Abstract
RNA viruses encode the information required to usurp cellular metabolism and gene regulation and to enable their own replication in two ways: in the linear sequence of their RNA genomes and in higher order structures that form when the genomic RNA strand folds back on itself. Application of high-resolution SHAPE structure probing to viral RNA genomes has identified numerous new regulatory elements, has defined new principles by which viral RNAs interact with the cellular host and evade host immune responses, and has revealed relationships between virus evolution and RNA structure. This review summarizes our current understanding of genome structure-function interrelationships for RNA viruses, as informed by SHAPE structure probing, and outlines opportunities for future studies.
Keywords: RNA structure, SHAPE, chemical probing, RNA viruses, functional validation
INTRODUCTION
RNA viruses encode the information required for their replication and that cause host pathogenesis in short genomes, roughly 10,000 nucleotides (nts) or fewer. Information is encoded both in the linear RNA sequence, such as regions that encode viral proteins, and in complex higher order structures. Higher order motifs include base-paired elements and tertiary structures characterized by closely packed RNA helices stabilized by numerous noncanonical interactions between specific nucleotides (1–3). Structured RNA elements are pervasive throughout viral genomes and have complex effects on replication, protein synthesis, packaging, evasion of host immune factors, and the hijacking of host cell machinery (4–17). These RNA elements mediate interactions with protein, RNA, and small-molecule ligands (17–21). A subset of viral RNA functional elements are well characterized. These elements include the human immunodeficiency virus (HIV) 5′ untranslated region (UTR) (22–24), the hepatitis C virus (HCV) internal ribosome entry site (IRES) (25–29), and flavivirus cis 5′-to-3′ cyclization elements such as those found in dengue (DENV) and Zika (ZIKV) viruses (30, 31), all of which fall in noncoding genome regions. Functional elements with complex structures are also found in viral protein-coding regions, including the HIV Rev response element (32), the severe acute respiratory syndrome (SARS) coronavirus ribosomal frameshifting signal-associated pseudoknot (12), and flavivirus capsid-coding region cis-acting elements (33–36).
Until recently, it was not possible to evaluate higher order RNA structure genome-wide. Over the past several years, high-throughput strategies based on chemical probing have been developed that enable investigation of structure across entire intact viral RNA genomes in highly efficient experiments. These strategies pair chemical structure probing, especially the SHAPE (selective 2’-hydroxyl acylation analyzed by primer extension) strategy, with massively parallel sequencing (Figures 1 and 2) (37–40). Studies of entire viral genomes have revealed that viral RNA genomes are highly structured and that many identified structural elements are critical for viral fitness (Table 1). In this review, we describe how SHAPE chemical probing technologies, as implemented by multiple laboratories and in diverse ways, have transformed our physical and functional understanding of viral RNA genomes. SHAPE chemical probing strategies have also been integrated with dimethyl sulfate (DMS) and psoralen crosslinking based RNA structure probing strategies, including RING-MaP (41), PARIS (42), and SPLASH (43), that specifically measure higher order interactions. Nucleotide-resolution structure probing experiments have identified and characterized extensive novel RNA regions and structures of interest and have revealed intriguing mechanisms by which RNA viruses use the information encoded in higher order RNA structures to facilitate and regulate replication.
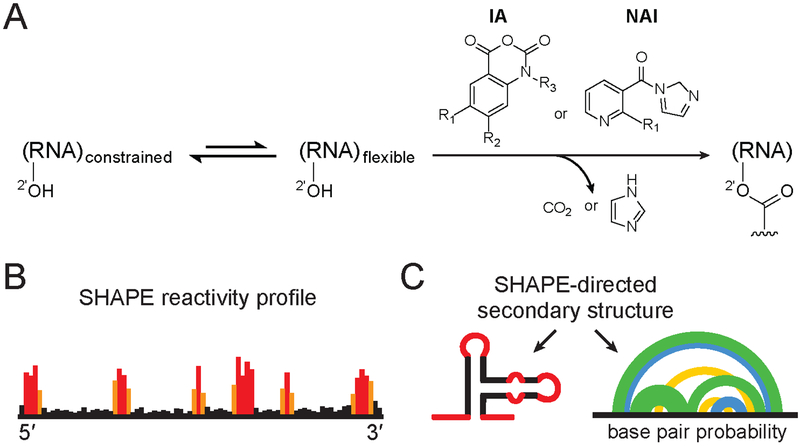
Figure 1.
SHAPE chemistry mechanism, reagents used to probe viral RNA structure, and SHAPE-directed structure modeling. (A) SHAPE reagents react with the 2′-hydroxyl group of conformationally flexible RNA nucleotides, yielding a covalent 2’-O-adduct. Widely used reagents are based on the isatoic anhydride (IA) or nicotinic acid imidazolide (NAI) scaffolds (47, 54, 115). (B) Representative SHAPE reactivity profile, with black, orange and red bars indicating low, medium and high nucleotide reactivities. (C) Use of SHAPE data to model a single minimum free energy structure (left) or characterize an ensemble consistent with SHAPE data (right). Arcs (right) connect base paired nucleotides with pairing probabilities illustrated (from highest to lowest) in green, blue and yellow. Data in panels B and C correspond to the same representative structural element; adapted from Siegfried et al. (37).
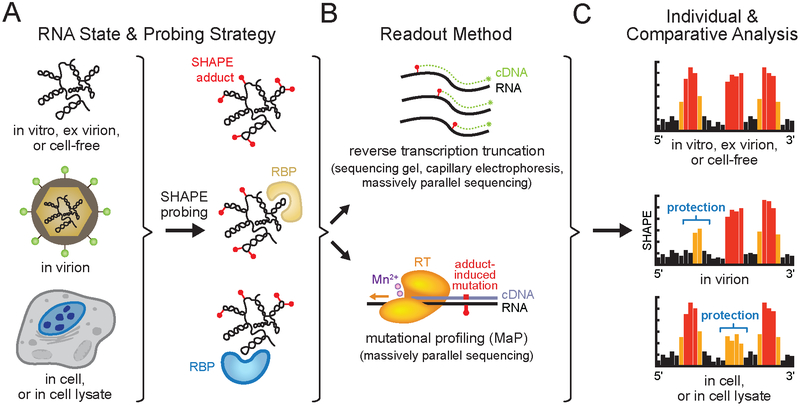
Figure 2.
SHAPE probing strategies. (A) Chemical probing of a viral RNA genome in an informative biological state (see Box 1 for definitions). Representative SHAPE adducts (red spheres) and RNA binding proteins (RBP) are shown. (B) Readout of per-nucleotide SHAPE reactivities by adduct-induced reverse transcription primer extension truncation or mutational profiling (MaP) and quantified by sequencing gel, capillary electrophoresis or massively parallel sequencing. (C) SHAPE reactivity profiles from distinct biological states can be compared to identify sites of SHAPE reactivity protections and enhancements, revealing state-specific RNA conformations, or protein or small molecule binding. Black, orange and red bars indicate low, medium and high nucleotide reactivities, respectively. Individual SHAPE reactivity profiles can also be incorporated into modeling algorithms to yield RNA secondary structure models (see Figure 1). Elements of figure adapted from Smola et al. (38, 116).
Table 1.
Whole viral genome studies by SHAPE.
| Virus Family/Genus | Virus Species | Ref. | Summary Statement |
|---|---|---|---|
| Retroviridae/Lentivirus | Human immunodeficiency virus (HIV-1) | (75) | The first whole viral genome interrogated by SHAPE reveals extensive secondary structure elements throughout the genome, many of which are functionally important for HIV-1. |
| Simian immunodeficiency virus (SIVmac239), HIV-1 | (76) | Many RNA structures previously identified in HIV-1 coding regions (75) are not conserved in the related SIVmac239 (~50% sequence similarity). | |
| HIV-1 | (37) | New SHAPE-MaP analysis strategy supports a refined genome structure model; low SHAPE/low Shannon entropy genome regions identify novel functional elements. | |
| SIVcpzMB897, SIVmac239, HIV-1 | (79) | Correlation of SHAPE reactivity patterns across the genomes of three lentivirus species pinpoint multiple novel functional structural elements. | |
| Picornaviridae/Enterovirus | Poliovirus | (78) | Second human virus family interrogated by SHAPE identifies novel functional elements, including the 3Dpol RNA element which may mediate an interaction with the viral 3Cpro protein. |
| Flaviviridae/Hepacivirus | Hepatitis C virus (HCV) | (65) | SHAPE analyses of genomes of three HCV genotypes reveals many conserved highly structured regions, including four novel conserved structures required for optimal viral fitness. |
| (69) | SHAPE-derived RNA structure models melded with covariation models identifies novel conserved stem-loops across the HCV genome, including four motifs required for viral fitness. | ||
| Togaviridae/Alphavirus | Sindbis virus (SINV), Venezuelan equine encephalitis virus (VEEV) | (73) | Structure-first SHAPE analysis identifies functional RNA elements only present in individual alphavirus species, and not detectable by covariation and comparative structure analyses. |
| Flaviviridae/Flavivirus | Dengue virus 2, (DENV2) | (58) | Higher order RNA tertiary structures are pervasive across the DENV2 genome, and promote a compact global genome architecture and enhance viral fitness. |
| DENV1, DENV2, DENV3, DENV4, Zika Virus (ZIKV) | (74) | Highly structured regions are conserved across four DENV serotypes and four ZIKV strains, and five long-range ZIKV RNA-RNA interactions are important for viral fitness. | |
| ZIKV | (72) | A long-range RNA-RNA interaction between sequences in the 5′-UTR and the Env-coding region occurs exclusively in epidemic ZIKV strains and is important for viral fitness. | |
| Orthomyxoviridae/ Alphainfluenzavirus | Influenza A Virus (IAV) | (64) | Inter-segment interactions between the eight IAV genomic segments are both redundant and important for viral packaging and growth. |
| IAV Segments 8, 7, 5(+) | (68, 70, 71) | IAV RNA genome segments 7 and 8 form highly structured conserved domains. Single-stranded regions in segment 5(+) can be targeted with antisense oligonucleotides to inhibit replication. | |
| Tombusviridae/Tombusvirus | Tomato bushy stunt virus (TBSV) | (63) | SHAPE and atomic force microscopy reveal a compact genome structure; local RNA structures provide a structural scaffold for the formation of functional long-range interactions. |
| Bromoviridae/Cucumovirus | Cucumber mosaic virus (CMV) Segment 3 | (81) | Covariation analysis of SHAPE-defined structures identifies four novel functional structural elements; SHAPE analysis in infected cell lysate reveals a protein binding site. |
| Plant satellite virus Virgaviridae/Tobamovirus | Satellite-Tobacco mosaic virus (STMV) | (62, 77, 80) | SHAPE-directed genome structure models from independent groups are in excellent agreement, revealing a three-domain genome architecture where each domain corresponds to a viral function. |
| Plant satellite virus Tombusviridae/Carmovirus | Satellite-Turnip crinkle virus | (66) | SHAPE structure probing of this 356-nt satellite viral genome identifies an extended functional hairpin, H2. |
| Plant satellite virus Tombusviridae/Tombusvirus | Satellite-Cymbidium ringspot virus | (67) | SHAPE structure probing of this 619-nt satellite viral genome identifies several functional genome secondary and tertiary structures, including an RNA switch. |
Studies are listed chronologically by date within each genus in the order: human viruses, plant viruses, and satellite viruses; with studies of whole genomes preceding genome segments.
SHAPE STRUCTURE PROBING
Chemical probing technologies, first developed 40 years ago, are powerful approaches for examining RNA structure (44–46). Chemical probing strategies have radically evolved in the last ten years with the advent of new chemistries that allow nearly every nucleotide in a complex RNA to be structurally interrogated in a single experiment. In addition, chemical probing experiments can now be read out by high-throughput strategies using massively parallel sequencing. These advances mean that increasingly complex systems can be studied and that these experiments can be undertaken by a broader set of investigators. The most widely used and information-rich approach involves SHAPE chemistry.
SHAPE chemistry exploits small hydroxyl-selective electrophilic reagents that react with the ribose 2′-hydroxyl group of conformationally flexible RNA nucleotides, yielding a covalent 2’-O-adduct (Figure 1A) (47). Reagents have been developed that are largely insensitive to nucleotide identity, and SHAPE experiments therefore yield single-nucleotide resolution information about local nucleotide flexibility and dynamics in a rapid and unbiased fashion (Figure 1B,C) (48, 49). Several strategies have been developed to quantify SHAPE reactivity of individual nucleotides (Figure 2). In the original methods, reverse transcriptase-mediated primer extension creates a cDNA which truncates at sites of adducts. cDNAs were initially analyzed using sequencing gel electrophoresis (48); subsequently, semiautomated capillary electrophoresis was implemented (48, 50). More recently, SHAPE data have been read out on genome-wide virus scales using massively parallel sequencing (reviewed in (51–54)). The mutational profiling (or MaP) strategy (37, 38) makes use of specialized conditions that allow the reverse transcriptase (RT) to read through chemically modified positions. The enzyme incorporates a noncomplementary nucleotide at the site of a chemical adduct. The locations of the SHAPE adducts are thus recorded in the resulting cDNA as internal mutations relative to the parent RNA sequence (37, 38). Although SHAPE data can also be read out by reverse transcription truncation and sequencing library-ligation strategies (39, 54–57), the high-throughput MaP strategy (SHAPE-MaP) is simpler to implement, allows targeted and rare RNAs to be examined, and is readily implemented on long RNA molecules (for example, DENV is 11 kb (58)).
SHAPE reactivity can be incorporated as restraints in RNA modeling algorithms to generate RNA secondary structure models with high accuracies (Figure 1B,C) (37, 59, 60) and to detect regions that form well-determined structures or that are likely to sample multiple conformations (Figure 1C). The SHAPE strategy has been applied to RNA molecules ranging in size from 75 to 18,000 nucleotides (37, 38, 61). SHAPE has been implemented by diverse laboratories to study viral genomes in a variety of both simplified and complex biologically relevant states. These states include RNA transcribed in vitro and refolded (referred to as in vitro RNA) (62–72), RNA gently extracted from virus particles (ex virion) (37, 58, 64, 73–81) or from infected cells (cell-free) (78), RNA in native virus particles (in virion) (58, 64, 74, 80), RNA in infected cell lysates (81), and RNA in infected cells (72, 74) (Box 1, Figure 2A). Comparisons of SHAPE reactivity profiles obtained for viral RNA molecules probed in different biological states reveal state-specific RNA conformations (31, 58, 64, 72, 74, 80–85) as well as sites occupied by RNA-binding proteins (RBP) (58, 64, 72, 80–83) or small molecules (86, 87) (Figure 2C).
BOX 1: DEFINITIONS OF USEFUL STATES FOR STUDYING VIRAL RNA STRUCTURE.
in vitro RNA: in vitro transcribed RNA, refolded in buffer; does not contain post-transcriptional modifications (114)
ex virion RNA: RNA (gently) extracted from virions, often the best state for de novo modeling RNA secondary structure
in virion RNA: RNA encapsulated within virions
cell-free RNA: RNA gently extracted from infected cells
infected cell lysate RNA: RNA in protease- and RNase-inhibited cell lysate
infected cell RNA: RNA in live cells
SHAPE structure probing has revealed a wealth of novel RNA structures across viral genomes. Many of these structures play significant roles in viral replication cycles, but some of these structures simply reflect the tendency of an RNA molecule to fold back on itself and do not appear to have a functional role. Multiple strategies have thus been developed to identify those viral RNA structural motifs that play functional roles in viral replication. In this review, we first give a broad overview of the results obtained in SHAPE-based studies of entire RNA genomes. To date, this list of examined viral genomes includes eleven viruses and three satellite viruses (Table 1, an example is shown for DENV in Figure 3). We then review the strategies used to analyze the SHAPE structure probing data, often in combination with other types of information, to identify and validate functional elements in these viral RNAs.
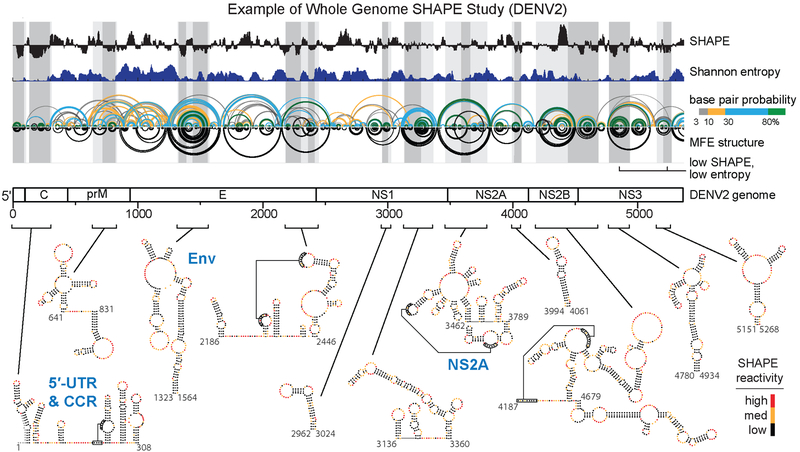
Figure 3.
Whole viral genome studies and well-determined structures. Genome wide SHAPE structure probing from a representative study of the DENV2 RNA genome. The 5’ half of the ~11,000 nucleotide long RNA genome is shown. Median ex virion 1M7 SHAPE reactivities (black) and Shannon entropies (dark blue) are plotted over centered 55-nt windows. Regions with both low SHAPE and low Shannon entropy, termed low SHAPE, Shannon entropy regions, are highlighted by dark-gray shading, with light-gray shading extended to encompass entire structures. Base pairing arcs are colored by probability (see scale), with green arcs indicating the most probable and well-determined base pairs. The minimum free-energy (MFE) secondary structure (inverted black arcs) was obtained using SHAPE reactivities as restraints (37, 38, 117). Secondary structures of the twelve boxed low SHAPE, low Shannon entropy elements are shown at bottom of image and are colored by SHAPE reactivity. Previously studied RNA structural elements in the 5′-UTR and capsid-coding region (CCR) and elements correctly modeled de novo by SHAPE-directed modeling are labeled in blue. Novel structural elements in the Env- and NS2A- coding regions identified by SHAPE structure probing and shown to be critical for viral fitness are also labeled in blue. Adapted from Dethoff et al. (58).
WHOLE VIRAL GENOME STUDIES
Lentiviruses
The first entire viral RNA genome structure to be investigated experimentally at single-nucleotide resolution by SHAPE chemical probing was HIV-1 (75). The resulting genome structure model revealed extensive RNA folding to form secondary structures across the length of the genome. In addition, all functional elements identified in prior work were modeled with good accuracy relative to accepted models. In some cases, the genome-wide SHAPE data clearly supported revisions to previously modeled structures, especially the HIV-1 frameshift element (75, 88). One intriguing observation to emerge from genome-wide structure interrogation of HIV-1 was that the RNA regions encoding junctions separating viral protein domains are highly structured. These structured elements were proposed to slow translation to promote folding of multi-domain proteins. This model has subsequently gained significant support (74, 89, 90) but does not appear to occur for all viruses (78).
Splice site acceptors and hypervariable regions across the HIV-1 genome were generally found to be unstructured and insulated from folding with neighboring regions by conserved helices. Subsequent second-generation structural studies of the HIV-1 genome using SHAPE-MaP identified three functional RNA pseudoknots (37), and multiple novel structures conserved across diverse lentivirus genomes (79). The second-generation studies supported a revised model in which only a subset of a large RNA genome forms stably folded structures (analogous to the example of a DENV genome study, shown in Figure 3). Regions with low SHAPE reactivities and low Shannon entropy (indicative of well-determined stably folded structures) are enriched with functional elements (37).
Structural studies comparing HIV-1NL4–3 and two simian immunodeficiency virus (SIV) strains, SIVcpzMB897 (the progenitor of HIV-1) and SIVmac239 (the progenitor of HIV-2), demonstrated that only a small subset of structured elements are conserved (76, 79). These conserved RNA structures are involved in protein binding, regulation of the reverse transcription step in retroviral replication, and splicing (79), and have higher G/C nucleotide content than nonconserved structures, suggesting pressure to retain stable RNA base pairing (76). A structure-based alignment of the three diverse lentivirus genomes, incorporating SHAPE data and nucleotide covariation information, revealed that regions with strongly correlated patterns of SHAPE reactivities were specifically enriched with functional structural elements (79).
Poliovirus
Another important early whole-genome SHAPE structure probing study identified numerous highly structured regions across the poliovirus genome (78). In poliovirus, unlike HIV, no significant correlation was observed between highly structured regions and protein domain junctions, suggesting that distinct RNA viruses – in these cases, a retrovirus and a positive-strand RNA virus – use RNA structure to regulate viral processes in distinct ways. Thirteen regions were identified in poliovirus with stable SHAPE-defined secondary structures and secondarily showing an evolutionarily conserved pattern of base pairing. These regions included known functional elements and eight novel structured RNA regions.
The most distinctive element, centered at position 7000 in the coding sequence for the viral RNA-dependent RNA polymerase (3Dpol), is conserved at the levels of sequence and structure among poliovirus and human enterovirus C sequences (Figure 4A). Viruses with mutations that disrupt this structure have defects in viral replication and infectivity (78). Interestingly, the functional effects of the structure-perturbing mutations were partially suppressed by mutations in the viral protease 3C (3Cpro), suggesting a potential direct or indirect interaction between this RNA element and 3Cpro (78, 91). Thus, a subset of mutations in the region encoding 3Dpol, known to compromise poliovirus infectivity, might do so by altering RNA secondary structure rather than protein structure.
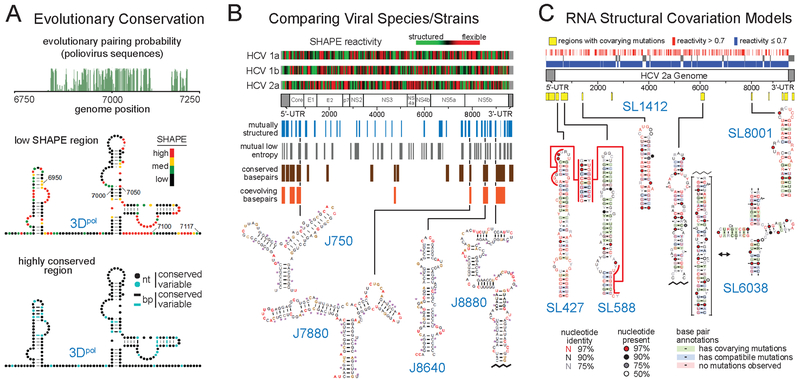
Figure 4.
Representative methods for melding sequence covariation information with SHAPE-directed structure models to identify functional RNA elements. (A) Coupling evolutionary pairing probability analysis of viral sequence alignments (green) with SHAPE structure probing to identify the 3Dpol functional element in the poliovirus genome. RNA nucleotides are labeled with SHAPE reactivities or nucleotide (nt) and base pair (bp) conservation. Adapted from Burrill et al. (78) with permission from American Society for Microbiology. (B) Comparison of sequences, SHAPE reactivities, and structure models across three HCV genotype subtypes to identify conserved functional structural elements. RNA structures are labeled by position in the JFH1 strain and residues are colored by SHAPE reactivity. Adapted from Mauger et al. (65). (C) RNA structural covariation models, built from a SHAPE-directed HCV secondary structure model and >1000 divergent HCV sequences. Functional motifs are colored by structural consensus and covariation. Conserved elements are located at different positions in distinct HCV genome sequences. Adapted from Pirakitikulr et al. (69) with permission from Elsevier.
Hepatitis C Virus
SHAPE structure probing of HCV genomes was undertaken by two independent groups and revealed themes that appear to be common to many RNA viruses: highly structured and well-defined RNA motifs are pervasive across both protein-coding and nonprotein-coding regions (Figure 4B,C) (65, 69). These studies accurately recovered most previously characterized structured RNA elements and identified a large number of novel well-structured elements. One study compared the SHAPE reactivity patterns of three distinct HCV molecular clones (H77c, Con1, and JFH1, corresponding to HCV genotypes 1a, 1b, and 2, respectively) and searched for regions with shared features (Figure 4B) (65). The second study focused on a single serotype, Jc1 (a chimera of genotype 2), identified stable stem-loop elements, and then sought evidence of their conservation based on sequence covariation models derived from diverse HCV sequences (Figure 4C) (69). Both studies identified structures with functional consequences for viral replication. Interestingly, the specific elements identified only partially overlapped, indicating potential complementary advantages of both strategies.
The studies on HCV also demonstrate the challenge of validating the functions of identified structural elements. For example, study one compared three HCV genotypes identified nine structural elements likely to have functional roles, four of which were previously uncharacterized (Figure 4B) (65). Mutations designed to disrupt these structures resulted in deficiencies in either genome replication or infectious virus production in the JFH1 genotype, but none resulted in large defects in both assays. The mutations that decreased infectious virus production (in J750 and J8640) may involve structures important in late-stage events in the virus replication cycle like viral assembly or packaging. Two of the structure-disrupting mutations were also tested in the much slower growing H77 strain and resulted in no phenotype. Study two, focused on the single Jc1 strain, identified seventeen well-conserved stem-loops, six of which were previously uncharacterized (69). Mutations in four of these regions (Figure 4C) decreased viral fitness. Notably, SL427/SL588 and SL6038 mutants abolished viral replication and infectivity; SL6038 functionally was interpreted as involving a dynamic switch between stem-loop and cloverleaf conformations. The SL1412 mutant affected viral infectivity without affecting replication, similar to elements in the study that compared three HCV genotypes.
An example of a subtle effect of RNA structure was revealed by comparison of structures likely to engage the innate immune system. These sites include UU and UA dinucleotides, the targets of RNase L (92), and helices of 16 base pairs or greater, which are detected by PKR (93), a human protein kinase inhibitory to viral replication. UU and UA dinucleotides occur overwhelmingly in well-structured motifs in the virus, and the very small number of strong RNase L cleavage sites identified in previous studies (65, 94, 95) fall in conformationally flexible single-stranded loops. Thus, the virus appears to have evolved such that the vast majority of potential RNase L cleavage sites are located in RNA structural contexts unfavorable for cleavage. Furthermore, most helices in the HCV genomes are short, with a median length of 4 base pairs (65). These studies of HCV genomes emphasize that structured RNA elements play diverse roles in viruses and that multiple assays are often needed to functionally characterize many of these viral RNA motifs.
Alphaviruses
A recent study comparing the Alphavirus species sindbis virus (SINV) and Venezuelan equine encephalitis virus (VEEV) revealed that nonconserved, species-specific RNA structural elements can modulate viral infectivity (73). SINV and VEEV share 50–60% sequence identity and were found to contain highly structured regions at many similar genome locations. SHAPE-directed structure models recapitulated known conserved RNA motifs including the 5′-conserved sequence element (5′-CSE) and the packaging signal element (96, 97). However, most of the well-determined structural elements are poorly conserved across alphavirus species. To assess the functional importance of both conserved and novel nonconserved structural elements in SINV, structure-disrupting mutations were introduced in the 5′-CSE and packaging signal elements and also in two SHAPE-identified, but nonconserved, structural elements in the nsP1 and nsP3 coding regions of SINV. Mutations in the 5′-CSE, packaging signal element, and nsP1 element all caused defects in viral infectivity, indicating that both conserved and nonconserved RNA structures can be important for viral fitness. These findings emphasize that there are important functional species-specific RNA structural motifs that cannot be identified by (or do not have) covariation and comparative structural conservation.
Dengue Virus
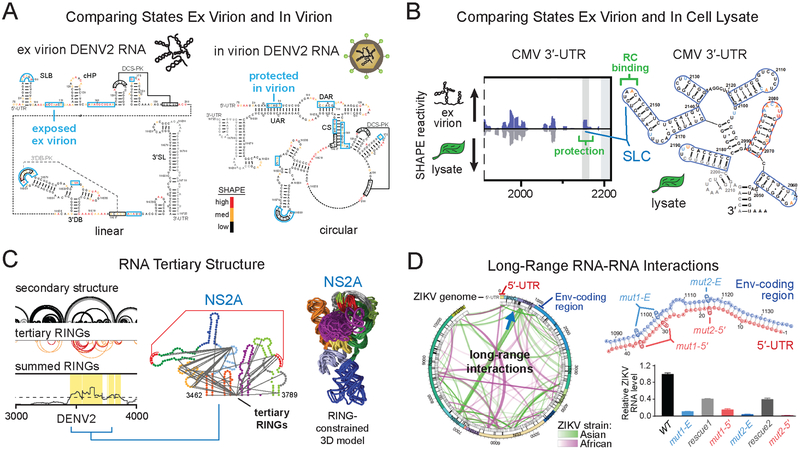
Two groups recently independently investigated the entire DENV2 genome using SHAPE (58, 74). All previously characterized RNA structural elements were identified along with numerous novel well-defined motifs (Figure 3). Linear and circular forms of the DENV2 RNA genome, which involve base pairing between the 5′ and 3′ ends of the genome, regulate replication and translation and are conserved between DENV and ZIKV (7, 18, 31, 98). The first SHAPE-based study detected the circular form in the virion but found that the linear form predominated in a capsid-free environment (Figure 5A) (58). Outside this circularization element, comparisons of in virion and ex virion SHAPE reactivities revealed additional limited, but significant, differences likely reflecting interactions between the RNA with the viral capsid. Twenty-four elements with well-defined structures were identified (as regions of low SHAPE reactivity and low Shannon entropy) (Figure 3), many of which showed clear evidence of evolutionary conservation (58). This study also employed the RING-MaP strategy, which detects through-space RNA tertiary interactions (41), to identify eight regions with dense internucleotide correlations, all of which overlapped with the SHAPE-defined well-determined secondary structure elements, consistent with RNA tertiary folds featuring closely packed RNA helices (58). Remarkably, the higher order RNA structures involve more than one-third of nucleotides in the genome. Structure-disrupting mutations in two RNA tertiary structure elements, in the Env and NS2A coding regions (Figure 3, 5C), disrupted the global RNA genome architecture and reduced viral fitness. These mutants maintained low viral fitness over more than 60 days of cell passage, suggesting that the disruption of RNA tertiary structures could guide the development of vaccines based on attenuated viruses.
Figure 5.
Methods for melding orthogonal structural data with SHAPE probing to identify candidate functional RNA elements. (A) Comparison of RNA genome SHAPE reactivities, ex virion and in virion, to identify functional conformations. Data indicate that the DENV2 genome forms the circularized state specifically in the in virion state. DENV2 nucleotides protected in virion (blue boxes) are shown on both linear and circular genome structures. Adapted from Dethoff et al. (58). (B) Comparison of ex virion and in cell lysate biological states to identify a functional CMV genome RNA-protein interaction (green) between stem-loop C (SLC) and the viral replicase (RC) in plant cell lysates. Adapted from Watters et al. (81) by permission of Oxford University Press. (C) Locating DENV2 genome regions with dense internucleotide correlations (boxed in yellow; based on tertiary RINGs) to identify the functional NS2A RNA tertiary structure element. Tertiary RINGs arcs are colored by correlation coefficient (from highest to lowest) in red and orange. The RING-constrained NS2A RNA element model (10 aligned lowest free-energy models shown) is colored in the same way as the NS2A secondary structure model. Adapted from Dethoff et al. (58). (D) PARIS-detected long-range RNA-RNA interactions in two ZIKV strains. Through-space interactions are shown as green and pink arcs. A functional interaction between Env-coding region (blue) and 5′-UTR (red) sequences, specific to the epidemic Asian ZIKV lineage, is indicated by a green arc and blue arrow (at left); secondary structure (at right) is shown in blue and red. Individual mutations that disrupt long-range interaction Env-coding region (mut1-E and mut2-E, blue) or 5′-UTR (mut1–5′ and mut2–5′, red) sequences yield attenuated viruses, while compensatory mutations introduced to restore base pairing (combined mut1-E and mut1–5′, combined mut2-E and mut2–5′) partially rescue viral fitness (gray). Adapted from Li et al. (72) with permission from Elsevier.
The second study independently identified many of the same highly structured regions across the DENV2 genome (74) but focused on structured regions that were conserved across the four main DENV serotypes (DENV1–4) and ZIKV strains. Sixteen structured regions were conserved in DENV1–4 (74), twelve of which overlapped structural elements identified in the first DENV2 study (58). One of these regions was found to create a strong ribosome pause site in ribosome profiling experiments in DENV1-infected human cells, suggesting that the structure may be involved in translational regulation. As observed in models of the HCV RNA (65), the DENV structure models had median helix lengths of four base pairs, likely important for evasion of the host immune response.
This study also used the psoralen-based SPLASH (43) crosslinking method to identify long-range interactions. Many of these interactions, including the known flavivirus circularization element, were observed in virions of two or more DENV serotypes or ZIKV strains. Most interactions were not present in infected cells (74). Viral or host helicases may actively unwind viral RNA structures in cells or these long-range interactions may only occur in the physically-constrained encapsidated genome state. Four long-range interactions in ZIKV observed in virions, in cells, or in both contexts were conserved among strains and were found to be important for viral fitness. Disruptions of the two long-range interactions present in both virions and infected cells resulted in the most severe attenuating phenotypes. Some of these long-range interactions may be dynamic. For example, an element in the Env-coding region of ZIKV forms long-range base pairing interactions with three other genome regions that appear to be mutually exclusive. Mutations individually disrupting each alternate long-range conformation compromised viral fitness, suggesting that each of the three conformations is functionally important.
Zika Virus
Recently, two groups independently investigated the ZIKV RNA genome using SHAPE (72, 74). All previously characterized RNA structural elements were identified along with numerous novel well-defined motifs. Twelve structured regions were conserved among four ZIKV strains analyzed in study one (74), a finding also largely consistent with study two focusing on two ZIKV strains belonging to the ancestral African lineage and the, currently epidemic, Asian lineage (72). Most specific individual RNA secondary structures in these structured regions were not conserved between two strains analyzed in study two (outside of the 5’- and 3’-UTRs); however, the sizes and locations of many structured RNA domains were conserved (72). Study two also examined long-range RNA-RNA interactions using a psoralen-based crosslinking method termed PARIS (42). The known flavivirus genome circularization element was detected, but only two of the detected interactions (72) overlapped with the ten interactions identified as conserved or functional in the psoralen crosslinking study of four ZIKV strains (74), discussed above. The lack of overlapping results may reflect real structural differences in the ZIKV strains or may reflect experimental idiosyncrasies resulting from differences in sequence coverage, library preparation biases, and other experimental features.
RNA structures unique to the Asian lineage may contribute to the higher pathogenicity of this ZIKV strain as compared to the African strain. One long-range interaction stood out as occurring exclusively in the epidemic Asian ZIKV strain (Figure 5D) (72). Mutations that disrupt this long-range interaction connecting the 5′-UTR and the Env coding region created attenuated viruses; compensatory mutations that restored base pairing partially rescued viral fitness (72). A separate study of another epidemic ZIKV strain, using a similar psoralen-based (COMRADES) crosslinking method, independently identified this same functional 5′-UTR–Env coding region interaction (99). This interaction overlaps with the known flavivirus genome circularization element and may affect the balance between translation, replication, and packaging.
Influenza A Virus
Influenza A virus (IAV) has a genome consisting of eight individual genomic segments. Structural interrogation by SHAPE has been undertaken by several groups (64, 68, 70, 71). Recently, a single group investigated the viral RNA negative sense strands for all eight genomic segments in virion and isolated from virions or based on RNAs transcribed in vitro (64). SHAPE-directed structure models recapitulated most previously identified functional structures (100, 101) and additionally revealed multiple novel highly structured regions (64). The eight genomic segments probed in the virion contained many highly structured regions; these regions were less structured than IAV RNAs that were refolded in vitro and probed, suggesting that binding of the nucleocapsid protein partially remodels RNA structures. Individual high-probability secondary structures are largely shared between in virion and in vitro states (64), and many are conserved across IAV strains (100, 101).
SHAPE structure probing of the IAV segment 5 sense RNA was specifically used to find single-stranded or loop regions with conserved sequences, and several of these single-stranded elements were successfully targeted by antisense oligonucleotides to inhibit IAV replication (71). Long-range intra- and intersegment base-pairing interactions between the eight IAV genomic segments were examined using SPLASH (64). Numerous intersegment interactions were detected. Five high-confidence interactions were selected for follow-up functional studies; each was shown to be critical for viral packaging and maintenance of normal growth kinetics, and intersegment genome ratios in virions.
Plant Viruses
A study of the tomato bushy stunt virus (TBSV) paired SHAPE structure probing with atomic-force microscopy (AFM) to reveal that the genome is highly folded and compact, with multiple RNA domains extending from a central hub (52, 63). Similar compact global genomic architectures have been visualized by AFM for HCV, hepatitis G virus, and satellite tobacco mosaic virus (STMV) (77, 102, 103) (previously reviewed and illustrated in Nicholson et al. (52)). This organization contrasts with those of poliovirus and rubella virus, which have extended genomic architectures as visualized by AFM (102). Long-range interactions within the TBSV RNA regulate genome replication, translation, and the production of subgenomic RNAs (30, 63). The SHAPE-directed structural model of the TBSV genome directly supports the formation of several of these known long-range interactions, and also suggests that local secondary structure places functional long-range interactions in close proximity. Two highly structured elements, SL27 and S31 are conserved in genomes of other members of the Tombusvirus genus. Mutations that disrupt these structures reduced viral accumulation in protoplast competition assays.
A recent genome-wide SHAPE chemical probing study investigated one of the three genome segments of cucumber mosaic virus (CMV), a segmented positive-strand RNA plant virus with the widest known host range of any virus (81). This 2200-nt genome segment has a highly branched structure both in infected plant cell lysates and as an RNA isolated from virions. SHAPE-based structure models accurately recovered five previously characterized stem-loops in the 3′-UTR that form a tRNA-like structure that binds the viral replicase (Figure 5B). Covariation analysis of SHAPE-supported structures led to the identification of six novel elements in the coat protein-coding region and the 3′-UTR.
A distinctive feature of CMV is its high adaptability to new hosts and environments (104), thus making it relatively easy to select CMV mutants for reversion in passaged populations in plant cells. Structure-disrupting mutations were introduced into four of the identified novel elements in CMV (81), and mutants were serially passaged in Nicotiana tabacum plants to identify biologically selected changes to the mutated sequences. After passaging, these four mutants showed partial reversion to regain sequences or structures similar to native sequences or structures, while mutations made outside of selected conserved structural elements did not revert, suggesting that these four viral RNA elements are functionally important. This study emphasizes the usefulness of functionally investigating RNA structural elements by serial cell passage and reversion analysis.
Plant Satellite Viruses
SHAPE structure probing has also been used to study several plant satellite viral genomes, which require coinfection with a helper virus to replicate (62, 77). The 1058-nt STMV genome was investigated independently by two groups yielding in vitro and ex virion SHAPE-directed genome structure models, with outstanding agreement between studies (62, 77, 80). The SHAPE-based STMV structure models reveal a multi-domain architecture supported by AFM and cryo-electron microscopy studies (77, 103) (previously reviewed and illustrated in Nicholson et al. (52)). The SHAPE-directed structure models have three structural domains, each corresponding to a distinct viral function: capsid-coding, translation regulation, and genome synthesis (77, 80). The modular organization of these functional domains may reflect distinct evolutionary origins of the domains and subsequent recombination from multiple viruses to form modern STMV. Comparing the STMV genome structure under in virion and ex virion conditions demonstrated that the RNA genome undergoes well-defined structural changes when released from its capsid environment (62, 77, 80). SHAPE probing studies of two other noncoding satellite plant viral RNAs, satellite-turnip crinkle virus and satellite-cymbidium ringspot virus, also revealed secondary and tertiary structures critical for replication and packaging (66, 67).
STRATEGIES FOR IDENTIFYING FUNCTIONAL ELEMENTS
SHAPE structure probing of numerous intact viral genomes has defined a wealth of novel RNA structures across coding and noncoding regions, linked by relatively unstructured regions. In general, previously characterized functional elements are consistently included among structured regions identified de novo by SHAPE. However, simply considering highly structured elements tends to overpredict the number of functional elements in an RNA. A major challenge therefore is identification of those RNA structures with functions important to viral fitness. In the following sections, we consider the strategies that have been used to identify functional viral RNA structural motifs.
Evaluation of Global SHAPE Reactivities
A common first step in the analysis of whole-genome SHAPE datasets involves identification of relatively highly structured regions. This is usefully done by plotting median SHAPE reactivities over 30-nt to 75-nt windows (37, 58, 63, 65, 72, 73, 75, 76) and identifying those regions with low median reactivities (Figure 3) relative to the global median. The median SHAPE reactivity that identifies a highly structured region can be calibrated by comparison to the medians of well-characterized structured RNA elements. Highly structured regions serve as a starting place for the identification of functional viral RNA structural motifs.
Identification of Evolutionarily Conserved Structures
Studies of HIV (75), SIV (76), poliovirus (78), and TBSV (63) combined SHAPE-based detection of highly structured regions with a method for assessing evolutionary conservation to identify regions of potential functional importance. For HIV, SIV, and poliovirus (Figure 4A), RNA secondary structure conservation was analyzed by evaluating the rate and pattern of nucleotide changes in viral species-specific sequence alignments of protein-coding regions to yield a predicted evolutionary base-pairing probability for each nucleotide (75, 76, 78, 105, 106). This general approach has proven useful in almost every SHAPE-based analysis of viral genomes performed to date.
Identification of Low SHAPE/low Shannon Entropy RNA Regions
An important analytical development for the discovery of functional RNA motifs combines identification of regions with low SHAPE reactivity with identification of regions with low entropy – these elements are likely to form a single stable structure (Figure 3). In this data analysis strategy, SHAPE reactivities are first used to constrain prediction of thermodynamically stable secondary structures, genome-wide, based on nearest-neighbor rules (59, 107). For a long viral RNA, this results in thousands of possible conformations. The probability of formation of each base pair is then calculated across all possible structures in the ensemble (37, 108). These base-pairing probabilities are used to calculate a Shannon entropy value for each nucleotide. Regions with low Shannon entropy are either likely to form a single stable structure or are unlikely to base pair at all. Identification of regions in viral genomes that are both highly structured (low SHAPE reactivity) and that have a well-determined structure (low Shannon entropy) have led to the discovery of novel well-defined RNA structures that are critical for viral fitness in HIV (37), HCV (65), SINV (73), and DENV (Figure 3) (58). The low SHAPE/low Shannon entropy metric also identifies functional elements in diverse cellular coding and noncoding RNAs (61, 109). Regions with low SHAPE reactivities but high entropy may still be functional but are likely to experience dynamic conformational changes.
Comparison of Structure in Related Viral Species and Strains
Functional RNA motifs in viral genomes can also be discovered by identifying highly structured regions or specific motifs conserved in two or more related viral species (of the same genus) or viral strains (subspecies). To date, comparative whole-genome SHAPE studies have been performed on lentivirus species HIV-1, SIVmac239, and SIVcpzMB897 (76, 79); HCV genotype subtypes 1a, 1b, and 2 (65); alphavirus species SINV and VEEV (73); and DENV and ZIKV flavivirus species (72, 74). These studies have led to the discovery of multiple novel functional motifs and have also revealed principles of viral RNA structure evolution.
In the study of three lentivirus species (79), SHAPE-directed structure models were used in combination with covariation analysis to create a first-of-its-kind structure-dependent genome-long sequence alignment. This alignment was then used to discover regions with statistically correlated SHAPE reactivity profiles and to construct consensus secondary structure models.
The study of the three HCV genotype subtypes paired identification of low SHAPE/low Shannon entropy regions with several metrics for evaluating evolutionary conservation (Figure 4B). Metrics for evaluating evolutionary conservation included measuring conserved base pairs, synonymous substitution rates, and coevolving base pairs. These metrics were also useful in a later study of the DENV2 genome (58).
The study of alphavirus species SINV and VEEV (73) employed several new analytical methods to identify functional elements in each individual species before comparing the two to evaluate structural conservation. Sequence conservation across the individual SINV and VEEV genomes was evaluated using a multiple sequence alignment representing the entire alphavirus genus phylogeny. Regions with low SHAPE/low Shannon entropy and sequence conservation were evaluated using a structural significance score (110) that compared the free energy for these identified regions with what would be expected if their sequences were shuffled at random. This combined analysis identified four known and thirteen novel significantly structured regions in SINV and four known and eleven novel significantly structured regions in VEEV. Nine of these structured regions were present in both viruses, however, most specific secondary structure motifs within these regions were not conserved. Structure-disrupting mutations in the only two conserved elements and in one novel nonconserved element reduced viral fitness. This work emphasizes the functional importance of both conserved and nonconserved RNA secondary structure elements and the value of structure-first SHAPE analysis for the discovery of functional species-specific elements.
Another study looked for structural conservation across the flavivirus family, specifically investigating the DENV1–4 serotypes and four ZIKV strains. This study compared regions of conserved sequence, well-defined structures as defined by low SHAPE/low Shannon entropy analysis, similar SHAPE reactivity profiles, and low mutation rates within and across serotypes and strains to identify potentially functional regions (74). A separate study used SHAPE analysis to identify RNA structural domains of similar size and genome location conserved in two ZIKV strains; intriguingly, while domain locations were conserved, specific RNA secondary structures within these domains were not conserved. The functional relevance of the structural elements discovered in the comparative analysis of DENV1–4 and multiple ZIKV strains remains to be investigated.
RNA Structural Covariation Models
Another important method for identifying functional elements was developed in a study investigating HCV (69). A preliminary covariation model was built from a SHAPE-directed HCV secondary structure model of a single strain coupled with a sequence-based alignment of multiple HCV genotype 2 viruses. Additional divergent HCV sequences with lower levels of sequence conservation were then incorporated into this preliminary covariation model (111) using algorithms developed to identify highly structured RNA motifs in shorter RNAs like riboswitches and ribozymes (112). This resulting covariation model aligned divergent HCV sequences based on structure and thus allowed the identification of conserved RNA motifs located at different positions in the primary sequences. Seventeen stem-loops containing multiple consecutive covarying nucleotides were identified (113) and, of the five novel elements evaluated, four were found to be important for viral fitness (69), emphasizing the power of this strategy for finding novel functional stem-loops in viral genomes (Figure 4C).
Comparison of RNA Structural States Under Different Conditions
Another important approach for the discovery of functional RNA motifs in viral genomes involves comparison of RNA structural elements in different biological states or contexts of a virus (Box 1). Comparison of the DENV2 genome in the ex virion and in virion states led to the discovery that flavivirus genomes are in their circularized form when packaged in virions (58) and are in their linear form in the absence of viral proteins (Figure 5A). Genome circularization, which is involved in regulating flavivirus replication and translation (18, 31, 98), may thus also play a role in viral packaging. Comparisons of the in virion, ex virion, and in vitro states have revealed potential RNA binding sites for capsid protein components of DENV (58) and IAV (64). Comparing the CMV genome segment 3 structure in virion and in infected cell lysates revealed an RNA motif likely bound by the viral replicase in infected cells (Figure 5B) (81). Flaviviral (72, 74), IAV (64), and STMV (77, 80) RNAs have higher SHAPE reactivities (are less structured) when analyzed in infected cells or in virions than are the in vitro or ex virion RNAs. These results are consistent with the idea that RNA in cells or in virions may be bound by proteins or unwound by viral or cellular helicases. Functional studies of the ZIKV genome found that RNA motifs present only in virions, only in cells, or both in virions and in cells each play critical roles in viral fitness (74).
Strategies Exploiting Analysis of RNA Tertiary Structures
Some RNA regions fold back on themselves to form complex higher order tertiary folds with closely packed base-paired helices. The evolutionary requirements to retain a complex higher order structure are likely substantial; thus, by finding motifs that form tertiary interactions, it might also be possible to identify novel functional elements. A recent study of the DENV2 RNA genome (58) used RING-MaP analysis to examine through-space RNA interactions throughout the genome (Figure 5C). This analysis identified regions with dense internucleotide correlations (RINGs) consistent with RNA tertiary folds. RING-MaP involves the chemical probing of RNA with DMS such that each RNA strand contains multiple structure-specific modifications. Nucleotide pairs involved in tertiary interactions are protected when the interaction is stably formed but are more likely to be modified in a correlated manner during structural “breathing” (41, 58). The eight regions that contained dense through space interactions all overlapped with highly structured elements identified by low SHAPE/low Shannon entropy (Figure 3) and evolutionary conservation analyses. Mutations were introduced into three of these eight elements. Two of the three mutants disrupted the global RNA genome architecture, reduced viral fitness, and created stable attenuated viruses, emphasizing that viral RNA regions that form tertiary interactions are also likely to contain functional RNA motifs.
Psoralen-based crosslinking methods SPLASH (43) and PARIS (42) have also been paired with SHAPE analysis to identify potential functional RNA structures in IAV, DENV, and ZIKV (Figure 5D) (64, 72, 74). These crosslinking methods can detect both short and long-range intramolecular (64, 72, 74) and intermolecular (64) viral RNA genome interactions. Genome interactions identified by these methods complement SHAPE-directed structure models by providing additional confidence for short-range base pairing interactions and by identifying long-range base pairing interactions that cannot be identified by current SHAPE-directed folding methods. SPLASH and PARIS yield complex data and additional filters are needed to identify the functional interactions from among the many interactions that are identified. These crosslinking methods have recovered known long-range interactions between the 5′ and 3′ cyclization sequences critical for flaviviral fitness (72, 74, 99). Melding SHAPE and crosslinking-based methods has also thus far identified six long-range ZIKV interactions required for viral fitness (72, 74), including one present exclusively in the epidemic Asian ZIKV lineage strains (Figure 5D) (72).
PERSPECTIVE
SHAPE structure probing of viral RNA is now a broadly used and well-validated strategy for discovering and characterizing the structures of viral RNA genomes. SHAPE-directed secondary structure models consistently recover nearly all known functional elements identified prior to SHAPE probing and also identify many novel elements. SHAPE interrogation of eleven full-length viral RNA genomes from diverse viral families has revealed a plethora of well-defined internal structures (Table 1), many which have been shown to be critical for viral fitness. RNA structure is pervasive across viral genome coding regions; these RNA structures involve motifs from simple helices and hairpins to complex pseudoknots, conformational switches, long-range RNA-RNA interactions, and tertiary structures that rival the complexity of previously characterized functional structures in viral UTRs. Research to date indicates that complex and dynamic RNA genome architectures govern and modulate interactions with functionally critical protein, RNA, and small-molecule ligand partners to regulate the balance between viral replication, protein synthesis, packaging, and evasion of host immune responses.
A diverse suite of bioinformatics and functional analysis tools have been developed by different laboratories and, collectively, these efforts support validation of numerous functional RNA structural elements across viral genomes. Nonetheless, it remains challenging to identify the subset of functionally important motifs in the context of extensive secondary structure folding. Implementing virus functional assays is time consuming and often inefficient, which has limited the number of elements that have been functionally investigated. New higher throughput strategies for viral functional analysis are critically needed. Also needed are new strategies for more efficiently paring down initial lists of potentially functional and well-structured elements by computational approaches. We anticipate that exploration of the structural complexity of viral genomes in distinct biological contexts and pairing SHAPE analysis with new methods for mapping higher order RNA structures will continue to unveil mechanistic details of viral replication cycles. Full-length RNA genomes are very complex structurally and likely form three-dimensional entities. If the extent to which viruses use RNA structure-based mechanisms to enable and regulate their replication is as broad and comprehensive as studies to date suggest, there may be opportunities to target these elements in the development of antiviral therapies and vaccination strategies.
SUMMARY POINTS.
SHAPE-guided structure probing serves as a “molecular microscope” yielding high-resolution models of structured RNA elements across viral genomes.
Bioinformatics analysis and functional assays are important for characterizing and validating novel functional structures in viral RNA genomes.
SHAPE structure probing has revealed that structured RNA elements are pervasive throughout viral genomes. Models and images of viral RNA genomes as long “single-stranded” entities should be abandoned.
Viral genomes form higher order structures in which a subset of secondary structures fold into tertiary structures and organize into complex and dynamic global genome architectures.
RNA structures regulate viral replication and fitness with often complex effects on virus pathogenicity.
The mechanisms by which structured elements in viral RNA genomes modulate viral replication are diverse and involve enabling, mediating, and fine-tuning interactions with RNA, protein, and small-molecule partners.
FUTURE ISSUES.
Current SHAPE-based structure interrogation methods have had a transformative effect on understanding how RNA genomes encode information but are not perfect. Ongoing innovations to increase confidence in structure models are needed.
Most structure models for viral genomic RNA have focused on and have been interpreted in the context of a single predominant conformation. Many RNA elements likely sample multiple structures, form defined ensembles, or function as switches. New strategies are needed to identify and characterize these dynamic elements.
High-throughput structure probing strategies are powerful for defining structured RNA elements, but new high-throughput strategies for functional analysis are needed to validate the importance of these elements.
Structures of most viral RNA genomes have been interrogated in only one or two biological states, leaving mechanistic details of viral replication cycles undiscovered. There is a broad opportunity to probe viral RNA genomes in virions, and in infected cells, infected cell nuclei or cytoplasm, and to analyze viral replication intermediates, negative-strands of positive-strand RNA viruses (and vice versa), and subgenomic viral RNA molecules.
Methods are emerging for mapping three-dimensional and through-space interactions in viral RNA genomes, but these methods are under active development, are supported by relatively modest validation, and merit additional effort to address challenges in reproducibility and information content.
Application of existing and novel methods to characterize interactions between structured elements in RNA genomes with protein, other RNAs, and small-molecule ligands represents an exciting frontier.
ACKNOWLEDGMENTS
This work in our laboratory is supported by the US National Institutes of Health (R35 GM122532 and R01 AI068462 to K.M.W.). M.A.B. is a Ruth L. Kirschstein Postdoctoral Fellow (F32 GM128330). We thank Parth Jariwala for helpful discussions in the early-stage planning of this review.
Footnotes
DISCLOSURE STATEMENT
K.M.W. serves as an advisor to and holds equity in Ribometrix.
LITERATURE CITED
- 1.Brion P, Westhof E. 1997. Hierarchy and Dynamics of RNA Folding. Annu. Rev. Biophys. Biomol. Struct 26(1):113–37 [DOI] [PubMed] [Google Scholar]
- 2.Batey RT, Rambo RP, Doudna JA. 1999. Tertiary motifs in RNA structure and folding. Angew. Chem. - Int. Ed 38(16):2326–43 [DOI] [PubMed] [Google Scholar]
- 3.Butcher SE, Pyle AM. 2011. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res 44(12):1302–11 [DOI] [PubMed] [Google Scholar]
- 4.Desselberger U, Racaniello VR, Zazra JJ, Palese P. 1980. The 3’ and 5’-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene 8(3):315–28 [DOI] [PubMed] [Google Scholar]
- 5.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516(7531):355–60 [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, Silverman RH, Jeang KT. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59(2):273–82 [DOI] [PubMed] [Google Scholar]
- 7.Ortín J, Parra F. 2006. Structure and function of RNA replication. Annu. Rev. Microbiol 60:305–26 [DOI] [PubMed] [Google Scholar]
- 8.Brierley I, Digard P, Inglis SC. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 57(4):537–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S-J, Kim Y-G, Park H-J. 2011. Identification of RNA Pseudoknot-Binding Ligand That Inhibits the −1 Ribosomal Frameshifting of SARS-Coronavirus by Structure-Based Virtual Screening. J. Am. Chem. Soc 133(26):10094–100 [DOI] [PubMed] [Google Scholar]
- 10.Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol 64(5):1920–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu K, Heng X, Summers MF. 2011. Structural Determinants and Mechanism of HIV-1 Genome Packaging. J. Mol. Biol 410(4):609–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, et al. 2008. A Highly Structured, Nuclease-Resistant, Noncoding RNA Produced by Flaviviruses Is Required for Pathogenicity. Cell Host Microbe 4(6):579–91 [DOI] [PubMed] [Google Scholar]
- 13.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, et al. 2014. The Structural Basis of Pathogenic Subgenomic Flavivirus RNA (sfRNA) Production. Science 344(6181):307–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, et al. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354(6316):1148–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. 2016. RNA Structure Duplications and Flavivirus Host Adaptation. Trends Microbiol. 24(4):270–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol 66(3):1476–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K-M, Chen C-J, Shih S-R. 2017. Regulation Mechanisms of Viral IRES-Driven Translation. Trends Microbiol. 25(7):546–61 [DOI] [PubMed] [Google Scholar]
- 18.Gebhard LG, Filomatori CV, Gamarnik AV. 2011. Functional RNA elements in the dengue virus genome. Viruses 3(9):1739–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, et al. 2014. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol 21(8):721–27 [DOI] [PubMed] [Google Scholar]
- 20.Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, et al. 2013. An Unusual Topological Structure of the HIV-1 Rev Response Element. Cell 155(3):594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. 2014. Functional conservation despite structural divergence in ligand-responsive RNA switches. PNAS 111(45):15952–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel AD, Young JAT. 1998. HIV-1: Fifteen Proteins and an RNA. Annu. Rev. Biochem 67(1):1–25 [DOI] [PubMed] [Google Scholar]
- 23.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. 2004. RNA Interactions in the 5′ Region of the HIV-1 Genome. J. Mol. Biol 336(2):369–79 [DOI] [PubMed] [Google Scholar]
- 24.Goff SP. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol 5(4):253–63 [DOI] [PubMed] [Google Scholar]
- 25.Fraser C, Doudna J. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol 5(1):29–38 [DOI] [PubMed] [Google Scholar]
- 26.Kieft JS. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci 33(6):274–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komar AA, Mazumder B, Merrick WC. 2012. A new framework for understanding IRES-mediated translation. Gene 502(2):75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, et al. 2014. Hepatitis C Virus Translation Inhibitors Targeting the Internal Ribosomal Entry Site. J. Med. Chem 57:1694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khawaja A, Vopalensky V, Pospisek M. 2015. Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdiscip. Rev. RNA 6(2):211–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson BL, White KA. 2014. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat. Rev. Microbiol 12(7):493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV. 2015. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol 89(6):3430–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjems J, Brown M, Chang DD, Sharp PA. 1991. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. PNAS 88(3):683–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379(2):314–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. 2012. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology 432(2):511–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z-Y, Li X-F, Jiang T, Deng Y-Q, Zhao H, et al. 2013. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J. Virol 87(12):6804–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, et al. 2016. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. eLife 5:e17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11(9):959–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. 2015. Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 10(11):1643–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, et al. 2011. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). PNAS 108(27):11063–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loughrey D, Watters KE, Settle AH, Lucks JB. 2014. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 42(21):e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homan PJ, Favorov OV, Lavender CA, Kursun O, Ge X, et al. 2014. Single-molecule correlated chemical probing of RNA. PNAS 111(38):13858–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, et al. 2016. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165(5):1267–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, et al. 2016. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 62(4):603–17 [DOI] [PubMed] [Google Scholar]
- 44.Peattie DA, Gilbert W. 1980. Chemical probes for higher-order structure in RNA. PNAS 77(8):4679–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehresmann B 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15(22):9109–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern S, Moazed D, Noller HF. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481–89 [DOI] [PubMed] [Google Scholar]
- 47.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc 127(12):4223–31 [DOI] [PubMed] [Google Scholar]
- 48.Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol 20(3):295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenyon J, Prestwood L, Lever A. 2014. Current perspectives on RNA secondary structure probing. Biochem. Soc. Trans 42(4):1251–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape : Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19(1):63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. 2015. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci 40(4):221–32 [DOI] [PubMed] [Google Scholar]
- 52.Nicholson BL, White KA. 2015. Exploring the architecture of viral RNA genomes. Curr. Opin. Virol 12:66–74 [DOI] [PubMed] [Google Scholar]
- 53.Rausch JW, Sztuba-Solinska J, Le Grice SFJ. 2018. Probing the Structures of Viral RNA Regulatory Elements with SHAPE and Related Methodologies. Front. Microbiol 8:2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strobel EJ, Yu AM, Lucks JB. 2018. High-throughput determination of RNA structures. Nat. Rev. Genet 19(10):615–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hector RD, Burlacu E, Aitken S, Le Bihan T, Tuijtel M, et al. 2014. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 42(19):12138–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seetin MG, Kladwang W, Bida JP, Das R. 2014. Massively parallel RNA chemical mapping with a reduced bias MAP-seq protocol. Methods Mol. Biol 1086:95–117 [DOI] [PubMed] [Google Scholar]
- 57.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519(7544):486–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dethoff EA, Boerneke MA, Gokhale NS, Muhire BM, Martin DP, et al. 2018. Pervasive tertiary structure in the dengue virus RNA genome. PNAS 115(45):11513–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. PNAS 106(1):97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. PNAS 110(14):5498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, et al. 2016. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. PNAS 113(37):10322–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Athavale SS, Gossett JJ, Bowman JC, Hud NV, Williams LD, Harvey SC. 2013. In Vitro Secondary Structure of the Genomic RNA of Satellite Tobacco Mosaic Virus. PLOS ONE 8(1):e54384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu B, Grigull J, Ore MO, Morin S, White KA. 2013. Global Organization of a Positive-strand RNA Virus Genome. PLOS Pathog. 9(5):e1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dadonaite B, Barilaite E, Fodor E, Laederach A, Bauer DL. 2017. The structure of the influenza A virus genome. bioRxiv 236620. 10.1101/236620 [DOI] [Google Scholar]
- 65.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, et al. 2015. Functionally conserved architecture of hepatitis C virus RNA genomes. PNAS 112(12):3692–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murawski AM, Nieves JL, Chattopadhyay M, Young MY, Szarko C, et al. 2015. Rapid evolution of in vivo-selected sequences and structures replacing 20% of a subviral RNA. Virology 483:149–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashton P, Wu B, D’Angelo J, Grigull J, White KA. 2015. Biologically-supported structural model for a viral satellite RNA. Nucleic Acids Res. 43(20):9965–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lenartowicz E, Kesy J, Ruszkowska A, Soszynska-Jozwiak M, Michalak P, et al. 2016. Self-Folding of Naked Segment 8 Genomic RNA of Influenza A Virus. PLOS ONE 11(2):e0148281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. 2016. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol. Cell 62(1):111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruszkowska A, Lenartowicz E, Moss WN, Kierzek R, Kierzek E. 2016. Secondary structure model of the naked segment 7 influenza A virus genomic RNA. Biochem. J 473(23):4327–48 [DOI] [PubMed] [Google Scholar]
- 71.Soszynska-Jozwiak M, Michalak P, Moss WN, Kierzek R, Kesy J, Kierzek E. 2017. Influenza virus segment 5 (+)RNA - secondary structure and new targets for antiviral strategies. Sci. Rep 7(1):15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li P, Wei Y, Mei M, Tang L, Sun L, et al. 2018. Integrative Analysis of Zika Virus Genome RNA Structure Reveals Critical Determinants of Viral Infectivity. Cell Host Microbe 24(6):875–86.e5 [DOI] [PubMed] [Google Scholar]
- 73.Kutchko KM, Madden EA, Morrison C, Plante KS, Sanders W, et al. 2018. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res. 46(7):3657–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huber RG, Lim XN, Ng WC, Sim A, Poh HX, et al. 2018. Structure mapping of dengue and Zika viruses reveals new functional long- range interactions. bioRxiv 381368. 10.1101/381368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460(7256):711–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, et al. 2013. Comparison of SIV and HIV-1 Genomic RNA Structures Reveals Impact of Sequence Evolution on Conserved and Non-Conserved Structural Motifs. PLOS Pathog. 9(4):e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Archer EJ, Simpson MA, Watts NJ, O’Kane R, Wang B, et al. 2013. Long-range architecture in a viral RNA genome. Biochemistry 52(18):3182–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burrill CP, Westesson O, Schulte MB, Strings VR, Segal M, Andino R. 2013. Global RNA structure analysis of poliovirus identifies a conserved RNA structure involved in viral replication and infectivity. J. Virol 87(21):11670–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lavender CA, Gorelick RJ, Weeks KM. 2015. Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes. PLOS Comput. Biol 11(5):e1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Larman BC, Dethoff EA, Weeks KM. 2017. Packaged and Free Satellite Tobacco Mosaic Virus (STMV) RNA Genomes Adopt Distinct Conformational States. Biochemistry 56(16):2175–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watters KE, Choudhary K, Aviran S, Lucks JB, Perry KL, Thompson JR. 2018. Probing of RNA structures in a positive sense RNA virus reveals selection pressures for structural elements. Nucleic Acids Res. 46(5):2573–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sztuba-Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, et al. 2017. Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 45(11):6805–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Filbin ME, Kieft JS. 2011. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA 17(7):1258–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Romero-López C, Barroso-delJesus A, García-Sacristán A, Briones C, Berzal-Herranz A. 2012. The folding of the hepatitis C virus internal ribosome entry site depends on the 3′-end of the viral genome. Nucleic Acids Res. 40(22):11697–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diaz-Toledano R, Lozano G, Martinez-Salas E. 2016. In-cell SHAPE uncovers dynamic interactions between the untranslated regions of the foot-and-mouth disease virus RNA. Nucleic Acids Res. 45(3):1416–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sztuba-Solinska J, Shenoy SR, Gareiss P, Krumpe LRH, Le Grice SFJ, et al. 2014. Identification of Biologically Active, HIV TAR RNA-Binding Small Molecules Using Small Molecule Microarrays. J. Am. Chem. Soc 136(23):8402–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abulwerdi FA, Shortridge MD, Sztuba-Solinska J, Wilson R, Le Grice SFJ, et al. 2016. Development of Small Molecules with a Noncanonical Binding Mode to HIV-1 Trans Activation Response (TAR) RNA. J. Med. Chem 59(24):11148–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Low JT, Garcia-Miranda P, Mouzakis KD, Gorelick RJ, Butcher SE, Weeks KM. 2014. Structure and dynamics of the HIV-1 frameshift element RNA. Biochemistry 53(26):4282–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang Y, Assmann SM, Bevilacqua PC. 2016. Protein Structure Is Related to RNA Structural Reactivity In Vivo. J. Mol. Biol 428(5):758–66 [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Chen J, Nikolaitchik OA, Desimmie BA, Busan S, et al. 2018. The roles of five conserved lentiviral RNA structures in HIV-1 replication. Virology 514:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tellez AB, Wang J, Tanner EJ, Spagnolo JF, Kirkegaard K, Bullitt E. 2011. Interstitial Contacts in an RNA-Dependent RNA Polymerase Lattice. J. Mol. Biol 412(4):737–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han J-Q, Barton DJ. 2002. Activation and evasion of the antiviral 2’–5’ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8(4):512–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng X, Bevilacqua PC. 2004. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10(12):1934–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Floyd-Smith G, Slattery E, Lengyel P. 1981. Interferon action: RNA cleavage pattern of a (2’−5’)oligoadenylate–dependent endonuclease. Science 212(4498):1030–32 [DOI] [PubMed] [Google Scholar]
- 95.Han J-Q, Wroblewski G, Xu Z, Silverman RH, Barton DJ. 2004. Sensitivity of Hepatitis C Virus RNA to the Antiviral Enzyme Ribonuclease L Is Determined by a Subset of Efficient Cleavage Sites. J. Interf. Cytokine Res 24(11):664–76 [DOI] [PubMed] [Google Scholar]
- 96.Fayzulin R, Frolov I. 2004. Changes of the secondary structure of the 5’ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations. J. Virol 78(10):4953–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I. 2011. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J. Virol 85(16):8022–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alvarez DE, Lodeiro MF, Ludueña SJ, Lía I, Gamarnik AV, et al. 2005. Long-Range RNA-RNA Interactions Circularize the Dengue Virus Genome Long-Range RNA-RNA Interactions Circularize the Dengue Virus Genome. J. Virol 79(11):6631–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, et al. 2018. COMRADES determines in vivo RNA structures and interactions. Nat. Methods 15(10):785–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kobayashi Y, Dadonaite B, van Doremalen N, Suzuki Y, Barclay WS, Pybus OG. 2016. Computational and molecular analysis of conserved influenza A virus RNA secondary structures involved in infectious virion production. RNA Biol. 13(9):883–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gultyaev AP, Tsyganov-Bodounov A, Spronken MIJ, Van Der Kooij S, Fouchier RAM, Olsthoorn RCL. 2014. RNA structural constraints in the evolution of the influenza A virus genome NP segment. RNA Biol. 11(7):942–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. 2008. Bioinformatic and Physical Characterizations of Genome-Scale Ordered RNA Structure in Mammalian RNA Viruses. J. Virol 82(23):11824–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garmann RF, Gopal A, Athavale SS, Knobler CM, Gelbart WM, Harvey SC. 2015. Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA 21(5):877–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thompson JR, Doun S, Perry KL. 2006. Compensatory capsid protein mutations in cucumber mosaic virus confer systemic infectivity in squash (Cucurbita pepo). J. Virol 80(15):7740–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J. 2004. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 32(16):4925–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Westesson O, Holmes I. 2012. Developing and Applying Heterogeneous Phylogenetic Models with XRate. PLOS ONE 7(6):e36898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. PNAS 101(19):7287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mathews DH. 2004. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10(8):1178–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, et al. 2018. Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 173(1):181–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soldatov RA, Vinogradova SV, Mironov AA. 2014. RNASurface: fast and accurate detection of locally optimal potentially structured RNA segments. Bioinformatics 30(4):457–63 [DOI] [PubMed] [Google Scholar]
- 111.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.El Korbi A, Ouellet J, Naghdi MR, Perreault J. 2014. Finding Instances of Riboswitches and Ribozymes by Homology Search of Structured RNA with Infernal In Therapeutic Applications of Ribozymes and Riboswitches: Methods and Protocols, ed. Lafontaine D, Dubé A, 1103:113–26. New York: Humana Press; [DOI] [PubMed] [Google Scholar]
- 113.Weinberg Z, Breaker RR. 2011. R2R - software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinform. 12(3):1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, et al. 2016. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20(5):654–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. 2013. RNA SHAPE analysis in living cells. Nat. Chem. Biol 9(1):18–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smola MJ, Calabrese JM, Weeks KM. 2015. Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry 54(46):6867–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rice GM, Leonard CW, Weeks KM. 2014. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA 20(6):846–54 [DOI] [PMC free article] [PubMed] [Google Scholar]