Abstract
Background:
The human leukocyte antigen (HLA) locus includes several genes with key roles in antigen presentation and immune response, some of them inclusively found to be associated with non-obstructive azoospermia. Still, HLA connections to other infertility phenotypes such as semen hyperviscosity (SHV), asthenozoospermia (AST) and oligozoospermia (OLI) have been often neglected.
Objectives:
In this work, we aimed to evaluate the association of HLA class I and II genes with SHV, AST and OLI phenotypes while exploring a possible role in an adaptive immune response to sexually transmitted diseases (STD).
Materials and methods:
Whole exome sequencing was performed in a Portuguese cohort of 71 infertility cases and 68 controls, followed by HLA typing using a specific software - HLA*PRG:LA tool. Molecular screenings of seven STD were carried out in a subset of 72 samples (30 cases and 42 controls).
Results:
Statistical tests uncovered three protective alleles: HLA-A*11:01, associated with all forms of male infertility (P=0.0006); HLA-DQB1*03:02 with SHV and OLI (PSHV=0.0303, POLI=0.0153) and HLA-A*29:02 with OLI (P=0.0355), which was found to interfere in sperm number together with HPV (P=0.0313). Five risk alleles were also identified: two linked with SHV (HLA-B*50:01, P=0.0278; and HLA-C*06:02, P=0.0461), another one with both SHV and OLI (HLA-DQA1*05:01, PSHV=0.0444 and POLI=0.0265) and two with OLI (HLA-C*03:03, P=0.0480; and HLA-DQB1*03:01, P=0.0499). Here, HLA-C*03:03 carriers tend to be HPV infected.
Conclusions:
The application of HLA*PRG:LA tool to the study of male infertility, provided novel insights for an HLA correlation with semen quality, namely among SHV and OLI phenotypes. The discovery of an HLA-A*29:02/HPV crosstalk, together with former reports of HLA alleles conferring resistance-susceptibility to diverse human pathogens raises the hypothesis of a mechanistic link between male infertility, HLA polymorphism and host response to STD.
Keywords: male infertility, semen hyperviscosity, oligozoospermia, association study, HLA-A, HPV
INTRODUCTION
Infertility is a major health issue that affects 10–20% of couples within reproductive age and in which male factors, either alone or in combination with female causes, are known to contribute to its etiology in ~50% of cases 1. More specifically, male infertility is a multifactorial disorder with a large phenotypic variability in semen quality, including: azoospermia (AZO) and oligozoospermia (OLI), for spermatozoa absence or low counts, respectively; asthenozoospermia (AST), for reduced spermatozoa motility; and semen hyperviscosity (SHV), for the persistence of semen viscous proprieties after its expected liquefaction time 2. Although such abnormalities may in some instances be correlated with congenital defects, endocrine dysfunction, urogenital infections and genetic anomalies, in a high proportion of cases the underlying causes for male infertility remain unknown 3.
In the last decade, several genome-wide association studies (GWAS) shed some light into the genetic makeup of this disorder, mainly concerning non-obstructive azoospermia (NOA), by uncovering novel susceptibility genes like PDE3A (phosphodiesterase 3A), SOX5 (SRY-box 5), DMRT1 (doublesex and mab-3 related transcription factor 1) and HLA-DRA (major histocompatibility complex, class II, DR alpha) 4–6. Aside from these GWAS, some candidate gene approaches centered on HLA classes I and II also reported several associations with specific HLA alleles (HLA-A*26, HLA-A*28, HLA-A*33, HLA-B*18, HLA-B*44, HLA-DPB1*04:01, HLA-DQB1*06:04 and HLA-DRB1*13:02) and idiopathic male infertility 7–10. However, once again these studies were predominantly focused on NOA patients and in most instances in men of East Asian ancestry.
In overview, the human leukocyte antigen (HLA) locus (6p21.32–22.1) is one of the most peculiar regions of our genome, characterized by high levels of polymorphism and heterozygosity, as well as extended linkage disequilibrium (LD) across different alleles. Briefly, the HLA system contains several genes and pseudogenes that can be divided into class I, II and III 11,12 (Supplementary Fig. S1A). Even though all three classes are involved in the immune response, HLA class I and II molecules are the ones in charge of antigen presentation to T cells. Classical HLA class I genes, HLA-A, -B, and -C, are expressed in most cells, where they code for distinct α-chains of HLA class I heterodimers and where the β-chain is encoded by β2-microglobulin (B2M; 15q21.1; Supplementary Fig. S1B). On the other hand, classical HLA class II genes are expressed on specific immune cells, such as B cells, activated T cells, macrophages and dendritic cells, and are organized in three families: HLA-DP, -DQ and -DR. Each family comprises at least two genes coding for α- and β-chain molecules (Supplementary Fig. S1). Here, HLA-DR family accommodates a copy number variation (CNV), determining the origin of β-chains in HLA-DRB3, -DRB4 or -DRB5 genes (Supplementary Fig. S1C)13.
Importantly, several alleles of HLA class I and II genes have already been correlated with sexually transmitted disease (STD) caused by Neisseria gonorrhoeae (gonorrhea) 14,15, Chlamydia trachomatis 14,16, Treponema pallidum (syphilis) 17,18, human immunodeficiency virus (HIV) 19,20, human papilloma virus (HPV) 21, hepatitis B virus (HBV)22 and hepatitis C virus (HCV) 23–25. Such microbial agents per se and together with activated immunological processes can lead to anatomical obstruction, compromise normal gland function, affect the availability of diverse seminal components and interfere in spermatozoa viability 26–30. To be more precise, HIV, HPV and HBV, can be internalized by spermatozoa causing DNA damage and altering sperm number, motility and morphology, while in case of HBV, it can also induce the production of antisperm antibodies (ASA) 29,31–37. Likewise, C. trachomatis and N. gonorrhoeae infections, as well as other bacteriospermias in general, may reduce semen quality by multiple mechanisms such as genital tract dysfunction, impaired spermatogenesis and bacterial cellular interactions 38–42.
In this work, we investigated the association of HLA class I and II genes to SHV, AST (two less explored infertility phenotypes) and OLI, by applying for the first time the HLA*PRG:LA tool 43 to a whole exome sequencing (WES) panel generated in a Portuguese cohort of patients and controls. The algorithm implemented through HLA*PRG:LA tool has the advantage over standard alignment methods used for the analysis of next-generation sequencing data, since it compares collected reads against multiple available HLA sequences instead of a single human reference. Thus, this method allows to circumvent the mapping problems arising from high polymorphism levels and from the presence of multiple paralog genes within the HLA locus. Then, for those HLA alleles showing significant associations with male infertility, we screened seven well known pathogens to test the hypothesis of increased susceptibility/resistance to STD.
Overall, our results provide further evidence for a possible association of HLA locus variability with male infertility, which we propose to be connected with an adaptive immune response to sexually transmitted pathogens often underlying asymptomatic urogenital infections or on the other hand, with an autoimmune reaction leading to the production of anti-sperm antibodies.
MATERIAL AND METHODS
Ethical approval
This study has been conducted in accordance with the ethical standards of the involved institutions and with the Helsinki Declaration. All participants provided written informed consent.
Patient recruitment and classification
A total of 139 samples were selected from a previous cohort of Portuguese men undergoing spermiogram analysis, for which peripheral blood and semen were collected 44,45. Since our infertility patients (N=71; mean age=37.3±4.33) showed several abnormal parameters in spermiogram, samples were stratified into three non-mutually exclusive phenotypes: SHV (semen drops form a thread >2cm long; N=61), AST (rapid progressive motility <25%; N=45) and OLI (sperm counts <20million/mL; N=28) 2. Samples that did not fit any of these infertility criteria were considered as controls (N=68; mean age=36.6±4.29). This cohort was excluded for male accessory gland infections (negative results in seminal culture), four sexually transmitted virus (HIV; HBV; HCV; and human T lymphotropic virus - HTLV) and syphilis (T. pallidum) 44,45. Individuals with known infertility causes, including chromosome anomalies and Yq microdeletions, were not considered in this study.
Whole exome sequencing (WES)
DNA was extracted from peripheral blood leukocytes using Citogene Blood Kit (Citomed). For cases, exonic and UTR regions were captured by SureSelect Human All Exon V5+UTR enrichment kit and paired-end sequenced on an Illumina HiSeq 2000 (Macrogen Inc, Seoul, South Korea). Raw reads were mapped to the human reference genome (GRCh37) using BWA-MEM v.0.7.15 with default parameters 46. Picard v.1.138 (http://broadinstitute.github.io/picard/) and Samtools v.1.2 47 were applied for post-alignment processes such as indexing, sorting and marking duplicates. The alignments were further submitted to indel realignment and base quality score recalibration using the Genome Analysis Toolkit (GATK) v. 3.4–0-g7e26428 48.
Control samples were analyzed in the framework of the Genetics of Male Infertility Initiative (GEMINI; http://gemini.wustl.edu/), aiming to map the genetic architecture of severe male fertility. Up to 39.1 Mb of exonic regions were captured using in-house exome targeting reagent and submitted to paired-end sequencing on Illumina HiSeq 4000 at the McDonnell Genome Institute of Washington University (genome.wustl.edu). The mapping and alignment procedures were similar to the approach applied to cases. In summary, raw reads were processed with Picard v.2.10.0 and samtools v.1.6 and mapped to the human reference genome assembly GRChg38 in an alternate contig-aware manner using BWA-MEM v.0.7.17 followed by INDEL recalibration and base quality score recalibration using GATK v.3.6.0. These WES data were extracted from two larger and ongoing works yet to be published.
HLA type inference
HLA typing using WES data was performed with HLA*PRG:LA program 43,49, a bioinformatics tool published only on bioRxiv (not peer-reviewed). This tool has the advantage of automatically detecting the genome version of inputted BAM files and using a population reference graph (PRG) framework to impute HLA alleles for 10 genes (HLA-A, -B, -C, -DQA1, -DQB1, -DPA1, -DPB1, -DRB1, -DRB3 and -DRB4). Two caveats of HLA*PRG:LA is that the model does not take into account the number of copies of HLA-DRB paralogs nor analyzes HLA-DRB5. Hence, we followed HLA*PRG:LA method recommendations and used the obtained coverage of HLA-DRB3 and -DRB4 together with HLA-DRB1 alleles to infer the CNV: HLA-DRB3, -DRB4, or -DRB5 13,43. Moreover, the presence/absence of HLA-DRB5 was confirmed in all individuals by two amplicons. Positive samples were sequenced with BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI 3130 automated sequencer. PCR conditions for HLA-DRB5 are described in Supplementary Table S1.
Molecular screening of sexually transmitted diseases (STD)
A subset of 72 samples (42 controls and 30 cases, distributed by phenotypes as 29 SHV, 20 AST, 16 OLI) was evaluated for seven well-known agents of STD: C. trachomatis, N. gonorrhoeae, Ureaplasma urealyticum, Mycoplasma genitalium, herpes simplex virus −1 and −2 (HSV-1; HSV-2) and HPV. To this end, total DNA was extracted from seminal plasma using QIAamp DNA mini kit (Qiagen) and several duplex and multiplex PCR reactions were carried out using primers reported in the literature 50–53 (for PCR conditions see Supplementary Table S2). All samples were amplified in triplicate and all PCR reactions were performed in the presence of negative and positive controls for each one of the tested STD. To confirm PCR specificity, obtained amplicons were submitted to Sanger sequencing (BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI 3130 automated sequencer) and inspected using BLAST tool and NCBI database (https://www.ncbi.nlm.nih.gov/).
Statistical analysis
To consider all possible associations to male infertility, four sets of comparisons were carried out in this study – all cases, SHV, AST or OLI.
Hardy–Weinberg equilibrium (HWE)
The HWE was evaluated at gene-level for controls and cases, separately, using PyPop v0.7.0 54 and assuming Guo and Thompson’s test 55. Then, Chen’s measure was used to identify those genotypes explaining the observed departures from HWE 56.
Association tests for HLA alleles
Identified HLA alleles (4-digit resolution, corresponding the first 2-digits to the allele group and the remaining ones to specific HLA proteins) were divided into common or rare according to their frequency (f) in the control sample (f ≥ 0.01 and f < 0.01, respectively). For common alleles, differences between cases and controls were estimated using Fisher’s exact test implemented in PyHLA package 57. Next, a Bonferroni correction was used to adjust P-values for multiple comparisons in which the number of observed common alleles per gene was used as the number of tests. Welch’s ANOVA was also performed for those alleles showing associations with either AST or OLI and nominal P-values < 0.05 (IBM SPSS Statistics v.25). For these tests, the entire cohort was used to evaluate the effect of HLA alleles on quantitative spermiogram parameters: sperm motility (fast forward progression) and sperm number, respectively. Taking into account the limited statistical power of these approaches to detect rare variant associations, an enrichment analysis of HLA alleles with f < 0.01 was assessed by a simplified CAST test 58,59. Essentially, the number of chromosomes carrying rare alleles was compared between cases and controls using a 2 × 2 contingency table and Fisher’s exact test.
Haplotype analysis
To evaluate the hypothesis of a non-random association of HLA alleles due to local high LD levels, pairwise D’ statistics were calculated using Pypop software 54. For allele pairs found to be strongly correlated (D’ > 0.85), the corresponding haplotypes were inferred using the bigdawg R package 60, and their association to different infertility phenotypes was addressed by Fisher’s exact tests. Moreover, to assess if those allele pairs contribute to a phenotype in a combined manner or if one of them results from a spurious association caused by LD, we used the statistical genetic approach called “interaction analysis” from PyHLA software 57.
Amino acid sharing analysis
To circumvent a possible lack of statistical power due to a reduced sample size or because of multiple comparisons, an amino acid sharing approach was implemented in our analysis. Specifically, association tests at residue level were performed for both common and rare alleles with the bigdawg R package using only HLA regions known to encode antigen recognition sites (exon 2 and 3 for class I and exon 2 for class II genes) 60. Contingency tables and χ2 tests were first performed for each polymorphic amino acid position, followed by contingency tables and Fisher’s exact test for each residue with significantly associated positions. For variants with expected counts below five (cases or controls), comparisons were done in a single combined category. The results are expressed according to the mature protein numbering.
Copy number variation (CNV) association tests
In the association analysis of HLA-DRB3, -DRB4 and -DRB5, we used two approaches. In the first one, HLA-DRB3, -DRB4, -DRB5 or none of the previous were treated as alternative alleles of a single CNV locus (a 2 × 4 contingency table and χ2 test was used). In the second approach, three bi-allelic CNV loci corresponding to the presence or absence of HLA-DRB3, -DRB4 and -DRB5 were considered (three 2 × 2 contingency tables and Fisher’s exact test were employed). In all instances, GraphPad Prism 5 software (GraphPad Software) was used to evaluate statistical significance of association.
Association tests for STD
To evaluate the strength of STD association to male infertility, we used the data from our molecular screening to perform Fisher’s exact tests (2 × 2 contingency tables for cases vs. controls). In addition, Welch’s ANOVA was carried out for sperm number and sperm motility variables. Then, to address whether detected HLA associations could be related with a host response to STD, two statistical approaches were used: Fisher’s exact tests were employed to compare presence/absence of HLA allele with STD; and two-way ANOVAs were carried out to accommodate the interaction between HLA allele, STD and the semen parameters “sperm number” or “sperm motility”. These analyses were performed for STD as a whole and for C. trachomatis and HPV separately. In all instances, IBM SPSS Statistics v.25 was used to evaluate statistical significance.
RESULTS
Fine-mapping of HLA alleles in male infertility
To assess a possible genetic influence of classical HLA class I and II genes into abnormal semen parameters, we analyzed the HLA allelic variation in 71 infertility cases and 68 controls genotyped as part of a whole-exome study (unpublished data). A large number of alleles were found in the full cohort: 113 common (f ≥ 0.01) and 63 rare (f < 0.01) (Supplementary Table S3). Globally, no violation of HWE was observed in our control dataset for HLA-A, -B, -C, -DQA1, -DQB1, -DRB1, -DPA1 and -DPB1 eliciting its usage in the following disease association tests.
Among common HLA alleles, eight showed significant differences (P < 0.05) between cases and controls (Table 1 and Fig. 1). Remarkably, seven of these associations could be correlated with one or two male infertility phenotypes. For instance, HLA-C*03:03 and HLA-C*06:02 were both detected as risk alleles for OLI and SHV, respectively. On the other hand, in HLA-DQB1 gene, whereas HLA-DQB1*03:01 was identified as an OLI risk allele, HLA-DQB1*03:02 was classified as a protective allele for OLI and SHV. The single association that appears to be transversal to male infertility (all cases) is correlated with HLA-A*11:01 allele that is absent from patients, while presenting a 7.4% frequency in controls. Even though this was the only allele reaching statistical significance after Bonferroni correction (P < 0.0029), other alleles displaying nominal P-values < 0.05 were correlated as well with the quantitative variables sperm number or sperm motility (Table 2).
Table 1 -.
Significant case-control associations for common HLA alleles.
| Gene | Allele | Frequency |
||||
|---|---|---|---|---|---|---|
| Controls | All cases | SHV cases | AST cases | OLI cases | ||
| HLA-A | 11:01 | 0.074 | 0 | 0 | 0 | 0 |
| 29:02 | 0.081 | 0.028 | 0.025 | 0.022 | 0 | |
| HLA-B | 50:01 | 0.015 | 0.063 | 0.074 | 0.056 | 0.071 |
| HLA-C | 03:03 | 0.022 | 0.042 | 0.041 | 0.056 | 0.089 |
| 06:02 | 0.052 | 0.113 | 0.123 | 0.067 | 0.089 | |
| HLA-DQA1 | 05:01 | 0.199 | 0.289 | 0.312 | 0.289 | 0.357 |
| HLA-DQB1 | 03:01 | 0.169 | 0.197 | 0.197 | 0.222 | 0.304 |
| 03:02 | 0.132 | 0.063 | 0.049 | 0.056 | 0.018 | |
Significant nominal P-values (P < 0.05) for Fisher’s exact test are underlined.
Significant P-value after adjustment for multiple testing (Bonferroni correction) are shown in bold.
Figure 1 -.
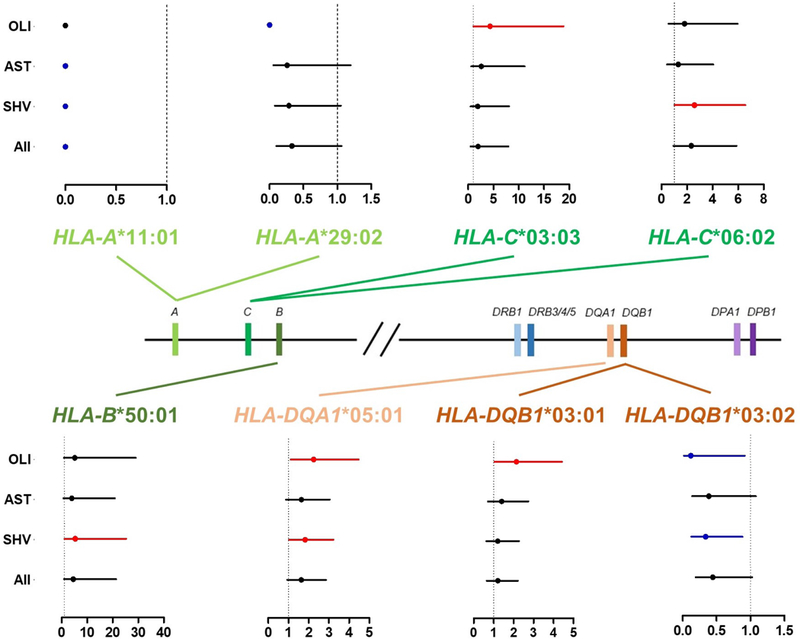
Risk effect analysis for HLA alleles in male infertility. The relative position of HLA genes (central diagram) is shown along with forest plots for alleles presenting significant associations with male infertility (P < 0.05). Each allele plot shows the Odd Ratios (OR) for four comparisons performed: Controls vs. oligozoospermia (OLI); Controls vs. asthenozoospermia (AST); Controls vs. semen hyperviscosity (SHV) and Controls vs. all cases. The dashed black line represents OR = 1. For each comparison OR values and confidence intervals are shown by a dot and a line, respectively, and where color denotes the allele effect: black – none (non-significant); blue – protective (significant); red – risk (significant).
Table 2 -.
Quantitative analysis of HLA alleles associated with male infertility (P<0.05).
| Gene | Allele | Welch’s ANOVA
P-value |
|
|---|---|---|---|
| Sperm number | Sperm motility | ||
| HLA-A | 11:01 | – | 0.0097 |
| 29:02 | 0.0186 | – | |
| HLA-C | 03:03 | 0.0360 | – |
| HLA-DQA1 | 05:01 | 0.0511 | – |
| HLA-DQB1 | 03:01 | 0.0218 | – |
| 03:02 | 0.2657 | – | |
Significant nominal P-values (P < 0.05) are underlined.
not performed
To overcome the low power of standard statistical methods to detect HLA rare allele associations, we performed an approach similar to the burden tests currently recommended for low-frequency variants with f < 0.01 58,61. Overall, a significant enrichment of rare alleles was identified for the entire HLA locus and across all four comparisons (Table 3). However, in a later gene-based analysis, the higher burden of rare alleles was found to be mainly attributed to HLA-A and -B, and SHV and/or AST phenotypes. These results also seem to explain the significant HWE departures detected for HLA-A in the full case set and in SHV patients (P = 0.0418 and P = 0.0486, respectively), which are probably connected with the occurrence of several unique genotypes (data not shown).
Table 3 -.
Enrichment analysis of rare HLA alleles among male infertility phenotypes.
| Locus |
P-value |
|||
|---|---|---|---|---|
| All cases vs Controls |
SHV cases vs Controls |
AST cases vs Controls |
OLI cases vs Controls |
|
| HLAa | 1.0443 × 10−5 | 8.0640 × 10−6 | 0.0003 | 0.0003 |
| HLA-A | 0.0179 | 0.0085 | 0.0660 | 0.0570 |
| HLA-B | 0.0220 | 0.0268 | 0.0418 | 0.0997 |
| HLA-C | 1 | 1 | 1 | 1 |
| HLA-DPA1 | 1 | 0.4729 | 0.3982 | 0.2917 |
| HLA-DPB1 | 1 | 1 | 1 | 1 |
| HLA-DQB1 | 0.6226 | 0.6041 | 0.3039 | 0.2042 |
| HLA-DRB1 | 0.0814 | 0.0714 | 0.1904 | 0.1219 |
all HLA genes covered in this study except HLA-DRB3, -DRB4 and -DRB5.
Significant nominal P-values (P < 0.05) for Fisher’s exact test are underlined.
Haplotype analysis
The pairwise LD analysis performed for the eight studied HLA genes uncovered nine regions with unusual LD levels (Supplementary Table S4). Among these, three could be connected with previously identified signals of association to male infertility (Table 1). A first one spanning HLA-A and -B (D’SHV = 0.86), a second comprising HLA-B and -C (D’SHV = 0.91) and a last one encompassing HLA-DQA1 and -DQB1 genes (D’SHV =0.87; D’OLI =0.87). Indeed, HLA-B*50:01 and HLA-C*06:02 were found to co-segregate in one of the most prevalent HLA-B~HLA-C haplotypes in SHV cases (fcontrols = 0.007 and fSHV= 0.066; P = 0.0118 and OR = 9.47). A similar finding was obtained for HLA-DQA1*05:01 and HLA-DQB1*03:01 alleles in the most common OLI haplotype (fcontrols = 0.125 and fOLI= 0.250; P = 0.0298 and OR = 2.33). On the other hand, the associations of HLA-A*11:01 and HLA-DQB1*03:02 (Table 1) to infertility phenotypes appear to be independent and not a by-product of the high LD levels observed for HLA-A~HLA-B and HLA-DQA1~HLA-DQB1 (Supplementary Table S4), given that neither of these alleles were linked to other significantly associated alleles.
In order to assess if co-segregating HLA alleles were both playing a role in SHV and OLI, or if one of them was a spurious association caused by LD, we tested the hypothesis of a combined allele contribution to the disease. According to our results, in both instances, the two alleles are likely to confer an increased infertility risk (HLA-B*50:01 / HLA-C*06:02 pair P = 0.0117 and OR = 7.53; HLA-DQA1*05:01 / HLA-DQB1*03:01 pair P = 0.032 and OR = 3.24).
Amino acid association analysis
To unravel functionally important domains of HLA molecules that otherwise may not emerge as associated to disease, we performed an analysis based on the amino acid variation. This approach led to similar results as the previous tests with HLA-A, -B, -DQA1 and -DQB1 showing associations to different infertility phenotypes (Table 1 and Supplementary Table S5). Interestingly, the protective effect of HLA-A*11:01 and HLA-A*29:02 to male infertility in general and to OLI, respectively, seems to be explained by the occurrence of a tyrosine (p.Y9 in HLA-A*11:01) or threonine (p.T9 in HLA-A*29:02) at residue 9 in prejudice of the phenylalanine (p.F9) (Fig. 2 and Supplementary Tables S5–S10). Concerning HLA-B*50:01 association with SHV, it is probably correlated with the increment of p.E152 in SHV over p.V152 (Fig. 2 and Supplementary Tables S6 and S10). In addition, for HLA-B the myriad of associations observed in AST is likely to be connected with the aforementioned enrichment of rare alleles and to a sharing of specific amino acid residues (Table 3 and Supplementary Tables S5, S8 and S10). Regarding HLA-DQA1*05:01, it appears to be related with the prevalence of p.S75 in OLI cases in contrast to p.I75 in controls (Fig. 2 and Supplementary Tables S9 and S10). Finally, the HLA-DQB1*03:01 allele conferring risk to OLI is possibly connected with p.A13, p.Y26 and p.45E in contrast to p.G13, p.L26 and p.G45 residues that are more common in controls and associated to the protective allele HLA-DQB1*03:02 (Fig. 2 and Supplementary Tables S9 and S10).
Figure 2 -.
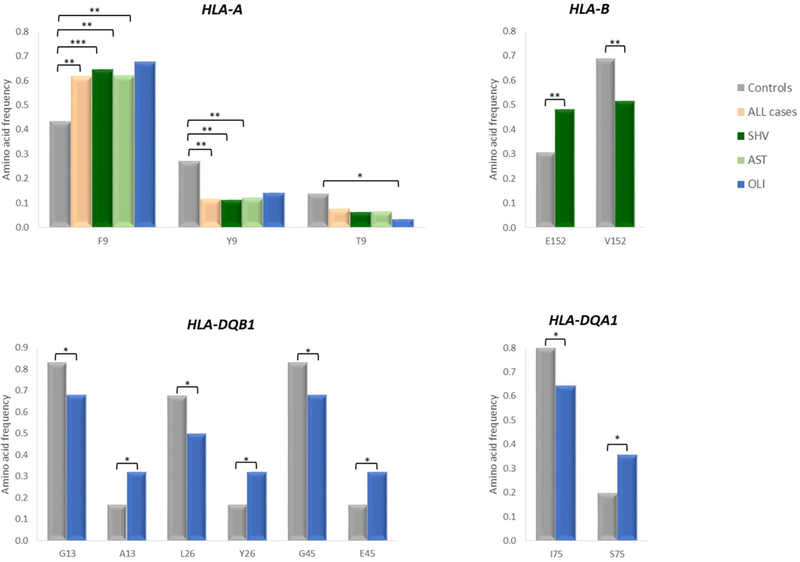
Evaluation of amino acid composition of HLA-A, HLA-B, HLA-DQA1 and HLA-DQB1 alleles associated to male infertility (P < 0.05). Frequencies of the different residues encoded by HLA alleles showing significant results for semen hyperviscosity (SHV), asthenozoospemia (AST) or oligozoospermia (OLI). *P < 0.05; **P < 0.01; ***P < 0.001.
Copy number variation of HLA-DRB3, -DRB4 and -DRB5
We started our analysis by treating the CNV as a single locus (Table 4), which uncovered a significant result between OLI and controls (χ2 = 10.4559 and P = 0.0151). Then, further tests were performed for the presence or absence of each paralog in a multiple loci approach that disclosed the association of HLA-DRB3 with SHV and OLI (P = 0.0454 and P = 0.0023, respectively) and of HLA-DRB4 with OLI alone (P = 0.0123; Table 4).
Table 4 -.
Association analysis for HLA-DRB3, -DRB4 and -DRB5 copy number polymorphism.
| Gene |
Frequencies |
||||
| Controls | All cases | SHV cases | AST cases | OLI cases | |
| HLA-DRB3 | 0.375 | 0.472 | 0.500 | 0.511 | 0.625 |
| HLA-DRB4 | 0.412 | 0.310 | 0.295 | 0.289 | 0.214 |
| HLA-DRB5 | 0.096 | 0.099 | 0.098 | 0.078 | 0.071 |
| NULLa | 0.118 | 0.120 | 0.107 | 0.122 | 0.089 |
|
P-value |
|||||
| Gene |
All
cases vs Controls |
SHV
cases vs Controls |
AST
cases vs Controls |
OLI
cases vs Controls |
|
| Single locus | 0.3145 | 0.1835 | 0.1874 | 0.0151 | |
| Multiple locus | HLA-DRB3 | 0.1152 | 0.0454 | 0.0545 | 0.0023 |
| HLA-DRB4 | 0.0815 | 0.0525 | 0.0671 | 0.0123 | |
| HLA-DRB5 | 1.0000 | 1.0000 | 0.8118 | 0.7817 | |
The NULL designation corresponds to chromosomes lacking any copy of HLA-DRB3, -DRB4 or -DRB5.
Significant nominal P-values (P < 0.05) for Fisher’s exact test or χ2 test are underlined.
STD screening in seminal plasma
To investigate whether HLA associations to male infertility could be related with susceptibility or resistance to STD, we screened seven well-known pathogens in a subset of 30 cases and 42 controls. Among tested STD, only C. trachomatis, N. gonorrhoeae, U. urealyticum and HPV were detected in the seminal plasma either alone or as co-infections (Supplementary Table S11). The most prevalent pathogens were C. trachomatis and HPV, but no differences were observed between cases and controls (C. trachomatis: fcontrols = 0.429 vs. fcases = 0.300; HPV: fcontrols = 0.286 vs. fcases = 0.400). Still, for HPV, previous reported associations with lower sperm numbers 29,30 were confirmed in this study when this parameter was considered as a quantitative trait (Fig. 3). No similar association was observed for C. trachomatis or STD in general (Fig. 3).
Figure 3 -.
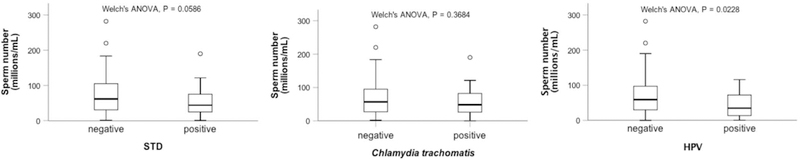
Effect of STD agents in sperm concentration. Group differences between sperm number and STD (all identified pathogens), Chlamydia trachomatis or human papilloma virus (HPV) presence/absence are shown.
Next, we examined whether STD prevalence diverged between carriers and non-carriers of HLA alleles associated to male infertility (nominal P < 0.05). For HLA-A locus, this revealed two times less STD affected subjects in the presence of the protective allele HLA-A*11:01 (P = 0.1096; OR = 0.2341; 95% CI = 0.0421–1.3017; Supplementary Fig. S2). On contrary, for HLA-C*03:03 a two-fold increase in HPV infected individuals was connected with this risk allele (P = 0.0906; OR = 4.6000; 95% CI = 0.7783–27.1885; Supplementary Fig. S2). Finally, statistical models of interaction were taken into account in the analysis of HLA allele, STD and infertility outcome. Notably, these tests showed that HLA-A*29:02 and HPV had an effect in sperm concentration (P = 0.0313), in which the highest sperm counts, even within the normal range, were observed in the presence of HLA-A*29:02 without HPV, differing from any other combination of analyzed variables (Fig. 4).
Figure 4 -.
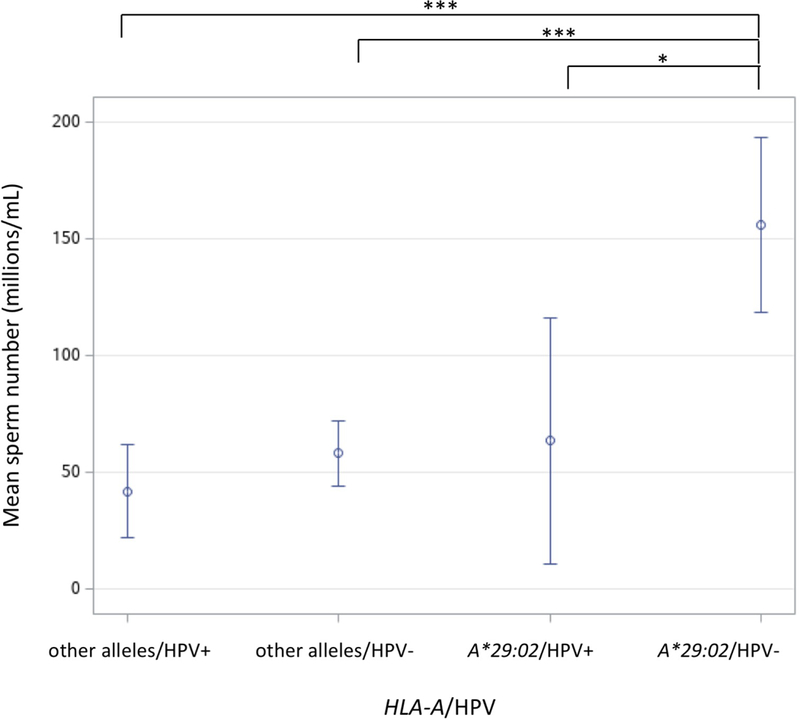
Interaction analysis of HLA-A*29:02 allele and human papilloma virus (HPV) in sperm concentration. Mean sperm number and 95% confidence intervals are shown for four combinations according to allele presence or absence and to HPV infection status (HPV+ or HPV-). *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
In this work, we evaluated the possible contribution of classical HLA variability into the etiology of different male infertility phenotypes, SHV, AST and OLI, through the analysis of a WES panel of Portuguese patients and controls. We identified three protective (HLA-A*11:01, HLA-A*29:02 and HLA-DQB1*03:02) and five risk alleles (HLA-B*50:01, HLA-C*03:03, HLA-C*06:02, HLA-DQA1*05:01 and HLA-DQB1*03:01). Notably, none of these have been previously linked to male infertility, neither former described associated alleles were replicated in this study 4,7–10,62,63. This is not entirely unexpected given that the analyzed populations and phenotypes are quite distinct. Indeed, except for the work of Aleksovski et al. that analyzed men from the former Yugoslavia with impaired spermatogenesis (OLI or AZO), all the remaining studies were performed in NOA patients with East-Asian ancestry 4,7–10,62,63. Moreover, in many instances, HLA alleles linked to male infertility are found at contrasting frequencies in different human populations 64, indicating that previous findings might have been influenced by the ethnicity of analyzed samples and thus restricted to those populations. Interestingly, as pivotal players in the immune response, HLA molecules are thought to have been shaped by natural selection, and their polymorphism is known to correlate with geography (distance from Africa) and pathogen richness 65. In this sense, the selective pressures exerted by a higher burden of infectious agents might have favored HLA diversification, especially in certain continental regions, to recognize a broader repertoire of pathogenic peptides 65–68.
Despite the absence of a prevailing rule on how HLA classes access antigens, class I molecules are believed to mainly bind intracellular peptides of viral origin, triggering a cytotoxic response by their presentation to CD8+ T cells. Conversely, class II molecules are thought to bind mostly to extracellular antigens of bacterial origin that, once presented to CD4+ T cells, stimulate antibody production by B cells 11,66,69,70. Therefore, HLA classes I and II would be expected to have dissimilar outcomes in STD like gonorrhea (N. gonorrhoeae) and syphilis (T. pallidum), and infections caused by C. trachomatis, HPV, HSV, HIV, HBV and HCV, which are all accepted risk factors for male infertility 29. However, according to our study results, only class I genes (HLA-A and -C) appear to influence the outcomes of HPV infection and STD in semen quality.
Notably, some of our candidate alleles for male infertility were already associated in the context of other disorders with several of these pathogens. For example, HLA-A*11:01 and HLA-DQB1*03:02 were both described to confer resistance against HIV and in the case of HLA-DQB1*03:02 it was also reported to protect from HBV in chronic hepatitis 19,20,22. Consistently, in our cohort, HLA-A*11:01 and HLA-DQB1*03:02 were correlated with a lower overall risk to male infertility and to SHV and OLI, respectively. These findings are therefore suggestive of a protective role through an improved immune response against HIV and HBV, but also possibly to other viruses, given that the presence of HIV and HBV was excluded in our samples 45. Indeed, this assumption seems to apply to another class I allele, HLA-A*29:02, for which we found evidence of a relationship with HPV infection in sperm counts.
Concerning HLA-DQB1*03:01 allele, identified as a risk factor for OLI, previous studies showed conflicting results: whereas in chronic hepatitis this allele has been correlated in some instances with HCV clearance 23–25, other authors have reported a link between HLA-DQB1*03:01 and HPV6 infection 21. Notably, this HPV subtype, despite being considered as low-risk for cervical cancer, is established to cause genital lesions and to be found in the semen 71,72. In our cohort, no association of HLA-DQB1*03:01 with HPV was not detected. However, it is important to note that no discrimination of HPV subtypes was done in our STD screening. Given that some HPV subtypes have more severe outcomes in semen quality than others, our molecular analysis might have hampered the detection of any association signal 73.
Less is known about bacterial agents, but for HLA-DQA1*05:01 allele correlated with OLI and SHV, a previous work showed contrasting results among women infected by C. trachomatis and N. gonorrhoeae. While in positive C. trachomatis cases HLA-DQA1*05:01 had an increased frequency, in N. gonorrhoeae patients this allele displayed only reduced counts 14,29. However, as far as we could evaluate, no association of HLA-DQA1*05:01 with C. trachomatis, nor with N. gonorrhoeae (found in a single case), was detected in our sample.
Noteworthy, our cohort overlaps with the one used for a global characterization of seminal microbiota, in which pooled samples of controls, asthenoteratozoospermia, oligoasthenoteratozoospermia (OAT) and SHV were screened by sequencing 16S ribosomal RNA gene 45. Again, according to Monteiro et al., the above association of HLA-DQA1*05:01 to Chlamydia alone seems improbable, given that its relative abundance compared to other bacterial taxa only reaches very low frequencies (f<0.1%). On contrary, Neisseria and other infectious agents known to cause urogenital infections, namely Haemophilus, Pseudomonas, Klebsiella, and Serratia, were found to be augmented in OAT and/or SHV samples 45. Although the different methodological approaches and sample subsets used in these studies might have prevented a throughout appreciation of their findings, the complexity of seminal microbiota still suggests the existence of multiple targets for HLA antigenic presentation in the male urogenital system.
Taking into account the above considerations, it is tempting to hypothesize HLA alleles as critical immunological players in STD susceptibility. Supporting this conjecture is the evidence for an activation of the immune system in asymptomatic OAT patients as shown by their infiltrations of HLA-DR positive macrophages in the ejaculate and also the expression of HLA class II genes in spermatozoa of infertile men 74,75. Nevertheless, we cannot rule out an autoimmunity cause for our findings, since HLA alleles can be associated with ASA production and male infertility as well 76,77.
In conclusion, we were able to apply for the first time a population reference graph approach implemented through HLA*PRG:LA tool to the study of male infertility. This allowed us to infer HLA genotypes from a panel of WES data as well as to perform a preliminary study of HLA variability in this disease. Even though our cohort may be considered as small sized it was enough to provide novel hints for a possible role of HLA locus in this complex disorder. We demonstrate that both HLA class I and II alleles can be correlated with SHV and OLI phenotypes and that these also appear to be implicated in the susceptibility/resistance to different pathogens known to cause male urogenital infections and to affect reproductive fitness. Future studies in extended WES cohorts, followed by seminal transcriptomics research of human and pathogens will be fundamental to assess the proposed host-pathogen interaction in male infertility.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank all patients for donating their samples and for collaborating in this study, Don Conrad for providing access to exome sequencing under the auspices of the GEMINI consortium and Maria José Borrego (National Institute of Health, Lisbon, Portugal) for supplying DNA samples as positive controls for STD. Ipatimup integrates the i3S Consortium, which is partially supported by the Portuguese Foundation for Science and Technology (FCT). This work was supported by Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER) - project NORTE-01–0145-FEDER-000029. This work was also financed by FEDER funds through COMPETE 2020 (Operacional Programme for Competitiveness and Internationalization - POCI) and by Portuguese funds through FCT - project POCI-01–0145-FEDER-007274, grant PTDC/BEX- GMG/0242/2012 to S.S. and fellowship SFRH/BPD/120777/2016 to P.I.M. A.M.L is funded by the Portuguese Government through the FCT research contract IF/01262/2014. L.N. was supported by grant R01HD07864 from the Eunice Shriver Kennedy National Institute of Child Health and Human Development of the United States.
BIBLIOGRAPHY
- 1.Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13(37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. 4th edn. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- 3.Jungwirth A, Diemer T, Dohle G., Giwercman A, Kopa Z, Krausz C European Association of Urology Guidelines on Male Infertility: The 2012 Update. Eur Urol. 2012;62:324–32. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z, Li Z, Yu J, Tong C, Lin Y, Guo X, et al. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat Commun. 2014;5:3857. [DOI] [PubMed] [Google Scholar]
- 5.Ramasamy R, Bakircioʇlu ME, Cengiz C, Karaca E, Scovell J, Jhangiani SN, et al. Whole-exome sequencing identifies novel homozygous mutation in NPAS2 in family with nonobstructive azoospermia. Fertil Steril. 2015;104(2):286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aston KI. Genetic susceptibility to male infertility: News from genome-wide association studies. Andrology. 2014;2(3):315–21. [DOI] [PubMed] [Google Scholar]
- 7.Jinam TA, Nakaoka H, Hosomichi K, Mitsunaga S, Okada H, Tanaka A, et al. HLA-DPB104:01 allele is associated with non-obstructive azoospermia in Japanese patients. Hum Genet. 2013;132(12):1405–11. [DOI] [PubMed] [Google Scholar]
- 8.Miura H, Tsujimura A, Nishimura K, Kitamura M, Kondoh N, Takeyama M, et al. Susceptibility to idiopathic azoospermia in Japanese men is linked to HLA class I antigen. J Urol. 1998;159(6):1939–41. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzaka Y, Makino S, Okamoto K, Oka A, Tsujimura A, Matsumiya K, et al. Susceptibility locus for non-obstructive azoospermia is localized within the HLA-DR/DQ subregion: Primary role of DQB1*0604. Tissue Antigens. 2002;60:53–63. [DOI] [PubMed] [Google Scholar]
- 10.Aleksovski D, Todorov Z, Chilimanov Z. Possibilities of association of azoospermia and oligospermia with some HLA system antigens. Hum Reprod. 1988;3(1):83–4. [DOI] [PubMed] [Google Scholar]
- 11.Klein J, Sato A. The HLA System. New Engl J Med Rev. 2000;343(10):702–9. [DOI] [PubMed] [Google Scholar]
- 12.Mosaad YM. Clinical Role of Human Leukocyte Antigen in Health and Disease. Scand J Immunol. 2015;82:283–306. [DOI] [PubMed] [Google Scholar]
- 13.Bergström TF, Erlandsson R, Engkvist H, Josefsson A, Erlich HA, Gyllensten U. Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol Rev. 1999;167:351–65. [DOI] [PubMed] [Google Scholar]
- 14.Ness RB, Brunham RC, Shen C, Bass DC . Associations among Human Leukocyte Antigen (HLA) Class II DQ Variants, Bacterial Sexually Transmitted Diseases, Endometritis, and Fertility among Women with Clinical Pelvic Inflammatory Disease. Sex Transm Dis. 2004;31(5):301–4. [DOI] [PubMed] [Google Scholar]
- 15.GEISLER WM, WANG C, TANG J, WILSON CM, CROWLEY-NOWICK PA, KASLOW RA Immunogenetic Correlates of Neisseria gonorrhoeae Infection in Adolescents. Sex Transm Dis. 2008;35(7):656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson KM, Tang J, Brown L, Press CG, Geisler WM. HLA-DQB1*06 is a risk marker for chlamydia reinfection in African American women. Genes Immun. 2019;20:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang Y-L, Ren G-J, Tian K-L, Li X-Y, Zhu Y-B, Liu J-L, et al. Human leukocyte antigen-C and killer cell immunoglobulin-like receptor gene polymorphisms among patients with syphilis in a Chinese Han population. APMIS. 2012; [DOI] [PubMed] [Google Scholar]
- 18.Jiang H-W, Tian H-Q, Liu H, Li N, Zhao Y, Zhang F-R. Association of the HLA-DRB1 locus with syphilis in a Chinese population. Int J Infect Dis. 2011;15(5):e342–5. [DOI] [PubMed] [Google Scholar]
- 19.Raghavan S, Selvaraj P, Swaminathan S, Narendran G. Short communication: association of HLA-A*1101 with resistance and B*4006 with susceptibility to HIV and HIV-TB: an in silico analysis of promiscuous T cell epitopes. AIDS Res Hum Retroviruses. 2009;25(10):1023–8. [DOI] [PubMed] [Google Scholar]
- 20.Rallón N, Restrepo C, Vicario JL, del Romero J, Rodríguez C, García-Samaniego J, et al. Human leucocyte antigen (HLA)-DQB1*03:02 and HLA-A*02:01 have opposite patterns in their effects on susceptibility to HIV infection. HIV Med. 2017;18(8):587–94. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Gaborieau V, Zhao Y, Chabrier A, Wang H, Waterboer T, et al. A systematic investigation of the contribution of genetic variation within the MHC region to HPV seropositivity. Hum Mol Genet. 2015;24(9):2681–8. [DOI] [PubMed] [Google Scholar]
- 22.Nishida N, Ohashi J, Khor SS, Sugiyama M, Tsuchiura T, Sawai H, et al. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep. 2016;6:24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Huang K, Xu R, Wang M, Liao Q, Xiong H, et al. The Associations of HLA-A∗02:01 and DRB1∗11:01 with Hepatitis C Virus Spontaneous Clearance Are Independent of IL28B in the Chinese Population. Sci Rep. 2016;6:31485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thio C, Thomas D, Goedert J, Vlahov D, Nelson K, Hilgartner M, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. [DOI] [PubMed] [Google Scholar]
- 25.El-Bendary M, Neamatallah M, Esmat G, Kamel E, Elalfy H, Besheer T, et al. Associations of human leucocyte antigen class II-DQB1 alleles with hepatitis C virus infection in Egyptian population: a multicentre family-based study. J Viral Hepat. 2016;23:961–70. [DOI] [PubMed] [Google Scholar]
- 26.du Plessis SS, Gokul S, Agarwal A. Semen hyperviscosity-causes, consequences, and cures. Front Biosci (Elite Ed). 2013;5:224–31. [DOI] [PubMed] [Google Scholar]
- 27.Flint M, du Plessis S, Menkveld R. Revisiting the assessment of semen viscosity and its relationship to leucocytospermia. Andrologia. 2014;46:837–41. [DOI] [PubMed] [Google Scholar]
- 28.Weidner W, Pilatz A, Diemer T, Schuppe HC, Rusz A, Wagenlehner F. Male urogenital infections: Impact of infection and inflammation on ejaculate parameters. World J Urol. 2013;31:717–23. [DOI] [PubMed] [Google Scholar]
- 29.Gimenes F, Souza RP, Bento JC, Teixeira JJ V, Maria-Engler SS, Bonini MG, et al. Male infertility: a public health issue caused by sexually transmitted pathogens. Nat Rev Urol. 2014;11(12):672–87. [DOI] [PubMed] [Google Scholar]
- 30.Bezold G, Politch JA, Kiviat NB, Kuypers JM, Wolff H, Anderson DJ. Prevalence of sexually transmissible pathogens in semen from asymptomatic male infertility patients with and without leukocytospermia. Fertil Steril. 2007;87(5):1087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garolla A, Pizzol D, Bertoldo A, Menegazzo M, Barzon L, Foresta C. Sperm viral infection and male infertility: focus on HBV, HCV, HIV, HPV, HSV, HCMV, and AAV. J Reprod Immunol. 2013;100:20–9. [DOI] [PubMed] [Google Scholar]
- 32.Lai YM, Soong YK, Lee JF, Yang FP, Huang HY, Pao CC. The effect of human papillomavirus infection on sperm cell motility. Fertil Steril. 1997;67(6):1152–5. [DOI] [PubMed] [Google Scholar]
- 33.Moretti A, Palù G, Foresta C, Pizzol D, Barzon L, Garolla A. Clinical and prognostic significance of human papillomavirus DNA in the sperm or exfoliated cells of infertile patients and subjects with risk factors. Fertil Steril. 2010;94(5):1723–7. [DOI] [PubMed] [Google Scholar]
- 34.Palù G, Zuccarello D, Pizzol D, Foresta C, Moretti A, Barzon L, et al. Human papillomavirus found in sperm head of young adult males affects the progressive motility. Fertil Steril. 2008;93(3):802–6. [DOI] [PubMed] [Google Scholar]
- 35.Ceballos A, Lenicov FR, Sabatté J, Rodrígues CR, Cabrini M, Jancic C, et al. Spermatozoa capture HIV-1 through heparan sulfate and efficiently transmit the virus to dendritic cells. J Exp Med. 2009;206(12):2717–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorusso F, Palmisano M, Chironna M, Vacca M, Masciandaro P, Bassi E. Impact of chronic viral diseases on semen parameters. Andrologia. 2010;42:121–6. [DOI] [PubMed] [Google Scholar]
- 37.Kang X, Xie Q, Zhou X, Li F, Huang J, Liu D, et al. Effects of hepatitis B virus S protein exposure on sperm membrane integrity and functions. PLoS One. 2012;7(3):e33471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eley A, Hosseinzadeh S, Hakimi H, Geary I, Pacey AA. Apoptosis of ejaculated human sperm is induced by co-incubation with Chlamydia trachomatis lipopolysaccharide. Hum Reprod. 2005;20(9):2601–7. [DOI] [PubMed] [Google Scholar]
- 39.Hosseinzadeh S, Brewis IA, Eley A, Pacey AA. Co-incubation of human spermatozoa with Chlamydia trachomatis serovar E causes premature sperm death. Hum Reprod. 2001;16(2):293–9. [DOI] [PubMed] [Google Scholar]
- 40.Satta A, Stivala A, Garozzo A, Morello A, Perdichizzi A, Vicari E, et al. Experimental Chlamydia trachomatis infection causes apoptosis in human sperm. Hum Reprod. 2006;21(1):134–7. [DOI] [PubMed] [Google Scholar]
- 41.Domes T, Lo KC, Grober ED, Mullen JBM, Mazzulli T, Jarvi K. The incidence and effect of bacteriospermia and elevated seminal leukocytes on semen parameters. Fertil Steril. 2012;97(5):1050–5. [DOI] [PubMed] [Google Scholar]
- 42.Mackern-Oberti JP, Motrich RD, Breser ML, Sánchez LR, Cuffini C, Rivero VE. Chlamydia trachomatis infection of the male genital tract: An update. J Reprod Immunol. 2013;100(1):37–53. [DOI] [PubMed] [Google Scholar]
- 43.Dilthey AT, Gourraud PA, Mentzer AJ, Cereb N, Iqbal Z, McVean G. High-Accuracy HLA Type Inference from Whole-Genome Sequencing Data Using Population Reference Graphs. PLoS Comput Biol. 2016;12(10):e1005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marques PI, Fonseca F, Carvalho AS, Puente DA, Damião I, Almeida V, et al. Sequence variation at KLK and WFDC clusters and its association to semen hyperviscosity and other male infertility phenotypes. Hum Reprod. 2016;31(12):2881–91. [DOI] [PubMed] [Google Scholar]
- 45.Monteiro C, Marques PI, Cavadas B, Damião I, Almeida V, Barros N, et al. Characterization of microbiota in male infertility cases uncovers differences in seminal hyperviscosity and oligoasthenoteratozoospermia possibly correlated with increased prevalence of infectious bacteria. Am J Reprod Immunol. 2018;79:e12838. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Depristo MA, Banks E, Poplin R, Garimella KV., Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dilthey AT, Mentzer AJ, Carapito R, Cutland C, Cereb N, Madhi SA, et al. HLA*PRG:LA - HLA typing from linearly projected graph alignments. bioRxiv. 2018;453555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abusarah EA, Awwad ZM, Charvalos E, Shehabi AA. Molecular detection of potential sexually transmitted pathogens in semen and urine specimens of infertile and fertile males. Diagn Microbiol Infect Dis. 2013;77:283–6. [DOI] [PubMed] [Google Scholar]
- 51.Mahony JB, Jang D, Chong S, Luinstra K, Sellors J, Tyndall M, et al. Detection of chlamydia trachomatis, neisseria gonorrhoeae, ureaplasma urealyticum, and mycoplasma genitalium in first-void urine specimens by multiplex polymerase chain reaction. Mol Diagnosis. 1997;2(3):161–8. [DOI] [PubMed] [Google Scholar]
- 52.Gimenes F, Medina FS, De Abreu ALP, Irie MMT, Esquica̧ IB, Malagutti N, et al. Sensitive Simultaneous Detection of Seven Sexually Transmitted Agents in Semen by Multiplex-PCR and of HPV by Single PCR. PLoS One. 2014;9(6):e98862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Roda Husman AM, Walboomers JMM, Van den Brule AJC, Meijer CJLM, Snijders PJF. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–62. [DOI] [PubMed] [Google Scholar]
- 54.Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G. PyPop update - A software pipeline for large-scale multilocus population genomics. Tissue Antigens. 2007;69(SUPPL. 1):192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo S, Thompson E. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48(2):361–72. [PubMed] [Google Scholar]
- 56.Chen JJ, Hollenbach JA, Trachtenberg EA, Just JJ, Carrington M, Rønningen KS, et al. Hardy-Weinberg testing for HLA class II (DRB1, DQA1, DQB1, AND DPB1) loci in 26 human ethnic groups. Tissue Antigens. 1999;54:533–42. [DOI] [PubMed] [Google Scholar]
- 57.Fan Y, Song YQ. PyHLA: Tests for the association between HLA alleles and diseases. BMC Bioinformatics. 2017;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dering C, Hemmelmann C, Pugh E, Ziegler A. Statistical Analysis of Rare Sequence Variants: An Overview of Collapsing Methods. Genet Epidemiol. 2011;35(Suppl 1):S12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: A cohort allelic sums test (CAST). Mutat Res. 2007;615:28–56. [DOI] [PubMed] [Google Scholar]
- 60.Pappas DJ, Marin W, Hollenbach JA, Mack SJ. Bridging Immunogenomic Data Analysis Workflow Gaps (bigdawg): An Integrated Case-Control Analysis Pipeline. Hum Immunol. 2016;77(3):283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kosmicki JA, Churchhouse CL, Rivas MA, Neale BM. Discovery of rare variants for complex phenotypes. Hum Genet. 2016;135:625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao H, Xu J, Zhang H, Sun J, Sun Y, Wang Z, et al. A genome-wide association study reveals that variants within the HLA region are associated with risk for nonobstructive azoospermia. Am J Hum Genet. 2012;90:900–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet. 2011;44(2):183–6. [DOI] [PubMed] [Google Scholar]
- 64.Gourraud PA, Khankhanian P, Cereb N, Yang SY, Feolo M, Maiers M, et al. HLA diversity in the 1000 genomes dataset. PLoS One. 2014;9(7):e97282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15:1022–7. [DOI] [PubMed] [Google Scholar]
- 66.Meyer D, Vitor VR, Bitarello BD, Débora DY, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70(1):5–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penman BS, Ashby B, Buckee CO, Gupta S. Pathogen selection drives nonoverlapping associations between HLA loci. Proc Natl Acad Sci. 2013;110(48):19645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bitarello BD, Francisco R dos S, Meyer D. Heterogeneity of dN/dS Ratios at the Classical HLA Class I Genes over Divergence Time and Across the Allelic Phylogeny. J Mol Evol. 2016;82(1):38–50. [DOI] [PubMed] [Google Scholar]
- 69.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Helper T Cells and Lymphocyte Activation In: Molecular Biology of the Cell. 4th editio New York: Garland Science; 2002. [Google Scholar]
- 70.Blum JS, Wearsch PA, Cresswell P. Pathways of Antigen Processing. Annu Rev Immunol. 2013;31:443–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lyu Z, Feng X, Li N, Zhao W, Wei L, Chen Y, et al. Human papillomavirus in semen and the risk for male infertility: A systematic review and meta-analysis. BMC Infect Dis. 2017;17(1):714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flores-Díaz E, Sereday KA, Ferreira S, Sirak B, Sobrinho JS, Baggio ML, et al. HPV-6 Molecular Variants Association With the Development of Genital Warts in Men: The HIM Study. J Infect Dis. 2017;215:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y, Jia CW, Ma YM, Zhou LY, Wang SY. Correlation between HPV sperm infection and male infertility. Asian J Androl. 2013;15:529–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paradisi R, Neri S, Pession A, Magrini E, Bellavia E, Ceccardi S, et al. Human leukocyte antigen II expression in sperm cells: comparison between fertile and infertile men. Arch Androl. 2000;45(3):203–13. [DOI] [PubMed] [Google Scholar]
- 75.Pelliccione F, D’Angeli A, Cinque B, Falone S, Micillo A, Francavilla F, et al. Activation of the immune system and sperm DNA fragmentation are associated with idiopathic oligoasthenoteratospermia in men with couple subfertility. Fertil Steril. 2011;95(8):2676–9. [DOI] [PubMed] [Google Scholar]
- 76.Omu AE, al-Qattan F, Ismail AA, al-Taher S, al-Busiri N. Relationship between unexplained infertility and human leukocyte antigens and expression of circulating autogeneic and allogeneic antisperm antibodies. Clin Exp Obstet Gynecol. 1999;26(3–4):199–202. [PubMed] [Google Scholar]
- 77.Bohring C, Krause W. Immune infertility: Towards a better understanding of sperm (auto)-immunity: The value of proteomic analysis. Hum Reprod. 2003;18(5):915–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


