most often display tetrasomy of chromosomes 14, 18, 21, and X/Y. Doubled low-hypodiploid karyotypes most often show tetrasomy for chromosomes 1, 8, 10, 11, 18, 19, 21, and 22. We found a doubled clone in some or all cells in 76 (66%) of 115 patients with hypodiploidy. The frequency of doubling was higher among patients with near-haploidy (47/65 [72%]) than those with low hypodiploidy (29/50 [58%]; Table 3). Also, patients with near-haploidy were more likely to display exact or near-exact doublings of the hypodiploid clone than those with low hypodiploidy, whose less-exact doublings resulted in more trisomies (Table 2). Notably, as the doubling became less exact with increasing numbers of trisomies rather than tetrasomies, it became less obvious that karyotypes arose through doubling, and the karyotypes were increasingly reminiscent of the hyperdiploid karyotype associated with favorable risk. We presume that the doubling is a result of failure during mitosis of daughter cells to separate at telophase. It is not known if doubling provides any proliferative advantage to a cell nor is it known if retention of specific chromosomes is advantageous.
Of 28 patients with hypodiploidy whose karyotype displayed only the hyperdiploid (doubled) clone, 21 (75%) were classified as having near-haploidy and only 7 (25%) were classified as having low hypodiploidy. The distribution was more even among 48 patients who displayed mosaicism with both hypodiploid and hyperdiploid (doubled) clones, with 26 (54%) having near-haploidy and 22 (46%) having low hypodiploidy. It is possible that doublings of hypodiploid clones are more common among patients with near-haploidy; however, it is also possible that doublings are just as common among patients with low hypodiploidy, but they are recognized less often because of less-exact doublings, resulting in trisomies, which mask characteristic features of the doubling process.
For 12 of 28 patients displaying only the hyperdiploid (doubled) clone, DI and/or FISH analyses showed evidence of a small population of hypodiploid cells. This confirms that hyperdiploid cells were the result of doubling. These patients were considered to be “partially masked.” In the 16 remaining patients with only a hyperdiploid (doubled) clone, DI and FISH analyses showed no evidence of a measurable population of hypodiploid cells, and they were considered “completely masked.” Microsatellite panel analysis confirmed LOH for loci on several different disomic chromosomes for all 16 patients. The coverage of the microsatellite panel used for this study was limited to 1 or 2 loci on 13 chromosomes, and future studies will benefit from the more extensive coverage offered by chromosomal microarray (CMA) platforms with single nucleotide polymorphism (SNP) probes. It seems likely that in our study, additional patients with masked hypodiploidy, particularly those with low hypodiploidy and inexact doublings, might have been missed because their karyotypes were not recognized to suggest masked hypodiploidy. Thus, estimates from this study should be considered as the minimum estimate of the true frequency of masked hypodiploid B-ALL in children. In adults, near-triploid karyotypes are considered to be in the same subgroup as hypodiploid karyotypes (21–23). Although near-triploid karyotypes are uncommon in childhood ALL, they should be investigated by SNP microarray or microsatellite analysis to determine whether doubling underlies the high chromosome number.
The 3 largest previous studies of hypodiploidy in children with ALL (10–12) reported a combined total of 110 patients with 24–39 chromosomes. Many of the patients exhibited mosaicism for a hypodiploid and a hyperdiploid (doubled) clone, but none of them had partially or completely masked hypodiploidy. We conclude that a considerable proportion (25% or higher) of blast cell hypodiploidy in children with B-ALL may have been overlooked by previous studies due to the presence of only a doubled hypodiploid clone mistakenly thought to represent typical hyperdiploidy associated with favorable risk. Cytogeneticists and clinicians need to have a heightened awareness of the unique karyotypic “signature” of a doubled hypodiploid clone. Also, the coordinated use of DI and FISH, as well as LOH and/or CMA studies when indicated for suspicious karyotypes is important to identify patients with masked hypodiploidy so that they can be appropriately administered treatment for very high-risk disease.
Acknowledgements
The authors thank the patients and parents who participated in the AALL03B1 protocol and the cytogeneticists at the COG-approved laboratories who contributed to this study. MLL is the UCSF Benioff Chair of Children’s Health and Deborah and Arthur Ablin Endowed Chair in Pediatric Molecular Oncology. EAR is a KiDS of NYU Foundation Professor at NYU Langone Health. SPH is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics at the Children’s Hospital of Philadelphia.
Sources of Funding;
Supported by grants U10 CA98543 and U10 CA180886 from the National Institutes of Health.
Footnotes
Conflict of Interest:
The authors declare no conflict of interest. All authors read and approved the first version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hunger SP, Lu X, Devidas M, Camitta BM,Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(14):1663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Möricke A, Zimmermann M, Reiter A, Henze G, Schrauder A, Gadner H, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–84. [DOI] [PubMed] [Google Scholar]
- [4].Silverman LB, Stevenson KE, O’Brien JE, Asselin BL, Barr RD, Clavell L, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000) Leukemia. 2010;24:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kamps WA, van der Pal-de Bruin KM, Veerman AJ, Fiocco M, Bierings M, Pieters R. Long-term results of Dutch Childhood Oncology Group studies for children with acute lymphoblastic leukemia from 1984 to 2004. Leukemia. 2010;24:309–19. [DOI] [PubMed] [Google Scholar]
- [6].Schultz KR, Pullen DJ, Sather HN, Shuster JJ, Dévidas M, Borowitz MJ, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood. 2007;109:926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heerema NA, Raimondi SC, Anderson JR, Biegel J, Camitta BM, Cooley LD, et al. Specific extra chromosomes occur in a modal number dependent pattern in pediatric acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46:684–93. [DOI] [PubMed] [Google Scholar]
- [8].Pui CH, Carroll AJ, Raimondi SC, Land VJ, Crist WM, Shuster JJ, et al. Clinical presentation, karyotypic characterization, and treatment outcome of childhood acute lymphoblastic leukemia with a near-haploid or hypodiploid less than 45 line. Blood. 1990;75:1170–7. [PubMed] [Google Scholar]
- [9].Heerema NA, Nachman JB, Sather HN Sensel MG, Lee MK, Hutchinson R, et al. Hypodiploidy with less than 45 chromosomes confers adverse risk in childhood acute lymphoblastic leukemia: a report from the children’s cancer group. Blood. 1999;94:4036–46. [PubMed] [Google Scholar]
- [10].Raimondi SC, Zhou Y, Mathew S, Shurtleff SA, Sandlund JT, Rivera GK, et al. Reassessment of the prognostic significance of hypodiploidy in pediatric patients with acute lymphoblastic leukemia. Cancer. 2003;98:2715–22. [DOI] [PubMed] [Google Scholar]
- [11].Harrison CJ, Moorman AV, Broadfield ZJ, Cheung KL, Harris RL, Reza Jalali G, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol. 2004;125:522–9. [DOI] [PubMed] [Google Scholar]
- [12].Nachman JB, Heerema NA, Sather H, Camitta B, Forestier E, Harrison CJ, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110:1112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nature Genetics. 2013;45:242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].ISCN (1995): An International System for Human Cytogenetic Nomenclature, Mitelman F (ed); S. Karger, Basel, 1995. [Google Scholar]
- [15].Dressler LG. Controls, standards and histogram interpretation in DNA flow cytometry. Methods Cell Biol. 1990;33:157–71. [DOI] [PubMed] [Google Scholar]
- [16].Hiddeman W, Schumann J, Andreeff M, Barlogie B, Herman CJ, Leif RC, et al. Convention on nomenclature for DNA cytometry. Cytometry. 1984;5:445–6. [DOI] [PubMed] [Google Scholar]
- [17].Larsen EC, Devidas M, Chen S, Salzer WL, Raetz EA, Loh ML, et al. Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: A report from Children’s Oncology Group study AALL0232. J Clin Oncol 2016;34:2380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, et al. Improved early event-free survival with imatinab in Philadelphia chromosome-positive acute lymphoblastic leukemia: A Children’s Oncology Group study. J Clin Oncol 2009;27:5175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schultz KR, Devidas M, Bowman WP, Aledo A, Slayton WB, Sather H, et al. Philadelphia chromosome-negative very high-risk acute lymphoblastic leukemia in children and adolescents: Results from Children’s Oncology Group study AALL0031. Leukemia 2014;28:964–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slaton WB, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia 2014;28:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Charrin C, Thomas X, Ffrench M, Le QH, Andrieux J, Mozziconacci MJ, et al. A report from the LALA-94 and LALA-SA groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2 possible expressions of a sole entity conferring poor prognosis in adult acute lymphoblastic leukemia (ALL). Blood. 2004; 104(8):2444–51. [DOI] [PubMed] [Google Scholar]
- [22].Moorman AV, Harrison CJ, Buck GA, Richards SM, Secker-Walker LM, Martineau M, et al. ; Adult Leukaemia Working Party, Medical Research Council/National Cancer Research Institute. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109(8):3189–97. [DOI] [PubMed] [Google Scholar]
- [23].Muhlbacher V, Zenger M, Schnittger S, Weissmann S, Kunze F, Kohlmann A, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer. 2014;53:524–6. [DOI] [PubMed] [Google Scholar]