Abstract
Like their host cells, many viruses express noncoding RNAs. Despite the technical challenge of ascribing function to ncRNAs, diverse biological roles for virally-expressed ncRNAs have been described, including regulation of viral replication, modulation of host gene expression, host immune evasion, cellular survival and cellular transformation. Insights into conserved interactions between viral ncRNAs and host cell machinery frequently lead to novel findings concerning host cell biology. In this review, we discuss the functions and biogenesis of ncRNAs produced by animal viruses.
Keywords: viruses, noncoding RNA, virus-host interactions
Introduction
High-throughput RNA sequencing has broadened appreciation of the prevalence and diversity of noncoding RNAs (ncRNAs) expressed by pathogens and their host cells. Although these studies have advanced our observational understanding of infectious disease states, mechanistic details lag behind. Viruses are parasites that habitually exchange genetic material with their hosts. However, the limited size of viral genomes means that coding potential is carefully rationed. RNAs are less immunogenic than proteins and therefore provide an advantageous means to commandeer host cell functions. As a result, RNA and DNA viruses regularly express and optimize ncRNAs for their own benefit. Therefore, even though a viral ncRNA may exhibit characteristics typical of a particular host cell ncRNAs, viral transcripts may have non-canonical functions. Investigating the complex interplay of viral ncRNAs and host cell activities has provided many insights into host cell biology.
The best studied viral ncRNAs are produced by herpesviruses, which are a sizable family of vertebrate viruses characterized by large double-stranded DNA genomes and the ability to cause life-long infections. Members of this virus family are notorious human pathogens, including Herpes simplex virus 1 (HSV-1; oral and genital herpes), Epstein-Barr virus (EBV; mononucleosis and Burkitt’s lymphoma), Kaposi’s sarcoma-associated herpesvirus (KSHV; Kaposi’s sarcoma) and varicella zoster virus (VZV; chicken pox and shingles). Herpesvirus-infected cells exist in two broadly defined states: the latent phase when the double-stranded circular viral genome is unobtrusively maintained in the host nucleus and the lytic phase when active replication produces new virions. Because of their large genomes, complex lifecycle and medical relevance, the majority of well-studied viral ncRNAs, as well as the greatest number of ncRNAs per viral genome, have been described in herpesviruses.
Some of the most abundant viral ncRNAs are transcribed by RNA polymerase III (Pol III). Such RNAs include the cytoplasmic VA (virus-associated) RNAs from adenovirus and the nuclear EBER1 and EBER2 from EBV. VA RNAs inhibit protein kinase R (PKR) to counteract the host cell antiviral defense (1; 2) and competitively block microRNA (miRNA) biogenesis by the RNase III enzyme Dicer (3; 4). Both VA RNAs and EBERs bind the autoantigen La, which recognizes the U-rich 3′ termini of RNA Pol III transcripts (5; 6). Beyond its binding partners La, RPL22 and AUF1, almost nothing is known about the function of EBER1 (7; 8). In contrast, the role of EBER2 is better understood; it recruits PAX5 to nascent transcripts emanating from the viral terminal repeat region to regulate EBV DNA replication (9), which is known to be critical for virally-induced tumorigenesis. Detailed reviews of EBERs and VA RNAs are provided elsewhere (10–12).
In addition to discrete noncoding transcripts produced by viral genomes, cis-acting structures in RNA viral genomes and messenger RNAs (mRNAs) are required for viral replication and protein production. For instance, Picornavirus cis-acting RNA structures present in the 5′ and 3′ non-translated regions and open reading frame (ORF) together facilitate replication of the RNA viral genome by mediating interactions with viral trans-acting factors (13). Mechanisms for non-canonical translation of viral transcripts enable viruses to customize translation to their own needs. No virus is known to encode its own ribosome; yet, evolutionary pressure to optimize viral protein expression has driven the evolution of internal ribosome entry sites and frameshifting pseudoknots. These fascinating structures are discussed in depth in other reviews (13–15).
In this review, we highlight a selection of viral ncRNAs that have either an established function or the potential to provide insights into an important aspect of ncRNA biology (Table 1). As in previous reviews, we will not discuss the many viral ncRNAs of low abundance, which have been identified by high-throughput sequencing efforts, but are as yet uncharacterized. We look forward to new mechanisms of host cell manipulation that may be uncovered by future studies.
TABLE 1.
Viral ncRNAs, excluding miRNAs
| Viral family (subgroup) |
virus | RNA | Abundance (copies per cell) |
Length (nt) |
RNA polymerase |
Associated proteins |
Function | References |
|---|---|---|---|---|---|---|---|---|
| Adenoviridae | Human adenovirus | VAI | 107 | ~160 | III | La, PKR, Dicer | Inhibition of PKR | (1; 3; 4; 143) |
| VAII | 5×106 | ~160 | III | La, Dicer | Inhibition of PKR | (4; 143) | ||
| Alpha-Herpesviridae | HSV-1 | LAT | ? | ~2000 | II | ? | Repression of HSV-1 gene expression | (113; 144) |
| MDV | vTR | ~385 | II | TERT | Induction of lymphomas | (99) | ||
| Beta-Herpesviridae | HCMV | 5-kb immediate-early sisRNA | ? | 5000 | II | ? | ? | (112) |
| MCMV | 7.2-kb sisRNA | ? | 7200 | II | ? | Promotes progression from acute to persistent infection | (111) | |
| Gamma-Herpesviridae | EBV | EBER1 | 106 | 167 | III | La, L22, hnRNPD | ? | (5; 7; 110; 145) |
| EBER2 | 2–5 × 105 | 172 | III | La, nucleolin, PAX5 | Regulation of viral gene expression | (5; 7; 9; 110) | ||
| ebv-sisRNA-1 | 106 | 81 | II | ? | ? | (110) | ||
| ebv-sisRNA-2 | ? | 2971 | II | ? | ? | (110) | ||
| circRNAs | ? | Variable | II | ? | ? | (119) | ||
| HVS | HSUR1 | 2 × 104 | 144–143 | II | Sm, Ago2, HuR, hnRNPD | miRNA degradation | (62; 69; 146-148) | |
| HSUR2 | 2 × 103 | 144–143 | II | Sm, HuR, Ago2, hnRNPD | miRNA-mRNA adaptor | (62; 72; 146-148) | ||
| HSUR5 | 2 × 103 | 111–114 | II | Sm, Ago2, HuR, hnRNPD | ? | (62; 146-148) | ||
| HSURs3, 4, 6, 7 | 103–104 | 75–106 | II | Sm | ? | (146; 149) | ||
| KSHV | PAN | 5 × 105 | 1060 | II | hnRNPC1, PABPC1, LANA, ORF57 | Promotion of late lytic viral gene expression and release of progeny virions | (76; 78; 150-152) | |
| circvIRF4 | ? | Variable | II | ? | ? | (118) | ||
| circPAN/K7.3 | ? | Variable | II | ? | ? | (118) | ||
| MHV68 | tRNA1–7 | ? | 72–84 | III | ? | ? | (153) | |
| Flaviviridae | Flaviviruses | sfRNA | ? | 520 | Viral | XRN1 | inhibition of interferon response and RNA decay pathways | (124; 134-138; 154; 155) |
| Retroviridae | HIV-1 | ASP RNA | ? | 2600 | II | PRC2 | Promotes viral latency | (89; 90) |
Viral miRNA are processed by non-canonical pathways of biogenesis
MiRNAs are small, ~22-nt ncRNAs that post-transcriptionally downregulate gene expression (16). A single miRNA can regulate hundreds of different mRNAs; indeed, they play roles in almost every cellular pathway. MiRNAs are distinguished from other small ncRNAs by their biogenesis, evolutionary conservation, and function (17). Namely, miRNAs are processed from hairpin stem-loop structures by Drosha and Dicer RNase III-family enzymes, and they guide Argonaute (AGO) proteins – as part of the RNA-induced silencing complex (RISC) – to imperfectly complementary sequences in mRNA 3′ untranslated regions (UTRs) (18). The first viral miRNAs were identified in EBV (19). Since then, hundreds of viral miRNAs have been described (20) – with roles in viral replication, pathogenesis and immune evasion [reviewed in (21; 22)]. Here, we highlight new insights into non-canonical pathways for viral miRNA production and novel methodologies for viral miRNA identification.
Canonical miRNA biogenesis begins with RNA Polymerase II (Pol II) transcription of primary miRNAs (pri-miRNAs). These capped and polyadenylated transcripts can be several kilobases long and feature distinct secondary structure characteristics that allow recognition and cleavage by the Microprocessor complex, comprised of the endonuclease Drosha and the RNA binding protein DGCR8 (23–25). The excised ~65-nt stemloop known as the precursor miRNA (pre-miRNA) is exported to the cytoplasm where it is further processed by Dicer to form one or two mature miRNAs (26–28). Viral miRNA biogenesis frequently deviates from this canonical miRNA production pathway as viruses often evolve creative methods to hijack host cellular machinery [reviewed in (29)]. Recently, even more examples of viral miRNA precursors transcribed by Pol III, or processed by Drosha- or Dicer-independent means have been discovered (30–42). These unconventional mechanisms expand our conception of the miRNA biogenesis machinery and virus-host coordination.
The ideal human pri-miRNA substrate for Drosha comprises a ~65-nt long pre-miRNA hairpin featuring a 10-nt loop, internal bulges in the stem (i.e. ~11-nt or one helical turn apart) and flanking unstructured RNA sequences (24; 25; 43). Roughly half of all human pri-miRNAs also exhibit specific sequence motifs at the 5′ base and loop of the hairpin that are required for efficient processing (44; 45). Bovine Foamy Virus (BFV) generates three mature viral miRNAs from a Pol III-transcribed bicistronic pri-miRNA cluster that harbors two hairpins separated by a single unpaired nucleotide (35; 36). How this pri-miRNA is processed is unknown; the dumbbell-shaped secondary should be a poor substrate for the Microprocessor because of the lack of unstructured regions at the base of each pre-miRNA stem. Mutational studies of the BFV pri-miRNA reveal that flanking sequences and local secondary structure contribute to proper expression of these BFV miRNAs, thereby suggesting secondary structural constraints similar to those for canonical pri-miRNA processing (35).
The first miRNA discovered in a single-stranded RNA virus came from a Flavivirus, West Nile Virus (WNV) (38). Prior to this discovery, one dilemma concerning miRNAs expressed by RNA viruses was how miRNAs could be excised without destruction of the RNA viral genome. To overcome this, flaviviruses produce a noncoding subgenomic flavivirus RNA (sfRNA) in the course of viral replication, which will be discussed later in this review. Bioinformatic interrogation and Northern blot analyses of viral ncRNAs in WNV-infected mosquito cells identified a miRNA-like small RNA, KUN-miR-1, processed from a pre-miRNA hairpin present in the 3′ UTR of the WNV genome (38). Some putative viral miRNAs are referred to as “miRNA-like small RNAs” because they have no known functions or evolutionary conservation, and rarely fully follow the canonical miRNA biogenesis pathway. In WNV, knockdown experiments determined that Dicer-1 is required for KUN-miR-1 expression, providing evidence that it is a true miRNA (38). Dengue virus 2 (DENV-2)-infected mosquito cells similarly express a miRNA-like small RNA, DENV-vsRNA-5, which maps to a stemloop located in the 3′ UTR of the DENV-2 viral genome (37). DENV-vsRNA-5 expression is reduced by half in Dicer-1 or AGO1 depleted DENV-2-infected mosquito cells, but is even more reduced in AGO2-depleted cells. In addition, the mature and precursor miRNA as well as a larger (240-nt) miRNA-containing transcript were present in RNA associated with AGO2, but not AGO1, which only associates with mature miRNA. These results strongly suggest AGO2 processing of this putative viral miRNA. In H5N1 influenza-infected human cell lines, RNAi of Dicer, Drosha or AGO-2 reduced expression of the miRNA-like small RNA miR-HA-3p significantly only after AGO-2 knockdown, but not Dicer or Drosha, again implicating AGO2-mediated miRNA maturation (39). Whether these AGO2-dependent putative viral miRNAs follow a biogenesis pathway similar to the well-studied example of host AGO2-dependent miRNA processing of vertebrate miR-451 requires further study (46–48).
Subgenomic or non-processive transcription is another method viruses exploit for miRNA generation. Human immunodeficiency virus 1 (HIV-1) possesses a 57-nt trans-activation response (TAR) RNA element which folds into a hairpin at the 5′ end of all HIV-1 transcripts (42; 49-51). The viral transactivator protein TAT interacts with TAR on nascent viral RNAs to promote HIV-1 transcription and replication (52; 53). Total small RNA sequencing and AGO2 HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) identified a miRNA-like viral small RNA, miR-TAR-3p, that maps to the HIV-1 TAR sequence. AGO-bound miR-TAR-3p is present at higher levels in infected 293T cells expressing a mutant virus engineered to block TAT-TAR transcriptional activation than those expressing wildtype HIV-1 (42). This phenotype was attributed to an increase in production of non-processive TAR transcripts from the mutant virus. Finally, investigation of Dicer-depleted and TAT-mutant cells revealed that TAT binding rearranges the TAR hairpin into a better substrate for Dicer processing. Overall, miR-TAR-3p is the product of short, paused or prematurely stopped RNA Pol II transcripts that are bound by TAT to allow processing by Dicer (42).
Another strategy used by viruses to generate miRNAs involves inclusion of other viral ncRNAs in the pri-miRNA transcript. Six miRNA sequences appear immediately downstream of the Herpesvirus saimiri (HVS) RNA Pol II viral ncRNA transcripts known as HSURs (Herpesvirus saimiri U-rich RNAs) (41). HSURs resemble the Sm-class of cellular small nuclear RNAs (snRNAs) that are essential for pre-mRNA splicing and histone pre-mRNA 3′ end processing (48; 49). Cleavage at the 3′ ends of HSUR2, 4 or 5 sequences by host Integrator complex releases a colinear downstream pre-miR-HSUR hairpin, each of which includes two viral miRNAs (41). Alignment of pre-miR-HSUR sequences revealed a second processing signal called the miR 3′ box, which is also recognized and cleaved by the host Integrator complex (54). Therefore, the Integrator complex directly interacts with HVS pri-miRNA to excise pre-miRNAs by cleaving both at the 5′ and 3′ sides of the hairpin.
Murine gamma-herpesvirus 68 (MHV68) miRNAs are cotranscribed by Pol III as pre-miRNA stemloops located immediately downstream of a transfer RNA (tRNA)-like structure. The tRNA processing enzyme, RNaseZ, cleaves at the 3′ end of the tRNA-like structure, releasing viral pre-miRNAs in a Drosha-independent manner (31; 32). Genetic studies using TMER (tRNA-miRNA-encoded RNAs) knockout viruses provide evidence that these transcripts are multifunctional – the tRNA-like ncRNA has an important role in infection and virulence equal to that of the viral miRNA in vivo (55; 56). It will be interesting to see how those roles relate to the emerging research on tRNA fragments, which have been implicated in regulation of gene expression, apoptosis and epigenetic inheritance [reviewed in (57; 58)].
Viral miRNAs are detected primarily by high-throughput small RNA sequencing coupled to computational tools that predict pre-miRNA hairpins (19; 37-39; 41; 59). Identifying viral miRNAs in this manner has been challenging given that the vast majority (>98%) of sequencing reads map to the human genome and viral miRNAs can be moderately or lowly expressed compared to other abundant viral transcripts. The hunt for more viral miRNAs, particularly from retroviruses, led to innovative bioinformatic and synthetic gene sequencing strategies (33; 34; 60). Candidate miRNA precursors (pre-miRNAs), identified by algorithms that predict RNA Pol III viral transcripts, which form RNA hairpin secondary structures, were transiently expressed in cells and subjected to Northern blot analysis to confirm production of mature miRNA sequences. Using this approach, viral miRNAs produced by bovine leukemia virus (BLV) and Simian foamy virus (SFV) were identified and characterized (33; 34). Discovery of five miRNAs expressed by four different papillomaviruses (PV) was accomplished using a new approach known as miDGE (miRNA discovery by forced genomic expression) (60). In this methodology, a group of PV genomes were fragmented, cloned into expression libraries and transfected into cells. High-throughput small RNA sequencing reads obtained from these cells were mapped to the initial pool of PV genomes and miRNAs were identified by the accumulation of sequencing reads at predicted pre-miRNA hairpins (61).
With the discovery of each new viral miRNA comes insight into the innovative approaches viruses use to manipulate cellular machinery. MiRNAs are powerful gene regulators that are compact: an ideal situation for any virus. Conversely, it is in the best interest of a virus to defend itself against host miRNAs – an undertaking also accomplished by viral ncRNAs.
Modulation of host microRNA stability and target selection by viral ncRNAs
HSUR1 from Herpesvirus saimiri was the first example of an RNA that binds to host miRNA and, instead of undergoing RISC-mediated degradation, induces decay of the bound miRNA (62). Normally, specific binding to target mRNAs is conferred by the seed sequence of the miRNA (nts 2–7) and results in RISC-mediated inhibition of translation, induction of mRNA decay by decapping and deadenylation, or both (16). HSUR1 binds to miR-27 family members in a unique fashion: in addition to base-pairing via the miRNA seed, HSUR1 exhibits extensive complementarity to the 3′ portion of the miRNA (Fig. 2A) (62). This manner of interaction induces miR-27 degradation. Decreased levels of miR-27 in T cells cause prolonged T cell activation and correlate with virus-induced oncogenic transformation (63). The discovery of HSUR1-mediated miR-27 degradation was followed by two reports of other herpesviral RNAs that also selectively induce host miRNA degradation: the m169 RNA from murine cytomegalovirus (CMV), which mediates degradation of host miR-27 family members, and a transcript from human CMV that targets host miR-17 family members (5). This target-directed miRNA degradation (TDMD) is believed to be a widespread cellular mechanism for regulating miRNA populations (64–66). Crystallization of AGO2-miR-27a-HSUR1 complexes has shed light on the molecular mechanism of TDMD. HSUR1 complementarity to the 3′ portion of miR-27a dislodges the miRNA 3′ end from its binding pocket in AGO2, thereby rendering the miRNA susceptible to host cell enzymatic attack (J. Sheu-Gruttadauria, P. Pawlica, J.A. Steitz and I.J. MacRae, submitted). Ongoing efforts to identify nucleotidyl transferases and exonucleases mediating TDMD have been challenging due to the redundancy and promiscuity of these cellular enzymes (67; 68). Bioinformatic discovery of novel TDMD effector RNAs awaits better definition of TDMD requirements, beyond finding that such ncRNAs must be devoid of internal structure (65; 66; 69; 70).
FIGURE 2. Modulation of miRNA stability and target selection by HSURs.
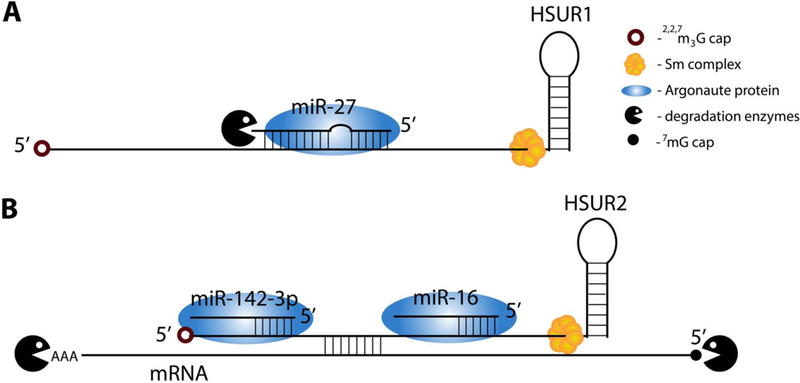
Schematized interactions between Sm-bound HSURs and host RNAs. (A) HSUR1 binding to miR-27a dislodges the miRNA 3′ end from its pocket in the Argonaute protein followed by miRNA degradation by yet unknown enzymes. (B) HSUR2 interaction with miR-142–3p and/or miR-16 recruits Argonaute protein to target mRNAs — also tethered by HSUR2 — for RNA-induced silencing complex-mediated mRNA decay.
The 116-nt HVS HSUR2 has evolved to mediate miRNA target selection (71). HSUR2 base-pairs with miR-142–3p and miR-16 (Fig. 2B), as well as with ~70 host mRNAs. HSUR2 tethering of these two miRNAs destabilizes target mRNAs, which likely occurs via recruitment of RISC. MRNA targets of miR-142–3p and miR-16 modulate hematopoietic lineage development and the G1-S cell cycle transition, respectively. In the presence of HSUR2, additional downregulation of mRNAs encoding proteins involved in cell cycle regulation (RB1), apoptosis (FAS) and the immune response (JAK1) are observed: these processes are relevant to viral maintenance during latency. It is unclear how HSUR2 targets multiple mRNAs; however, unpublished evidence suggests that different segments of HSUR2 base pair with different mRNA targets ((72), Demian Cazalla, personal communication).
Deciphering the roles of HSURs 1 and 2 led to discovery of two novel mechanisms of miRNA regulation, but the roles of the other five HSURs remain unknown. These advances highlight the potential for other viral ncRNAs to modulate miRNA activities and biogenesis in ways that have yet to be imagined.
Functions of viral long ncRNAs remain elusive
Long ncRNAs (lncRNAs) are arbitrarily defined as RNA transcripts longer than 200 nts that are transcribed by RNA Pol II and lack identifiable protein coding potential (73). According to their genomic positions relative to protein-coding genes, lncRNAs are categorized as antisense lncRNAs, bidirectional lncRNAs, enhancer RNAs (eRNAs), intronic lncRNAs, long intergenic ncRNAs (lincRNAs) or transcribed pseudogene lncRNAs. In contrast to host lncRNAs that tend to be expressed in low copy numbers, viral lncRNAs can be among the most abundant transcripts in the cell. This disparity in expression levels hints at essential roles for lncRNAs during viral infection. High expression levels facilitate biochemical detection of these RNAs, yet deciphering their functions is far from easy.
In lytic KSHV-infected cells, the most abundant polyadenylated transcript is a nuclear lncRNA called PAN RNA. The best-studied aspect of PAN RNA is a triple-stranded 3′ end element called the element for nuclear expression (ENE) that robustly inhibits nuclear RNA degradation and is required to maintain PAN RNA at elevated levels (74–77). PAN RNA homologues in other gammaherpesviruses have been identified through bioinformatic searches for ENE structures (77). Loss of KSHV PAN RNA expression, either through knockout from the viral genome or antisense depletion of the transcript, results in misregulation of late lytic viral genes and host immune response genes (78; 79). Characterization of PAN RNA interaction partners has led to hypotheses that PAN RNA epigenetically regulates host and viral gene expression, promotes viral lytic reactivation or sequesters host proteins [reviewed in (10; 80)]. Chemical probing of PAN RNA identifies three branched domains, but local structural details within each domain change depending on the biological source of PAN RNA: nucleus, cytoplasm, virion (81). This structural variability suggests that PAN RNA may associate with, and potentially sequester (81) or concentrate (82), different protein partners during the viral lytic cycle. Delineating salient interaction partners and functions of PAN RNA will continue to illuminate how this viral lncRNA acts as a global regulator of the herpesvirus lytic phase.
During the latent phase, one of the few transcripts expressed from the HSV-1 genome is LAT (latency-associated transcript). Mutational and genetic ablation studies of LAT have implicated this viral lncRNA in preventing apoptosis (83), supporting neuronal tropism (84), establishing latency (85), promoting lytic reactivation (86) and regulating histone methylation (87; 88). More studies are needed to tease apart the temporal and spatial factors that allow this lncRNA to influence so many aspects of HSV-1 latency.
HIV-1, in addition to the full-length genomic RNA and spliced mRNAs, produces antisense RNAs (89; 90). One such transcript, the HIV-1 antisense protein (ASP) RNA, is a 2.6-kb, nuclear lncRNA transcribed from the 3′ long terminal repeat (LTR) U3 region of integrated proviral DNA (89). ASP RNA lacks a poly(A) tail and interacts with and recruits polycomb repressor complex 2 (PRC2) to the HIV-1 5′ LTR. Subsequently, PRC2 trimethylates histone H3 lysine 27 (H3K27me3), which promotes nucleosome assembly and suppresses viral gene expression. Therefore, ASP RNA facilitates proviral latency through accumulation of the suppressive epigenetic mark H3K27me3 and reduction of proviral transcription (90). Although ASP RNA is readily detectable in chronically infected cell lines and latently infected cells from clinical samples (~5–25 copies per cell) (90), it has been hypothesized that the recruitment of PRC2 complexes to HIV-1 proviral loci might be mediated through other, yet unknown, lncRNAs (91).
The sometimes contradictory list of phenotypes and functions associated with each viral RNA highlights how difficult it is to ascribe a mechanism of action to a lncRNA. Further complicating is the fact that many lncRNAs may be multifunctional. Advances in biochemical and high-throughput techniques are needed to devise unbiased approaches to decipher the pertinent roles that viral lncRNAs play in infection.
Viral telomerase RNA is essential for oncogenesis
Telomeres are repetitive sequences at the ends of chromosomes that prevent loss of genetic content through cycles of DNA replication. Telomeres are maintained by telomerase, consisting of two essential components: a reverse transcriptase (TERT) and an RNA molecule known as telomerase RNA (TR) that serves as a template for telomere repeat addition by TERT (92). Heightened telomerase activity has been detected in many cancers and contributes to transformation by maintaining cells in a proliferative state (93; 94).
Oncogenic viruses take advantage of telomeres and telomerase using different strategies. Examples include upregulation of host mRNA encoding TERT, which promotes survival of the infected cell (95) and integration of the viral genome into the host genome mediated by telomeric repeat sequences in the viral genome, which promotes prolonged infection of the host cell (96; 97). Marek’s disease virus (MDV), an alphaherpesvirus that causes lymphomas in chickens (98), produces its own viral TR (vTR), which complements the function of the chicken TR (chTR) (98–100). Interestingly, vTR, relative to chTR, elevates telomerase activity more efficiently within the cell (101). The increased efficiency of vTR is attributed to sequence variations in the P2 and P3 helices of the pseudoknot domain (Fig. 3). Four nucleotides in the P2b stem of chTR, when inserted into vTR, diminish the capacity of vTR to promote telomerase activity (Fig. 3).
FIGURE 3. Mutational analyses reveal regions important for vTR function.
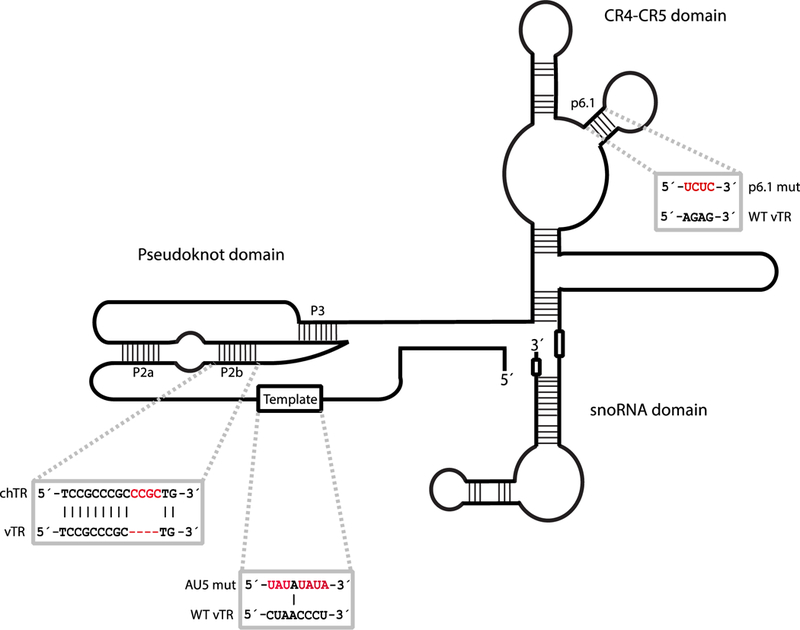
A comparison of chTR and vTR nucleotide sequences revealed additional nucleotides in the P2b region of the pseudoknot of chTR that reduce the ability of vTR to promote telomerase activity (101). Mutating the template region of TR (AU5 mutant) in the MDV genome prevents lymphomagenesis (103). Disruption of the p6.1 stem loop also precludes tumor formation (104). Mutational analysis of p6.1 and template sequences were performed in the context of vTR.
The high transformation rate of MDV-infected cells, along with the precedent of heightened telomerase activity in cancer, has prompted investigation into the role of vTR in lymphomagenesis. MDV mutants that lack vTR do not efficiently induce lymphomas in their hosts, but are able to replicate normally (102). Disrupting the interaction between vTR and chTERT by mutating the p6.1 stem of vTR (Fig. 3) delays, but does not eliminate, the onset of lymphomagenesis (103). This suggests that elevated telomerase activity is important for the rate of transformation, but is not essential for disease progression (104). Mutagenesis also revealed that an intact template region of vTR is necessary for transformation of T cells upon MDV infection (Fig. 3) (103). The reduced rate of tumorigenesis resulting from a vTR template mutant virus infection revealed the therapeutic efficacy of vTR: vaccinating chickens with a vTR template mutant MDV produced an efficient, protective immune response (98; 103).
The possibility that vTR has roles independent of its interaction with TERT is supported by the finding that vTR interacts with the ribosomal protein RPL22 (104). RPL22 also interacts with human telomerase (105) and with the EBER1 ncRNA, which is produced by the oncogenic gammaherpesvirus EBV and implicated in tumorigenesis (7). Expression of EBER1, vTR or chTR is required for relocalization of RPL22 from the cytoplasm and nucleoli to the nucleoplasm (7; 104). Furthermore, the interaction between EBER1 and RPL22 is important for proliferation (106). The relevance of the interaction between vTR and RPL22 in tumor progression upon MDV infection has yet to be addressed.
EBER2, which modulates lytic EBV replication by regulating latent gene expression (9), is able to drive oncogenesis in MDV-infected cells lacking vTR (107). It is unclear whether the mechanisms by which EBER2 promotes oncogenesis are similar to those employed by vTR. Mutational analysis and the identification of additional vTR interacting partners will further the understanding of this process. The role of EBERs and vTR in cancer development and progression highlights the functional relevance of viral ncRNAs.
Viral circular RNAs and stable intronic sequence RNAs: pervasive, abundant and enigmatic
Alternative splicing of nascent pre-mRNAs differentially joins exons, resulting in RNA isoforms that may have different cellular functions or encode different protein variants. A form of alternative splicing called backsplicing joins a 3′ splice donor to an upstream 5′ splice acceptor to generate a covalently-closed circular RNA (circRNA) (108). CircRNAs are remarkably stable due to the absence of exonuclease-susceptible termini. Most introns and intron fragments produced as a result of splicing are unstable; however, there are some notable exceptions including small nucleolar RNAs (snoRNAs) and stable intronic sequence RNAs (sisRNAs) (109; 110). SnoRNAs function in the modification or processing of ribosomal RNAs, small nuclear RNAs, mRNAs and tRNAs [reviewed in (109)]. Definitive biological functions for sisRNAs and circRNAs have yet to be elucidated, but since both classes of these ncRNAs are produced by viruses, they are potentially potent regulators of cellular function.
SisRNAs have been identified in at least four viruses: HCMV, MCMV, HSV-1 and EBV (110–113). EBV-encoded sisRNAs were discovered during analysis of a nuclear RNA library constructed from cultured EBV-infected B lymphocytes at the cancer-associated latency III stage. Ebv-sisRNA-1 and −2 are abundant nuclear transcripts expressed from the W-repeat region of the EBV internal repeat 1 and are produced as a result of EBNA-LP (Epstein-Barr nuclear antigen leader protein) pre-mRNA splicing (110). Transformation of B-cells by EBV requires EBNA-LP expression (114), and genetic mutation ebv-sisRNA-1 reduces EBNA-LP expression and inhibits the transformation of B-cells by EBV (115). Ebv-sisRNA-1 and −2 interact with as many as 81 human proteins, seven of which have been validated biochemically: FUS, hnRNPA1, hnRNPC, hnRNPL, HuR, LIN28 and hnRNPD/AUF1 (116). The viral ncRNA EBER1, which also binds hnRPNPD/AUF1 (9), may cooperate with ebv-sisRNA-1 to perturb the homeostasis of hnRNPD/AUF1-dependent mRNA regulation in EBV-infected cells. Interaction of the splicing repressor hnRNPL with ebv-sis-RNA may alter the nucleo-cytoplasmic distribution of hnRNPL, thereby influencing alternative splicing of host transcripts (117). Alternatively, association of ebv-sisRNA with hnRNPL might alter EBNA-LP pre-mRNA splicing to regulate sisRNA production (116). Further functional investigations are needed to establish the implications of sisRNA interactions in EBV infection and disease.
CircRNAs in EBV- and KSHV-infected cells were recently discovered by high-throughput RNA sequencing of ribosomal RNA-depleted and linear RNA-depleted (exonuclease RNase R-treated) libraries. At least 30 unique viral circRNAs were reported, including EBV-encoded circRNAs from the BART/RPMS1, BHLF1, and LMP-2 loci, and KSHV-encoded circRNAs from the vIRF4 and PAN/K7.3 loci (118; 119). Investigation of KSHV circRNAs is still in its infancy.
EBV circRNAs are expressed in both latent and lytic stages and some display extraordinarily high expression levels. Although mechanistic analyses of EBV circRNAs are as yet lacking, the literature hints at potential functions. CircBHLF1 may be the functional form of BHLF1 RNA that associates with the viral origin of replication, oriLyt, to facilitate lytic viral DNA replication (120). The EBV BART locus produces four abundant circRNAs; two nuclear intron-containing circBARTs and two exon-only circBARTs that are found in both the nucleus and the cytoplasm. Host RNA helicases UAP56 and URH49, which contribute to host circRNA nuclear export (121), may likewise control export of viral circRNAs. The circBARTs and BART miRNAs are generated from pre-mRNA transcripts produced by the same locus. Like EBV BART miRNAs, circBARTs are present in all forms of EBV tumor latency; however, BART deletion studies showed that neither circBARTs nor BART miRNAs are essential for the maintenance of the EBV genome in cell culture. Nonetheless, circBART expression was detected in all EBV-associated tumor samples tested, implicating these ncRNAs in tumorigenesis.
The field of circRNA biology is nascent. CircRNAs can sponge miRNAs, modulate RNA splicing, provide templates for cap-independent translation and act in competition with their equivalent linear mRNA [reviewed in (122)]. Given the broad spectrum of latent and lytic genes from which viral circRNAs originate, their diverse patterns of expression and their various sub-cellular distributions, viral circRNAs may contribute to a wide array of functions. Additionally, the extreme stability of latency-associated and cancer-associated viral circRNAs enables their use as biopsy markers and diagnostic reagents (118; 119).
Inhibition of 5′ to 3′ exonucleases by structured RNA elements generates viral ncRNAs
An unusual mode of ncRNA production is employed by RNA viruses of the genus Flavivirus, which includes such arthropod-borne human pathogens such as DENV, WNV, Zika virus (ZIKV) and yellow fever virus (YFV). An ~300–700-nt long subgenomic flaviviral RNA (sfRNA) is generated by incomplete degradation of the genomic RNA (gRNA) by the cellular 5′ to 3′ exonuclease XRN1 [reviewed in (123; 124)]. SfRNA is the most abundant viral RNA in flavivirus-infected cells.
The positive-sense gRNAs of Flaviviruses possess a single ORF preceded and followed by highly-structured UTRs [reviewed in (125)]. Some of the 3′ UTR structures, referred to as XRN1-resistant RNAs (xrRNAs), stall XRN1 progression to generate stable sfRNAs. Most Flavivirus gRNAs possess two adjacent xrRNA structures (Fig. 4), although virus gRNAs with only one xrRNA have been identified (126). The 5′-proximal xrRNA usually constitutes the major block to XRN1 progression and thus demarcates the 5′ end of the most abundant form of sfRNA. Secondary structure mapping and mutational analysis of DENV sfRNA revealed a three-way junction in xrRNA that is critical for halting XRN1 (127). Subsequently, the crystal structures of xrRNAs from Murray Valley encephalitis and Zika viruses revealed an elegant RNA structure-based mechanism for XRN1 stalling: the RNA fold organized around the three-way junction adopts a ring-like conformation with the 5′ end of the XRN1-resistant structure passing through the ring (Fig. 4) (128). The ring-like conformation is reinforced by a conserved pseudoknot and two long-range interactions of 5′-proximal nucleotides with residues at the three-way junction. Although the primary, and to some extent the secondary structure of xrRNAs from distant branches of Flaviviruses differ, based on nucleotide conservation and multiple compensatory changes within the long-range interactions, the tertiary fold is believed to be similar (129). Interestingly, subgenomic ncRNAs produced by incomplete degradation of the 3′ UTR of genomic RNA by cellular 5′ to 3′ exonucleases are found also in other animal and plant viral families (130–132). A crystal structure of xrRNA from a plant dianthovirus revealed a fold that is different from that of flavivarial xrRNAs (133). Nevertheless, both kinds of xrRNAs possess a common topological feature, i.e. a pseudoknot that creates a protective ring around the 5′ end of xrRNA to obstruct further degradation.
FIGURE 4. Tertiary structure of Zika virus xrRNA.
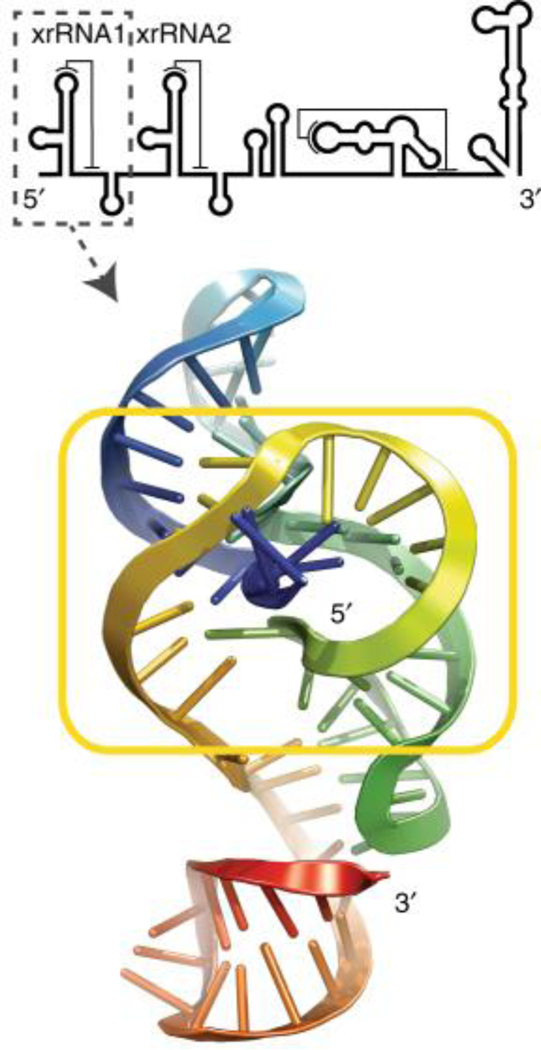
The yellow box marks a unique ring-like structure through which the 5′ end (blue) passes. The location of this xrRNA in ZIKV sfRNA is shown above. Adapted from (129).
SfRNA is required for Flavivirus cytopathicity and pathogenicity. It has been demonstrated to i) dampen the antiviral activity of type I interferon [reviewed in (124)], ii) interfere with cellular 5′ to 3′ RNA decay by inhibiting XRN1 (134), and iii) inhibit the RNAi pathway in vertebrate and arthropod species, most likely by serving as a decoy substrate for Dicer (135; 136). Inhibition of the host interferon response appears to be, at least in some Flaviviruses like DENV-2, achieved by binding and inactivating cellular regulators of interferon-stimulated mRNA translation (137). Moreover, DENV-2 sfRNA binds TRIM25 to prevent TRIM25-mediated ubiquitination of the cytosolic RNA sensor RIG-I, thus averting viral RNA detection and interferon production (138). The RNAi inhibitory activity of sfRNA appears to be especially important for counteracting a potent antiviral RNAi response during the insect phase of the Flavivirus life cycle.
Conclusions and Prospects
Powerful sequencing technologies present unprecedented opportunities to investigate questions in virology and RNA biology. The combined expertise of bioinformaticians and virologists will identify new viruses and viral ncRNAs, facilitate prediction of RNA structures and enable increasingly penetrating transcriptomic analyses. Each newly discovered ncRNA has its own customized function. The peculiar ncRNA functions highlighted in this review are likely a small sampling of potential viral control mechanisms. Giant viruses, such as mimiviruses, are conceivably an untapped resource for viral ncRNA biology. These viruses typically have genomes larger than 200 kb and therefore possess substantial potential for expressing ncRNAs (139). Although giant viruses are studied primarily as infectious agents of single-celled eukaryotes, they have been isolated from human samples indicating potential roles in human health (140; 141). Finally, our understanding of the influence of RNA modifications on ncRNA function is only nascent (142). Post-transcriptional RNA modifications may prove to be potent modulators of the battle between viral ncRNAs and host cell defenses.
FIGURE 1. MiRNA biogenesis pathways.
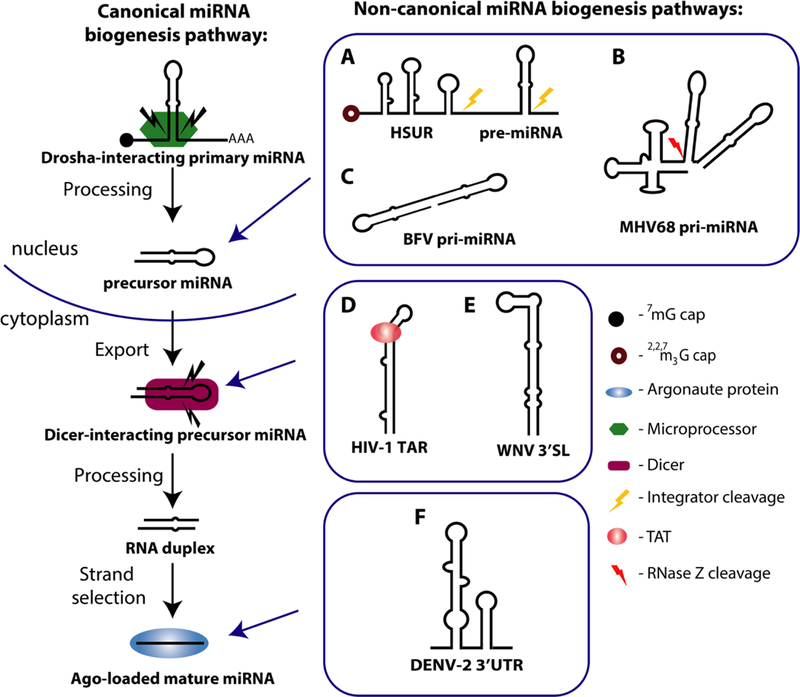
Canonical pathway: A capped and polyadenylated primary miRNA (pri-miRNA) undergoes cleavage by the Microprocessor complex. The ~65-nt precursor miRNA (pre-miRNA) is transported to the cytoplasm, where it is subsequently cleaved by Dicer to produce a ~21 nt double-stranded product that features 2 nt overhangs at both ends. The mature miRNA strand is then loaded into Argonaute (Ago), where it works as part of the RNA induced silencing complex (RISC). Non-canonical viral miRNA biogenesis pathways include: (A) Herpesvirus saimiri U-rich RNAs (HSUR) are noncoding messages colinear with a viral pre-miRNA hairpin. Integrator processes the precursor by cleaving at the 3′ ends of the HSUR and of the pre-miRNA. (B) Murine gamma-herpesvirus 68 (MHV68) encodes eight small ncRNAs known as TMERS (transfer RNA-miRNA-encoded RNAs). RNase Z cleavage at the 3′ end of the tRNA-like structure releases the co-transcribed pre-miRNA hairpin. (C) Bovine Foamy Virus (BFV) primary miRNA (pri-miRNA) harbors two pre-miRNA hairpins that bypass Drosha processing in an unknown manner. (D) Human immunodeficiency virus 1 (HIV-1) has a miRNA-containing trans-activation response (TAR) RNA element at the 5′ end of all HIV-1 transcripts. The miRNA is excised when viral transactivator protein, TAT, interacts with stalled transcripts to allow Dicer processing of the stem-loop. (E) The 3′ terminal stemloop (3′SL) of West Nile Virus (WNV) is recognized and processed by Dicer. (F) The 3′ untranslated region (3′ UTR) of Dengue Virus 2 (DENV) consist of stem-loop structures, one of which associates with and is processed by AGO2 to release a miRNA.
Acknowledgements
We thank Angie Miccinello for editorial expertise and all Steitz laboratory members for stimulating discussions. Our work is supported by NIH grants (##). NARM is supported by a Ford Foundation Predoctoral Fellowship in combination with a Yale University fellowship. PP is supported by a K99 grant (K99GM129412). SFT is a Damon Runyon postdoctoral fellow. JAS is an investigator of the Howard Hughes Medical Institute.
Contributor Information
Johanna B Withers, Email: johannawithers@yale.edu.
Vanessa Mondol, Email: vanessa.mondol@yale.edu.
Paulina Pawlica, Email: paulina.pawlica@yale.edu.
Nicolle A Rosa-Mercado, Email: nicolle.rosa-mercado@yale.edu.
Kazimierz T Tycowski, Email: ktt@yale.edu.
Salehe Ghasempur, Email: salehe.ghasempur@yale.edu.
Seyed F Torabi, Email: seyed.torabi@yale.edu.
Joan A Steitz, Email: joan.steitz@yale.edu.
References
- 1.Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J Virol 65:5657–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson JL, Vachon VK, Sunita S, Schwartz SL, Conn GL. 2014. Dissection of the adenoviral VA RNAI central domain structure reveals minimum requirements for RNA-mediated inhibition of PKR. The Journal of biological chemistry 289:23233–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu S, Cullen BR. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 78:12868–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol 79:9556–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerner MR, Andrews NC, Miller G, Steitz JA. 1981. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A 78:805–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francoeur AM, Mathews MB. 1982. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci U S A 79:6772–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toczyski DP, Matera AG, Ward DC, Steitz JA. 1994. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci U S A 91:3463–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N, Pimienta G, Steitz JA. 2012. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 18:2073–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, Moss WN, Yario TA, Steitz JA. 2015. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160:607–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tycowski KT, Guo YE, Lee N, Moss WN, Vallery TK, et al. 2015. Viral noncoding RNAs: more surprises. Genes Dev 29:567–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vachon VK, Conn GL. 2016. Adenovirus VA RNA: An essential pro-viral non-coding RNA. Virus Res 212:39–52 [DOI] [PubMed] [Google Scholar]
- 12.Moss WN, Lee N, Pimienta G, Steitz JA. 2014. RNA families in Epstein-Barr virus. RNA Biol 11:10–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steil BP, Barton DJ. 2009. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res 139:240–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93:1385–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. 2009. Structural and functional diversity of viral IRESes. Biochim Biophys Acta 1789:542–57 [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. 2018. Metazoan microRNAs. Cell 173:20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, et al. 2003. A uniform system for microRNA annotation. RNA 9:277–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509. [DOI] [PubMed] [Google Scholar]
- 19.Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, et al. 2004. Identification of virus-encoded microRNAs. Science 304:734–6 [DOI] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8:e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernier A, Sagan S. 2018. The diverse roles of microRNAs at the host–virus interface. Viruses 10:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen Tuan A, Jo Myung H, Choi Y-G, Park J, Kwon CS, et al. 2015. Functional anatomy of the human microprocessor. Cell 161:1374–87 [DOI] [PubMed] [Google Scholar]
- 24.Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, et al. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901 [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y, Cullen BR. 2005. Efficient processing of primary microRNA hairpins by drosha requires flanking nonstructured RNA sequences. J Biol Chem 280:27595–603 [DOI] [PubMed] [Google Scholar]
- 26.Wilson Ross C, Tambe A, Kidwell Mary A, Noland Cameron L, Schneider Catherine P, Doudna Jennifer A. 2015. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell 57:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint É, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–8 [DOI] [PubMed] [Google Scholar]
- 28.Ketting RF, Fischer SEJ, Bernstein E, Sijen T, Hannon GJ, Plasterk RHA. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15:2654–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie M, Steitz JA. 2014. Versatile microRNA biogenesis in animals and their viruses. RNA Biol 11:673–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diebel KW, Smith AL, van Dyk LF. 2010. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16:170–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. 2010. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol 84:10344–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell 37:135–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A 109:3077–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS. 2014. Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao W, Heit A, Hotz-Wagenblatt A, Löchelt M. 2018. Functional characterization of the bovine foamy virus miRNA expression cassette and its dumbbell-shaped pri-miRNA. Virus Genes 54:550–60 [DOI] [PubMed] [Google Scholar]
- 36.Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, et al. 2014. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J Virol 88:4679–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain M, Asgari S. 2014. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc Natl Acad Sci U S A 111:2746–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, et al. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res 40:2210–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Fu Z, Liang H, Wang Y, Qi X, et al. 2018. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell Res 28:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diebel KW, Claypool DJ, van Dyk LF. 2014. A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene 544:8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cazalla D, Xie M, Steitz Joan A. 2011. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell 43:982–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harwig A, Jongejan A, van Kampen Antoine HC, Berkhout B, Das AT 2016. Tat-dependent production of an HIV-1 TAR-encoded miRNA-like small RNA. Nucleic Acids Res 44:4340–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Ahn C, Han J, Choi H, Kim J, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–9 [DOI] [PubMed] [Google Scholar]
- 44.Conrad T, Marsico A, Gehre M, Orom UA. 2014. Microprocessor activity controls differential miRNA biogenesis In Vivo. Cell Rep 9:542–54 [DOI] [PubMed] [Google Scholar]
- 45.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. 2013. Beyond Secondary Structure: Primary-Sequence Determinants License Pri-miRNA Hairpins for Processing. Cell 152:844–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, et al. 2010. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A 107:15163–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465:584–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328:1694–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennasser Y, Le S-Y, Yeung ML, Jeang K-T. 2004. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 1:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, et al. 2007. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, et al. 2008. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 36:2353–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng S, Holland EC. 1988. HIV-1 tat trans-activation requires the loop sequence within tar. Nature 334:165–7 [DOI] [PubMed] [Google Scholar]
- 53.Garcia JA, Harrich D, Soultanakis E, Wu F, Mitsuyasu R, Gaynor RB. 1989. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J 8:765–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie M, Zhang W, Shu M-D, Xu A, Lenis DA, et al. 2015. The host Integrator complex acts in transcription-independent maturation of herpesvirus microRNA 3′ ends. Genes Dev 29:1552–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldman ER, Kara M, Oko LM, Grau KR, Krueger BJ, et al. 2016. A gammaherpesvirus noncoding RNA is essential for hematogenous dissemination and establishment of peripheral latency. mSphere 1:00105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diebel KW, Oko LM, Medina EM, Niemeyer BF, Warren CJ, et al. 2015. Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo. mBio 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schorn AJ, Martienssen R. 2018. Tie-break: host and retrotransposons play tRNA. Trends Cell Biol [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar P, Kuscu C, Dutta A. 2016. Biogenesis and function of transfer RNA related fragments (tRFs). Trends Biochem Sci 41:679–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2:269. [DOI] [PubMed] [Google Scholar]
- 60.Chirayil R, Kincaid RP, Dahlke C, Kuny CV, Dälken N, et al. 2018. Identification of virus-encoded microRNAs in divergent Papillomaviruses. PLoS Pathog 14:e1007156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cazalla D, Yario T, Steitz JA. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo YE, Riley KJ, Iwasaki A, Steitz JA. 2014. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell 54:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bitetti A, Mallory AC, Golini E, Carrieri C, Carreno Gutierrez H, et al. 2018. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 25:244–51 [DOI] [PubMed] [Google Scholar]
- 65.Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghini F, Rubolino C, Climent M, Simeone I, Marzi MJ, Nicassio F. 2018. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat Commun 9:3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174:350–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, et al. 2016. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res 44:2873–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pawlica P, Moss WN, Steitz JA. 2016. Host miRNA degradation by Herpesvirus saimiri small nuclear RNA requires an unstructured interacting region. RNA 22:1181–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S, Song J, Kim S, Kim J, Hong Y, et al. 2013. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13:678–90 [DOI] [PubMed] [Google Scholar]
- 71.Gorbea C, Mosbruger T, Cazalla D. 2017. A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis. Nature 550:275–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cazalla D 2018. Novel roles for Sm-class RNAs in the regulation of gene expression. RNA Biol:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cech TR, Steitz JA. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94 [DOI] [PubMed] [Google Scholar]
- 74.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330:1244–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. 2012. Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A 109:19202–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conrad NK, Steitz JA. 2005. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J 24:1831–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. 2012. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep 2:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7:e1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossetto CC, Pari GS. 2011. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol 85:13290–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conrad NK. 2016. New insights into the expression and functions of the Kaposi’s sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res 212:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sztuba-Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, Le Grice SFJ. 2017. Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res 45:6805–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vallery TK, Withers JB, Andoh JA, Steitz JA. 2018. KSHV mRNA accumulation in nuclear foci is influenced by viral DNA replication and the viral noncoding PAN RNA. J Virol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, et al. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500–3 [DOI] [PubMed] [Google Scholar]
- 84.Margolis TP, Imai Y, Yang L, Vallas V, Krause PR. 2007. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J Virol 81:1872–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson ZL, Washington SD, Phelan DM, Lewin AS, Tuli SS, et al. 2018. In vivo knockdown of the herpes simplex virus 1 latency-associated transcript reduces reactivation from latency. J Virol 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, et al. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saayman S, Ackley A, Turner AW, Famiglietti M, Bosque A, et al. 2014. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 22:1164–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zapata JC, Campilongo F, Barclay RA, DeMarino C, Iglesias-Ussel MD, et al. 2017. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 506:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mbonye U, Karn J. 2017. The molecular basis for human immunodeficiency virus latency. Annu Rev Virol 4:261–85 [DOI] [PubMed] [Google Scholar]
- 92.Blackburn EH, Collins K. 2011. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3:a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernardes de Jesus B, Blasco MA. 2013. Telomerase at the intersection of cancer and aging. Trends Genet 29:513–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hackett JA, Greider CW. 2002. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene 21:619–26 [DOI] [PubMed] [Google Scholar]
- 95.Van Doorslaer K, Burk RD. 2012. Association between hTERT activation by HPV E6 proteins and oncogenic risk. Virology 433:216–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greco A, Fester N, Engel AT, Kaufer BB. 2014. Role of the short telomeric repeat region in Marek’s disease virus replication, genomic integration, and lymphomagenesis. J Virol 88:14138–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wallaschek N, Sanyal A, Pirzer F, Gravel A, Mori Y, et al. 2016. The telomeric repeats of human herpesvirus 6A (HHV-6A) are required for efficient virus integration. PLoS Pathog 12:e1005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kheimar A, Previdelli RL, Wight DJ, Kaufer BB. 2017. Telomeres and telomerase: role in Marek’s disease virus pathogenesis, integration and tumorigenesis. Viruses 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fragnet L, Blasco MA, Klapper W, Rasschaert D. 2003. The RNA subunit of telomerase is encoded by Marek’s disease virus. J Virol 77:5985–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brown AC, Nair V, Allday MJ. 2012. Epigenetic regulation of the latency-associated region of Marek’s disease virus in tumor-derived T-cell lines and primary lymphoma. J Virol 86:1683–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fragnet L, Kut E, Rasschaert D. 2005. Comparative functional study of the viral telomerase RNA based on natural mutations. The Journal of biological chemistry 280:23502–15 [DOI] [PubMed] [Google Scholar]
- 102.Trapp S, Parcells MS, Kamil JP, Schumacher D, Tischer BK, et al. 2006. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J Exp Med 203:1307–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaufer BB, Arndt S, Trapp S, Osterrieder N, Jarosinski KW. 2011. Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog 7:e1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaufer BB, Trapp S, Jarosinski KW, Osterrieder N. 2010. Herpesvirus telomerase RNA(vTR)-dependent lymphoma formation does not require interaction of vTR with telomerase reverse transcriptase (TERT). PLoS Pathog 6:e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Le S, Sternglanz R, Greider CW. 2000. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell 11:999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Houmani JL, Davis CI, Ruf IK. 2009. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J Virol 83:9844–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kheimar A, Kaufer BB. 2018. Epstein-Barr virus-encoded RNAs (EBERs) complement the loss of Herpesvirus telomerase RNA (vTR) in virus-induced tumor formation. Sci Rep 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, et al. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Falaleeva M, Stamm S. 2013. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 35:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moss WN, Steitz JA. 2013. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics 14:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulesza CA, Shenk T. 2006. Murine cytomegalovirus encodes a stable intron that facilitates persistent replication in the mouse. Proc Natl Acad Sci U S A 103:18302–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kulesza CA, Shenk T. 2004. Human cytomegalovirus 5-kilobase immediate-early RNA is a stable intron. J Virol 78:13182–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Farrell MJ, Dobson AT, Feldman LT. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci U S A 88:790–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kelly GL, Milner AE, Tierney RJ, Croom-Carter DS, Altmann M, et al. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, −3B, and −3C expression in Burkitt’s lymphoma cells and with increased resistance to apoptosis. J Virol 79:10709–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szymula A, Palermo RD, Bayoumy A, Groves IJ, Ba Abdullah M, et al. 2018. Epstein-Barr virus nuclear antigen EBNA-LP is essential for transforming naive B cells, and facilitates recruitment of transcription factors to the viral genome. PLoS Pathog 14:e1006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tompkins VS, Valverde DP, Moss WN. 2018. Human regulatory proteins associate with non-coding RNAs from the EBV IR1 region. BMC Res Notes 11:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15:689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, et al. 2018. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 115:E8737–E45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, et al. 2018. The Epstein Barr virus circRNAome. PLoS Pathog 14:e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rennekamp AJ, Lieberman PM. 2011. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J Virol 85:2837–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang C, Liang D, Tatomer DC, Wilusz JE. 2018. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32:639–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li X, Yang L, Chen LL. 2018. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–42 [DOI] [PubMed] [Google Scholar]
- 123.Bidet K, Garcia-Blanco MA. 2014. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J 462:215–30 [DOI] [PubMed] [Google Scholar]
- 124.Slonchak A, Khromykh AA. 2018. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antiviral Res [DOI] [PubMed] [Google Scholar]
- 125.Fernandez-Sanles A, Rios-Marco P, Romero-Lopez C, Berzal-Herranz A. 2017. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front Microbiol 8:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kieft JS, Rabe JL, Chapman EG. 2015. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol 12:1169–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 3:e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, et al. 2014. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344:307–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.MacFadden A, O’Donoghue Z, Silva P, Chapman EG, Olsthoorn RC, et al. 2018. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat Commun 9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Charley PA, Wilusz CJ, Wilusz J. 2018. Identification of phlebovirus and arenavirus RNA sequences that stall and repress the exoribonuclease XRN1. The Journal of biological chemistry 293:285–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flobinus A, Chevigny N, Charley PA, Seissler T, Klein E, et al. 2018. Beet necrotic yellow vein virus noncoding RNA production depends on a 5’-->3’ Xrn exoribonuclease activity. Viruses 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, et al. 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5’->3’ RNA decay represses both cap-independent and cap-dependent translation. J Virol 82:10162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Steckelberg AL, Akiyama BM, Costantino DA, Sit TL, Nix JC, Kieft JS. 2018. A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc Natl Acad Sci U S A 115:6404–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, et al. 2012. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 18:2029–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, et al. 2012. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol 86:13486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J. 2015. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 485:322–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bidet K, Dadlani D, Garcia-Blanco MA. 2014. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10:e1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, et al. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wilhelm SW, Bird JT, Bonifer KS, Calfee BC, Chen T, et al. 2017. A student’s guide to giant viruses infecting small eukaryotes: from acanthamoeba to zooxanthellae. Viruses 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yolken RH, Jones-Brando L, Dunigan DD, Kannan G, Dickerson F, et al. 2014. Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc Natl Acad Sci U S A 111:16106–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Popgeorgiev N, Boyer M, Fancello L, Monteil S, Robert C, et al. 2013. Marseillevirus-like virus recovered from blood donated by asymptomatic humans. J Infect Dis 208:1042–50 [DOI] [PubMed] [Google Scholar]
- 142.Jacob R, Zander S, Gutschner T. 2017. The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs. Int J Mol Sci 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. 2006. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80:1376–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–9 [DOI] [PubMed] [Google Scholar]
- 145.Fok V, Friend K, Steitz JA. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. The Journal of cell biology 173:319–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. 1988. Four novel U RNAs are encoded by a herpesvirus. Cell 54:599–607 [DOI] [PubMed] [Google Scholar]
- 147.Myer VE, Lee SI, Steitz JA. 1992. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc Natl Acad Sci U S A 89:1296–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cook HL, Mischo HE, Steitz JA. 2004. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Molecular and cellular biology 24:4522–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Albrecht JC, Fleckenstein B. 1992. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res 20:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun R, Lin SF, Gradoville L, Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 93:11883–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sahin BB, Patel D, Conrad NK. 2010. Kaposi’s sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog 6:e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Campbell M, Kim KY, Chang PC, Huerta S, Shevchenko B, et al. 2014. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol 88:1843–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bowden RJ, Simas JP, Davis AJ, Efstathiou S. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol 78 ( Pt 7):1675–87 [DOI] [PubMed] [Google Scholar]
- 154.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, et al. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–91 [DOI] [PubMed] [Google Scholar]
- 155.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. 2014. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6:404–27 [DOI] [PMC free article] [PubMed] [Google Scholar]


