Abstract
Three-channel 3D printed centerpieces with two sample sectors next to a joint solvent reference sector were recently described as a strategy to double the throughput of sedimentation velocity analytical ultracentrifugation experiments [Anal. Chem. 91 (2019) 5866-5873]. They are compatible with Rayleigh interference optical detection in commercial analytical ultracentrifuges, but require the rotor angles of data acquisition to be repeatedly adjusted during the experiment to record from the two sample sectors. Here we present an approach to automate this data acquisition mode through the use of a secondary, general-purpose automation software, and an accompanying data pre-processing software for scan sorting.
Keywords: analytical ultracentrifugation, 3D printing, laboratory automation, sedimentation velocity
Nearly a century after the initial development of analytical ultracentrifugation (AUC) [1,2], this classical technique of physical biochemistry is again in rapid development [3-5]. This is rooted in the powerful and simple concept of observing particle redistribution in solution after application of a gravitational force, which can be adapted to modern problems. Taking advantage of new theoretical, computational, and experimental capabilities [6,7], the traditional areas of application have been expanded into a variety of fields such as food sciences [8], archeology [9], characterization of nanoparticles [10-14], biotechnology [15-18], and the study of extremely strong, and ultra-weak macromolecular interactions [19-22]. In the last few years, interest in instrumental developments has included new detectors [23,24], calibration and alignment procedures [25-29], and strategies for sample handling [30-32].
A long-standing limitation of AUC, however, is the relatively low throughput, which allows only 7–8 samples to be studied in a single run that may take several hours or days. This can pose a problem, in particular, for screening applications comparing many different molecules or solvents, protein interaction studies requiring titration series spanning a wide range of concentration, and studies of multi-protein interactions that require multiple control samples [7,10,15,33-37].
To increase the sample throughput, our laboratory has previously introduced an alternative strategy of using the absorbance detection system [38]. The absorbance detection system was originally designed to function like a dual-beam spectrophotometer with one reference and one sample solution [39]. However, we have shown that the logarithm of the recorded light intensity in each sector exhibits a data structure equivalent to that of interference optical data (the time-invariant radially-dependent baseline offset playing the same role as the local detector sensitivity profile), and therefore data from each sector can be analyzed separately using the same algebraic noise decomposition method [40,41]. This allows a standard double sector centerpiece to be operated with two independent samples, and thereby run SV experiments with twice the throughput [34,38]. Unfortunately, this popular strategy is limited to absorbance detection.
A key part of AUC instrumentation is the ‘centerpiece’, which holds a sector-shaped liquid volume between optical windows and is sealed against high vacuum during ultracentrifugation. Recently we have demonstrated how 3D printing technology liberates the experimenter from the constraints of commercial centerpieces that use designs going back half a century or more [30,31]. Current 3D printing allows the inexpensive fabrication of centerpieces that are watertight and provide a seal against the windows during exposure to centrifugal forces in the AUC, which in sedimentation velocity (SV) experiments can reach up to ~300,000 g at 60,000 rpm. Similarly, suitable spacers can be printed to elevate the centerpiece to the filling port of existing cell assembly barrels [31]. While they can be printed in a variety of materials [30,31], our work has mostly focused on epoxy-like photopolymer ‘MicroFine Green’ (ProtoLabs Inc., Maple Plain, MN) that can be printed by micro-stereolithography with sufficiently high resolution such that no separate gaskets are required for the vacuum seal. We have demonstrated that such centerpieces allow unhindered macromolecular sedimentation at a precision that is on par with commercial Epon epoxy centerpieces [31]. Various centerpiece designs can be created with OpenSCAD [42], a parametric scripting language for the creation of 3D objects from mathematically defined primitives. This flexibility, combined with quick turnaround of commercial 3D printing, opens new experimental possibilities in AUC. This includes centerpieces that require significantly lower sample volumes, provide short optical pathlengths for work at high concentrations, and/or provide longer observable solution columns for higher precision sedimentation velocity experiments [20,22,31].
3D printing has enabled a different strategy for increasing sample throughput, as it is possible to accommodate three sectors in the centerpieces instead of only two [31], as shown in Figure 1. Whereas previous designs featured sector angles of 1° and 1.5° to reduce sample volume, in the current work we have created sector angles of 2.2° to allow easier filling of the sectors, and to provide more tolerance for the adjustment of detection angles. The sectors have a dome-shaped ceiling [31] to raise the meniscus and extend the observable radial solution column lengths. As any other design, they can be printed with different optical pathlengths as needed.
Figure 1.
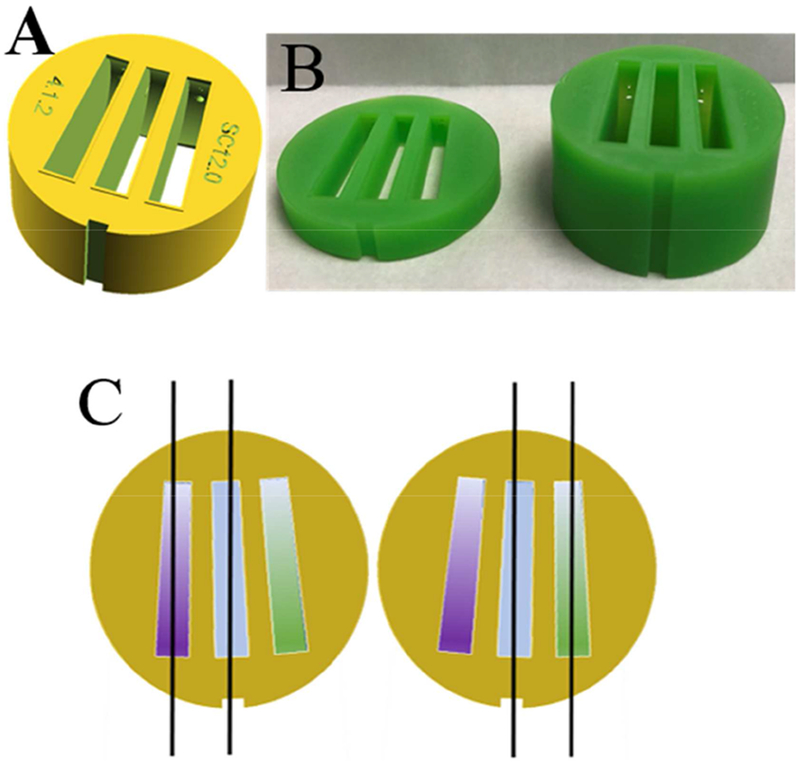
(A) Design of 3D printed three-channel centerpiece with sector angles of 2.2°, filling and venting holes, dome-shaped ceilings, and embossed rims surrounding the sectors that serve as integrated gaskets for vacuum seal. (B) Photographs of centerpieces 3D printed in an epoxy-like photopolymer “MicroFine Green” (ProtoLabs, Maple Plain, MN) using modification of a design file 3DPX-010758 of the NIH 3D Print Exchange (3dprint.nih.gov). Shown are centerpieces with 4 mm (left) and 12 mm pathlength (right), respectively. (C) Principle of illumination modes of three-channel centerpieces with Rayleigh interference optical detection. Two coherent parallel planar beams traverse either the left and the middle sector (left sketch), or the right and the middle sector (right sketch). The middle sector contains a joint reference buffer (blue), such that in the left configuration the purple sample can be observed, and in the right configuration the green sample can be observed. The two configurations have a 3-4° different rotation angle at the time of data acquisition, and due to the reversed beams they differ in the sign of the fringe shift signal.
While three sectors should, in principle, enable absorbance detection in transmitted intensity mode for three samples per cell, unfortunately this is not possible with the current commercial AUC operating software, which assumes the standard double-sector cell configuration in the synchronization of data acquisition with the rotation of the rotor. There is no obvious reason for this other than the custom of using double sector centerpieces, which has been hard-wired into the AUC operation software. Fortunately, however, the Rayleigh interference optical system in the most widely used ProteomeLab and XLA/I instruments allows the user to set the data acquisition angles. (In the most recently presented AUC model of this manufacturer this opportunity has been removed.) This detection system is based on phase differences of coherent light traversing either a sample or a reference solution, measured through shifts in their interference fringe pattern recorded in a camera [43]. Three sectors offer the possibility of measuring two samples if they share the same reference solution, as depicted in Figure 1C.
The two configurations can be selected through different rotor angles for data acquisition. In ProteomeLab and XLA/I instruments the rotor angles of data acquisition are determined by the user during the run setup [4]. They depend on the exact position of the magnet underneath the overspeed disc of the rotor, which serves as an index for the rotor angle. Optimal angles of sequential rotor positions differ by 90° or 45° for 4-hole or 8-hole rotors, respectively, and follow almost precisely αn= α0 + n×45 (or αn= α0 + n×90 for 4-hole rotors). When using the three-channel centerpieces this adjustment can proceed as usual, but must be followed by exploring adjustment of the alternate configuration approximately Δα = 3-4° distant from the first. The angular difference between the two configurations will be the same for all cells αnA= α0 + n×45 and αnB= α0 + Δα + n×45 (assuming 8-hole rotors, with n denoting the rotor hole numbers containing three-channel centerpieces), since it is determined solely by the geometry of the 3D printed centerpiece. Therefore, after initial adjustment, the two configurations can be recalled simply by (re)starting data acquisition after shifting the set of data acquisition angles alternately by +Δα and −Δα, with Δα being the initially defined and constant angle shift. Proof of principle of this data acquisition strategy was shown previously [31].
In this form, the run provides higher throughput, but is also much more labor intensive, since it requires periodic manual switching of the acquisition angles for the two samples in each cell. The motivation of the present communication is to alleviate this problem and to switch rotor angles automatically during the SV run. Even though this has not been envisioned in the AUC user interface, and the latter does currently not offer any support in this regard, it is possible to use general-purpose automation software to activate the necessary AUC user interface functions. To this end, we employed the freeware scriptable software AutoIt (version 3.3.14.5, AutoIt Consulting Ltd.; https://www.autoitscritp.com), which has been used previously in other scientific context to expand capabilities of commercial software and for laboratory automation [44-46]. AutoIt is a scripting language and scheduler for mouse and keyboard inputs, and can therefore be used effectively as meta-program using functions of commercial software as building blocks.
A sample script can be found in the Supplementary Material for an experiment with eight 3-channel cells in an 8-hole rotor. (It can be easily adapted to other rotor/cell configurations.) It consists of a timed sequence of mouse clicks at positions that correspond to menu items on the AUC user interface. Since they are specified in units of screen pixels, the positions must be recorded prior to the SV run for the specific computer screen and display resolution used. The script relies on the reproducibility of screen positioning of the AUC user interface program. Short wait times of 100 msec are included between clicks to allow execution of the invoked AUC user interface functions.
In more detail, the script will be activated after the start of the AUC run and after Rayleigh interferometric scans have been set to the angles αnA. Execution of the script will bring the AUC operating program into the foreground and carry out a loop for changing scan settings, in this example 600 times, corresponding to running the script for approximately 16 hours. In each iteration of this loop, a first block defines the clicks needed to stop the current scan. (When the script is initiated, this step will be included but there is no method scan to stop; this will not affect the following steps.) Next are the specific clicks needed to adjust the laser settings for all eight cells, one at a time: 1) select Detail; 2) select Laser Setup; 3) clicking on the Laser delay slider, adjust the laser settings by Δα (in the present example 3°) if the loop number is even, or - Δα if it is odd; 4) after closing the details, move the slider of the cell selection down (in the present script using two clicks) to view the next cell to adjust. After all cells are adjusted, the last block moves the cell selection slider in the methods window up to the top and presses the Start Method Scan button. Finally, each iteration of the loop contains a pause (here for 85 sec) to allow data acquisition with the current settings before starting over with the adjustments.
In contrast to standard SV runs where the scan files are consecutively numbered and located in a single folder representing the start time of the scan set, after execution of the script the scan files have the same filenames but can be found in consecutive folders. They alternate between the two samples. Thus, we created a software program 3ChannelSorter that renames all scans (using different identifiers for the two samples), multiplies the scan data from the left sample with −1 to restore positive signals, and saves the scan files in a single folder for conventional data analysis. This software is available for download from our laboratory sharepoint site sedfitsedphat.nibib.nih.gov.
Figure 2A-D shows sedimentation boundary data recorded from left and right sectors that were filled with identical bovine serum albumin solutions. In contrast to the previous proof-of-principle data with sparse, uneven manual switching [31], the scans are now automatically acquired with high frequency for each sample sector, evenly spanning the entire SV experiment. A superposition of the sedimentation coefficient distributions c(s) is shown in Figure 2E. The standard deviation of the BSA monomer peak is 0.02 S, consistent with experimental error [26]. Slight variation of the BSA trimer peak s-value is observed, consistent with known limitations in the precision of measured sedimentation coefficient of trace components [18,47]. Neither the data acquisition mode nor the 3D printed centerpiece has any discernable impact on the data quality.
Figure 2.
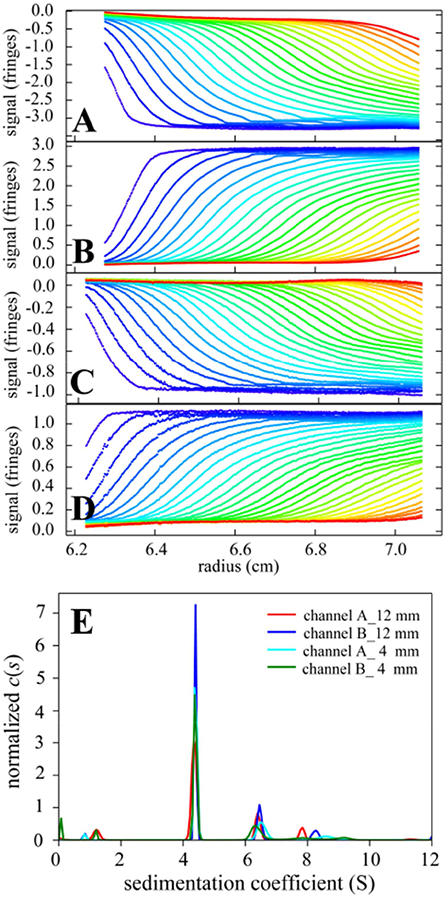
(A-D) Sedimentation boundaries recorded in the left (A, C) and right (B, D) sample sectors of two three-channel centerpieces with 12 mm (A and B) and 4 mm (C and D) pathlength, respectively. (The 4 mm centerpiece was centered in height relative to the filling port, which allows measurement of gradients of up to ~100 fringes/cm [48].) Both sample sectors were filled with 1 mg/ml bovine serum albumin in phosphate buffered saline, with the buffer filled into the middle sector. SV experiments were carried out at 20° C and 50,000 rpm with 8 three-channel centerpieces in an 8-hole rotor following standard protocols [4,49] except for centerpieces and data acquisition script. Raw data in the left configuration (A, C) of Figure 1C are inverted. (E) Superposition of sedimentation coefficient distributions c(s) [6] calculated from the sedimentation boundaries. The s-value of the BSA monomer peak is 4.353 S (red), 4.393 S (blue), 4.402 S (cyan), 4.384 S (green).
In summary, in the present brief communication we show how one hurdle toward doubling the sample numbers in analytical ultracentrifugation using Rayleigh interference optics can be overcome. In practice, the remaining limitation is that sample loading is still laborious, and that we found three channel centerpieces in the current design not to seal as well as double sector centerpieces. However, the latter may be solved by slight changes in the embossed seals, or effectively addressed through the use of separate gaskets. The key point of the present work is to demonstrate how some restrictions in the use of AUC that are built into its current operating software may be easily lifted using freely available computer automation and simple scripting, thus motivating further developments of this technique. Exploiting similar strategy, user interface automation may be useful for the next generation of ultracentrifuges, and other computer-controlled laboratory instruments, to extend manufacturer envisaged instrument use.
Supplementary Material
3-channel centerpieces increase throughput in analytical ultracentrifugation
The required new data acquisition mode can be implemented with automation software
Computer user interface functions of instrumentation can be scripted and scheduled
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering. KJM and TV-J acknowledges support from the Aarhus University Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Svedberg T, Rinde H, The determination of the distribution of size of particles in disperse systems, J. Am. Chem. Soc 45 (1923) 943–954. doi: 10.1021/ja01657a012. [DOI] [Google Scholar]
- [2].Svedberg T, Rinde H, The ultra-centrifuge, a new instrument for the determination of size and distribution of size of particle in amicroscopic colloids, J. Am. Chem. Soc 46 (1924) 2677–2693. doi: 10.1021/ja01677a011. [DOI] [Google Scholar]
- [3].Unzai S, Analytical ultracentrifugation in structural biology, Biophys. Rev 10 (2018) 229–233. doi: 10.1007/s12551-017-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schuck P, Zhao H, Brautigam CA, Ghirlando R, Basic Principles of Analytical Ultracentrifugation, CRC Press, Boca Raton, FL, 2015. [Google Scholar]
- [5].Schuck P, Analytical ultracentrifugation as a tool for studying protein interactions, Biophys. Rev 5 (2013) 159–171. doi: 10.1007/s12551-013-0106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schuck P, Sedimentation Velocity Analytical Ultracentrifugation: Discrete Species and Size-Distributions of Macromolecules and Particles, CRC Press, Boca Raton, FL, 2016. [Google Scholar]
- [7].Schuck P, Zhao H, Sedimentation Velocity Analytical Ultracentrifugation: Interacting Systems, CRC Press, Boca Raton, FL, 2017. [Google Scholar]
- [8].Morris GA, Adams GG, Harding SE, On hydrodynamic methods for the analysis of the sizes and shapes of polysaccharides in dilute solution: A short review, Food Hydrocoll. 42 (2014) 318–334. doi: 10.1016/j.foodhyd.2014.04.014. [DOI] [Google Scholar]
- [9].Harding SE, The Svedberg Lecture 2017. From nano to micro: the huge dynamic range of the analytical ultracentrifuge for characterising the sizes, shapes and interactions of molecules and assemblies in Biochemistry and Polymer Science, Eur. Biophys. J (2018). doi: 10.1007/s00249-018-1321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lam S, Zheng M, Fagan JA, Characterizing the Effect of Salt and Surfactant Concentration on the Counterion Atmosphere around Surfactant Stabilized SWCNTs Using Analytical Ultracentrifugation, Langmuir. 32 (2016) 3926–3936. doi: 10.1021/acs.langmuir.6b00605. [DOI] [PubMed] [Google Scholar]
- [11].Sousa AA, Morgan JT, Brown PH, Adams A, Mudiyanselage P, Zhang G, Ackerson CJ, Kruhlak MJ, Leapman RD, Synthesis, Characterization, and Direct Intracellular Imaging of Ultrasmall and Uniform Glutathione-Coated Gold Nanoparticles., Small. (2012) 1–10. doi: 10.1002/smll.201200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carney RP, Kim JY, Qian H, Jin R, Mehenni H, Stellacci F, Bakr OM, Determination of nanoparticle size distribution together with density or molecular weight by 2D analytical ultracentrifugation., Nat. Commun 2 (2011) 335. doi: 10.1038/ncomms1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wawra SE, Pflug L, Thajudeen T, Kryschi C, Stingl M, Peukert W, Determination of the two-dimensional distributions of gold nanorods by multiwavelength analytical ultracentrifugation, Nat. Commun (2018) 1–11. doi: 10.1038/s41467-018-07366-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mehn D, Rio-Echevarria IM, Gilliland D, Kaiser M, Vilsmeier K, Schuck P, Wohlleben W, Identification of nanomaterials: A validation report of two laboratories using analytical ultracentrifugation with fixed and ramped speed options, NanoImpact. 10 (2018) 87–96. doi: 10.1016/j.impact.2017.12.005. [DOI] [Google Scholar]
- [15].Chaturvedi SK, Schuck P, A Reappraisal of Sedimentation Nonideality Coefficients for the Analysis of Weak Interactions of Therapeutic Proteins, AAPS J. 21 (2019) 35. doi: 10.1208/s12248-019-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berkowitz SA, Philo JS, Characterizing biopharmaceuticals using analytical ultracentrifugation, in: Houde DJ, Berkowitz SA (Eds.), Biophys. Charact. Proteins Dev. Biopharm, Elsevier, Amsterdam, 2015: pp. 211–260. doi: 10.1016/B978-0-444-59573-7.00009-9. [DOI] [Google Scholar]
- [17].Gabrielson JP, Randolph TW, Kendrick BS, Stoner MR, Sedimentation velocity analytical ultracentrifugation and SEDFIT/c(s): Limits of quantitation for a monoclonal antibody system, Anal. Biochem 361 (2007) 24–30. [DOI] [PubMed] [Google Scholar]
- [18].Wafer L, Kloczewiak M, Luo Y, Quantifying trace amounts of aggregates in biopharmaceuticals using analytical ultracentrifugation sedimentation velocity: Bayesian analyses and F statistics, AAPS J. (2016). doi: 10.1208/s12248-016-9925-y. [DOI] [PubMed] [Google Scholar]
- [19].Zhao H, Mayer ML, Schuck P, Analysis of protein interactions with picomolar binding affinity by fluorescence-detected sedimentation velocity, Anal. Chem 86 (2014) 3181–3187. doi: 10.1021/ac500093m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chaturvedi SK, Sagar V, Zhao H, Wistow G, Schuck P, Measuring Ultra-Weak Protein Self-Association by Nonideal Sedimentation Velocity, J. Am. Chem. Soc (2019) jacs.8b11371. doi: 10.1021/jacs.8b11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chaturvedi SK, Ma J, Zhao H, Schuck P, Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions, Nat. Protoc 12 (2017) 1777–1791. doi: 10.1038/nprot.2017.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chaturvedi SK, Ma J, Brown PH, Zhao H, Schuck P, Measuring macromolecular size distributions and interactions at high concentrations by sedimentation velocity, Nat. Commun 9 (2018) 4415. doi: 10.1038/s41467-018-06902-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schilling K, Krause F, Analysis of antibody aggregate content at extremely high concentrations using sedimentation velocity with a novel interference optics, PLoS One. 10 (2015) e0120820. doi: 10.1371/journal.pone.0120820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pearson J, Cölfen H, LED based near infrared spectral acquisition for multiwavelength analytical ultracentrifugation: A case study with gold nanoparticles, Anal. Chim. Acta (2018). doi: 10.1016/j.aca.2018.07.022. [DOI] [PubMed] [Google Scholar]
- [25].LeBrun T, Schuck P, Wei R, Yoon JS, Dong X, Morgan NY, Fagan J, Zhao H, A radial calibration window for analytical ultracentrifugation, PLoS One. 13 (2018) e0201529. doi: 10.1371/journal.pone.0201529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao H, Ghirlando R, Alfonso C, Arisaka F, Attali I, Bain DL, Bakhtina MM, Becker DF, Bedwell GJ, Bekdemir A, Besong TMD, Birck C, Brautigam CA, Brennerman W, Byron O, Bzowska A, Chaires JB, Chaton CT, Cölfen H, Connaghan KD, Crowley KA, Curth U, Daviter T, Dean WL, Diez AI, Ebel C, Eckert DM, Eisele LE, Eisenstein E, England P, Escalante C, Fagan JA, Fairman R, Finn RM, Fischle W, de la Torre J. García, Gor J, Gustafsson H, Hall D, Harding SE, Hernández Cifre JG, Herr AB, Howell EE, Isaac RS, Jao S-C, Jose D, Kim S-J, Kokona B, Kornblatt JA, Kosek D, Krayukhina E, Krzizike D, Kusznir EA, Kwon H, Larson A, Laue TM, Le Roy A, Leech AP, Lilie H, Luger K, Luque-Ortega JR, Ma J, May CA, Maynard EL, Modrak-Wójcik A, Mok Y-F, Mücke N, Nagel-Steger L, Narlikar GJ, Noda M, Nourse A, Obsil T, Park CK, Park J-K, Pawalek PD, Perdue EE, Perkins SJ, Perugini MA, Peterson CL, Peverelli MG, Piszczek G, Prag G, Prevelige PE, Raynal BDE, Rezabkova L, Richter K, Ringel AE, Rosenberg R, Rowe AJ, Rufer AC, Scott DJ, Seravalli JG, Solovyova AS, Song R, Staunton D, Stoddard C, Stott K, Strauss HM, Streicher WW, Sumida JP, Swygert SG, Szczepanowski RH, Tessmer I, Toth RT IV, Tripathy A, Uchiyama S, Uebel SFW, Unzai S, Vitlin Gruber A, von Hippel PH, Wandrey C, Wang S-H, Weitzel SE, Wielgus-Kutrowska B, Wolberger C, Wolff M, Wright E, Wu Y-S, Wubben JM, Schuck P, A multilaboratory comparison of calibration accuracy and the performance of external references in analytical ultracentrifugation, PLoS One. 10 (2015) e0126420. doi: 10.1371/journal.pone.0126420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Channell G, Dinu V, Adams GG, Harding SE, A simple cell-alignment protocol for sedimentation velocity analytical ultracentrifugation to complement mechanical and optical alignment procedures, Eur. Biophys. J 1 (2018) 3. doi: 10.1007/s00249-018-1328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Doyle BL, Budyak IL, Rauk AP, Weiss WF, An optical alignment system improves precision of soluble aggregate quantitation by sedimentation velocity analytical ultracentrifugation, Anal. Biochem (2017). doi: 10.1016/j.ab.2017.05.018. [DOI] [PubMed] [Google Scholar]
- [29].Zhao H, Ghirlando R, Piszczek G, Curth U, Brautigam CA, Schuck P, Jakoby W, Zhao H, Ghirlando R, Piszczek G, Curth U, Brautigam CA, Schuck P, Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation, Anal. Biochem 437 (2013) 104–108. doi: 10.1016/j.ab.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Desai A, Krynitsky J, Pohida TJ, Zhao H, Schuck P, 3D-Printing for Analytical Ultracentrifugation, PLoS One. 11 (2016) e0155201. doi: 10.1371/journal.pone.0155201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].To SC, Brautigam CA, Chaturvedi SK, Bollard MT, Krynitsky J, Kakareka JW, Pohida TJ, Zhao H, Schuck P, Enhanced Sample Handling for Analytical Ultracentrifugation With 3D-Printed Centerpieces, Anal. Chem 91 (2019) 5866–5873. doi: 10.1021/acs.analchem.9b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schneider CM, Haffke D, Cölfen H, Band Sedimentation Experiment in Analytical Ultracentrifugation Revisited, Anal. Chem (2018) acs.analchem.8b02768. doi: 10.1021/acs.analchem.8b02768. [DOI] [PubMed] [Google Scholar]
- [33].Manna A, Zhao H, Wada J, Balagopalan L, Tagad HD, Appella E, Schuck P, Samelson LE, Cooperative assembly of a four-molecule signaling complex formed upon T cell antigen receptor activation, Proc. Natl. Acad. Sci 115 (2018) 201817142. doi: 10.1073/pnas.1817142115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rosovitz MJ, Schuck P, Varughese M, Chopra AP, Mehra V, Singh Y, McGinnis LM, Leppla SH, Alanine scanning mutations in anthrax toxin protective antigen domain 4 reveal residues important for binding to the cellular receptor and to a neutralizing monoclonal antibody, J. Biol. Chem 278 (2003) 30936–30944. doi: 10.1074/jbc.M301154200. [DOI] [PubMed] [Google Scholar]
- [35].Pavlov GM, Rowe AJ, Harding SE, Conformation zoning of large molecules using the analytical ultracentrifuge zones, Trends Anal. Chem 16 (1997) 401–405. doi: 10.1016/S0165-9936(97)00038-1. [DOI] [Google Scholar]
- [36].Esfandiary R, Hayes DB, Parupudi A, Casas-Finet J, Bai S, Samra HS, Shah AU, Sathish HA, A systematic multitechnique approach for detection and characterization of reversible self-association during formulation development of therapeutic antibodies, J. Pharm. Sci 102 (2013) 3089–3099. doi: 10.1002/jps.23654. [DOI] [PubMed] [Google Scholar]
- [37].Schönfeld HJ, Pöschl B, Müil F, Quasi-elastic light scattering and analytical ultracentrifugation are indispensable tools for the purification and characterization of recombinant proteins, Biochem. Soc. Trans 26 (1998) 753–758. [DOI] [PubMed] [Google Scholar]
- [38].Kar SR, Kingsbury JS, Lewis MS, Laue TM, Schuck P, Analysis of transport experiments using pseudo-absorbance data, Anal. Biochem 285 (2000) 135–142. doi: 10.1006/abio.2000.4748. [DOI] [PubMed] [Google Scholar]
- [39].Hanlon S, Lamers K, Lauterbach G, Johnson R, Schachman HK, Ultracentrifuge studies with absorption optics. I. An automatic photoelectric scanning absorption system, Arch. Biochem. Biophys 99 (1962) 157–174. doi: 10.1016/0003-9861(62)90258-8. [DOI] [PubMed] [Google Scholar]
- [40].Schuck P, Demeler B, Direct sedimentation analysis of interference optical data in analytical ultracentrifugation, Biophys. J 76 (1999) 2288–2296. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schuck P, Some statistical properties of differencing schemes for baseline correction of sedimentation velocity data, Anal. Biochem 401 (2010) 280–287. doi: 10.1016/j.ab.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kintel M, OpenSCAD, (2019). http://www.openscad.org/. [Google Scholar]
- [43].Richards EG, Schachman HK, Ultracentrifuge Studies with Rayleigh Interference Optics. I. General Application, J. Phys. Chem 63 (1959) 1578–1591. doi: 10.1021/j150580a008. [DOI] [Google Scholar]
- [44].Shivelley AR, Shade PA, Pilchak AL, Tiley JS, Kerns R, A novel method for acquiring large-scale automated scanning electron microscope data, J. Microsc 244 (2011) 181–186. doi: 10.1111/j.1365-2818.2011.03524.x. [DOI] [PubMed] [Google Scholar]
- [45].Scherholz ML, Forder J, Androulakis IP, A framework for 2-stage global sensitivity analysis of GastroPlus™ compartmental models, J. Pharmacokinet. Pharmacodyn 45 (2018) 309–327. doi: 10.1007/s10928-018-9573-1. [DOI] [PubMed] [Google Scholar]
- [46].Carvalho MC, Practical Laboratory Automation, Wiley, Weinheim, Germany, 2017. [Google Scholar]
- [47].Brown PH, Balbo A, Schuck P, A Bayesian approach for quantifying trace amounts of antibody aggregates by sedimentation velocity analytical ultracentrifugation, AAPS J. 10 (2008) 481–493. doi: 10.1208/s12248-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brown PH, Balbo A, Schuck P, On the analysis of sedimentation velocity in the study of protein complexes, Eur. Biophys. J 38 (2009) 1079–1099. doi: 10.1007/s00249-009-0514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhao H, Brautigam CA, Ghirlando R, Schuck P, Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation, Curr. Protoc. Protein Sci 7 (2013) 20.12.1. doi: 10.1002/0471140864.ps2012s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


